Abstract
Embryonic stem cells (ESCs), due to their intrinsic capability to generate somatic cells of all three germ layers, are the potential sources of neural cells for cell replacement therapies. However, the empirical differentiation protocols and the lack of mechanistic understanding of the neural differentiation of ESCs has limited the utility of ESCs as a developmental model or as a cell source for neural cell population for replacement therapies. Coculturing ESCs with stromal cells is one of the extensively used methods to induce neural differentiation. Despite several studies attempting to identify neural inducing factors in stromal cell induced neural differentiation, self-regulatory effects of ESCs in the neural differentiation process remains unexplored. For the first time, we elucidate the self-regulatory role of mESCs on their neural cell differentiation by supplementing conditioned media from differentiating mESCs to the mESCs-PA6 cocultures and quantitatively evaluating the change in neural differentiation. Moreover, we use statistical tools to analyze the expressions of various growth and trophic factors and distinguish the factors produced primarily by PA6 cells versus mESCs in coculture. We observed that addition of the medium containing mESCs-secreted factors to the single mESCs colony cocultured with PA6 cells significantly enhanced the neural differentiation of mESCs compared to the medium extracted from the stromal cells only. Hierarchical clustering of gene expression data from PA6 and cocultured mESCs segregated two group of factors that are produced by the stromal cells and differentiating mESCs. Identifying the major soluble factors that drive and regulate the neural differentiation process in mESCs-PA6 coculture niche will help understand the molecular mechanisms of neural development. Moreover, it can be the first step toward developing novel protocols to differentiate stem cells with mESCs derived factors supplementation without using feeder cells with greater efficiency compared to existing approaches.
Keywords: embryonic stem cells, self-regulating factors, stromal signaling, neural differentiation, co-culture
Graphical Abstract
INTRODUCTION
Embryonic stem cells (ESCs) provide a potential cell source for neural tissue repair and cell replacement therapies due to their infinite self-renewal, expansion, and multi-lineage differentiation potential.1,2 In addition, ESCs enable in vitro modeling of embryonic organogenesis to help understand normal brain development and neural cell diseases.3,4 There are several methods to derive neural cells from ESCs. ESCs may be cultured as compact, three-dimensional (3D) aggregates known as embryoid bodies and differentiated using specific chemicals such as retinoic acid.5 However, differentiation of ESCs with the embryoid body culture is not exclusive to neural cells and mesodermal and endodermal cells also occur.6 ESCs cultured as a monolayer in defined media supplemented with specific growth factors also undergo neural differentiation but with a relatively low efficiency. Subsequent growth of differentiated cells followed by dissociation and re-plating on adherent surfaces is required to derive neuronal cells.7,8 Alternatively, neural differentiation can result from co-culturing of ESCs with certain stromal cells.9,10 ESCs grow in clumps over the stromal cells layer and undergo differentiation without any additional differentiation-inducing agents or re-plating of differentiated cells. This method mimics intercellular signaling during embryonic neurogenesis and effectively results in dopaminergic neuronal differentiation of ESCs by soluble and surface-bound factors collectively referred to as “stromal cell derived inducing activity (SDIA)”.9 However, a major limitation of this method is the use of animal feeder cells to induce differentiation. Moreover, the underlying mechanism of differentiating ESCs is poorly understood. Hayashi et al. showed that conditioned medium of PA6 stromal cells induced moderate levels of dopaminergic neuronal differentiation of mouse ESCs.11 Swistowska et al. reported that conditioned medium of PA6 cells induced dopaminergic differentiation in neuronal precursors isolated from a human embryonic carcinoma cell line as well as neural stem cells derived from different human ESC lines.12 Vazin et al. observed that fixing or mitotically inactivating PA6 cells minimally affects the neural inducing effect of the stromal cells but significantly decreases differentiation to dopaminergic neurons.13 Stromal cell-derived factors such as SHH, FGF8, and Wnt5a were suggested to play a major role in SDIA-mediated neural differentiation of ESCs.12,14
Despite extensive use of the stromal PA6 cells-ESCs co-culture method, self-regulatory effects of ESCs in the neural differentiation process remains unexplored. Metabolites of ESCs contain soluble factors such as morphogens and activins that can directly influence differentiation of ESCs.15,16 This is supported by our previous study that standalone mESC colonies in co-culture with a monolayer of PA6 cells show disproportionately greater neural differentiation by increase in the mESC colony size.10 We hypothesize that in addition to SDIA, soluble factors secreted by differentiating mESCs additionally regulate the neural differentiation phenotype. Here, we conduct a comprehensive study to test this hypothesis through addition of mediua containing PA6-secreted factors only and both PA6 cells- and mESCs-secreted factors to the mESCs-PA6 cells co-cultures, and quantitative evaluation of neural differentiation of mESCs using protein and gene expression analyses. Considering that quantification of differentiation of randomly-seeded ESCs in the co-culture system is difficult and subjective, we use a non-contact cell microprinting technology to localize a defined number of mESCs over a layer of supporting stromal cells and allow the mESCs to form a single colony of defined size.10,17 This approach enables convenient imaging and quantification of protein expression of differentiated cells.
Our data indicate that addition of the medium containing mESCs-secreted factors to the co-culture of a single mESCs colony on PA6 cells significantly enhances neural differentiation of mESCs compared to the effect of the medium extracted from the stromal cells only. To elucidate the self-regulatory role of mESCs on their neural cell differentiation, we use statistical tools to analyze the expression of various growth and trophic factors in PA6 and mESCs. We identify two group of factors that are produced by the stromal cells only or primarily by the differentiating mESCs. Understanding molecular mechanisms of neural differentiation of ESCs in co-culture with stromal cells including identification of major soluble factors that drive and regulate this process will advance the development of ESCs-based in vitro model systems and cell replacement therapy strategies for neurodegenerative diseases, and may result in novel protocols to differentiate stem cells without using feeder cells but with greater efficiency compared to existing approaches.18
MATERIALS AND METHODS
Cell maintenance
Mouse embryonic stem cells, mESCs, (EB5, Riken) were cultured in a maintenance medium in flasks coated with a 0.1% gelatin solution (Sigma) for 1 hour. The maintenance medium consisted of Glasgow Minimum Essential Medium (GMEM, Life Technologies) supplemented with 10% knockout serum replacement (KSR, Life Technologies), 2 mM glutaMAX (Life Technologies), 1% fetal bovine serum, FBS (Sigma), 0.1 mM non-essential amino acids (NEAA, Life Technologies), 1 mM sodium pyruvate (Life Technologies), 0.1 mM 2-mercaptoethanol (Life Technologies), and 2000 units/ml leukemia inhibitory factor (Millipore). The cells were kept in a humidified incubator at 37°C and 5% CO2. The stromal PA6 cells (Riken), derived from newborn mouse calvaria, were cultured on gelatin-coated flasks in a medium consisting of αMEM (Life Technologies), 10% FBS (Sigma), and 1% antibiotic-antimycotic (Life Technologies). All experiments were conducted within the first ten passages of both cell types.
