Abstract
This study evaluated the prognostic roles of murine double minute 2 (MDM2) and p53 in pancreatic cancer patients treated with gemcitabine-based chemotherapy. A total of 137 advanced or recurrent adenocarcinoma patients who were treated with gemcitabine-based palliative chemotherapy were reviewed, selected from 957 patients with pancreatic malignancy between 2008 and 2013 at our hospital. Immunohistochemical staining for MDM2 and p53 with formalin-fixed, paraffin-embedded tumor tissues was independently reviewed. Nuclear or cytoplasmic expression of MDM2 and p53 was found in tumor cells of 30 (21.9%) and 71 (51.8%) patients, respectively. Patients with MDM2 expression had shorter median overall survival (OS) (3.7 vs 5.8 mo; P = .048) and median progression-free survival (PFS) (1.5 vs 2.5 mo; P < .001); by contrast, p53 expression was not correlated with OS or PFS. In the multivariate analysis, MDM2 expression (hazard ratio = 1.731; P = .025) was an independent and unfavorable prognostic factor of OS. Additionally, MDM2 expression was significantly associated with progressive disease (PD) and death (P = .015) following first-line gemcitabine-based therapy. In advanced pancreatic cancer patients, MDM2 expression is associated with shorter OS and PFS after gemcitabine-based chemotherapy.
Introduction
Pancreatic cancer is one of the leading causes of cancer-related mortalities in the world, resulting in more than 330000 deaths per year [1]. The 5-year overall survival (OS) rate is only 20% among patients receiving curative surgery and adjuvant gemcitabine, and patients with advanced diseases face even lower (< 5%) OS [2, 3]. Gemcitabine has been the most crucial element in the development of first-line chemotherapy since 1997 [3–6]. Following FOLFIRINOX establishing the role in first-line therapy for advanced pancreatic cancer [7], gemcitabine plus nab-paclitaxel also has become a new treatment standard for patients with favorable performance status (PS) [8]. Regarding the mechanisms of gemcitabine activation and metabolism, human equilibrative nucleoside transporter 1 represents the most consistent predictive biomarker for the efficacy of gemcitabine; however, data on other markers, such as deoxycytidine kinase and ribonucleotide reductase subunits 1 and 2, are heterogeneous [9]. The complex genetic background may largely contribute to the biology of pancreatic cancer and limit the utility of any single biomarker for drugs [10].
Gemcitabine, a nucleoside analogue, incorporates with DNA after activation, subsequently terminating DNA elongation [11]. After gemcitabine-induced DNA damage, p53 is activated and may contribute to apoptosis or cell cycle arrest [12, 13]. The chemosensitivity of gemcitabine in pancreatic cancer is enhanced after the restoration of p53 function [14]. However, p53 is mutated in more than 50% of pancreatic cancer cases [15], and MDM2, the negative regulator of p53, is induced and overexpressed by Ras signaling in pancreatic cancer [16]. MDM2 suppresses the transcriptional activity of p53 by binding to the transactivation domain of p53 [17]. In addition, MDM2 is an E3 ubiquitin ligase for p53 to mediate its degradation [18]. Therefore, functional p53-mediated apoptosis and cell cycle regulation may be inefficient, thus contributing little to gemcitabine-mediated cytotoxicity in pancreatic cancer patients. Furthermore, the status of p53 is not prognostic for pancreatic cancer [19–22], and the prognostic significance of MDM2 in resected pancreatic cancer is inconsistent [21, 22].
MDM2 exerts numerous other biological effects unrelated to p53, such as the regulation of p21, E2F1, XIAP, p73, and NF-κB/p65 [23–27]. In addition, the association between chemotherapy and MDM2 status in pancreatic cancer is largely unknown. In this study, we evaluated the prognostic values of MDM2 and p53 expression in advanced pancreatic cancer patients receiving gemcitabine-based palliative chemotherapy.
Methods and materials
The cancer registry database of the Medical Information Management Office at National Taiwan University Hospital was screened for primary pancreatic malignancy diagnoses between 2008 and 2013. The patients selected for this study were required to have received palliative treatment with gemcitabine-containing chemotherapy (S1 Table) for advanced or recurrent pancreatic cancer; complete available medical records and histopathological archival tissues were also obtained. Patients with benign tumors, neuroendocrine tumors, solid pseudopapillary neoplasm, or pancreatic malignancies other than adenocarcinoma were excluded. In total, 137 patients who met our inclusion criteria were selected for analysis (S1 Fig). This study was approved by the Research Ethics Committee of National Taiwan University Hospital (approval number: 201309033RINB). Written consents were waived by the Research Ethics Committee. The dataset generated and/or analyzed during the current study was de-identified and available in the supplement.
Immunohistochemistry
We applied immunohistochemical (IHC) staining to formalin-fixed, paraffin-embedded tumor tissue sections (4-μm thick), using the OptiView DAB IHC Detection Kit (Roche) and Ventana automated slide strainers (Roche). The primary antibodies and their dilutions comprised anti-MDM2 diluted to 1:100 (#33–7100, Invitrogen Corporation) and anti-p53 diluted to 1:50 (M 7001, Dako). Stained tissue sections were reviewed and scored by a pathologist (Jen-Chieh Lee) who is an expert in the interpretation of MDM2 expression [28] and was blinded to the patients’ demographic data and clinical outcomes. Expression was defined as positive when at least 10% of the tumor cells had positive staining [21]. The positive controls of p53 and MDM2 staining were colon adenoma and liposarcoma, respectively.
