Abstract
KCNE1 is known to modulate the voltage-gated potassium channel α subunit KCNQ1 to generate slowly activating potassium currents. This potassium channel is essential for the cardiac action potential that mediates a heartbeat as well as the potassium ion homeostasis in the inner ear. Therefore it is important to know the structure and dynamics of KCNE1 to better understand its modulatory role. Previously the Sanders group solved the three-dimensional structure of KCNE1 in LMPG micelles, which yielded a better understanding of this KCNQ1/KCNE1 channel activity. However, research in the Lorigan group showed different structural properties of KCNE1 when incorporated into POPC/POPG lipid bilayers as opposed to LMPG micelles. It is hence necessary to study the structure of KCNE1 in a more native-like environment such as multi-lamellar vesicles (MLVs). In this study, the dynamics of lipid bilayers upon incorporation of the membrane protein KCNE1 were investigated using 31P solid-state NMR spectroscopy. Specifically, the protein/lipid interaction was studied at varying molar ratios of protein to lipid content. The static 31P NMR and T1 relaxation time were investigated. The 31P NMR powder spectra indicated significant perturbations of KCNE1 on the phospholipid headgroups of MLVs as shown from the changes in the 31P spectral line shape and the chemical shift anisotropy (CSA) line width. 31P T1 relaxation times were shown to be reversely proportional to the molar ratios of KCNE1 incorporated. The 31P NMR data clearly indicate that KCNE1 interacts with the membrane.
Keywords: KCNE1, 31P Solid-state NMR, Multi lamellar vesicles, Lipid bilayer
Graphical abstract
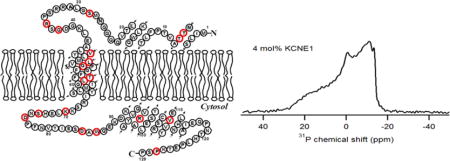
1. Introduction
KCNE1, also known as MinK, is one of the five members of the KCNE family that modulate the voltage-gated potassium channel α subunit KCNQ1 to generate slowly activating potassium currents[1, 2]. This ion channel is essential to the cardiac action potential that mediates heartbeat and the ion homeostasis in the inner ear[1–3]. Moreover, dominant mutations in either KCNE1 or KCNQ1 can cause congenital long-QT syndrome and deafness[4–7]. To better understand the mechanism of this important voltage-gated potassium channel, especially how KCNE1 modulates KCNQ1 to form the slowly activating potassium currents, tremendous efforts on functional and structural studies have been devoted and significant progress has been achieved[4, 8–11].
To fully understand how membrane protein functions, it is essential to know its structure in its native-like environment. The three-dimensional structure of KCNE1 was determined by solution NMR in LMPG detergent micelles by the Sanders group previously[8]. It has significantly enhanced our understanding of KCNE1 and helped to build a reasonable model for the mechanism of the modulatory role of KCNE1 on KCNQ1. However, the structure of KCNE1 was determined in LMPG micelles, which may not represent the native environment for proper function of KCNE1. A big distinction between a soluble protein and a membrane protein is that membrane proteins require a specific hydrophobic environment to fold and function correctly. Due to the ease and good solubility of detergent micelles, most structural information obtained so far has been conducted in a micellar environment. However, it has been shown that micelles are not an ideal membrane mimetic due to the lack of a lipid bilayer and curvature of the micelles which potentially could alter the folding of the membrane proteins[12–15]. The work from the Lorigan group has shown different secondary structural properties of KCNE1 when incorporated into POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine)/POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt)) multi-lamellar vesicles (MLVs) as opposed to LMPG detergent micelles using Circular Dichroism (CD) and Electron Paramagnetic Resonance (EPR) spectroscopic studies[12, 15–17]. It is hence necessary to investigate KCNE1 in a lipid bilayer environment such as MLVs in contrast to traditional detergent micelles to gain a better understanding of this important modulatory protein.
In this study, membrane mimetic MLVs and 31P solid-state NMR spectroscopy were utilized to probe the interaction between KCNE1 and the membrane. MLVs composed of multi-layers of lipid bilayers are commonly used to study membrane proteins and proven to be an excellent membrane mimetic[18, 19]. Solid-state NMR spectroscopy is one of the few spectroscopic techniques that are capable of obtaining pertinent structural information of membrane proteins reconstituted into lipid bilayers. It is widely used to study the structural and dynamic properties of proteins and lipids[20–22]. By incorporating KCNE1 into MLVs, KCNE1 is stabilized in its native-like environment retaining its functionality and thus provide more accurate structural property. 31P is a useful native probe for studying the interaction of the protein with the lipid headgroup region from the perspective of lipids. The conformation and lipid phase can be obtained with the use of native 31P probe. Static 31P NMR experiment was conducted to probe the structural and dynamic changes of the phospholipid bilayer induced by the incorporation of the KCNE1 protein[22–24].
