Abstract
Brain cytoplasmic 200 RNA (BC200 RNA) is a neuron-specific non-coding RNA, implicated in the inhibition of local synaptodendritic protein synthesis, and is highly expressed in some cancer cells. Although BC200 RNA has been shown to inhibit translation in vitro, the cellular location of this inhibition is unknown. In this study, we used a BC200 RNA-recognizing antibody to identify the cellular locations of BC200 RNA in HeLa cervical carcinoma cells. We observed punctate signals in both the cytoplasm and nucleus, and further discovered that BC200 RNA co-localized with the p-body decapping enzyme, DCP1A, and the heterogeneous nuclear ribonucleoprotein E2 (hnRNP E2). The latter is a known BC200 RNA-binding partner protein and a constituent of p-bodies. This suggests that BC200 RNA is localized to p-bodies via hnRNP E2.
Keywords: BC200 RNA, hnRNP E2, p-body, RNA-recognizing antibody
INTRODUCTION
In 1993, brain cytoplasmic 200 RNA (BC200 RNA) was first identified by Tiedge’s group as a non-coding RNA expressed in human neuronal cells (1). A possible correlation between BC200 RNA and cancer was reported by the same group in 1997 (2). The authors detected BC200 RNA in human germ cells and several immortalized cell lines that did not originate from neurons. Moreover, in situ hybridization experiments showed that BC200 RNA was abnormally overexpressed in various cancers, including breast, lung, tongue, esophagus, ovary, and cervix (2). A later study found that particularly high levels of BC200 RNA were expressed in invasive breast cancer cells (3). These findings suggest that BC200 RNA can contribute to tumorigenesis. Although there is some controversy (4), recent studies suggest that BC200 RNA helps cancer cells avoid apoptosis, and may enhance metastasis (5, 6). For example, BC200 RNA upregulates Bcl-xL (an isoform of Bcl-x) in breast cancer cells by altering its splicing pattern; this upregulation of Bcl-xL enabled cells to avoid apoptosis (5). Moreover, in non-small-cell lung cancers, BC200 RNA was induced by the transcription factor c-Myc, which is highly associated with cancer metastasis, and induced RNA enhanced the cellular motility, thereby possibly promoting metastasis (6).
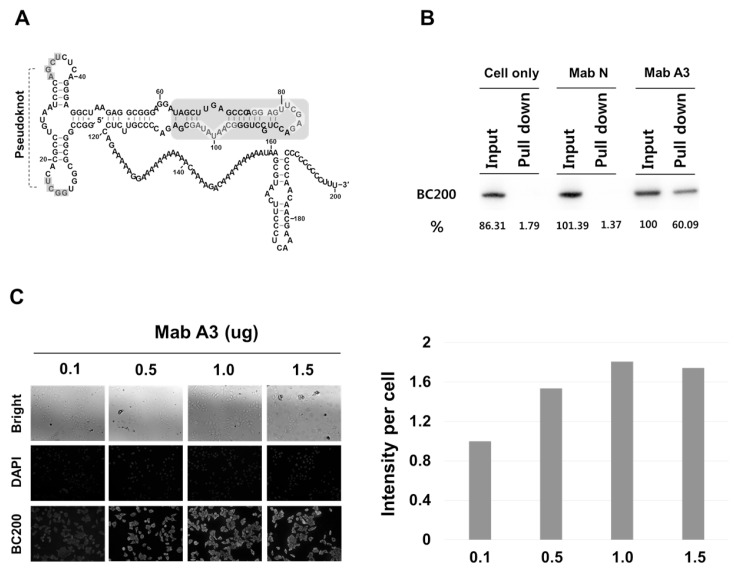
BC200 RNA inhibits the initiation of translation, both in vitro and in vivo (7–9), by interacting with components of the translational machinery, such as eIF4A, eIF4B, and PABP (7, 10). We recently found that only specific genes are down-regulated in BC200 RNA-knockdown HeLa cells (11), suggesting that BC200 RNA-mediated inhibition of translation is controlled within the cell. However, relatively little is known about how this process is regulated. Since the cellular localization of BC200 RNA potentially regulates its cellular function, we set out to determine the cellular localization of BC200 RNA in HeLa cervical cancer cells. In a previous study, we showed that the human monoclonal antibody, MabBC200-A3, recognizes a domain of BC200 (nts 63-107) in a structure- and sequence-dependent manner (Fig. 1A) (12). The BC200 RNA concentration-dependent immunoanalytical signals of MabBC200-A3 coincide with the corresponding conventional hybridization signals (12). Here, we first confirmed that MabBC200-A3 can be used to immunostain BC200 RNA in HeLa cells, and thereafter used it to study the cellular localization of BC200 RNA in these cells. We found that the antibody yielded concentration-dependent immunostaining signals for BC200 RNA in the tested cell line, and the BC200 RNA was localized as punctuates in both the cytoplasm and the nucleus of HeLa cells.