Statistical analysis
Most (n = 130) selected patients were dead before initiation of this study, and the other seven patients without the confirmation of death also had been selected. OS was the primary endpoint in this study and was defined from the first day of gemcitabine-based chemotherapy to the day of death or final follow-up. Tumor response was evaluated according to the Response Evaluation Criteria in Solid Tumors (RECIST), Version 1.1 [29]. PFS was defined as imaging-documented PD with RECIST or death after initiation of a gemcitabine-based chemotherapy; therefore, progression was defined as PD or death after a gemcitabine-based chemotherapy.
A Fisher’s exact test was used to analyze the correlations between the discrete clinicopathologic characteristics and the IHC expression of MDM2 and p53. The association between MDM2 and p53 IHC expression was analyzed using the Fisher’s exact test. The prognostic significance of OS among the clinicopathologic factors and the expression of MDM2 and p53 were evaluated using univariate analysis and Kaplan—Meier survival curves (i.e., log-rank test). The clinicopathologic factors with significance in the univariate analysis were subsequently introduced into a multivariate analysis (i.e., Cox regression model) for OS. The cutoff point of OS data follow-up was July 2015.
The SPSS statistical software system (IBM SPSS Statistics for Windows, Version 20.0; IBM Corp., Armonk, NY, USA) was employed for statistical analyses, and P < .05 was considered statistically significant.
Results
Patient characteristics
Our analysis included 137 patients. The median age was 62 years (range: 27–84 y), and male patients comprised 60.6% (n = 83) of the study population. Most of the patients had favorable PS according to the criteria of the Eastern Cooperative Oncology Group (ECOG) 0–1 (81.0%, n = 111), and most of them were at the advanced stages of disease (stages III or IV, according to the American Joint Committee on Cancer) (74.5%, n = 102). Initially, of the 86 patients with stage IV disease, the most common metastatic sites were the liver (n = 69), peritoneum or omentum (n = 30), and lungs (n = 17). Patients in the first-line gemcitabine subgroup had comparable clinical characteristics comparing to the whole study cohort. The baseline patient characteristics are summarized in Table 1.
Table 1. Patient characteristics.
| Characteristics | All patients | 1st–line gemcitabine | Non-1st-line gemcitabine |
|---|---|---|---|
| n = 137 (%) | n = 121 (%) | n = 16 (%) | |
| Age | |||
| median | 62 | 62 | 63 |
| range | 27–84 | 27–84 | 46–73 |
| Sex | |||
| male | 83 (60.6) | 74 (61.2) | 9 (56.3) |
| female | 54 (39.4) | 47 (38.8) | 7 (43.8) |
| ECOG PS | |||
| 0–1 | 111 (81.0) | 98 (81.0) | 13 (81.3) |
| 2–3 | 26 (19.0) | 23 (19.0) | 3 (18.8) |
| Stage* | |||
| I | 2 (1.5) | 1 (0.8) | 1 (6.3) |
| II | 33 (24.1) | 23 (19.0) | 10 (62.5) |
| III | 16 (11.7) | 14 (11.6) | 2 (12.5) |
| IV | 86 (62.8) | 83 (68.6) | 3 (18.8) |
| T | |||
| 1–2 | 21 (15.3) | 18 (14.9) | 3 (18.8) |
| 3 | 65 (47.4) | 56 (46.3) | 9 (56.3) |
| 4 | 51 (37.2) | 47 (38.8) | 4 (25.0) |
| N | |||
| 0 | 60 (43.8) | 56 (46.3) | 4 (25.0) |
| 1 | 77 (56.2) | 65 (53.7) | 12 (75.0) |
| Diabetes | |||
| Yes | 57 (41.6) | 54 (44.6) | 3 (18.8) |
| No | 80 (58.4) | 67 (55.4) | 13 (81.3) |
| Cigarette smoking | |||