2. Materials and Methods
2.1. Expression and purification of KCNE1
The expression and purification of KCNE1 were described previously[9, 12]. In brief, the plasmids containing wild-type KCNE1 cDNA were transformed into E. coli BL21-CodonPlus(DE3)-RP competent cells. A single colony was inoculated into 5 mL of LB (Luria broth) containing 50 μg/mL ampicillin for overnight growth. The pre-culture was then transferred into 1 L of sterile M9 minimal medium containing 50 μg/mL ampicillin. The large culture was allowed to grow at 37 °C and 250 rpm. When the OD600 reached about 0.8, the culture was induced with 1 M IPTG to a final concentration of 1 mM. Cells were collected by centrifugation after overnight induction. The cell pellets were resuspended in 40 mL of lysis buffer (70 mM Tris-Cl, 300 mM NaCl, pH 8.0). After the addition of 0.5 mg of DNase I (bovine pancreas, Fischer Scientific), 1 mg of lysozyme (egg white, Fischer Scientific), 40 μL of RNase A (20 mg/mL, Thermo Scientific), the cells were disrupted with Fisher Scientific Sonic Dismembrator (5 seconds on, 5 seconds off, 10 minutes total on time, 40% amplitude). The lysates were centrifuged at 40,000 g for 20 minutes. The resulting pellet was resuspended in 30 mL of urea buffer (8 M urea, 20 mM Tris-Cl, 150 mM NaCl, 0.2% SDS, 2 mM β-ME, pH 8.0) and rotated overnight at room temperature. Overnight suspensions were again centrifuged at 40,000 g for 20 minutes. The resulting supernatant was mixed with 4 mL of Ni-NTA resin (Thermo Scientific) that had been previously equilibrated with the urea buffer. Samples were then rotated at room temperature for 2 hours. The resin suspension was washed with 100 mL of TS/SDS buffer (20 mM Tris-Cl, 200 mM NaCl, 0.2% SDS (sodium dodecyl sulfate), 2 mM β-ME, pH 8.0). KCNE1 was then eluted with 6 mL of elution buffer (250 mM imidazole, 0.2% SDS, 2 mM β-ME, pH 7.0). The protein concentration was obtained by measuring the OD280 with the extinction coefficient of 1.2 mg/mL per 1.0 absorbance value. The purity of the KCNE1 protein was confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
2.2. Reconstitution into lipid bilayers
A POPC:POPG lipid bilayer was used to mimic phospholipids typically found in mammalian membranes[9, 12, 15–17, 25, 26]. POPC and POPG powdered lipids (Avanti, Alabaster, AL) were dissolved to a total final concentration of 5 mM (9/1 POPC/POPG molar ratio) in 5 mM HEPES and 0.5% DM (n-Dodecyl-β-D-maltoside) (pH 7.0) by repeating at least 10 freeze/thaw cycles until the solutions were clear[12]. SM2 Bio Beads (Bio-Rad) were washed with 8× 20 mL volumes of methanol followed by 8× 20 mL volumes of water prior to use. Samples were reconstituted by mixing 1 mL of the lipid slurry with 157 μg of KCNE1 protein in 0.2% SDS (500/1 lipid/protein molar ratio) and repeating 2 freeze/thaw cycles. The lipid/protein mixture was then added to 300 mg of wet Bio Beads and nutated at room temperature for 2 hours. Previous studies showed that this amount of Bio Beads effectively removes all detergent and forms MLVs[27]. The resulting sample was centrifuged at 2,000 g for 5 minutes to remove excess Bio Beads. The supernatant was concentrated by ultracentrifugation at 300,000 g for 30 minutes. The proteoliposome pellet was thoroughly resuspended in 50 μL of 100 mM NaH2PO4 (pH 7.2) and immediately used in spectroscopic studies.
