Abstract
Purpose
To examine predictors of understanding preemptive CYP2D6 pharmacogenomic test results and to identify key features required to improve future educational efforts of preemptive pharmacogenomic testing.
Methods
1010 participants were surveyed after receiving preemptive CYP2D6 pharmacogenomic test results.
Results
Eighty-six percent (n=869) of patients responded. Responders were 98% white, 55% female, 57% had four or more years of post-secondary education, and aged 58.9±5.5 years on average. Among responders, 26% reported they only somewhat understood their results and 7% reported they did not understand them at all. Only education predicted understanding. The most common suggestion for improvement was the use of layperson's terms when reporting results. In addition, responders suggested results should be personalized by referring to medications that they were currently taking. Of those reporting imperfect drug adherence in the past, most (91%) reported they would be more likely to take medication as prescribed if pharmacogenomic information was used to help select the drug or dose.
Conclusion
Despite great efforts to simplify pharmacogenomic results (or because of them), about one-third of responders did not understand their results. Future efforts need to provide more examples and tailor results to the individual. Incorporation of pharmacogenomics is likely to improve medication adherence.
Keywords: Pharmacogenomics, Return of Results, CYP2D6, preemptive genotyping, patient understanding
Introduction
Preemptive pharmacogenomics testing is drug-gene testing done prior to a clinical need so that the data are already available at the point of care. A recent publication outlined implementation of preemptive pharmacogenomic testing at five clinical centers and reported the types of genotype platforms used, the numbers of genes assayed, and clinical decision support used to aid prescribers 1. However, little research has been done to determine how best to return and explain test results to patients prior to actual time of prescription.
Learning how to best return and explain preemptive pharmacogenomics test results has become an especially critical issue with the advent of national interest in tailoring all types of clinical care to each patient (“Personalized Medicine”). In particular, pharmacogenomics and return of research results are two key components of the new Precision Medicine Initiative (PMI) proposed by the National Institutes of Health in 2015 2. The PMI Cohort Program has a goal of developing a cohort of more than one million people in the United States in order to improve our knowledge of the “biological, environmental, and behavioral influences ….of diseases”2. The current plan is to allow the PMI Cohort Program participants to access their own genomic data, including pharmacogenomic results. However, there is scarce knowledge around returning such results outside of the clinical setting. Traditionally, pharmacogenomic tests have been ordered by clinicians at the time a new medication was being considered. If results were returned to patients at all, such results were interpreted by the provider in the context of the medication being considered. However, the PMI cohort members, like patients receiving preemptive pharmacogenomic results, will receive their pharmacogenomics results independent of medication prescriptions. It is currently unclear how such results should be returned in order to ensure that patients find the results of preemptive pharmacogenomics testing useful and easy to understand.
The Mayo Clinic Right Drug, Right Dose, Right Time Protocol (RIGHT Protocol) is a study of preemptive pharmacogenomics in 1013 patients 3. CYP2D6 genotypes were among the first clinically-validated pharmacogenomic results that were deposited into participants' electronic health records (EHR).
The CYP2D6 gene encodes for an important enzyme that is involved in the metabolism of xenobiotics, including up to 25% of all prescribed medications 4. However, the standard clinical genetic test report can be difficult to comprehend, as it includes technical jargon and unfamiliar terms. Complicating this is the effects of various CYP2D6 variants vary depending upon the form of the medication (e.g. pro-drug vs. active form) at time of administration. Some medications, such as codeine, require activation by CYP2D6 before they have their intended pharmacologic effect. A patient with an “ultra-rapid metabolizer” phenotype would rapidly convert codeine to morphine and experience overdose symptoms. Other medications, such as Prozac, are administered in an active form and are then deactivated by CYP2D6 prior to elimination from the body. The same patient with the “ultra-rapid metabolizer” phenotype would experience lack of efficacy as they rapidly deactivated the medication. Such patients require higher than normal doses to achieve therapeutic effectiveness.
To call attention to the availability of the results in the EHR and provide help in the interpretation of those results, we mailed a letter with a summary of results and educational materials. The aims of this study were to: 1) understand the variation in the perceived understanding of results, including potential demographic differences in perceived understanding; 2) investigate attitudes and beliefs about sharing and using CYP2D6 test results; 3) explore patient ideas for improving materials to present and interpret results.