Feeder layer preparation for differentiation experiments
For preparation of a confluent stromal feeder layer, 4×104 and 4×103 PA6 cells were seeded into a gelatin-coated 35 mm Petri dish and per well of a 96-well plate, respectively. The selected density was to ensure that after two days, PA6 cells spread well on the culture surface, display a normal morphology, and form a confluent layer. At this point, the cells were mitotically arrested using 10 μg/ml mitomycin-c (Sigma) for 2 hrs. The mitomycin-c solution was removed and cells were washed three times with PBS. The mitotically inactivated PA6 cells were incubated overnight at 37°C and 5% CO2 in a standard medium (SM) containing GMEM, 10% KSR, 2 mM glutaMAX, 0.1 mM NEAA, 1 mM sodium pyruvate, and 0.1 mM 2-mercaptoethanol.
Printing of mESC colonies using an aqueous two-phase system technology
Two aqueous polymer solutions were prepared by dissolving polyethylene glycol (PEG, Mw: 35 kDa) (Sigma) and dextran (DEX, Mw: 500 kDa) (Pharmacosmos) in the SM at the concentrations of 5.0% (w/v) and 12.8% (w/v), respectively.19 mESCs were suspended in an equal volume of the SM and the DEX phase solution to yield a cell density of 104 cells/μl. This also reduced the polymer concentration to 6.4% (w/v). The suspension was transferred into wells of a 384-well plate (source plate). A liquid handler (SRT Bravo, Agilent) was used to load 50 nl of the suspension into hydrophobic slot pins (VP Scientific). The pins were then slowly lowered into a 35 mm Petri dish or a 96-well plate containing a monolayer of mitotically-inactivated PA6 cells immersed in the PEG phase solution. Pins were kept in the close vicinity of the PA6 layer to allow the aqueous DEX phase containing 500 mESCs autonomously dispense from each pin and form a drop on the monolayer of the PA6 cells. mESCs remained confined in the DEX phase drop and adhered to the PA6 cells layer. Polymeric solutions were replaced with the SM after 3 hrs of incubation. Printed mESCs proliferated to form an individual colony and differentiated over time. Colonies were cultured for 8 days in the SM and for an additional 6 days in the SM containing 1X N2 (Life Technologies). Media exchange was performed on days 4 and 6, and then every day until day 14.
Collection and supplementation of conditioned media
Mitotically-inactivated PA6 cells were kept in the SM in 35 mm Petri dishes to allow factors secreted by the PA cells accumulate. This culture was used as the source of stromal cells conditioned medium (SCM). In parallel, nine colonies of mESCs were generated on a layer of mitotically-inactivated PA6 cells and kept in the SM to allow soluble factors produced by the PA6 and mESCs accumulate. The resulting culture was used as the source for co-culture conditioned medium (CCM). Every day, 1 ml of the SCM and CCM was recovered from each Petri dish. The depleted medium that comprised one third of the total medium volume in each dish was replaced with fresh SM. The SCM and CCM were each separately supplemented to a single colony of mESCs on mitotically-inactivated PA6 cells in each well of a 96-well plate daily. The culture medium in each well was completely replaced with 100 μl of either SCM or CCM. In parallel, a negative control group, i.e., co-culture of a colony of mESCs on PA6 cells in a 96 well plate, received 100 μl of the SM. Media renewal in Petri dishes that were the sources for SCM and CCM was performed on days 4 and 6, and then every day until day 14.
Immunofluorescence and image analysis
Cells were fixed in 3.7% formaldehyde (Fisher Scientific) for 15 min followed by 3 washes with PBS, each for 5 min. The samples were blocked with a 5% donkey serum solution (Sigma) in PBS (Sigma) for one hour to prevent nonspecific binding. The samples were then incubated with a primary antibody solution at 4°C overnight. The primary antibody for TuJ was a rabbit monoclonal class III β-tubulin antibody (1:2000) (Biolegend). For Nestin, an affinity purified Nestin chicken antibody was used (1:200) (Neuromics). For glial fibrillary acidic protein (GFAP), an affinity purified chicken IgY (1:2000) (Neuromics) was used. Secondary antibodies to visualize protein expression were raised in donkey and fluorescently conjugated with Aminomethylcoumarin (AMCA) (1:100), Rhodamine red (1:100), and Alexa Fluor 488 (1:800) (Jackson ImmunoResearch). Sections of each immunostained mESC colony were imaged at 10X and merged (Adobe Photoshop CS) to generate a panorama of the entire colony. The exposure time for imaging of each protein was kept fixed. After subtracting the background signal from images using the “subtract background” tool in ImageJ (NIH), the expression of each protein was quantified by (i) measuring the net fluorescence intensity in each image and (ii) using adaptive thresholding in ImageJ. The adaptive thresholding technique involved conversion of images to black and white by calculating a threshold for each pixel based on its surrounding pixels. The resulting white pixels count corresponded to protein expression.
Quantitative real-time PCR (q-PCR) analysis
Cells were lysed using a solution of TRK lysis buffer (Omega Biotek) and mercaptoethanol (Sigma) at a 1:50 (v/v) ratio. The samples were homogenized by loading into homogenizer mini columns (Omega Biotek). RNA was isolated using a commercial kit (Omega Biotek). A RNase-free DNase kit was then used per the manufacturer’s instructions to remove DNase (Omega Biotek). The purity and concentration of RNA was determined using OD 260/280 spectrophotometry (Synergy H1M, Biotek instruments). cDNA was synthesized from 1 μg of total RNA using random hexamer primers according to the manufacturer’s instructions and a transcriptor reverse transcriptase cDNA synthesis kit (Roche). Reverse transcription reactions were incubated at 25°C for 10 min, at 55°C for 30 min, and at 85°C for 5 min. Resulting products were kept at 4°C.
q-PCR was performed on the resulting cDNA in a Roche LightCycler 480 II instrument using a SYBR Green Master Mix (Roche). Briefly, 50 ng of cDNA was combined with forward and reverse primers, and the SYBR green Master Mix was diluted to a final volume of 15 μl according to the manufacturer’s instructions. The reactions were pre-incubated at 95°C for 5 min followed by 45 cycles of amplification, i.e., at 95°C for 10 sec, at 60°C for 10 sec, and at 72°C for 10 sec. Melting curve analysis was conducted post-amplification at temperatures of 95°C for 5 sec and at 65°C for 1 min followed by continuous fluorescence reading at 97°C. Each sample was run in duplicates. Specific primer sequences for all the 25 genes in this study are listed in Table S1. Expression levels of mRNA for different marker genes were calculated relative to GAPDH and β-actin using the ΔΔCt method. Fold change in mRNA expression was determined as 2-ΔΔCt.19,20
Statistical analysis
All experiments were performed in triplicates for q-PCR and in duplicates for immunostaining. Statistical significance of protein expression levels was assessed using one-way ANOVA followed by Fisher’s LSD post hoc test in Minitab 16 software. Statistical significance of gene expression levels was assessed in Minitab using two-sample t-test performed on the -ΔΔCt (i.e., the log2 relative expression levels) values from days 4, 6, and 8 of cultures. Agglomerative hierarchical cluster analysis with the complete linkage method was also performed on the -ΔΔCt values on days 4, 6, and 8. This analysis helped segregate the factors expressed dominantly in stromal cells or differentiating mESCs. Agglomerative hierarchical cluster analysis was performed using the R statistical programming language.