2.3. 31P solid-state NMR measurements
All NMR data were recorded on a Bruker Avance 500 MHz WB NMR spectrometer equipped with a CPMAS probe (Bruker, Billerica, MA). 31P NMR spectra were acquired at 200 MHz with proton decoupling using a 4 μs 90° pulse and a 4 s recycle delay. For the 31P static NMR spectra, 512 scans were taken and the free induction delay was processed using 200 Hz of line broadening. The spectral width was set to 500 ppm. All 31P NMR spectra were referenced by assigning the 85% H3PO4 31P peak to 0 ppm. The 31P T1 relaxation times were measured using an inversion recovery pulse sequence (180°-τ-90°-acq) with CP-MAS[28]. For T1 calculations, the data were fit to a single-exponential function: I(t)=I(0)-Aexp(-t/T1)[29–31].
3. Results and Discussion
31P solid-state NMR spectroscopy is extensively utilized to study the interaction between the lipid bilayers and membrane proteins[22, 23]. There is a 100% natural isotopic abundance of 31P within the phospholipid headgroup, it is thus useful to observe changes in the headgroup region of the lipid bilayers through 31P solid-state NMR spectroscopy. Chemical shift anisotropy (CSA) and proton-phosphorus heteronuclear dipolar couplings are the two dominant interactions in the static solid-state 31P NMR spectroscopy, and the latter interaction is removed by high-power proton decoupling. The shape of the 31P powder pattern spectrum is defined by CSA[32]. Reduction of the CSA line width may result from increased membrane surface fluidity, increased dynamics of the vesicles, or decreased size of the vesicles induced by membrane proteins. 31P longitudinal relaxation time T1 indicates changes in the lipid motions including the long axis rotation and lipid diffusion upon protein interaction[23]. 31P T1 relaxation time is associated with the correlation time (τC) of fast molecular motion and the Larmor frequency (ω0). This correlation is described as 1/T1 ∝ τC/(1+ω02τC2). T1 is shortened and the relaxation mechanism of the phospholipids is most efficient when ω0τC ≈ 1[29–31].
To better understand the interaction between KCNE1 and the lipid bilayer, especially the influence which KCNE1 exerts on the membrane, traditional membrane mimetic POPC/POPG (9/1) multi lamellar vesicles (MLVs) was used[33]. A cartoon representation of the topology of KCNE1 in lipid bilayers is shown in Figure 1[8, 12, 15]. Static 31P NMR experiments were conducted to investigate the potential lamellar and/or non-lamellar phase transitions of the lipid bilayers upon incorporation of KCNE1.
Figure 1.
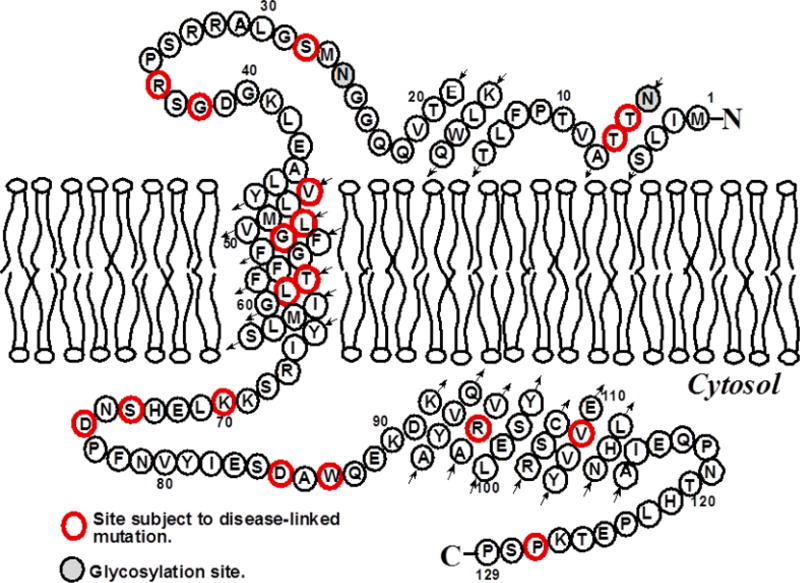
A predicted topology of KCNE1 in lipid bilayers based on previous solution NMR studies[8].