Methods
Setting and Patient Selection
The RIGHT Protocol 3 was designed to test whether health care providers can use preemptive pharmacogenomic test results to guide patient prescriptions at the point of care. Further, we wanted to study the effects of preemptive genotyping on the healthcare utilization and outcomes of primary care patients. Participants were selected from the Mayo Clinic Biobank 5 based on age, sex, and race 3. A total of 2000 subjects were identified; 1013 participants provided a blood sample and informed consent and were included in the RIGHT Protocol. We conducted clinical CYP2D6 genetic testing for all participants and deposited results in the EHR. Methods for interpreting pharmacogenomic variants have been previously described6. At the time of this study, Mayo Clinic used the following CYP2D6 nomenclature groups: ultra-rapid, extensive to ultra-rapid, extensive, intermediate to extensive, intermediate, poor to intermediate, and poor. In addition, one Mayo Clinic specific nomenclature category was used (i.e. intermediate to ultra-rapid) for 6 patients with indeterminant copy number variations. Of the 1013 participants, 1010 were alive and able to participate in the current study.
Study Setting
This study was approved by the Mayo Clinic Institutional Review Board. Study participants were mailed a packet of materials that included a results letter summarizing their phenotypic CYP2D6 metabolizer status. Metabolizer status was divided into two phenotypic categories: actionable and non-actionable. Each category was then further categorized into groups to describe the rate at which patients metabolize the CYP2D6 medication. For non-actionable or “normal” phenotypes, patients received a result summary that stated: “You process CYP2D6-related medications at a regular rate”. The pharmacogenomic-based recommendation for individuals with these non-actionable CYP2D6 phenotypes (Mayo Clinic nomenclature groups: extensive, intermediate to extensive, and intermediate) is to adhere to standard drugs and/or dosages. The second category included actionable CYP2D6 phenotypes (i.e. ultra-rapid, extensive to ultra-rapid, intermediate to ultra-rapid, poor to intermediate, and poor). At Mayo Clinic, individuals in this category typically are recommended to change dose or use an alternative medication. For individuals with these phenotypes, the summary letter stated “You process some medications differently”. Note that genotype and phenotype methods used at Mayo Clinic during the time of this study are not necessarily those used across the United States. For all RIGHT participants, the mailed packet also included 1) an educational brochure (See Supplemental materials) with one page of information about how to log into the Mayo Clinic Patient Portal to access results, 2) two pages of educational material about pharmacogenomic testing and CYP2D6, 3) a nine-page survey, and 4) a postage-paid return envelope.
Data collection
The mailed survey contained attitudinal questions about the result letter, educational materials, participants' beliefs about personal medication use, plans for using CYP2D6 results, and the usefulness of pharmacogenomic testing in general. Questions included in the survey were developed by the study investigators. Study material questions concerning perceived understanding allowed responders to report if they understood ‘completely’, ‘mostly’, ‘somewhat’, or ‘not at all’ (Supplemental Table 1). Space was provided for participants to write in things that were helpful and things that would have made the study materials more helpful. Questions about their confidence in their ability to explain their results to a friend or family member included categories of ‘extremely’, ‘somewhat’, ‘a little’ and ‘not at all’. Those who had not responded within 30 days of the initial mailing were sent a second survey. Data collection was closed 30 days after the second mailing. No incentives or telephone follow-up calls were included.
Data analyses
To assess potential non-response bias, demographic and CYP2D6 phenotype data were compared for survey responders and non-responders using a Chi-square or Fisher's exact test for categorical variables and a t-test for continuous variables, as appropriate. Between group variance was similar between compared groups.Associations between participant characteristics including age, sex, education, CYP2D6 phenotype, number of prescription medications, marital status, self-reported health status, and severity and age weighted sum of diseases and perceived understanding of the CYP2D6 results and their utility were assessed using logistic regression and summarized using the odds ratio (OR) and 95% confidence interval (CI). For these analyses, ‘completely understood’ and ‘mostly understood’ were combined and compared to a combined group of those who reported understanding only ‘somewhat’ or ‘not at all’. For attitudinal questions about encouraging others to consider getting pharmacogenomic testing, we used unadjusted logistic regression models.