RESULTS AND DISCUSSION
Microprinting of mESC to generate isolated colonies
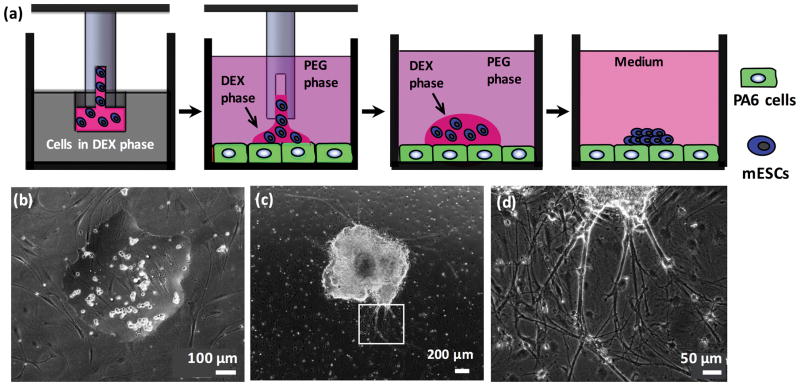
The aqueous two-phase system technology is a non-contact cell microprinting approach that uses biocompatible polymeric aqueous solutions to maintain viability and functionality of cells.10,17,20 mESCs microprinted onto a confluent layer of PA6 cells remained partitioned within the DEX phase drop due to an ultralow interfacial tension between the aqueous PEG and DEX phases.19,21 mESCs adhered to the stromal layer within 3 hrs of incubation (Fig 1a–b).19,21 The layer of mitotically inactive PA6 cells remained intact and viable during the 2-week culture period while mESCs rapidly proliferated and formed individual colonies (Fig 1c). A density of 500 cells per drop resulted in uniformly-sized colonies of 2.0±0.3mm in average diameter on day 8 of culture, irrespective of type of culture vessel. In addition to proliferation, differentiating mESCs migrated outward from the colonies,22,23 contributing to the measured size. Physical and biochemical interactions between mESCs and stromal cells were sufficient to induce neural cell differentiation, without a need for non-physiologic chemicals.24,25 Close examination of cultures showed thick neurite bundles and processes extending out from colonies (Fig 1d). A single colony was printed in a well of the 96-well plate and nine colonies with a center to center spacing of 9 mm were printed in a Petri dish.
Figure 1.
Aqueous two-phase system (ATPS)-mediated microprinting of mESCs to generate a single stem cells colony. (a) mESCs (blue) suspended in the aqueous DEX phase are aspirated using a slot pin from a microwell. The pin is immersed into the aqueous PEG phase and lowered close to the PA6 cells (green). The content of the pin dispenses onto the layer of PA6 cells in the aqueous PEG phase within a well of a 96-well plate or a Petri dish, forming a DEX phase drop containing mESCs. Stem cells attach to the PA6 cells and proliferate to form a colony. (b) Phase image of mESCs within a DEX phase drop on PA6 cells immersed in the aqueous PEG phase. (c) mESCs proliferate and form a single colony; the image shows a colony on day 8 of co-culture. (d) Magnified view from panel (c) shows that neurite processes emerge from differentiating mESCs in the colony; the underlying PA6 cells remain completely intact during the two-week co-culture.
Soluble factors effect on neural differentiation of mESCs
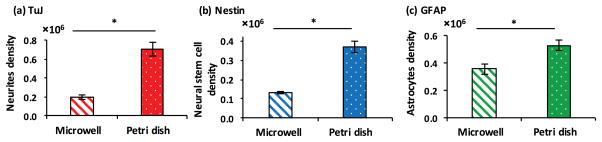
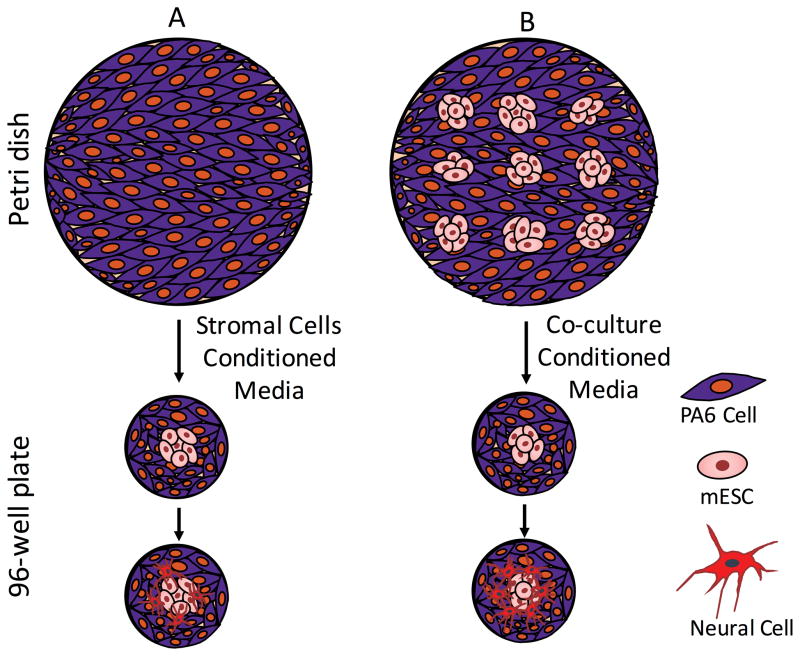
Despite the use of similar co-cultures, a single colony of mESCs in a well of a 96-well plate showed significantly lower neural differentiation compared to each of the nine isolated colonies in a Petri dish. We confirmed this through quantification of neural cell markers TuJ, Nestin, and GFAP (Fig 2a–c). The small diameter of a single colony (2.0±0.3 mm) compared to wells of 96-well plates (6.85 mm) excludes the possibility of limited space as the cause of poor neural differentiation and neurites extensions in microwells. Therefore, we hypothesized that differences in the total number of PA6 cells and/or mESCs between the two culture platforms and resulting difference in concentrations of cell-secreted soluble factors cause this effect. Previous studies have shown that soluble factors produced by PA6 cells induce neural differentiation of mESCs.9 However, the role of mESCs-derived soluble factors is unknown. To test the hypothesis, we harvested conditioned media from a culture of PA6 cells only in a Petri dish (SCM) and from the co-culture of nine colonies of mESCs on a layer of PA6 cells in a Petri dish (CCM), and investigated effects of these media on neural differentiation of a single colony of mESCs in co-culture with PA6 cells in 96-well plates. All cultures used mitotically-inactivated PA6 cells. This design is schematically shown in Fig 3. We maintained the cultures for two weeks with daily complete renewal with fresh SCM and CCM, and immunostained the mESCs on days 8 and 14 for protein expression analysis of specific neural cell markers.