Figure 2 showed the perturbations of KCNE1 on the dynamics of MLVs with varying protein/lipid molar ratios ranging from 0 mol% to 4 mol%. In the absence of KCNE1, the 31P NMR spectrum of MLVs showed a characteristic 31P powder pattern with a CSA line width of 42 ppm[34]. These spectra were acquired at room temperature which is above the melting transition temperature (−2 °C) for either POPC or POPG lipids, indicating a liquid-crystalline phase. In contrast to empty MLVs, MLVs containing 2 mol% KCNE1 has a CSA line width of 36 ppm, which showed a line width reduction of 6 ppm. For the sample of 4 mol% KCNE1, the CSA line width was further decreased to 34 ppm, which presented a significant 8 ppm smaller than the empty MLVs. These data are consistent with the previously published literature[35, 36]. This clear difference of CSA line width upon the addition of KCNE1 suggested that the fluctuations of the headgroup region of phospholipid bilayers were increased[34]. Reduction of the CSA line width may result from increased membrane surface fluidity, which might be the result of the interaction between the membrane surface and the helical components outside the transmembrane domain of KCNE1. Our previous published studies did show KCNE1 in liposomes containing helical structure in addition to TMD, which is consistent with the results observed in this study[8, 9, 12, 15–17]. The larger perturbation on the dynamics of MLVs was observed with the incorporation of higher amounts of proteins. Accordingly, an isotropic peak increased with increasing protein concentration, which indicated the presence of small rapidly tumbling MLVs or smaller fragments. The presence of additional helices other than TMD helix might cause perturbation on the dynamics of MLVs contributing towards the appearance of an isotropic peak in the 31P NMR spectra[8, 9, 12, 15–17]. The effect of KCNE1 on MLVs may be due to an increase in stochastic fluctuations of the lipid headgroups as a result of increased disorder in the individual lipids. The 31P solid-state NMR spectra provide evidence for dynamic changes on the membrane surface upon incorporation of KCNE1. And the association of KCNE1 and lipid bilayer dramatically decreases the 31P CSA line width, which revealed a dynamic increase in the 31P headgroup region on the membrane surface.
Figure 2.
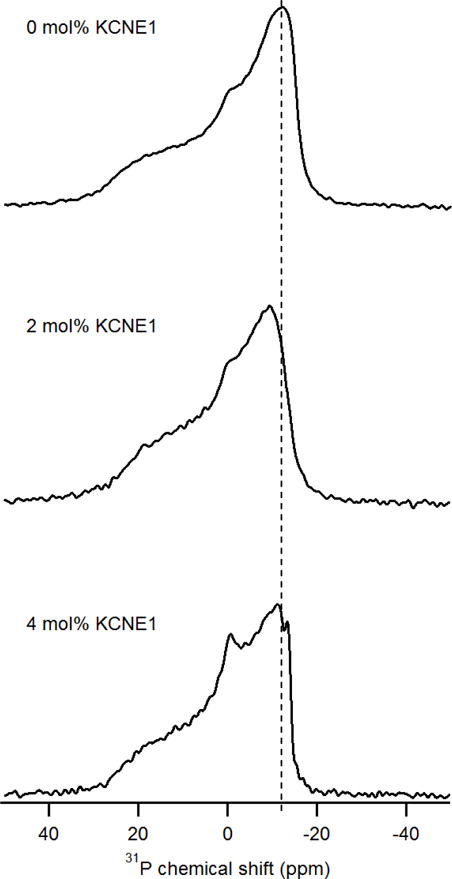
31P chemical shift of KCNE1 protein incorporated MLVs with varying protein/lipid molar ratios at room temperature.
31P NMR spin-lattice relaxation time T1 reflects the fast lipid headgroup rotational motions and the acyl-chain conformational changes, as well as the lipid diffusion in the presence of membrane proteins[20]. A plot of T1 as a function of protein/lipid molar ratios is shown in Figure 3. In the absence of KCNE1, the T1 value of 31P from empty MLVs was found to be (750±5) ms, which was the highest value among all samples. Upon incorporation of KCNE1 to 2 mol%, the T1 value was reduced to (600±10) ms. Increasing the molar ratio to 4 mol% further reduced the T1 value to (450±15) ms. The changes in lipid T1 values in this paper upon addition of protein are comparable to similar measurements in other membrane protein systems[34, 35]. The significant reduction in the T1 value upon the addition of KCNE1 suggested novel disturbances on the formation of MLVs when compared to the empty MLVs. This disturbance may be due to the presence of longer N- and C-termini of KCNE1 partially exposed to the aqueous region and partially interacting with the membrane surface[15]. This plot also revealed a near reversed linear correlation of T1 values to protein/lipid molar ratios. The minimum T1 value indicating the most efficient relaxation was seen at the presence of highest amount of KCNE1. These observations are likely due to the increasing of the stochastic fluctuations of the lipid headgroups as a result of the enhanced disorder of individual lipids.