Responses to the open-ended question, “What is the one thing that would have made your result letter more helpful?” were entered into a database, coded and analyzed for common themes by two independent reviewers (JEO, MEM). Discrepancies were reviewed and discussed until consensus was reached. If consensus could not be reached, then a third investigator (CRV) determined the final code.
Results
A total of 1010 participants were alive and eligible for the survey at the time of mailing. Of these, 869 (86%) responded to the survey (Table 1). Despite the high response rate of our survey, we observed differences between responders and non-responders in multiple characteristics, including sex, education level, and marital status (Table 1). Responders were more likely to be female, better educated, and married. Among responders, 98% were white, 55% were female, 57% had four or more years of post-secondary education, and mean age was 59 (±5.5) years (Table 1). The prominence of white race and high education level among responders is consistent with overall education levels in the Mayo Clinic Biobank, the source population from which this study sample was drawn 5. Eighty-seven percent had EHR data indicating use of at least one prescription medication in 2014. Over one-third (39%) of responders had CYP2D6 test results classified by the study team as actionable.
Table 1. Baseline characteristics.
| Characteristic | Non-responder (N=141) | Responder (N=869) | P‡ |
|---|---|---|---|
| Male, N (%) | 78 (55.3) | 394 (45.3) | 0.03 |
| White, N (%) | 135/139 (97.1) | 842/860 (97.9) | 0.53 |
| Age (years), mean (SD)* | 58.2 (5.0) | 58.9 (5.5) | 0.16 |
| Age (years), N (%)* | 0.18 | ||
| <55 | 35 (24.8) | 191 (22.0) | |
| 55-64 | 92 (65.2) | 540 (62.1) | |
| ≥65 | 14 (9.9) | 138 (15.9) | |
| CYP2D6 phenotype, N (%) | 0.03 | ||
| Ultra-rapid | 15 (10.6) | 68 (7.8) | |
| Extensive to ultra-rapid | 17 (12.1) | 141 (16.2) | |
| Extensive | 36 (25.5) | 169 (19.4) | |
| Intermediate to extensive | 36 (25.5) | 161 (18.5) | |
| Intermediate | 18 (12.8) | 200 (23.0) | |
| Poor to intermediate | 9 (6.4) | 66 (7.6) | |
| Poor | 10 (7.1) | 64 (7.4) | |
| CYP2D6 phenotype, N (%)† | 0.52 | ||
| Non-actionable | 90 (63.8) | 530 (61.0) | |
| Actionable | 51 (36.2) | 339 (39.0) | |
| Education, N (%) | 0.003 | ||
| High school graduate or less | 22 (15.6) | 80 (9.2) | |
| Some college/vocational/tech/associates degree including community college | 63 (44.7) | 293 (33.7) | |
| Four-year college graduate | 29 (20.6) | 273 (31.4) | |
| Graduate or professional school | 26 (18.4) | 218 (25.1) | |
| Unknown | 1 (0.7) | 5 (0.6) | |
| Prescription medications in 2014, N (%) | 0.02 | ||
| None | 32 (22.7) | 109 (12.5) | |
| 1 to 2 | 33 (23.4) | 207 (23.8) | |
| 3 to 5 | 32 (22.7) | 262 (30.1) | |
| 6 to 10 | 22 (15.6) | 177 (20.4) | |
| 11 to 20 | 17 (12.1) | 91 (10.5) | |
| ≥21 | 5 (3.5) | 23 (2.6) | |
| Marital status, N (%) | 0.01 | ||
| Single/divorced/widowed | 31 (22.0) | 122 (14.0) | |
| Married | 110 (78.0) | 747 (86.0) | |
| Health (self-reported) | - | ||
| Excellent/very good/good | - | 816 (93.9) | |
| Fair/poor | - | 42 (4.8) | |
| Not reported | - | 11 (1.3) | |
| Severity and age weighted sum of diseases, mean (SD) | 2.8 (2.2) | 2.9 (2.4) | 0.79 |
Abbreviations: SD, standard deviation
Chi-square or Fisher's exact P value presented for categorical variables and t-test P value presented for age and severity and age weighted sum of diseases
Age for non-responders was calculated as the patient's age when the survey was sent out; age for responders was calculated as the patient's age when the survey was filled out
Phenotypes categorized as non-actionable are extensive, intermediate to extensive, and intermediate, phenotypes categorized as actionable are ultra-rapid, extensive to ultra-rapid, intermediate to ultra-rapid, poor to intermediate, and poor
Perceived understanding of results
When asked, “How well do you feel you understand or don’t understand your CYP2D6 result?” the majority reported (67%) they either completely or mostly understood their results. Of concern, 33% reported that they either only somewhat understood (26%) or did not understand their results at all (7%). We examined the influence of multiple factors on failure to understand results (Table 2). Key variables included age, sex, education, CYP2D6 phenotype, and number of prescription medications in 2014. Other covariates examined include marital status, self-reported health status, and perceived health status (Table 2). Of these, lower education was associated with failure to understand results. Compared to those with a four-year college degree or greater, those with a high school education or less or those with some higher education (but not a four-year degree) were 1.6 times more likely to report understanding somewhat or not at all (p=0.006).