Figure 2.
Quantified protein expression of differentiated mESCs from immunostaining for (a) TuJ, (b) Nestin, and (c) GFAP. Each panel compares differentiation from a single mESC colony in co-culture with feeder PA6 cells in a microwell and each of nine isolated colonies of mESCs in co-culture with feeder PA6 cells in a Petri dish. *p<0.01. N=20.
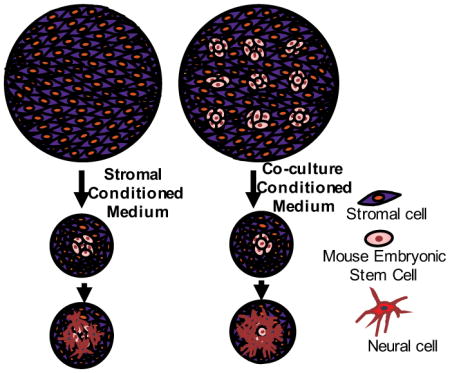
Figure 3.
Schematic representation of the experimental protocol. (A) Addition of stromal cells derived medium (SCM) from a Petri dish to a culture of a single colony of mESCs on PA6 cells in a well of 96-well plates moderately increases neural differentiation. (B) Addition of co-culture derived medium (CCM) from a Petri dish to a culture of a single colony of mESCs on PA6 cells in a well of 96-well plates Significantly enhances neural differentiation.
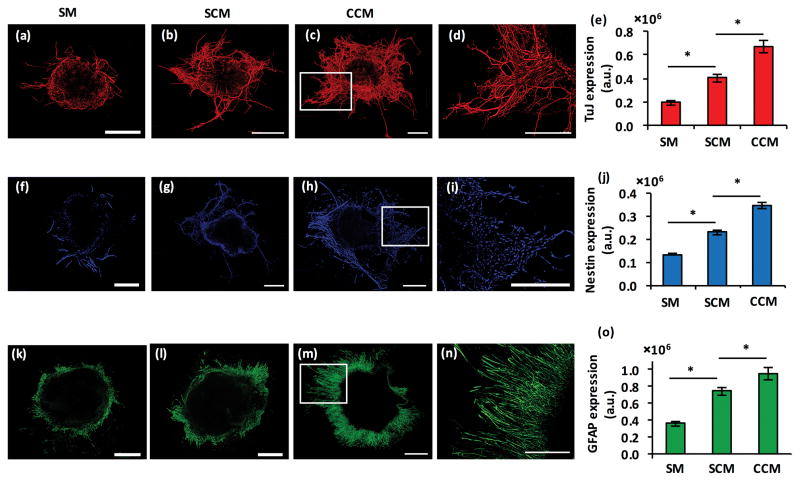
Addition of both SCM and CCM enhanced TuJ expression of differentiated mESCs by 2.0 and 3.3 folds, respectively (Fig 4a–e). Importantly, CCM addition enhanced TuJ expression of a single colony in the microwell to the level of nine colonies in co-culture with PA6 cells (cf. Fig 4e and 2a), suggesting that the added soluble factors of stromal and differentiating stem cells synergistically amplify the neural cell marker expression. Similar results were observed with the neural stem cell marker, Nestin. Both SCM and CCM significantly enhanced Nestin expression and caused a 1.7-fold and 2.6-fold increase in the protein levels, respectively (Fig 4f–j). The use of CCM with the single colony in a microwell increased Nestin expression to that of each of the nine colonies in co-culture with PA6 cells (cf. Fig 4j and 2b). The expression of a specific marker of astrocytes, GFAP, also showed enhanced protein expression by both conditioned media (Fig 4k–o). SCM and CCM addition to the single mESC colony in a microwell increased GFAP expression by 2.1 and 2.7 folds, respectively. Interestingly, both SCM and CCM elevated GFAP levels greater than that in the nine isolated colonies (cf. Fig 4o and 2c), indicating the important role of soluble factors produced by PA6 cells and differentiating stem cells on amplifying astrocytic differentiation. These results were also confirmed using total fluorescence signal measurements of immunostained samples (Fig S1).
Figure 4.
(a–c) TuJ-stained colonies of mESCs in co-culture with PA6 cells in a microwell under treatment with SM, SCM, and CCM, respectively. (d) Magnified view of neurite processes from the panel (c) image. (e) Quantified neurites pixel density is computed from TuJ+ mESC colonies. Data represent day 8 co-cultures. (f–h) Nestin staining of a single mESC colony in co-culture with PA6 cells in a microwell under treatment with SM, SCM, and CCM, respectively. (i) Magnified view of the boxed portion of the panel (h) image. (j) Pixel density computed from Nestin+ mESC colonies under different treatment conditions. Data represent day 8 cultures. (k–m) GFAP staining of a single mESC colony in co-culture with PA6 cells in a microwell under treatment with SM, SCM, and CCM, respectively. (n) Magnified view of the boxed portion of the panel (m) image. (o) Pixel density computed from GFAP+ mESC colonies under different treatment conditions. Data represent day 14 cultures. Scale bar is 500 μm. *p<0.01. The number of replicates for each treatment was N=20.