Fig. 1.
Specific recognition of BC200 RNA by the antibody, MabBC200-A3, in HeLa cells. (A) Possible secondary structures of BC200 RNA. The blue- shaded region is the domain, recognized by the antibody MabBC200-A3. Protected regions by the MabBC200-A3 antibody are highlighted in red letters. (B) HeLa cell lysates were immunoprecipitated with MabBC200-A3. RNAs were purified from the immunoprecipitates and subjected to Northern blot analysis. Cell only, without antibody. Mab N, a negative control antibody. Mab A3, MabBC200-A3. (C) Cells treated with increasing amounts of MabBC200-A3 were incubated with Cy™2 AffiniPure Donkey Anti-Human IgG and subjected to confocal microscopy. BC200 RNA is represented by green fluorescence. DAPI was used for nuclei staining.
The binding of proteins to BC200 RNA could play an important role in its subcellular localization. Recently, we identified heterologous nuclear ribonucleoprotein E2 (hnRNP E2) as a binding partner of BC200 RNA, as assessed using a yeast three-hybrid assay (13). hnRNP E2 is a multifunctional protein that participates in a variety of cellular processes, including RNA metabolism (14, 15) and translational enhancement (16). Although it is mainly located in the nucleus, a considerable portion of hnRNP E2 is found in the cytoplasm, enriched in the p-bodies and stress granules, where RNA-processing factors function to control the RNA metabolism (17). Since hnRNP E2 is a constituent of p-bodies, we suspect that BC200 RNA might be localized to p-bodies through its binding to hnRNP E2. Indeed, our immunostaining analysis with MabBC200-A3 shows that BC200 RNA and hnRNP E2 co-localized along with the p-body decapping enzyme, DCP1A.
RESULTS AND DISCUSSION
To investigate the localization of BC200 RNA, we first examined whether the MabBC200-A3 antibody (12) could immunostain the BC200 RNA in HeLa cells. When total cell lysates were treated with the antibody, about half of the cellular BC200 RNA molecules were immunoprecipitated by the antibody (Fig. 1B), suggesting that the antibody effectively recognizes the BC200 RNA in the cell. However, about 50% of the BC200 RNA molecules were not recovered by immunoprecipitation. This reflects that some proteins capable of interacting with the MabBC200-A3 binding motif of BC200 RNA (nts 63-107) compete with the antibody for RNA binding (12), enabling some BC200 RNA molecules to avoid interacting with the antibody.
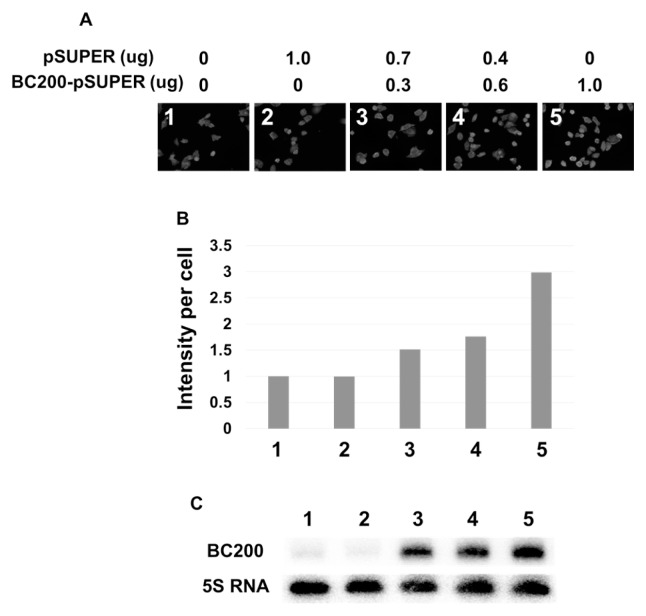
Next, we immunostained the cellular BC200 RNA and subjected the cells to confocal fluorescence microscopy. When permeabilized cells were treated with increasing amounts of MabBC200-A3, we found that the fluorescent signal increased dose-dependently, up to 1 μg (Fig. 1C). To examine whether this saturation point reflected a limited amount of cellular BC200 RNA available for antibody binding, we transfected HeLa cells with increasing amounts of a BC200 RNA-expressing plasmid (pSUPER-BC200), and examined whether the fluorescent signal increased with the amount of cellular BC200 RNA. Indeed, we found that the transfected cells showed dose-dependent increase in the fluorescent signal (Fig. 2A and B), proportional to the cellular content of BC200 RNA (Fig. 2C). Finally, we used the validated antibody to further investigate the subcellular localization of BC200 RNA. We observed a dispersed fluorescence throughout the cells, including both punctate staining in the nuclei and fine punctates throughout the cytoplasm (Fig. 3A).