Table 2. Associations with not understanding (somewhat or not at all) the CYP2D6 result returned in the results letter‡.
| Characteristic | Unadjusted OR (95% CI) | P | Adjusted* OR (95% CI) | P |
|---|---|---|---|---|
| Age | 0.58 | 0.52 | ||
| <55 | Reference | Reference | ||
| 55-64 | 1.08 (0.76, 1.54) | 1.13 (0.79, 1.62) | ||
| 65+ | 1.28 (0.80, 2.03) | 1.33 (0.82, 2.15) | ||
| Sex | 0.24 | 0.24 | ||
| Female | Reference | Reference | ||
| Male | 0.84 (0.63, 1.12) | 0.84 (0.62, 1.13) | ||
| Education | 0.003 | 0.006 | ||
| Four-year college graduate or greater | Reference | Reference | ||
| Some higher education | 1.67 (1.23, 2.27) | 1.63 (1.19, 2.22) | ||
| High school graduate or less | 1.62 (0.98, 2.66) | 1.58 (0.96, 2.60) | ||
| CYP2D6 phenotype† | 0.38 | 0.48 | ||
| Non-actionable | Reference | Reference | ||
| Actionable | 1.14 (0.85, 1.52) | 1.11 (0.83, 1.49) | ||
| Prescription medications in 2014 | 0.07 | 0.13 | ||
| None | Reference | Reference | ||
| 1 to 2 | 1.45 (0.85, 2.47) | 1.34 (0.78, 2.31) | ||
| 3 to 5 | 1.77 (1.06, 2.96) | 1.61 (0.96, 2.71) | ||
| 6 to 10 | 2.15 (1.25, 3.68) | 1.97 (1.14, 3.41) | ||
| 11 to 20 | 2.00 (1.08, 3.71) | 1.74 (0.92, 3.27) | ||
| ≥21 | 1.05 (0.35, 3.15) | 0.82 (0.27, 2.53) | ||
| Marital status | 0.60 | 0.54 | ||
| Married | Reference | Reference | ||
| Single/divorced/widowed | 0.89 (0.59, 1.36) | 0.88 (0.57, 1.34) | ||
| Health (self-reported) | 0.22 | 0.36 | ||
| Excellent/very good/good | Reference | Reference | ||
| Fair/poor | 1.49 (0.79, 2.82) | 1.36 (0.71, 2.60) | ||
| Severity and age weighted sum of diseases | 1.03 (0.97, 1.09) | 0.28 | 1.02 (0.96, 1.09) | 0.53 |
Abbreviations: CI, confidence interval; OR, odds ratio
Due to missing data, model N's ranged from 847 to 862
Age adjusted for sex and education, sex adjusted for age and education, education adjusted for age and sex, all other adjusted for age, sex, and education
Phenotypes categorized as non-actionable are extensive, intermediate to extensive, and intermediate; phenotypes categorized as actionable are ultra-rapid, extensive to ultra-rapid, intermediate to ultra-rapid, poor to intermediate, and poor
To understand patient perceived understanding of laboratory reports with CYP2D6 result, we asked them to indicate their level of agreement to the following statement: “It was easy to understand my pharmacogenomic result in the Patient Portal”. Among the 499 patients who said they had viewed their Patient Portal results, over half (53%) agreed (15% strongly and 38% somewhat), 12% neither agreed nor disagreed, 33% disagreed (21% somewhat and 12% strongly), and 1% did not specify.