Gene expression profiling of growth factors that mediate neural differentiation of mESCs
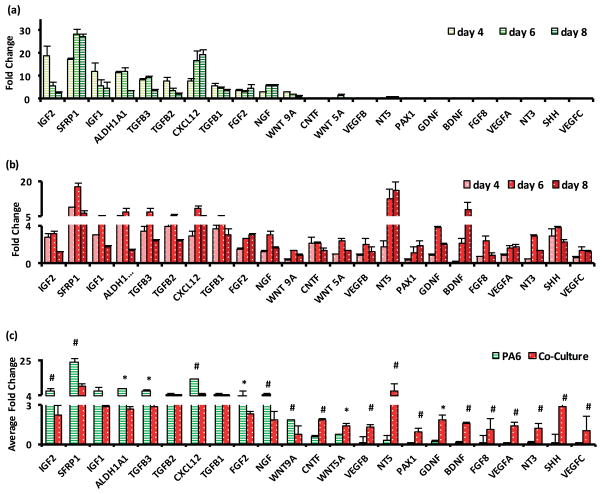
Our protein expression study indicates that soluble factors of stromal cells significantly enhance neural cell differentiation of mESCs, and that endogenous soluble factors of mESCs further amplify this phenotype. The main goal of this study was to discriminate between contributions of factors in SCM and CCM. Our preliminary study showed that ELISA assays were not sufficiently sensitive to detect all the soluble factors in the SCM and CCM; therefore, we evaluated gene level activities of 23 soluble factors (Table S1) in the cultures from which SCM and CCM were derived. These factors were selected for their known roles in ESCs differentiation and based on a thorough literature survey. For example, neurotrophins such as NT3 and NT5 enhance survival and neural differentiation of ESCs,26,27 ciliary neurotrophic factor (CNTF) and sonic hedgehog (SHH) promote stem cells survival and differentiation into specific neural cell lineages,12,14,28 and basic fibroblast growth factor-2 (FGF2) promotes generation of neural stem cells.29,30 We performed qPCR analysis with samples retrieved from mitotically-inactive PA6 cells only in a Petri dish, from co-cultures of nine isolated colonies of mESCs with PA6 cells in a Petri dish, and from a single colony of mESCs in co-culture with PA6 cells in a 96-well plate as control. Based on our previous work that showed the onset of neural differentiation of mESCs in this co-culture system was day 4, we harvested samples on days 4, 6, and 8 of cultures. Fold change in mRNA expression levels for different genes was calculated relative to the single colony in a 96-well plate, using the ΔΔCt method.
Our qPCR data showed that IGF2 and SFRP1 had an impressive 19- and 17-fold greater expression in PA6 cells on day 4 (Fig 5a). These factors also had larger expression in the co-culture group but only by 2.8 folds for IGF2 and 8.9 folds for SFRP1 (Fig 5b). Multiple factors with significant gene expression in PA6 cells on day 4 included IGF1, ALDHA1, TGF β1, TGF β2, TGF β3, CXCL12, FGF2, NGF, and WNT9A, all with greater than at least two folds. These factors also showed higher than 2-fold expression in the co-culture group on day 4; however, their expression levels always remained lower than that in the PA6 group (cf. Fig 5a and 5b). Considering that there are about the same number of PA6 cells in both cultures, these data indicates that PA6 cells produce the resulting growth factors.
Figure 5.
Fold change of mRNA levels of factors in (a) PA6 cells only and (b) co-culture of mESC colonies and PA6 cells in a Petri dish. (c) Average fold change over three days of measurements for each factor. * represents p < 0.05, # represents p < 0.01. The number of replicates was N=3.
Close scrutiny of Fig 5a shows that the expression levels of IGF1, IGF2, ALDHA1, TGF β1, TGF β2, TGF β3, and WNT9A in the PA6 group significantly decreased with time from day 4 to day 8. This result implies that PA6 cells-derived factors have a dominant role in survival of stem cells and guiding them toward a neural lineage. This is consistent with previous observations by Parmar et al. that stromal cell derived inducing activity was confined to a period before the expression of Sox1, which peaked on day 7 in their study. Therefore, PA6 cells primarily induce neural commitment of stem cells and have a limited role during the later phase of neuronal differentiation of neural precursor cells.31,32 Considering the crucial roles of IGFs, TGF β, and Wnt signaling in maintaining ESCs in a proliferative state and driving cell fate determination, PA6 cells-secreted factors support these activities. We note that despite reduced expression levels, these factors also support differentiation of ESCs to specific neural cells. For example, IGF1 regulates differentiation and maturation of neurons from neural stem and progenitor cells during embryonic development via PI3K/Akt pathway.33 IGF2 is a component of SDIA crucial for dopaminergic neuron differentiation of human ESC-derived progenitors.23 Similarly, TGF-β signaling regulates asymmetric cell division of neural stem cells with differentiation of one daughter cell into neuronal or glial cell and maintaining the neural stem cell population via the second daughter cell.34 Collectively, our data suggest reduced activities of this set of PA6 cells-derived factors with the culture time, consistent with a previous report.32 On the other hand, SFRP1, CXCL12, NGF, and FGF2 are a few PA6 cells-derived factors that maintain their high fold change until day 8. Several of these factors are established as components of SDIA in past studies. For example, Vazin et al. found that PA6 cells expressed high levels of CXCL12, IGF2 and SFRP1.23,35 Their results implicated potential roles of IGF2 in maintaining survival of differentiating stem cells and CXCL12 in inducing dopaminergic differentiation. Schwartz et al also found CXCL12 and SFRP1 as major dopaminergic neuron-inducing components in stromal cell derived media.36 Our data are consistent with the reduced fold change of IGFs and increased fold change for CXCL12 from day 4 to 8 in our study.
Remaining factors including CNTF, WNT5A, VEGF A, VEGF B, VEGF C, NT3, NT5, PAX1, GDNF, BDNF, FGF8, and SHH have negligible gene expression in PA6 cells but significant fold change in the co-culture group. The expression of these factors persisted, and in some cases increased, during the measurement window. These data strongly suggest that the factors are produced by differentiating mESCs in the co-culture group. In past studies, neurotrophins such as BDNF, NT3, NT5, and GDNF were found to play vital roles in nervous system development by regulating proliferation, differentiation, and maturity of neural cells.26 NT3 was also shown to enhance neuronal precursor cell migration and proliferation, neurites outgrowth, and regulate neuronal specification.26,27 CNTF promoted differentiation of ESCs into astrocytes as well as survival of matured neurons.28,37 Persistent and increasing gene expression of these factors in differentiating mESCs is consistent with their roles in development of specific neural cells that takes place during the second week of co-culture of mESCs and PA6 cells.38,39 In summary, the fold change data in Fig 5a–b suggests that a group of 12 factors self-regulate neural cell differentiation of mESCs in this co-culture system.
To make a more direct, statistical comparison of gene expression of these factors between the PA6 and co-culture groups, we averaged the temporal fold change data for each gene in each group and subjected the results to a two-sample t-test (Fig 5c). This analysis showed that the expression levels of all genes except for TGF β1, TGF β2, and IGF1 were statistically different between the PA6 and co-culture groups. Factors such as IGF2, FGF2, SFRP, ALDHA1, and CXCL12 showed significantly higher average fold change in PA6 cells, whereas factors including BDNF, GDNF, CNTF, NT3, NT5, and VEGFs showed higher average fold change in co-culture group. This analysis again emphasizes the presence of two cohorts of factors distinctly expressed by PA6 and differentiating mESCs.