| Yes | 44 (32.1) | 37 (30.6) | 7 (43.8) |
| No | 93 (67.9) | 84 (69.4) | 9 (56.3) |
| Primary | |||
| head | 66 (48.2) | 55 (45.5) | 11 (68.8) |
| neck or body | 40 (29.2) | 36 (29.8) | 4 (25.0) |
| tail | 31 (22.6) | 30 (24.8) | 1 (6.3) |
| Surgery | |||
| none | 79 (57.7) | 76 (62.8) | 3 (18.8) |
| curative | 26 (19.0) | 18 (14.9) | 8 (50.0) |
| bypass | 28 (20.4) | 23 (19.0) | 5 (31.3) |
| other | 4 (2.9) | 4 (3.3) | 0 |
| Radiotherapy | |||
| Yes | 20 (14.6) | 13 (10.7) | 7 (43.8) |
| No | 117 (85.4) | 108 (89.3) | 9 (56.3) |
| Differentiation | |||
| poor | 44 (32.1) | 42 (34.7) | 2 (12.5) |
| moderate | 72 (52.6) | 62 (51.2) | 10 (62.5) |
| good | 21 (15.3) | 17 (14.0) | 4 (25.0) |
| CA 19–9 (U/mL) | |||
| <500 | 59 (43.1) | 54 (44.6) | 5 (31.3) |
| ≥500 | 70 (51.1) | 60 (49.6) | 10 (62.5) |
| unknown | 8 (5.8) | 7 (5.8) | 1 (6.3) |
| CEA (ng/mL) | |||
| <3 | 49 (35.8) | 42 (34.7) | 7 (43.8) |
| ≥3 | 74 (54.0) | 68 (56.2) | 6 (37.5) |
| unknown | 14 (10.2) | 11 (9.1) | 3 (18.8) |
| Hematology and biochemistry¶# | |||
| WBC (per mm3) | |||
| median | 7,550 | 7,580 | 6,825 |
| range | 3,570–14,680 | 3,570–14,680 | 5,160–11,970 |
| PMN (per mm3) | |||
| median | 5,352 | 5,554 | 4,187 |
| range | 1,911–13,315 | 1,911–13,315 | 2,673–10,390 |
| Mono (per mm3) | |||
| median | 393 | 393 | 370 |
| range | 45–1,757 | 45–1,757 | 193–622 |
| Lym (per mm3) | |||
| median | 1,409 | 1,398 | 1,640 |
| range | 338–3,691 | 338–3,691 | 498–3,580 |
| Platelet (x103; per mm3) | |||
| median | 237 | 237 | 241 |
| range | 67–539 | 67–539 | 136–410 |
| CRP (mg/dL) | |||
| median | 1.93 | 1.93 | 2.48 |
| range | 0.07–21.21 | 0.07–21.21 | 0.56–4.40 |
| Albumin (g/dL) | |||
| median | 4.3 | 4.3 | 4.3 |
| range | 0.8–5.3 | 2.3–5.3 | 0.8–4.9 |
*Stage: TNM system of the American Joint Committee on Cancer (7th edition)
¶Hematology and biochemistry: WBC, white blood cell; PMN, polymorphonuclear granulocyte; Mono, monocyte; Lym, lymphocyte; CRP, C-reactive protein
#Missing data (patient number) in the whole study group: PMN, Mono, Lym (n = 8); CRP (n = 107); Albumin (n = 16)
In total, 26 patients received curative operations, 5 of whom were given adjuvant therapy with 5-FU-based chemotherapy and/or concurrent chemoradiotherapy; all patients experienced recurrence. As for palliative chemotherapy, gemcitabine had been used in the first-, second-, third-, or later-line therapy in 121, 39, 6, and 6 patients, respectively.
IHC expression of MDM2 and p53 versus clinical characteristics
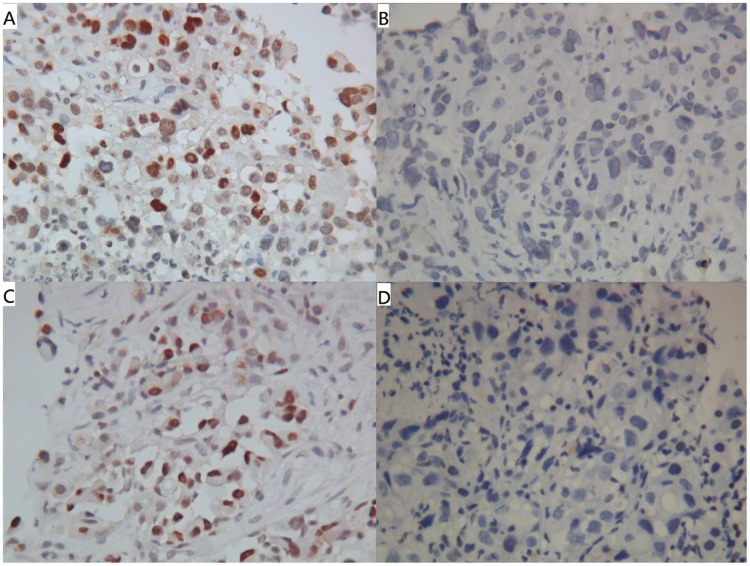
Nuclear or cytoplasmic expression of MDM2 and p53 was found in tumor cells of 30 (21.9%) and 71 (51.8%) cases, respectively (Fig 1A, 1B, 1C and 1D). The associations of MDM2 and p53 expression with patient clinical characteristics are presented in S2 Table. MDM2 or p53 expression was not significantly associated with any clinical factors. The association between the expression of MDM2 and p53 was not significant (P = .215). The positive rates of MDM2 and p53 expression stratified by the status of curative surgery did not show significant difference (S3 Table).
Fig 1. Representative cases of IHC expression (magnification 400X).
Cases of IHC expression with (A) MDM2+, (B) MDM2-, (C) p53+, and (D) p53- were demonstrated. The positive staining was predominantly nuclear for both MDM2 and p53.