Participants were also asked to indicate their confidence level in their ability to explain their results to a friend or family member. Thirty-eight percent responded “somewhat confident”. Responses, however, varied by education. Thirteen percent (10/77) of those with a high school education or less and 23% (48/211) of those with a graduate or professional degree responded that they were “extremely confident”. Conversely, 30% (23/77) of those with a high school education or less responded “not at all confident”, although even 18% (38/211) of those with a graduate or professional degree gave the same response.
Participant attitudes to Pharmacogenomics testing
In order to understand how much these participants valued pharmacogenomic testing of CYP2D6, we summarized responses to four questions including 1) whether they reported sharing or were planning to share their results with others, 2) whether they would encourage others to get pharmacogenomic testing, 3) how useful did they believe pharmacogenomic results will be, and 4) whether use of pharmacogenomics would affect their medication compliance.
Among the 69% (N=588) who had or planned to share their CYP2D6 test result, 83% planned to share their results with a health care provider, 60% planned to share with a spouse or partner, 30% with children, 20% with siblings, 10% with friends, 7% with parents, and 17% planned to share with a pharmacist. Some (17%) planned to discuss or share with no one; 14% were unsure if they would discuss or share results with anyone. Note that these categories were non-exclusive.
In general, 64% of the participants would encourage others to consider getting pharmacogenomic testing. The strongest predictor of who would encourage others to get tested was their reported level of understanding of their CYP2D6 result. Those who reported either completely or mostly understanding their result were more likely to encourage others to get tested than those who reported understanding their result only somewhat or not at all (OR=1.82, 95% CI 1.34 - 2.46). Other factors considered (age, sex, education, CYP2D6 phenotype, marital status, health status) were not associated with likelihood of encouraging others to get tested.
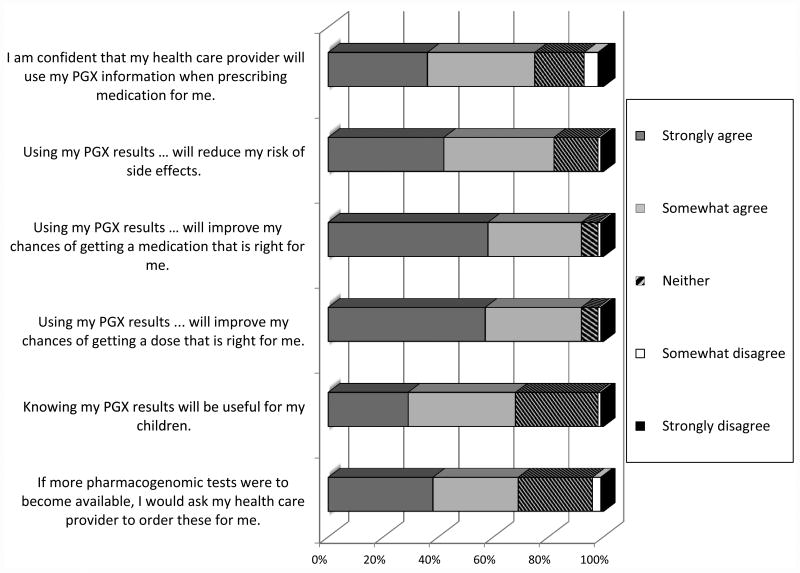
Participant attitudes on the usefulness of pharmacogenomic testing are summarized in Figure 1. Responders were generally positive concerning pharmacogenomics testing usefulness. Most participants (87%) strongly or somewhat agreed with the statement “Using my pharmacogenomic results when prescribing medications for me will improve my chances of getting a dose that is right for me.” In addition, 87% strongly or somewhat agreed with the statement “Using my pharmacogenomic results when prescribing medications for me will improve my chances of getting a medication that is right for me”. Participants were least likely to agree with the statement “Knowing my pharmacogenomic results will be useful for my children” (447/669, 67%), even after removing those without children.