Fig. 2.
Analysis of HeLa cells expressing increasing amounts of BC200 RNA. HeLa cells were transfected with increasing amounts of the BC200 RNA-expressing plasmid (pSUPER-BC200), and subjected to fluorescent signal analysis (A), and the relative signals are presented in arbitrary units (B). (C) Total RNAs were purified from the same cells indicated by the lane number subjected to Northern blot analysis.
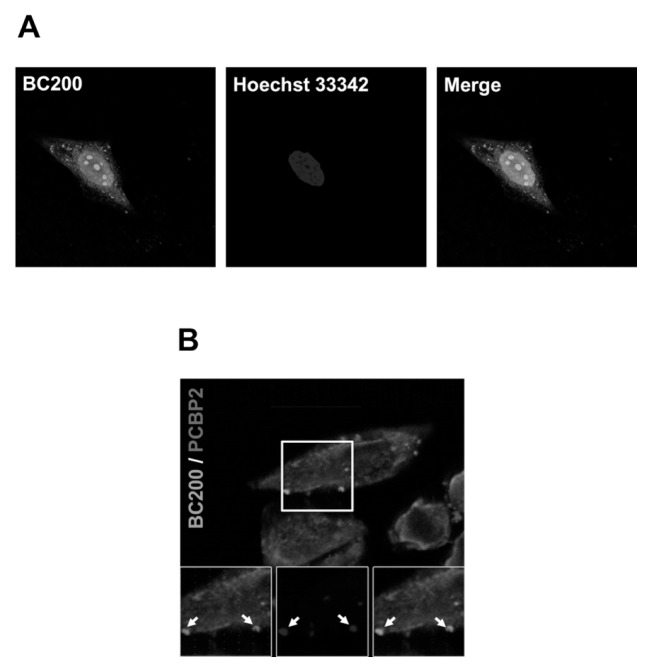
Fig. 3.
Localization of BC200 RNA and colocalization with hnRNP E2 in HeLa cells. (A) HeLa cells were treated with MabBC200-A3 and incubated with Cy™2 AffiniPure Donkey Anti-Human IgG, and the nuclei were stained with Hoechst 33342. BC200 RNA and the nuclei were visualized as green and blue fluorescence, respectively, under confocal microscopy. (B) HeLa cells were transfected with an hnRNP E2-eCFP-expressing construct, treated with MabBC200-A3, and incubated with Cy™2 AffiniPure Donkey Anti-Human IgG. BC200 RNA and hnRNP E2-eCFP were visualized as green and blue fluorescence, respectively, under confocal microscopy. Co-localization is shown by arrows.
We recently identified a heterogeneous ribonucleoprotein E2 (hnRNP E2, also known as PCBP2) as a BC200 RNA-interacting protein (13). hnRNP E2 binds to BC200 RNA mainly through the unique 3′ C-rich domain (nts 162-200) of BC200 RNA (Fig. 1A), which does not overlap with the antibody-binding motif. To evaluate whether the interaction of BC200 RNA and hnRNP E2 is detected by immunostaining with MabBC200-A3, we transfected cells with an hnRNP E2-eCFP-expressing plasmid, immunostained BC00 RNA with MabBC200-A3, and examined the distribution of BC200 RNA and the hnRNP E2-eCFP fusion protein by confocal microscopic analysis. As shown in Fig. 3B, the hnRNP E2-eCFP fusion protein exhibited both dispersed and punctate staining patterns. The punctate staining pattern overlapped with that of BC200 RNA, suggesting that BC200 RNA and hnRNP E2 co-localize in HeLa cells.