Prognostic analyses
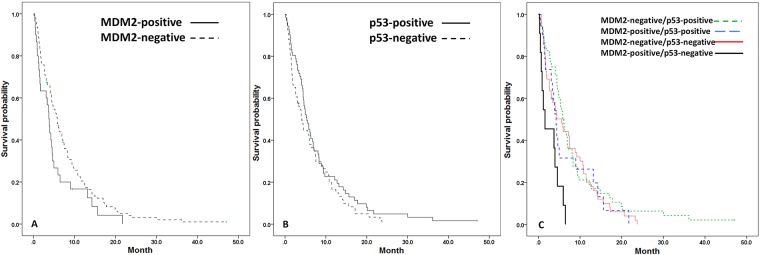
For the entire study group, patients with MDM2 expression had significantly poorer prognosis than those without MDM2 expression did as calculated from the start of a gemcitabine-based regimen (median OS = 3.7 vs 5.8 mo; P = .048) (Fig 2A). By contrast, p53 expression had no prognostic significance (median OS = 5.3 vs 4.1 mo; P = .192) (Fig 2B). After stratification of all the patients into four subgroups (MDM2+/p53-, MDM2+/p53+, MDM2-/p53+, and MDM2-/p53-), the median OS following the start of a gemcitabine-based regimen was 1.6, 4.2, 5.8, and 5.6 months, respectively (P = .003; Fig 2C); within the same subgroups, patients with unresectable diseases (n = 111) demonstrated a median OS of 0.9, 4.2, 5.8, and 7.4 months (P = .001), respectively, following the start of a gemcitabine-based regimen. The association between MDM2 and poor OS was similar irrespective of surgery status (S4 Table).
Fig 2. Survival curves in patients stratified with MDM2 and p53.
The OS curves stratified with (A) MDM2 expression, (B) p53 expression, and (C) MDM2 and p53 statuses were demonstrated. The dots represented censored observation. The OS was worse in patients with MDM2+ IHC staining (P = .048). The OS did not differ significantly between p53+ and p53- patients (P = .192). After stratification of MDM2 and p53 status, patients with MDM2+/p53- staining had the shortest OS (P = .003).
In patients with stage III or IV pancreatic cancer and receiving first-line gemcitabine monotherapy (n = 36), MDM2 but not p53 was a poor prognostic factor (S5 Table).
In addition to MDM2 expression, age (P = .032), ECOG PS (P < .001), initial carcinoembryonic antigen (CEA) level (P = .024), and initial albumin level (P = .038) were all significantly associated with median OS from the start of any gemcitabine-based regimen in the univariate analyses (Table 2). Notably, the poor prognostic factors in the univariate analysis were not associated with any MDM2 or p53 subgroup. Moreover, after the significant clinical characteristics were introduced into the multivariate analysis (Table 3), only ECOG PS (HR = 5.032; P < .001) and expression of MDM2 (HR = 1.731; P = .025) remained unfavorable prognostic factors for OS from the start of a gemcitabine-based regimen.
Table 2. Univariate analysis for OS.
| Characteristic | Value | Events | Median OS | SE* | P |
|---|---|---|---|---|---|
| Age (years) | ≥60 | 74 | 4.4 | 0.5 | 0.032 |
| <60 | 56 | 5.6 | 1.1 | ||
| Sex | Male | 77 | 5.0 | 0.9 | 0.395 |
| Female | 53 | 4.6 | 1.0 | ||
| ECOG PS | 0–1 | 104 | 6.0 | 0.8 | <0.001 |
| 2–3 | 26 | 1.6 | 0.1 | ||
| Stage | I/II/III | 47 | 5.8 | 0.6 | 0.191 |
| IV | 83 | 4.4 | 0.5 | ||
| T | 1–3 | 80 | 4.6 | 0.6 | 0.540 |
| 4 | 50 | 6.0 | 1.5 | ||
| N | 0 | 56 | 4.6 | 0.9 | 0.751 |
| 1 | 74 | 5.2 | 0.7 | ||
| Diabetes | No | 75 | 5.0 | 0.7 | 0.471 |
| Yes | 55 | 4.5 | 0.8 | ||
| Smoking | No | 89 | 5.6 | 0.6 | 0.756 |
| Yes | 41 | 4.2 | 0.3 | ||
| Primary | Tail | 28 | 4.6 | 1.3 | 0.190 |
| Others | 102 | 5.0 | 0.5 | ||
| Differentiation | Poor | 43 | 4.0 | 0.7 | 0.057 |
| Good/Moderate | 87 | 5.8 | 0.5 | ||
| CA 19–9 (U/mL) | ≥500 | 65 | 4.4 | 0.3 | 0.117 |
| <500 | 57 | 5.8 | 0.5 | ||
| CEA (ng/mL) | ≥3 | 71 | 4.6 | 0.3 | 0.024 |
| <3 | 46 | 5.6 | 1.5 | ||
| CRP (mg/dL) | ≥1.5 | 18 | 3.6 | 0.8 | 0.057 |
| <1.5 | 11 | 5.8 | 4.0 | ||
| Albumin (g/dL) | ≥4 | 83 | 6.0 | 0.8 | 0.038 |
| <4 | 31 | 3.4 | 0.6 | ||
| MDM2 | positive | 29 | 3.7 | 0.4 | 0.048 |
| negative | 101 | 5.8 | 0.6 | ||
| p53 | positive | 67 | 5.3 | 0.7 | 0.192 |
| negative | 63 | 4.1 | 0.6 | ||
| MDM2/p53 | +/+ (N = 19) | 18 | 4.2 | 0.5 | 0.003 |
| +/- (N = 11) | 11 | 1.6 | 1.6 | ||
| -/+ (N = 52) | 49 | 5.8 | 0.6 | ||
| -/- (N = 55) | 52 | 5.6 | 1.1 |
*SE: standard error
Table 3. Multivariate analysis for OS.