Figure 1. Participant attitudes on usefulness of pharmacogenomics (PGx) testing.
Patients were provided the following instructions for the statements above: “Please indicate your agreement or disagreement with the following statements”
*Among the 669 for which it applies (Those who marked “Does not apply” were not included in the presented figure results)
Finally, 513 participants (59%) reported that they did not always follow medication instructions as directed. However, most of these participants reported that they would either be “much more likely” (67%) or “somewhat more likely” (24%) to take the medication as prescribed if pharmacogenomic information was used to help select the medication and dose.
Participant suggestions for improving results
Using an open-ended question, we asked participants, “What is one thing that would have made your result letter more helpful?” Three overlapping themes emerged from the analysis of responses. First, explanations of results need to be translated into plain language and preferably in person. Second, results need to be personalized to be useful for patients. Third, layout and content of information could be simplified (Table 3).
Table 3. Sample responses out of 444 meaningful comments to “What is one thing that would have made your results letter more helpful?”.
| Primary theme | Sub theme | Count | Example Responses |
|---|---|---|---|
| Plain language from clinician | Simplify language | 297 | Use Layman's terms. The term intermediate was not explained well as being “typical” or “normal”. I showed this to my Mayo doctor, he said ignore it. |
| The letter still had a lot of medical detail that was difficult to understand for someone without a medical background | |||
| Name all genes. Not CYP2D6 – call it CARL. That, I can remember. | |||
| Clinical assistance needed | 14 | Given to me while at a Dr. appt to explain. | |
| If my doctor or nurse had gone over the results it would be more helpful | |||
| If the letter would have included “talk to your provider if you take… medications”. | |||
| I feel this information is for the doctor and not sure I need to know this. | |||
| Personalize results | Information for personal health | 59 | Really what does it mean for the drugs I take? |
| If I should contact my provider about medications I am currently taking. | |||
| What does this mean to me? Do I need to be concerned? | |||
| Medication info | 93 | List of medications affected by CYP2D6 would be valuable. | |
| Maybe reference to which medications my body may process differently and should a person mention this to our healthcare providers. | |||
| How are my current meds being metabolized? | |||
| Suggestions for improvements | Electronic Health Record suggestions | 9 | To explain each test when clicking on it in the online results. |
| The letter is ok. The online comments are very involved and difficult to understand. | |||
| Suggestion on layout | 62 | A small graph showing where my results were as to “normal”. | |
| Put test results in tabular form, ie: Gene CYP2D6 Result normal (extensive metabolizer) This would be particularly helpful if more than one result is available |
The most common responses suggested that results be translated into plain language and emphasized the need to simplify language and to have the information delivered by someone either in person or over the phone who could explain the results. The second was that results be personalized in a way that patients could better manage their care. Many responders requested a table or list of medications affected by CYP2D6 and comments frequently included requests for matching results with current medications. The third theme included suggestions for improving the presentation of the data. One person suggested that they wanted “comparative information in graph form – more visual understanding of my particular info”. Another made suggestions to personalize results in a clearer way, asking for “Information about the range of results. Where do I fall in a range as an ‘intermediate metabolizer?”
Discussion
The goal of this work was to understand the variation in the perceived understanding of results; 2) investigate attitudes and beliefs about sharing and using CYP2D6 test results; and 3) explore patient ideas for improving materials to present and interpret results. This information is especially important as plans unfold for participants in large research initiatives, including the Precision Medicine Initiative now underway. We found that our first attempt to return CYP2D6 phenotype results to participants in our study in an understandable way was only partially successful, but also identified key elements required for improving future effort to return pharmacogenomics results.
Most participants were enthusiastic about pharmacogenomics testing after receiving their results. Most had or planned to share their results with someone, would encourage others to get tested, and thought that the results would be useful for either their own health care or for their children.
Results show there is still significant work to provide patient-centered results that meet patient needs. Providing clear and concise results while trying to balance the limitations of current science, the complexity of test results, and patient needs will continue to be a challenge as national efforts move forward. Many participants had difficulty understanding their results, and those with less education were more likely to report problems with interpretation. Many participants also recommended that we improve our explanations using non-technical language. Avoiding the standard scientific categories used by the clinical lab, such as “intermediate to ultra-rapid metabolizer”, could significantly improve interpretation. While these are standard names for the CYP2D6 phenotypes, the actual effect of such phenotypes on a patient's response to a specific medication is difficult to interpret.