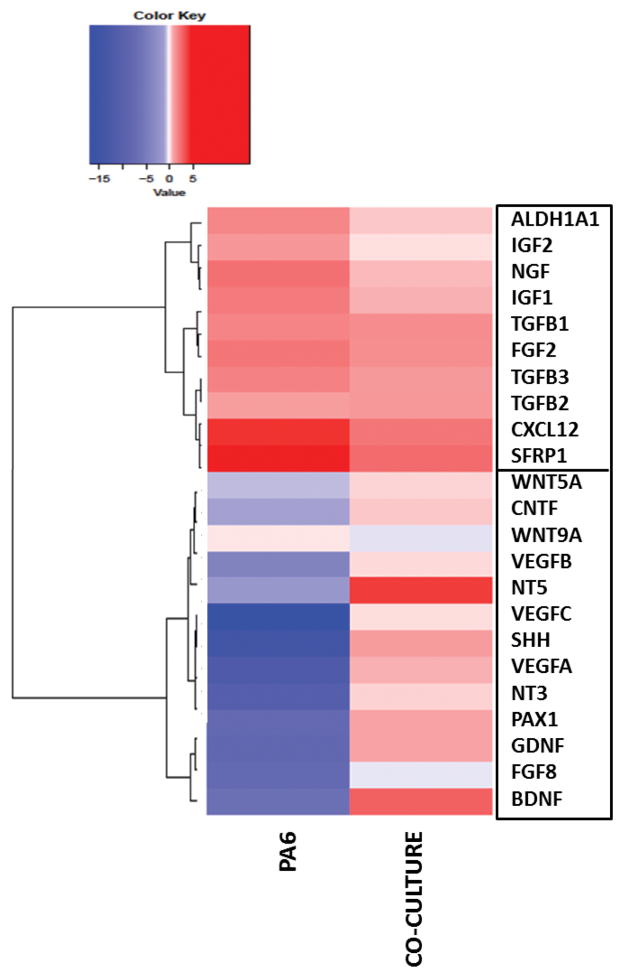
Hierarchical clustering identification of mESCs-contributing factors
To substantiate the validity of our gene expression analysis and data interpretation, we applied an agglomerative hierarchical cluster analysis to the data on all three days of analysis. On each day, only genes that exceeded the expression level of the control group by two folds or more in at least one group were included for the cluster analysis. A representative heatmap from day 8 data is shown in Fig 6. Hierarchical clustering segregated the genes into two distinct clusters on each day (Fig 6 for day 8 and Fig S2 for days 4 and 6). Examination of the results shows that the genes that are highly expressed in PA6 cells or differentiating mESCs in the co-culture group segregate into two clusters. The bottom cluster represents the collection of genes that are expressed in high levels in the co-culture group and with low or negligible expression in PA6 cells. This cluster consists of 12 factors that include neurotrophins such as BDNF, GDNF, CNTF, NT3, and NT5, VEGFs, WNTs, and transcription factors SHH and PAX1. All these factors are known to have definitive roles in induction, maintenance, or proliferation of differentiating neural lineage cells. For example, VEGFs have shown direct neurogenic, neuroprotective, proliferative, and anti-apoptotic effects on neural progenitor cells. VEGFA deficiency led to degeneration of cerebral cortex,40–42 whereas VEGFC stimulation of embryonic NPCs resulted in cellular proliferation.43,44 Our results show that the VEGF family factors are primarily active in differentiating mESCs in the co-culture group at a moderate, few fold change level. The transcription factor PAX1 maintains the neural progenitor pool and promotes cellular proliferation.45,46
Figure 6.
Heatmap and hierarchical clustering of 23 genes representing major soluble factors important in neural differentiation of stem cells. Data represent day 8 cultures.
The top cluster represents the genes expressed by PA6 cells in both groups. The expression levels of each gene in the two groups are comparable, although those in the PA6 group always show slightly higher expression indicated by the darker shade of red in the heatmap. This cluster includes IGFs, TGFs, ALDHA1, NGF, FGF2, CXCL12, and SFRP1. Expression of SFRP1 and ALDH1A1 was maximum on day 6 in the PA6 group with 28- and 12-fold change, respectively. ALDHA1 is a critical mediator of retinoic acid synthesis in vivo and suggested to promote neural patterning during embryonic brain development.47 Presence of high levels of ALDH1A1 in PA6 cells implies that neural fate regulation of mESCs by PA6-derived media is partly due to the endogenous retinoic acid. SFRP1, on the other hand, is a soluble modulator of Wnt signaling and previously known as a dominant neuron-inducing factor in PA6 conditioned media.35,36
To further validate that the factors segregated to the bottom cluster of the heatmap are indeed expressed by the differentiating mESCs, and not by the PA6 cells potentially over-stimulated by mESCs under the co-culture condition, we physically isolated mESC colonies from the culture vessel under a microscope on day 6. After lysing cells of the manually harvested colonies, extracting mRNA, and performing qPCR analysis, we subjected the data to agglomerative hierarchical clustering. The result is shown in Supplementary Fig S3. The major conclusion emerging from this analysis is that separating mESCs colonies and PA6 cells prior to performing qPCR still results in the segregation of two factors into two clusters identical to those generated on day 6 without cell separation (Supplementary Fig S2b). It is noteworthy from supplementary Fig S3 that factors in the bottom cluster show significantly lower expression in mESCs compared to PA6 cells, whereas the expression levels of these factors in both PA6 and co-culture groups are similar (Fig 2b). Again, this indicates that the factors in this cluster are primarily produced by PA6 cells in co-culture group as well.
In summary, our comprehensive gene expression and statistical analysis of growth factors and transcription factors from PA6 cells culture and PA6-mES cells co-culture helped identify and segregate factors that each cell type produces to induce or regulate neural differentiation of mESCs. Moreover, identifying differences in the temporal expression of these factors helped estimate their regulatory roles at different stages of differentiation. For the first time, we identified self-regulatory factors from differentiating mESCs that augment neural differentiation in this co-culture niche. Our data are consistent with studies that report self-regulatory roles of stem cells in their fate determination. For example, neural stem cells conditioned media induced nestin positive cells from mESCs,48 and secreted mitogens in media derived from an embryoid body culture showed wound repair and tissue remodeling effects.49 Nevertheless, our study identified a large number of these factors and that self-regulate and enhance of neural differentiation of stem cells. Further studies are required to establish the presence of stem cell derived factors at a protein level and to determine their concentration profiles in the co-culture conditioned medium. This study and its future extensions is expected to lead to the formulation of media compositions to maximize the yield of neural cells.
CONCLUSION
We establish that mESCs in co-culture with stromal cells produce self-regulatory factors that significantly augment their neural differentiation. This effect is beyond the previously-known stromal cell derived inducing activity (SDIA) effect of the stromal cells on differentiation of ESCs in this co-culture system. Our hierarchical clustering analysis segregates the collection of the secretory factors into two clusters and identifies secretory molecules of mESCs that self-regulate their neural differentiation. This study establishes a foundation for future investigations of combining factors derived from ES and PA6 cells as a potential strategy to formulate efficient neural differentiation protocols and eliminate the physical presence of stromal PA6 cells.