| Status | HR* (95% CI) |
P | ||
|---|---|---|---|---|
| Characteristic | Unfavourable | Favourable | ||
| Age (years) | ≥60 | <60 | 1.493 (0.967–2.304) |
0.070 |
| ECOG PS | 2–3 | 0–1 | 5.032 (2.687–9.421) |
<0.001 |
| CEA (ng/mL) | ≥3 | <3 | 1.455 (0.921–2.299) |
0.108 |
| Albumin (g/dL) | <4 | ≥4 | 0.989 (0.583–1.680) |
0.968 |
| MDM2 | positive | negative | 1.731 (1.070–2.798) |
0.025 |
*HR: hazard ratio
IHC expression of MDM2 and p53 versus chemotherapy outcomes
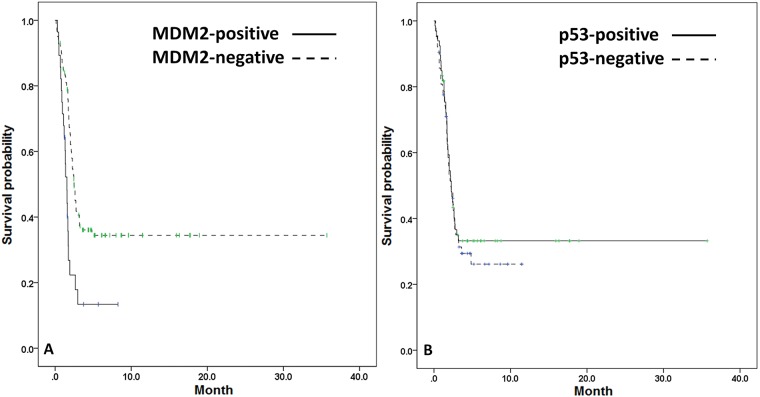
We analyzed the association of best response to gemcitabine-based regimens and the expression of MDM2 and p53, but no significant association was observed in the entire study population (Table 4). The median PFS after initiation of any gemcitabine-based therapy in the entire study population was 2.3 months; furthermore, MDM2 expression was significantly associated with shorter median PFS (positive vs negative = 1.5 vs 2.5 mo; P < .001; Fig 3A) but p53 expression was not (positive vs negative = 2.3 vs 2.2 mo; P = .630; Fig 3B).
Table 4. MDM2 and p53 status versus chemotherapy response and outcome.
| MDM2 | P | p53 | P | |||
|---|---|---|---|---|---|---|
| + | - | + | - | |||
| Best response (n = 107*) | 0.601 | 0.561 | ||||
| CR/PR/SD¶ | 6 | 38 | 25 | 19 | ||
| PD¶ | 12 | 51 | 32 | 31 | ||
| Progression | ||||||
| Progression-free survival (month) | ||||||
| All patients (n = 129*) | 1.5 | 2.5 | <0.001 | 2.3 | 2.2 | 0.630 |
| 1st line (n = 114*) | 1.4 | 2.5 | <0.001 | 2.1 | 2.2 | 0.940 |
| 2nd line (n = 37*) | 1.7 | 3.2 | 0.279 | 2.2 | 3.6 | 0.619 |
| 1st line (n = 114*) | 0.015 | 1.000 | ||||
| No progression (CR/PR/SD) ¶ | 3 | 34 | 19 | 18 | ||
| Progression (PD/death) ¶ | 22 | 55 | 38 | 39 | ||
| 2nd line (n = 37*) | 0.660 | 0.517 | ||||
| No progression (CR/PR/SD) ¶ | 2 | 16 | 8 | 10 | ||
| Progression (PD/death) ¶ | 4 | 15 | 11 | 8 | ||
*evaluable patients
¶CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease
Fig 3. PFS curves in patients stratified with MDM2 and p53.
The PFS curves were stratified with (A) MDM2 expression and (B) p53 expression. The median PFS was worse in patients with MDM2+ IHC staining (P < .001). The median PFS did not differ significantly between p53+ and p53- patients (P = .630). The dots represented censored observation.
We also stratified the patients according to their progression through gemcitabine-based therapy. MDM2 was significantly associated with progression after first-line gemcitabine-based therapy (P = .015) (Table 4).
Discussion
In this study, MDM2 was determined to be a prognostic factor for poor prognosis and progression under gemcitabine-based chemotherapy in addition to other poor prognostic factors identified in a previous study, such as old age, poor ECOG PS, high CEA level, and low albumin level [30]. Although it was not clearly linked to any baseline characteristics associated with poor prognosis, MDM2 had borderline significance associated with negative regional lymph node involvement, which is generally a favorable prognostic factor. However, the lymph node status was not indicative of prognostic significance, regardless of curative resection. Previous studies have suggested that E-cadherin is a target for MDM2-mediated ubiquitination and degradation in breast cancer cells [31]; additionally, overexpression of MDM2 can inhibit cell—cell contact and increase cell motility [31]. Thus, although MDM2 may mediate distant nonregional lymph node metastasis through the downregulation of E-cadherin, the poor efficacy of systemic chemotherapy in the palliative setting actually outweighs the significance of regional lymph node metastasis.
The percentages of MDM2 and p53 IHC expression in this study were similar to those from previous reports [19, 21, 22]. Under ordinary conditions, MDM2 and p53 form a negative regulation loop [23]. MDM2 expression has been activated through the Ras—Raf—MEK pathway [16, 32], but the inverse relationship between MDM2 and p53 levels has not been observed in our study and in pancreatic cancer cell lines with mutant p53 [16]. Although MDM2 regulates the stability of mutant p53 in transgenic animal models [33], it also ubiquitinates mutant p53 less efficiently [34]. Therefore, both active Ras signaling and p53 mutation may partially contribute to the protection and decoupling of mutant p53 from MDM2-mediated degradation.