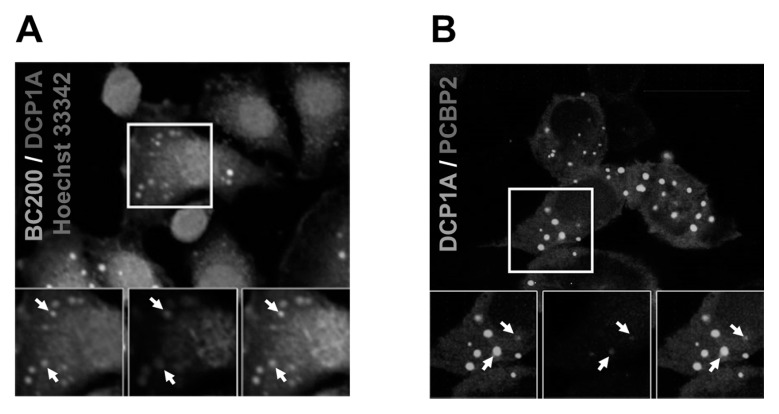
Since hnRNP E2 is known to be a constituent of p-bodies (17), we used DCP1A as a p-body marker protein (18), and examined whether BC200 RNA localizes to p-bodies. When HeLa cells were transfected with a DCP1A-mCherry fusion construct, the DCP1A fluorescence staining pattern overlapped with that of BC200 RNA (Fig. 4A). Additional transfection experiments with both hnRNP E2-eCFP and DCP1A-mCherry fusion constructs confirmed that hnRNP E2 co-localized with DCP1A (Fig. 4B).
Fig. 4.
Co-localization between BC200 RNA and DCP1A, and between hnRNP E2 and DCP1A. (A) HeLa cells were transfected with a DCP1A-mCherry-expressing construct, treated with MabBC200-A3, and incubated with Cy™2 AffiniPure Donkey Anti-Human IgG. The nuclei were stained with Hoechst 33342. BC200 RNA, DCP1A-mCherry, and the nuclei were visualized as green, red, and blue fluorescence, respectively, under confocal microscopy. Co-localization is shown by arrows. (B) HeLa cells were transfected with hnRNP E2-eCFP- and DCP1A-eGFP-expressing constructs, and the cells were subjected to confocal microscopic analysis. hnRNP E2-eCFP and DCP1A-eGFP were visualized as blue and green fluorescence, respectively. Co-localization is shown by arrows.
BC200 RNA acts as a general translation inhibitor in vitro, by interacting with components of the translational machinery (7–9). However, recent in vivo studies indicate that only a limited number of genes were altered by BC200 RNA (11), suggesting that one or more mechanisms govern the specificity of BC200 RNA-mediated inhibition of translation in vivo. In neuronal cells, BC200 RNA is normally restricted to dendrites, and thus acts on the dendritic mRNAs (19). In cancer cells, such as HeLa cells, BC200 RNA may also affect the specifically localized mRNAs. Thus, our present observation that BC200 RNA is localized in p-bodies suggests that BC200 RNA inhibits the translation of mRNAs localized in p-bodies. BC200 RNA is known to be targeted to dendrites by hnRNP A2/B1 in neuronal cells (19). Likewise, it is likely that BC200 RNA could be targeted to p-bodies by hnRNP E2 in HeLa cells because the p-body constituent (hnRNP E2) is a binding partner of BC200 RNA (13).
Although the cellular localization of BC200 RNA can be visualized by RNA fluorescence in situ hybridization (FISH) using antisense nucleic acids (20), the use of MabBC200-A3 has three major advantages over FISH. 1) Since both the sequence and structure of RNA are crucial for the antibody-RNA interaction, the recognition of RNA by an antibody is more specific than a complementarity-based RNA interaction, which is generally associated with nonspecific signals. 2) Whereas antisense nucleic acids are used under conditions that prevent the retention of structure, an antibody can be used to monitor complex and dynamic behaviors of RNAs that have various conformations and are involved in the formation of different ribonucleoproteins. 3) FISH is a multistep procedure that generally involves cell fixation and denaturation of target RNA (20), making it difficult (if not impossible) to detect RNA dynamics over time in living cells.
MATERIALS AND METHODS
Cell culture and transfection
HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 1% antibiotic-antimycotic (Gibco), at 37°C with 5% CO2. For transfections, cells were seeded and incubated for 24 h, and plasmid DNA transfections were performed using Lipofectamine 3000 (Invitrogen), according to the manufacturer’s protocol.