Figure 3.
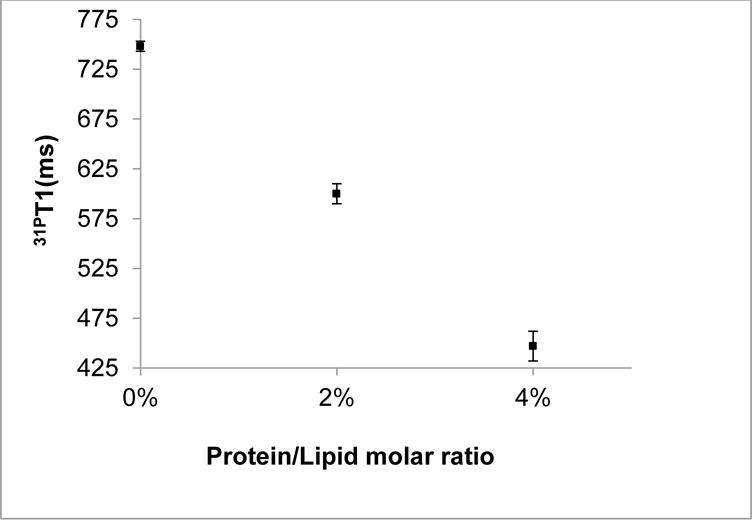
31P spin-lattice relaxation time T1 as a function of varying protein/lipid molar ratios at room temperature. The error bars were estimated from multiple batch of sample preparation.
4. Conclusion
The POPC/POPG (9/1) MLVs mimicked biological membranes providing a controlled environment to assay the interaction between protein and membrane. Both static 31P NMR powder pattern spectra and T1 relaxation time confirmed the membrane association of KCNE1 and the perturbations on the headgroup region of the phospholipids. These results revealed that the addition of higher protein causes a disturbance of stable lipid environment and hence it is important to have specific concentration for better mimic system. The previous studies on KCNE1 have been performed mainly from the perspective of protein in a less ideal membrane mimetic such as micelles, and in this study we focused on the behavior of membrane in the presence of KCNE1 protein. These experiments reveal the perturbation of the membrane resulting from the addition of KCNE1. Also, the addition of protein increases the disorderness of the lipid membrane and hence it is important to have specific concentration to obtain physically relevant environment for a functional membrane protein. Concentration below 2% is recommended for future structural studies of KCNE1. Future solid-state NMR experiments can be designed to investigate the spatial position and orientation of the KCNE1 with respect to the membrane utilizing site-specific 15N-labeled KCNE1 incorporated into proper membrane mimetic such as bicelles.
Acknowledgments
This work was supported by National Institutes of Health Grants R01 GM108026 (to GAL) and R01 DC007416 (to CRS), as well as National Science Foundation Grant CHE-1305664 (to GAL).
Footnotes
Notes
The authors declare no competing financial interests.