Furthermore, we determined that MDM2 expression, but not p53 expression, was associated with disease progression, poor PFS, and poor OS after gemcitabine-based chemotherapy. Notably, the shortest OS was observed in patients with MDM2+/p53- expression. Previous studies have also indicated that mutant p53 is associated with short OS, irrespective of curative resection [35, 36]. Recently, Fiorini et al observed that CDK1 and CCNB1 were induced after gemcitabine treatment in PANC1 cells expressing mutant p53 protein; however, they also noted that the effects were reversed after the downregulation of mutant p53 [37]. Conversely, in AsPC1 cells without expression of mutant p53 protein, the induction of CDK1 and CCNB1 expression occurred after the transfection of the mutant p53 (i.e., R273H) plasmid [37]. Therefore, the function gain that accompanies p53 mutation not only reverses cell cycle inhibition of wild-type p53 but also induces chemoresistance to gemcitabine in pancreatic adenocarcinoma cells.
In addition, p53 expression is not correlated with mutational status; this is also true of PANC1 and AsPC1 cells [37]. Because we did not incorporate p53 mutation analysis into the present study, we could not deduce the p53 mutation status of individual patients from the p53 IHC expression data. The four subgroups stratified by MDM2 and p53 IHC expression were not associated with poor prognostic factors. Notably, the two subgroups with extreme OS difference had opposite status of MDM2 and p53 expression. Therefore, we can assume that the balance between MDM2 and p53 mediates the tumor aggressiveness; as prior research similarly revealed, the downregulation of MDM2 in SW1990HM pancreatic adenocarcinoma cells increases levels of E-cadherin and decreases levels of matrix metallopeptidase 9 and Ki-67 [38]. Downregulation and induction of the autoubiquitination of MDM2 with SP141 inhibit pancreatic adenocarcinoma both in vitro and in vivo. Additionally, the induction of apoptosis, p21, and Bax, accompanied by a reduction of cyclin E and Bcl-2, occurs after SP141 treatment [39]. Although MDM2 amplification is among the mechanisms of MDM2 expression in specific malignancies [40], a typical pattern of the phenomenon was not found in this study (S2 Fig). Therefore, MDM2 amplification was not the major mechanism of MDM2 expression in our patient population, which aligns with previous studies reporting rare MDM2 amplification in pancreatic adenocarcinoma [41].
There were some missing data in Table 1 due to the limitation of retrospective study. Most patients did not have baseline data of CRP. Although the level of CRP may be associated with prognosis, it was not a routine test at the diagnosis of pancreatic cancer. The distribution of baseline characteristics, MDM2 and p53 expression was similar among the entire study population and subjects without missing data of CEA, CA 19–9, polymorphonuclear granulocyte (PMN) count, monocyte count, and lymphocyte count (S6 Table). In addition, the multivariate analysis of the subjects without missing data (S7 Table) was comparable to the original analysis (Table 3).
Conclusions
In summary, MDM2 expression is associated with poor prognosis and progression after gemcitabine-based chemotherapy in advanced pancreatic adenocarcinoma. The major limitation of our study was the heterogeneous patient population, comprising patients both with and without curative resection. However, all of these patients had been previously treated with gemcitabine. To the best of our knowledge, this is the first clinical study to evaluate the association of chemotherapy with MDM2 in pancreatic cancer. Future basic or clinical studies applying chemotherapy and MDM2-targeted therapy with a non-p53 dependent mechanism are warranted.
Supporting information
The process of patient selection was demonstrated.
(TIFF)
FISH patterns in the three patients with polysomy of chromosome 12 with concomitant increase numbers of the centromere and mdm2 staining [27] were demonstrated; red = MDM2; green = centromere 12.