Plasmids
For expression of hnRNP E2-eCFP, the hnRNP E2-encoding sequence was PCR amplified from a human cDNA library and cloned into the HindIII/BamHI sites of pECFP(n1) (Clontech). For expression of the DCP1A-eGFP and DCP1A-mCherry fusion proteins, the DCP1A-encoding sequence was PCR amplified and cloned into the XhoI/SacII sites of pEGFP(c1) (Clontech) or the HindIII/BamHI sites of pmCherry(c1) (Clontech). For ectopic expression of BC200 RNA, the BC200 RNA-encoding sequence was PCR amplified and cloned into pSUPER (Clonetech) to generate pSUPER-BC200.
RNA pull-down assay
Cells were washed, suspended in 10% FBS/PBS, permeabilized by 0.1% PBST, and incubated with MabBC200-A3 (5 μg) for 30 min at room temperature. MabBC200-A3 was prepared as previously described (12). The cells were washed twice with cold PBS, and lysed in cold RIPA buffer [50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Na-deoxycholate (m/v), 0.1% SDS (w/v), 1% Triton X-100 (v/v), 2 mM EDTA]. The cell lysates were incubated with protein G agarose beads (Invitrogen) for 1 h at room temperature. The beads were collected and washed three times with cold RIPA buffer, and the immunoprecipitated RNAs were purified by phenol extraction and examined by Northern blotting. As an input control, whole-cell lysates were prepared in parallel without any antibody.
Northern blot analysis
Northern blot analysis was performed as described previously (21). Total RNA was fractionated on a 6% polyacrylamide gel containing 7 M urea, and transferred to a Hybond-XL membrane (GE Healthcare). Antisense oligonucleotides were 5′ end labeled with [γ-32P] ATP (PerkinElmer Life Sciences) by T4 polynucleotide kinase (Enzynomics) and used as probes for hybridization. Hybridization was carried out in Rapid-Hyb buffer (GE Healthcare), and samples were incubated at 40°C overnight (for BC200 RNA) or at 42°C for 1 h (for the 5S RNA). The membrane was washed twice (20 min each time) at 25°C in 2× SSC buffer (20 mM sodium phosphate, pH 7.4, 0.3 M NaCl, 2 mM EDTA) containing 0.1% SDS, and twice (20 min each time) in 0.2× SSC buffer containing 0.1% SDS. The membrane was exposed to an imaging plate (Fuji BAS-IP), and the results were analyzed on a phospho-image analyzer (Fuji FLA-7000).
Confocal laser scanning microscopy
Confocal laser scanning was performed as described previously (22). Briefly, HeLa cells were seeded on sterile cover slips (Marienfeld) in a 24-well plate (100,000 cells/well), and transfected with or without fluorescent fusion protein-encoding plasmids. For immunostaining, cells were fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, followed by incubation with 1% BSA to block non-specific binding. The MabBC200-A3 antibody (500 ng/100 ul) was used to stain BC200 RNA, following which it was visualized using Cy™2 AffiniPure Donkey Anti-Human IgG (1:200, Jackson Immunoresearch). Imaging was performed using a confocal Zeiss microscope (Zeiss LSM780).
ACKNOWLEDGEMENTS
This study was supported by National Research Foundation of Korea (NRF) funded by the Korea government (MSIP) (2011-0020322 and 2017R1A6A3A11031308) and the Intelligent Synthetic Biology Center of Global Frontier Project funded by MSIP (2013M3A6A8073557).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting financial interests.