References
- 1.McCrossan ZA, Abbott GW. The MinK-related peptides. Neuropharmacology. 2004;47:787–821. doi: 10.1016/j.neuropharm.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 2.Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- 3.Sanguinetti MC, Curran ME, Zou A, Shen J, Spector PS, Atkinson DL, Keating MT. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 4.Abbott GW, Goldstein SA. Disease-associated mutations in KCNE potassium channel subunits (MiRPs) reveal promiscuous disruption of multiple currents and conservation of mechanism. Faseb j. 2002;16:390–400. doi: 10.1096/fj.01-0520hyp. [DOI] [PubMed] [Google Scholar]
- 5.Chen YH, Xu SJ, Bendahhou S, Wang XL, Wang Y, Xu WY, Jin HW, Sun H, Su XY, Zhuang QN, Yang YQ, Li YB, Liu Y, Xu HJ, Li XF, Ma N, Mou CP, Chen Z, Barhanin J, Huang W. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 6.Jespersen T, Grunnet M, Olesen SP. The KCNQ1 potassium channel: from gene to physiological function. Physiology (Bethesda) 2005;20:408–416. doi: 10.1152/physiol.00031.2005. [DOI] [PubMed] [Google Scholar]
- 7.Splawski I, Tristani-Firouzi M, Lehmann MH, Sanguinetti MC, Keating MT. Mutations in the hminK gene cause long QT syndrome and suppress IKs function. Nat Genet. 1997;17:338–340. doi: 10.1038/ng1197-338. [DOI] [PubMed] [Google Scholar]
- 8.Kang C, Tian C, Sonnichsen FD, Smith JA, Meiler J, George AL, Jr, Vanoye CG, Kim HJ, Sanders CR. Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel. Biochemistry. 2008;47:7999–8006. doi: 10.1021/bi800875q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian C, Vanoye CG, Kang C, Welch RC, Kim HJ, George AL, Jr, Sanders CR. Preparation, functional characterization, and NMR studies of human KCNE1, a voltage-gated potassium channel accessory subunit associated with deafness and long QT syndrome. Biochemistry. 2007;46:11459–11472. doi: 10.1021/bi700705j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panaghie G, Tai KK, Abbott GW. Interaction of KCNE subunits with the KCNQ1 K+ channel pore. J Physiol. 2006;570:455–467. doi: 10.1113/jphysiol.2005.100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panaghie G, Abbott GW. The role of S4 charges in voltage-dependent and voltage-independent KCNQ1 potassium channel complexes. J Gen Physiol. 2007;129:121–133. doi: 10.1085/jgp.200609612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coey AT, Sahu ID, Gunasekera TS, Troxel KR, Hawn JM, Swartz MS, Wickenheiser MR, Reid RJ, Welch RC, Vanoye CG, Kang C, Sanders CR, Lorigan GA. Reconstitution of KCNE1 into lipid bilayers: comparing the structural, dynamic, and activity differences in micelle and vesicle environments. Biochemistry. 2011;50:10851–10859. doi: 10.1021/bi2009294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cross TA, Murray DT, Watts A. Helical membrane protein conformations and their environment. Eur Biophys J. 2013;42:731–755. doi: 10.1007/s00249-013-0925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou HX, Cross TA. Influences of membrane mimetic environments on membrane protein structures. Annu Rev Biophys. 2013;42:361–392. doi: 10.1146/annurev-biophys-083012-130326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahu ID, Craig AF, Dunagan MM, Troxel KR, Zhang R, Meiberg AG, Harmon CN, McCarrick RM, Kroncke BM, Sanders CR, Lorigan GA. Probing Structural Dynamics and Topology of the KCNE1 Membrane Protein in Lipid Bilayers via Site-Directed Spin Labeling and Electron Paramagnetic Resonance Spectroscopy. Biochemistry. 2015;54:6402–6412. doi: 10.1021/acs.biochem.5b00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sahu ID, McCarrick RM, Troxel KR, Zhang R, Smith HJ, Dunagan MM, Swartz MS, Rajan PV, Kroncke BM, Sanders CR, Lorigan GA. DEER EPR measurements for membrane protein structures via bifunctional spin labels and lipodisq nanoparticles. Biochemistry. 2013;52:6627–6632. doi: 10.1021/bi4009984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahu ID, Kroncke BM, Zhang R, Dunagan MM, Smith HJ, Craig A, McCarrick RM, Sanders CR, Lorigan GA. Structural investigation of the transmembrane domain of KCNE1 in proteoliposomes. Biochemistry. 2014;53:6392–6401. doi: 10.1021/bi500943p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stockton GW, Polnaszek CF, Tulloch AP, Hasan F, Smith IC. Molecular motion and order in single-bilayer vesicles and multilamellar dispersions of egg lecithin and lecithin-cholesterol mixtures. A deuterium nuclear magnetic resonance study of specifically labeled lipids. Biochemistry. 1976;15:954–966. doi: 10.1021/bi00650a003. [DOI] [PubMed] [Google Scholar]