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(XLS)
Acknowledgments
Parts of the data in this manuscript were presented in the 2015 International Symposium of Cancer Center of Excellence: Frontiers on Cancer Research and Treatment, Taipei, Taiwan.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by MOHW103-TD-B-111-04 and MOHW104-TD-B-111-04, Ministry of Health and Welfare (http://www.mohw.gov.tw/EN/Ministry/).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2): 87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Oettle H, Neuhaus P, Hochhaus A, Hartmann JT, Gellert K, Ridwelski K, et al. Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: the CONKO-001 randomized trial. JAMA. 2013;310(14): 1473–1481. doi: 10.1001/jama.2013.279201 [DOI] [PubMed] [Google Scholar]
- 3.Burris HA 3rd, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15(6): 2403–2413. doi: 10.1200/JCO.1997.15.6.2403 [DOI] [PubMed] [Google Scholar]
- 4.Heinemann V, Quietzsch D, Gieseler F, Gonnermann M, Schönekäs H, Rost A, et al. Randomized phase III trial of gemcitabine plus cisplatin compared with gemcitabine alone in advanced pancreatic cancer. J Clin Oncol. 2006;24(24): 3946–3952. doi: 10.1200/JCO.2005.05.1490 [DOI] [PubMed] [Google Scholar]
- 5.Poplin E, Feng Y, Berlin J, Rothenberg ML, Hochster H, Mitchell E, et al. Phase III, randomized study of gemcitabine and oxaliplatin versus gemcitabine (fixed-dose rate infusion) compared with gemcitabine (30-minute infusion) in patients with pancreatic carcinoma E6201: a trial of the Eastern Cooperative Oncology Group. J Clin Oncol. 2009;27(23): 3778–3785. doi: 10.1200/JCO.2008.20.9007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ueno H, Ioka T, Ikeda M, Ohkawa S, Yanagimoto H, Boku N, et al. Randomized phase III study of gemcitabine plus S-1, S-1 alone, or gemcitabine alone in patients with locally advanced and metastatic pancreatic cancer in Japan and Taiwan: GEST study. J Clin Oncol. 2013;31(13): 1640–1648. doi: 10.1200/JCO.2012.43.3680 [DOI] [PubMed] [Google Scholar]
- 7.Conroy T, Desseigne F, Ychou M, Bouché O, Guimbaud R, Bécouarn Y, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364(19): 1817–1825. doi: 10.1056/NEJMoa1011923 [DOI] [PubMed] [Google Scholar]
- 8.Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18): 1691–1703. doi: 10.1056/NEJMoa1304369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei CH, Gorgan TR, Elashoff DA, Hines OJ, Farrell JJ, Donahue TR. A meta-analysis of gemcitabine biomarkers in patients with pancreaticobiliary cancers. Pancreas. 2013;42(8): 1303–1310. doi: 10.1097/MPA.0b013e3182a23ae4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540): 495–501. doi: 10.1038/nature14169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang P, Chubb S, Hertel LW, Grindey GB, Plunkett W. Action of 2',2'-difluorodeoxycytidine on DNA synthesis. Cancer Res. 1991;51(22): 6110–6117. [PubMed] [Google Scholar]
- 12.Achanta G, Pelicano H, Feng L, Plunkett W, Huang P. Interaction of p53 and DNA-PK in response to nucleoside analogues: potential role as a sensor complex for DNA damage. Cancer Res. 2001;61(24): 8723–8729. [PubMed] [Google Scholar]
- 13.Galmarini CM, Clarke ML, Falette N, Puisieux A, Mackey JR, Dumontet C. Expression of a non-functional p53 affects the sensitivity of cancer cells to gemcitabine. Int J Cancer. 2002;97(4): 439–445. doi: 10.1002/ijc.1628 [DOI] [PubMed] [Google Scholar]
- 14.Cascalló M, Calbó J, Capellà G, Fillat C, Pastor-Anglada M, Mazo A. Enhancement of gemcitabine-induced apoptosis by restoration of p53 function in human pancreatic tumors. Oncology. 2005;68(2–3): 179–189. doi: 10.1159/000086772 [DOI] [PubMed] [Google Scholar]
- 15.Redston MS, Caldas C, Seymour AB, Hruban RH, da Costa L, Yeo CJ, et al. p53 mutations in pancreatic carcinoma and evidence of common involvement of homocopolymer tracts in DNA microdeletions. Cancer Res. 1994;54(11): 3025–3033. [PubMed] [Google Scholar]
- 16.Sui X, Shin S, Zhang R, Firozi PF, Yang L, Abbruzzese JL, et al. Hdm2 is regulated by K-Ras and mediates p53-independent functions in pancreatic cancer cells. Oncogene. 2009;28(5): 709–720. doi: 10.1038/onc.2008.423 [DOI] [PubMed] [Google Scholar]
- 17.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362(6423): 857–860. doi: 10.1038/362857a0 [DOI] [PubMed] [Google Scholar]
- 18.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420(1): 25–27. doi: 10.1016/S0014-5793(97)01480-4 [DOI] [PubMed] [Google Scholar]
- 19.Ruggeri BA, Huang L, Berger D, Chang H, Klein-Szanto AJ, Goodrow T, et al. Molecular pathology of primary and metastatic ductal pancreatic lesions: analyses of mutations and expression of the p53, mdm-2, and p21/WAF-1 genes in sporadic and familial lesions. Cancer. 1997;79(4): 700–716. doi: 10.1002/(SICI)1097-0142(19970215)79:4<700::AID-CNCR7>3.0.CO;2-H [PubMed] [Google Scholar]