Supplementary Material
Supplementary Figure S1. Immunofluorescence quantification of neural cell differentiation of mESCs for (a) TuJ, (b) Nestin, and (c) GFAP. Data represent differentiation of a single mESC colony in co-culture with PA6 cells in a microwell under treatment with SM, SCM, and CCM. The number of replicates was N=20.
Supplementary Figure S2. Heatmap and hierarchical clustering of genes of representing specific soluble factors on (a) day 4 and (b) day 6 of cultures. Factors with fold change less than 2 are not included in the analysis.
Supplementary Figure S3. Heatmap and hierarchical clustering of genes representing specific soluble factors. Data are from qPCR experiments with mESCs performed on day 8 after manually separating the differentiated mES colonies from PA6 feeder layer.
Supplementary Table S1. List of primers, and forward and reverse sequence and length of the 26 primers used for qPCR.
Supplementary Table S2. List of the reagents used, with their vendors and catalog numbers.
INSIGHT, INNOVATION, INTEGRATION.
Mechanistic understanding of neural differentiation of embryonic stem cells (ESCs) will facilitate the use of ESCs as a developmental model and deriving specific neural cells for cell replacement therapies. Using a cell microprinting technology, we create standalone colonies of ESCs in co-culture with a stromal cell to differentiate ESCs to neural lineages. For the first time, we show that ESCs produce self-regulatory factors that significantly augment their neural differentiation. We use hierarchical clustering of gene expression data to identify major soluble factors that regulate the neural differentiation process in ESCs-stromal cells co-culture. This study lays a foundation toward developing novel protocols to differentiate stem cells with greater efficiency using ESCs-derived factors and without using feeder cells.
Acknowledgments
This research is supported by grants 1264562 from National Science Foundation, CA182333 from National Institutes of Health, and TECG20140954 from Ohio Third Frontier.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICT OF INTEREST
The authors declared no potential conflicts of interest with respect to the research, authorship and/or publication of this article.
References
- 1.Wobus AM, Boheler KR. In Vitro. 2005;85:635–678. doi: 10.1152/physrev.00054.2003. [DOI] [PubMed] [Google Scholar]
- 2.Liao MC, Diaconu M, Monecke S, Collombat P, Timaeus C, Kuhlmann T, Paulus W, Trenkwalder C, Dressel R, Mansouri A. World J Stem Cells. 2014;6:248–55. doi: 10.4252/wjsc.v6.i2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishikawa SI, Jakt LM, Era T. Nat Rev Mol Cell Biol. 2007;8:502–507. doi: 10.1038/nrm2189. [DOI] [PubMed] [Google Scholar]
- 4.Evans M. Nat Rev Mol Cell Biol. 2011;12:680–686. doi: 10.1038/nrm3190. [DOI] [PubMed] [Google Scholar]
- 5.Bain G, Ray WJ, Yao M, Gottlieb DI. Biochem Biophys Res Commun. 1996;223:691–4. doi: 10.1006/bbrc.1996.0957. [DOI] [PubMed] [Google Scholar]
- 6.Itskovitz-Eldor J, Schuldiner M, Karsenti D, Eden A, Yanuka O, Amit M, Soreq H, Benvenisty N. Mol Med. 2000;6:88–95. [PMC free article] [PubMed] [Google Scholar]
- 7.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 8.Qu Q, Li D, Louis KR, Li X, Yang H, Sun Q, Crandall SR, Tsang S, Zhou J, Cox CL, Cheng J, Wang F. Nat Commun. 2014;5:141–146. doi: 10.1038/ncomms4449. [DOI] [PubMed] [Google Scholar]
- 9.Kawasaki H, Mizuseki K, Nishikawa S, Kaneko S, Kuwana Y, Nakanishi S, Nishikawa SI, Sasai Y. Neuron. 2000;28:31–40. doi: 10.1016/s0896-6273(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 10.Tavana BH, Mosadegh B, Takayama S. Adv Mater. 2010;22:2628–2631. doi: 10.1002/adma.200904271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashi H, Morizane A, Koyanagi M, Ono Y, Sasai Y, Hashimoto N, Takahashi J. Eur J Neurosci. 2008;27:261–8. doi: 10.1111/j.1460-9568.2008.06027.x. [DOI] [PubMed] [Google Scholar]
- 12.Swistowska AM, da Cruz AB, Han Y, Swistowski A, Liu Y, Shin S, Zhan M, Rao MS, Zeng X. Stem Cells Dev. 2010;19:71–82. doi: 10.1089/scd.2009.0107. [DOI] [PubMed] [Google Scholar]
- 13.Vazin T, Chen J, Lee CT, Amable R, Freed WJ. Stem Cells. 2008;26:1517–25. doi: 10.1634/stemcells.2008-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vazin T, Ashton RS, Conway A, Rode Na, Lee SM, Bravo V, Healy KE, Kane RS, Schaffer DV. Biomaterials. 2014;35:941–948. doi: 10.1016/j.biomaterials.2013.10.025. [DOI] [PubMed] [Google Scholar]
- 15.Vermeren M, Keynes R. Encylopedia Life Sci. 2010:1–12. [Google Scholar]
- 16.Rhee YH, Yi SH, Kim JY, Chang MY, Jo AY, Kim J, Park CH, Cho JY, Choi YJ, Sun W, Lee SH. Sci Rep. 2016;6:32025. doi: 10.1038/srep32025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrak D, Atefi E, Yin L, Chilian W, Tavana H. Biotechnol Bioeng. 2014;111:404–412. doi: 10.1002/bit.25100. [DOI] [PubMed] [Google Scholar]
- 18.Martino G, Pluchino S. Nat Rev Neurosci. 2006;7:395–406. doi: 10.1038/nrn1908. [DOI] [PubMed] [Google Scholar]
- 19.Atefi E, Joshi R, Mann JA, Tavana H. ACS Appl Mater Interfaces. 2015;7:21305–14. doi: 10.1021/acsami.5b05757. [DOI] [PubMed] [Google Scholar]
- 20.Tavana H, Jovic A, Mosadegh B, Lee QY, Liu X, Luker KE, Luker GD, Weiss SJ, Takayama S. Nat Mater. 2009;8:736–741. doi: 10.1038/nmat2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atefi E, Mann JA, Tavana H. Langmuir. 2014;30:9691–9. doi: 10.1021/la500930x. [DOI] [PubMed] [Google Scholar]