REFERENCES
- 1.Tiedge H, Chen W, Brosius J. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J Neurosci. 1993;13:2382–2390. doi: 10.1523/JNEUROSCI.13-06-02382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W, Bocker W, Brosius J, Tiedge H. Expression of neural BC200 RNA in human tumours. J Pathol. 1997;183:345–351. doi: 10.1002/(SICI)1096-9896(199711)183:3<345::AID-PATH930>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 3.Iacoangeli A, Lin Y, Morley EJ, et al. BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis. 2004;25:2125–2133. doi: 10.1093/carcin/bgh228. [DOI] [PubMed] [Google Scholar]
- 4.Wu DI, Wang T, Ren C, et al. Downregulation of BC200 in ovarian cancer contributes to cancer cell proliferation and chemoresistance to carboplatin. Oncol Lett. 2016;11:1189–1194. doi: 10.3892/ol.2015.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh R, Gupta SC, Peng WX, et al. Regulation of alternative splicing of Bcl-x by BC200 contributes to breast cancer pathogenesis. Cell Death Dis. 2016;7:e2262. doi: 10.1038/cddis.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu T, Lu YR. BCYRN1, a c-MYC-activated long non-coding RNA, regulates cell metastasis of non-small-cell lung cancer. Cancer Cell Int. 2015;15:36. doi: 10.1186/s12935-015-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondrashov AV, Kiefmann M, Ebnet K, Khanam T, Muddashetty RS, Brosius J. Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)-binding protein (PABP) J Mol Biol. 2005;353:88–103. doi: 10.1016/j.jmb.2005.07.049. [DOI] [PubMed] [Google Scholar]
- 8.Lin D, Pestova TV, Hellen CU, Tiedge H. Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol Cell Biol. 2008;28:3008–3019. doi: 10.1128/MCB.01800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Iacoangeli A, Popp S, et al. Dendritic BC1 RNA: functional role in regulation of translation initiation. J Neurosci. 2002;22:10232–10241. doi: 10.1523/JNEUROSCI.22-23-10232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eom T, Berardi V, Zhong J, Risuleo G, Tiedge H. Dual nature of translational control by regulatory BC RNAs. Mol Cell Biol. 2011;31:4538–4549. doi: 10.1128/MCB.05885-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin H, Lee J, Kim Y, et al. Knockdown of BC200 RNA expression reduces cell migration and invasion by destabilizing mRNA for calcium-binding protein S100A11. RNA Biol. 2017 doi: 10.1080/15476286.2017.1297913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung E, Lee J, Hong HJ, Park I, Lee Y. RNA recognition by a human antibody against brain cytoplasmic 200 RNA. RNA. 2014;20:805–814. doi: 10.1261/rna.040899.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jang S, Shin H, Lee J, Kim Y, Bak B, Lee Y. Regulation of BC200 RNA-mediated translation inhibition by hnRNP E1 and E2. FEBS Lett. 2017;591:393–405. doi: 10.1002/1873-3468.12544. [DOI] [PubMed] [Google Scholar]
- 14.Chkheidze AN, Lyakhov DL, Makeyev AV, Morales J, Kong J, Liebhaber SA. Assembly of the alpha-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein alphaCP. Mol Cell Biol. 1999;19:4572–4581. doi: 10.1128/MCB.19.7.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blyn LB, Swiderek KM, Richards O, Stahl DC, Semler BL, Ehrenfeld E. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc Nat Acad Sci U S A. 1996;93:1115–1120. doi: 10.1073/pnas.93.20.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blyn LB, Towner JS, Semler BL, Ehrenfeld E. Requirement of poly (rC) binding protein 2 for translation of poliovirus RNA. J Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujimura K, Katahira J, Kano F, Yoneda Y, Murata M. Selective localization of PCBP2 to cytoplasmic processing bodies. Biochim Biophys Acta. 2009;1793:878–887. doi: 10.1016/j.bbamcr.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Kedersha N, Tisdale S, Hickman T, Anderson P. Real-Time and quantitative imaging of mammalian stress granules and processing bodies. Method Enzymol. 2008;448:521–552. doi: 10.1016/S0076-6879(08)02626-8. [DOI] [PubMed] [Google Scholar]
- 19.Muslimov IA, Patel MV, Rose A, Tiedge H. Spatial code recognition in neuronal RNA targeting: role of RNA-hnRNP A2 interactions. J Cell Biol. 2011;194:441–457. doi: 10.1083/jcb.201010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werner M, Wilkens L, Aubele M, Nolte M, Zitzelsberger H, Komminoth P. Interphase cytogenetics in pathology: principles, methods, and applications of fluorescence in situ hybridization (FISH) Histochem and Cell Biol. 1997;108:381–390. doi: 10.1007/s004180050179. [DOI] [PubMed] [Google Scholar]
- 21.Moon Y, Park B, Park H. Hypoxic repression of CYP7A1 through a HIF-1alpha- and SHP-independent mechanism. BMB Rep. 2016;49:173–178. doi: 10.5483/BMBRep.2016.49.3.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi SH, Park BK, Lee KW, Chang J, Lee Y, Kwon HJ. Effect of respiratory syncytial virus on the growth of hepatocellular carcinoma cell-lines. BMB Rep. 2015;48:565–570. doi: 10.5483/BMBRep.2015.48.10.268. [DOI] [PMC free article] [PubMed] [Google Scholar]