- 19.Jesorka A, Orwar O. Liposomes: technologies and analytical applications. Annu Rev Anal Chem (Palo Alto Calif) 2008;1:801–832. doi: 10.1146/annurev.anchem.1.031207.112747. [DOI] [PubMed] [Google Scholar]
- 20.Watts A. Solid-state NMR approaches for studying the interaction of peptides and proteins with membranes. Biochim Biophys Acta. 1998;1376:297–318. doi: 10.1016/s0304-4157(98)00012-4. [DOI] [PubMed] [Google Scholar]
- 21.Nakazawa Y, Asakura T. Structure determination of a peptide model of the repeated helical domain in Samia cynthia ricini silk fibroin before spinning by a combination of advanced solid-state NMR methods. J Am Chem Soc. 2003;125:7230–7237. doi: 10.1021/ja0300721. [DOI] [PubMed] [Google Scholar]
- 22.Abu-Baker S, Lorigan GA. Phospholamban and its phosphorylated form interact differently with lipid bilayers: a 31P, 2H, and 13C solid-state NMR spectroscopic study. Biochemistry. 2006;45:13312–13322. doi: 10.1021/bi0614028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dave PC, Tiburu EK, Damodaran K, Lorigan GA. Investigating structural changes in the lipid bilayer upon insertion of the transmembrane domain of the membrane-bound protein phospholamban utilizing 31P and 2H solid-state NMR spectroscopy. Biophys J. 2004;86:1564–1573. doi: 10.1016/S0006-3495(04)74224-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu S, Hawes JW, Lorigan GA. Solid-state NMR spectroscopic studies on the interaction of sorbic acid with phospholipid membranes at different pH levels. Magn Reson Chem. 2009;47:651–657. doi: 10.1002/mrc.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrett PJ, Song Y, Van Horn WD, Hustedt EJ, Schafer JM, Hadziselimovic A, Beel AJ, Sanders CR. The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science. 2012;336:1168–1171. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Y, Hustedt EJ, Brandon S, Sanders CR. Competition between homodimerization and cholesterol binding to the C99 domain of the amyloid precursor protein. Biochemistry. 2013;52:5051–5064. doi: 10.1021/bi400735x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambert O, Levy D, Ranck JL, Leblanc G, Rigaud JL. A new “gel-like” phase in dodecyl maltoside-lipid mixtures: implications in solubilization and reconstitution studies. Biophys J. 1998;74:918–930. doi: 10.1016/S0006-3495(98)74015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aussenac Fabien, Laguerre Michel, Schmitter a Jean-Marie, Dufourc* EJ. Detailed Structure and Dynamics of Bicelle Phospholipids Using Selectively Deuterated and Perdeuterated Labels. 2H NMR and Molecular Mechanics Study. 2003 [Google Scholar]
- 29.Lu JX, Blazyk J, Lorigan GA. Exploring membrane selectivity of the antimicrobial peptide KIGAKI using solid-state NMR spectroscopy. Biochim Biophys Acta. 2006;1758:1303–1313. doi: 10.1016/j.bbamem.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Pinheiro TJ, Watts A. Resolution of individual lipids in mixed phospholipid membranes and specific lipid-cytochrome c interactions by magic-angle spinning solid-state phosphorus-31 NMR. Biochemistry. 1994;33:2459–2467. doi: 10.1021/bi00175a014. [DOI] [PubMed] [Google Scholar]
- 31.Pinheiro TJ, Watts A. Lipid specificity in the interaction of cytochrome c with anionic phospholipid bilayers revealed by solid-state 31P NMR. Biochemistry. 1994;33:2451–2458. doi: 10.1021/bi00175a013. [DOI] [PubMed] [Google Scholar]
- 32.kiel ICPSaIH. Phosphorus-31 NMR: Principles and applications. Gorenstein, D. G.: Academic Press, INC; 1984. Chapter 15: Phosphorus-31 NMR of phospholipids in membranes. [Google Scholar]
- 33.Gennis RB. Biomembranes: Molecular Structure and Function. Springe-Verlag; New York: 1989. [Google Scholar]
- 34.Zhang L, Liu L, Maltsev S, Lorigan GA, Dabney-Smith C. Investigating the interaction between peptides of the amphipathic helix of Hcf106 and the phospholipid bilayer by solid-state NMR spectroscopy. Biochim Biophys Acta. 2014;1838 doi: 10.1016/j.bbamem.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X, Chu S, Hagerman AE, Lorigan GA. Probing the interaction of polyphenols with lipid bilayers by solid-state NMR spectroscopy. J Agric Food Chem. 2011;59:6783–6789. doi: 10.1021/jf200200h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dean RE, O’Brien LM, Thwaite JE, Fox MA, Atkins H, Ulaeto DO. A carpet-based mechanism for direct antimicrobial peptide activity against vaccinia virus membranes. Peptides. 2010;31:1966–1972. doi: 10.1016/j.peptides.2010.07.028. [DOI] [PubMed] [Google Scholar]