- 22.Tavana H, Mosadegh B, Zamankhan P, Grotberg JB, Takayama S. Biotechnol Bioeng. 2011;108:2509–2516. doi: 10.1002/bit.23190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vazin T, Chen J, Lee CT, Amable R, Freed WJ. Stem Cells. 2008;26:1517–1525. doi: 10.1634/stemcells.2008-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim M, Habiba A, Doherty JM, Mills JC, Mercer RW, Huettner JE. Dev Biol. 2009;328:456–471. doi: 10.1016/j.ydbio.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee LMY, Leung C-Y, Tang WWC, Choi H-L, Leung Y-C, McCaffery PJ, Wang C-C, Woolf aS, Shum aSW. Proc Natl Acad Sci. 2012;109:13668–13673. doi: 10.1073/pnas.1200872109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maisonpierre PC, Belluscio L, Friedman B, Alderson RF, Wiegand SJ, Furth ME, Lindsay RM, Yancopoulos GD. Neuron. 1990;5:501–509. doi: 10.1016/0896-6273(90)90089-x. [DOI] [PubMed] [Google Scholar]
- 27.Park S, Lee KS, Lee YJ, Shin HA, Cho HY, Wang KC, Kim YS, Lee HT, Chung KS, Kim EY, Lim J. Neurosci Lett. 2004;359:99–103. doi: 10.1016/j.neulet.2004.01.073. [DOI] [PubMed] [Google Scholar]
- 28.Sloan SA, Barres BA. Curr Opin Neurobiol. 2014;27:75–81. doi: 10.1016/j.conb.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okabe S, Forsberg-Nilsson K, Spiro aC, Segal M, McKay RDG. Mech Dev. 1996;59:89–102. doi: 10.1016/0925-4773(96)00572-2. [DOI] [PubMed] [Google Scholar]
- 30.Willerth SM, Faxel TE, Gottlieb DI, Sakiyama-Elbert SE. Stem Cells. 2007;25:2235–44. doi: 10.1634/stemcells.2007-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.James D, Levine AJ, Besser D, Hemmati-Brivanlou A. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 32.Parmar M, Li M. BMC Dev Biol. 2007;7:86. doi: 10.1186/1471-213X-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nieto-Estevez V, Defterali C, Vicario-Abejon C. Front Neurosci. 2016;10:1–9. doi: 10.3389/fnins.2016.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yun C, Mendelson J, Blake T, Mishra L, Mishra B. Dis Markers. 2008;24:251–255. doi: 10.1155/2008/747343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vazin T, Becker KG, Chen J, Spivak CE, Lupica CR, Zhang Y, Worden L, Freed WJ. PLoS One. 2009:4. doi: 10.1371/journal.pone.0006606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz CM, Tavakoli T, Jamias C, Park S, Martin B, Phillips TM, Yao PJ, Itoh K, Ma W, Rao MS, Arenas E, Mattson MP. J Neurosci Res. 2012;90:1367–1381. doi: 10.1002/jnr.23064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borsini A, Zunszain PA, Thuret S, Pariante CM. Trends Neurosci. 2015;38:145–57. doi: 10.1016/j.tins.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 38.Chen X, Gu Q, Wang X, Ma Q, Tang H, Yan X, Guo X, Yan H, Hao J, Zeng F. Int J Mol Med. 2013;32:25–34. doi: 10.3892/ijmm.2013.1372. [DOI] [PubMed] [Google Scholar]
- 39.Joshi R, Buchanan JC, Paruchuri S, Morris N, Tavana H. PLoS One. 2016;11:e0166316. doi: 10.1371/journal.pone.0166316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai C, Grabel L. Dev Dyn. 2007;236:3255–3266. doi: 10.1002/dvdy.21306. [DOI] [PubMed] [Google Scholar]
- 41.Haigh JJ, Morelli PI, Gerhardt H, Haigh K, Tsien J, Damert A, Miquerol L, Muhlner U, Klein R, Ferrara N, Wagner EF, Betsholtz C, Nagy A. Dev Biol. 2003;262:225–241. doi: 10.1016/s0012-1606(03)00356-7. [DOI] [PubMed] [Google Scholar]
- 42.Mackenzie F, Ruhrberg C. Development. 2012;139:1371–80. doi: 10.1242/dev.072348. [DOI] [PubMed] [Google Scholar]
- 43.Raab S, Plate KH. Acta Neuropathol. 2007;113:607–626. doi: 10.1007/s00401-007-0228-3. [DOI] [PubMed] [Google Scholar]
- 44.Le Bras B, Barallobre MJ, Homman-Ludiye J, Ny A, Wyns S, Tammela T, Haiko P, Karkkainen MJ, Yuan L, Muriel MP, Chatzopoulou E, Bréant C, Zalc B, Carmeliet P, Alitalo K, Eichmann A, Thomas JL. Nat Neurosci. 2006;9:340–348. doi: 10.1038/nn1646. [DOI] [PubMed] [Google Scholar]
- 45.Blake Ja, Ziman MR. Development. 2014;141:737–51. doi: 10.1242/dev.091785. [DOI] [PubMed] [Google Scholar]
- 46.Osumi N, Shinohara H, Numayama-Tsuruta K, Maekawa M. Stem Cells. 2008;26:1663–1672. doi: 10.1634/stemcells.2007-0884. [DOI] [PubMed] [Google Scholar]
- 47.Duester G. J Nutr. 1998;128:459S–462S. doi: 10.1093/jn/128.2.459S. [DOI] [PubMed] [Google Scholar]
- 48.Zhang JQ, Yu XB, Ma BF, Yu WH, Zhang AX, Huang G, Mao FF, Zhang XM, Wang ZC, Li SN, Lahn BT, Xiang AP. Neuroreport. 2006;17:981–986. doi: 10.1097/01.wnr.0000227977.60271.ca. [DOI] [PubMed] [Google Scholar]
- 49.Ngangan AV, Waring JC, Cooke MT, Mandrycky CJ, McDevitt TC. Stem Cell Res Ther. 2014;5:26. doi: 10.1186/scrt415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Immunofluorescence quantification of neural cell differentiation of mESCs for (a) TuJ, (b) Nestin, and (c) GFAP. Data represent differentiation of a single mESC colony in co-culture with PA6 cells in a microwell under treatment with SM, SCM, and CCM. The number of replicates was N=20.
Supplementary Figure S2. Heatmap and hierarchical clustering of genes of representing specific soluble factors on (a) day 4 and (b) day 6 of cultures. Factors with fold change less than 2 are not included in the analysis.
Supplementary Figure S3. Heatmap and hierarchical clustering of genes representing specific soluble factors. Data are from qPCR experiments with mESCs performed on day 8 after manually separating the differentiated mES colonies from PA6 feeder layer.
Supplementary Table S1. List of primers, and forward and reverse sequence and length of the 26 primers used for qPCR.
Supplementary Table S2. List of the reagents used, with their vendors and catalog numbers.