- 20.Gerdes B, Ramaswamy A, Ziegler A, Lang SA, Kersting M, Baumann R, et al. p16INK4a is a prognostic marker in resected ductal pancreatic cancer: an analysis of p16INK4a, p53, MDM2, an Rb. Ann Surg. 2002;235(1): 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong M, Ma G, Tu W, Guo KJ, Tian YL, Dong YT. Clinicopathological significance of p53 and mdm2 protein expression in human pancreatic cancer. World J Gastroenterol. 2005;11(14): 2162–2165. doi: 10.3748/wjg.v11.i14.2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermanova M, Karasek P, Nenutil R, Kyr M, Tomasek J, Baltasova I, et al. Clinicopathological correlations of cyclooxygenase-2, MDM2, and p53 expressions in surgically resectable pancreatic invasive ductal adenocarcinoma. Pancreas. 2009;38(5): 565–571. doi: 10.1097/MPA.0b013e31819fef8b [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Wang H, Li M, Agrawal S, Chen X, Zhang R. MDM2 is a negative regulator of p21WAF1/CIP1, independent of p53. J Biol Chem. 2004;279(16): 16000–16006. doi: 10.1074/jbc.M312264200 [DOI] [PubMed] [Google Scholar]
- 24.Martin K, Trouche D, Hagemeier C, Sørensen TS, La Thangue NB, Kouzarides T. Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature. 1995;375(6533): 691–694. doi: 10.1038/375691a0 [DOI] [PubMed] [Google Scholar]
- 25.Gu L, Zhang H, Liu T, Zhou S, Du Y, Xiong J, et al. Discovery of Dual Inhibitors of MDM2 and XIAP for Cancer Treatment. Cancer Cell. 2016;30(4): 623–636. doi: 10.1016/j.ccell.2016.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng X, Chen L, Jost CA, Maya R, Keller D, Wang X, et al. MDM2 suppresses p73 function without promoting p73 degradation. Mol Cell Biol. 1999;19(5): 3257–3266. doi: 10.1128/MCB.19.5.3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu L, Findley HW, Zhou M. MDM2 induces NF-kappaB/p65 expression transcriptionally through Sp1-binding sites: a novel, p53-independent role of MDM2 in doxorubicin resistance in acute lymphoblastic leukemia. Blood. 2002;99(9): 3367–3375. https://doi.org/10.1182/blood.V99.9.3367 [DOI] [PubMed] [Google Scholar]
- 28.Lee JC, Fletcher CD. Malignant fat-forming solitary fibrous tumor (so-called "lipomatous hemangiopericytoma"): clinicopathologic analysis of 14 cases. Am J Surg Pathol. 2011;35(8): 1177–1185. doi: 10.1097/PAS.0b013e318219cd0b [DOI] [PubMed] [Google Scholar]
- 29.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2): 228–247. doi: 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 30.Yang SH, Guo JC, Yeh KH, Tien YW, Cheng AL, Kuo SH. Association of radiotherapy with favorable prognosis in daily clinical practice for treatment of locally advanced and metastatic pancreatic cancer. J Gastroenterol Hepatol. 2016;31(12): 2004–2012. doi: 10.1111/jgh.13395 [DOI] [PubMed] [Google Scholar]
- 31.Yang JY, Zong CS, Xia W, Wei Y, Ali-Seyed M, Li Z, et al. MDM2 promotes cell motility and invasiveness by regulating E-cadherin degradation. Mol Cell Biol. 2006;26(19): 7269–7282. doi: 10.1128/MCB.00172-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ries S, Biederer C, Woods D, Shifman O, Shirasawa S, Sasazuki T, et al. Opposing effects of Ras on p53: transcriptional activation of mdm2 and induction of p19ARF. Cell. 2000;103(2): 321–330. http://dx.doi.org/10.1016/S0092-8674(00)00123-9 [DOI] [PubMed] [Google Scholar]
- 33.Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, et al. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 2008;22(10): 1337–1344. doi: 10.1101/gad.1662908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukashchuk N, Vousden KH. Ubiquitination and degradation of mutant p53. Mol Cell Biol. 2007;27(23): 8284–8295. doi: 10.1128/MCB.00050-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamori S, Yashima K, Murakami Y, Ishikawa O, Ohigashi H, Imaoka S, et al. Association of p53 gene mutations with short survival in pancreatic adenocarcinoma. Jpn J Cancer Res. 1995;86(2): 174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weyrer K, Feichtinger H, Haun M, Weiss G, Ofner D, Weger AR, et al. p53, Ki-ras, and DNA ploidy in human pancreatic ductal adenocarcinomas. Lab Invest. 1996;74(1): 279–289. [PubMed] [Google Scholar]
- 37.Fiorini C, Cordani M, Padroni C, Blandino G, Di Agostino S, Donadelli M. Mutant p53 stimulates chemoresistance of pancreatic adenocarcinoma cells to gemcitabine. Biochim Biophys Acta. 2015;1853(1): 89–100. doi: 10.1016/j.bbamcr.2014.10.003 [DOI] [PubMed] [Google Scholar]
- 38.Shi W, Meng Z, Chen Z, Hua Y, Gao H, Wang P, et al. RNA interference against MDM2 suppresses tumor growth and metastasis in pancreatic carcinoma SW1990HM cells. Mol Cell Biochem. 2014;387(1–2): 1–8. doi: 10.1007/s11010-011-1208-4 [DOI] [PubMed] [Google Scholar]
- 39.Wang W, Qin JJ, Voruganti S, Wang MH, Sharma H, Patil S, et al. Identification of a new class of MDM2 inhibitor that inhibits growth of orthotopic pancreatic tumors in mice. Gastroenterology. 2014;147(4): 893–902. doi: 10.1053/j.gastro.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimura H, Dobashi Y, Nojima T, Nakamura H, Yamamoto N, Tsuchiya H, et al. Utility of fluorescence in situ hybridization to detect MDM2 amplification in liposarcomas and their morphological mimics. Int J Clin Exp Pathol. 2013;6(7): 1306–1316. [PMC free article] [PubMed] [Google Scholar]
- 41.Birnbaum DJ, Adélaïde J, Mamessier E, Finetti P, Lagarde A, Monges G, et al. Genome profiling of pancreatic adenocarcinoma. Genes Chromosomes Cancer. 2011;50(6): 456–465. doi: 10.1002/gcc.20870 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The process of patient selection was demonstrated.
(TIFF)
FISH patterns in the three patients with polysomy of chromosome 12 with concomitant increase numbers of the centromere and mdm2 staining [27] were demonstrated; red = MDM2; green = centromere 12.
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.