Many patients also commented that they wanted more information about which medications are affected by CYP2D6, and some requested lists of medications to avoid or use with caution. More broadly, a theme that consistently emerged from the comments was that patients really wanted to know what the results meant for them personally. We tried to keep our results report simple and did not discuss all the possible ways that CYP2D6 might affect the metabolism of various medications, because the impact of CYP2D6 phenotype on response to a particular medication varies depending on whether the medication is delivered in an active state or if it needs to be activated to achieve its desired therapeutic benefit 4. We chose not to present the differential activity on various medications and thus may have inadvertently caused confusion in some of our participants. Interpreting the CYP2D6 phenotypes in lay language and indicating which drugs are likely to be affected by the phenotype could significantly improve patient satisfaction of return of results.
Finally, we found that over half of our participants reported that they did not take their medications routinely or as directed. Medication adherence varies depending on type of condition and medication, but has been reported to range from 17% to 80% 7. Therefore, our study population seems to be typical in their medication use behavior. However, a vast majority of those that were non-adherent to their medication regimens indicated that pharmacogenomic information tailoring their prescription would make them much more likely to take their medication. Lack of medication adherence has also been strongly associated with increases in health care utilization and cost 8. Our results suggest that, at least for a subset of the population, pharmacogenomic information, or more precise tailoring of prescription medications, could improve medication adherence.
Another interesting result pertains to respondant confidence that their healthcare provider will use pharmacogenomic information when prescribing medication for them in the future. Unlike other questions about the value of pharmacogenomics testing which showed extremely few respondants disagreeing, seven percent indicated that they either disagreed strongly (5%) or somewhat (2%) with the statement that they were confident that their provider would use pharmacogenomic information when prescribing their medications. We found this disconnect to be of interest and plan to explore this further in future studies of patient and physician attitudes toward pharmacogenomics. To date we have only been able to have explore this preliminarily9.
Limitations of our study include the potential lack of generalizability. Our study population was highly motivated and enthusiastic about the potential for implementing pharmacogenomics into routine patient care. In addition, they are mostly white, highly educated, and in very good health. Populations with different characteristics may be less interested in pharmacogenomic testing, and our results indicate that persons with less education were also less likely to understand study results. However, many people in our study population still had difficulty understanding what their pharmacogenomics results meant for their personal health. Therefore, future studies involving return of preemptive test results will need to ensure that patient materials related to delivery of pharmacogenomics results are tested and validated in multiple populations, with multiple education levels, to ensure that the results are simple and easy to comprehend in all populations. Another limitation is that we were unable to verify that patients had actually viewed their results in the Mayo Clinic Patient Portal. Further we were not able to ask specific questions about their knowledge of their results. Future research in this area is warranted.
In summary, we found that participants in a study of preemptive pharmacogenomics testing were enthusiastic about the potential benefits of such testing, but felt that it was difficult to interpret results, and the personal impact of such test results was unclear. Future studies focused on ensuring that pharmacogenomic results are clear and easy to interpret in multiple populations will be essential for all initiatives focused on delivery of personalized medicine.
Supplementary Material
Acknowledgments
This study was funded as part of the NHGRI-supported eMERGE (Electronic Records and Genomics) Network (HG04599 and HG006379). Additional support was provided by the Mayo Center for Individualized Medicine and the Mayo Clinic Robert D. and Patricia E. Kern Center for the Science of Health Care Delivery.
Footnotes
Conflict of Interest Notifications: DISCLOSURE: The authors declare no conflict of interest
References
- 1.Dunnenberger HM, Crews KR, Hoffman JM, et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu Rev Pharmacol Toxicol. 2015;55:89–106. doi: 10.1146/annurev-pharmtox-010814-124835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Precision Medicine Initiative Cohort Program – Building a Research Foundation for 21st Century Medicine. [Accessed March 17, 2016, 2016];Precision Medicine Initiative (PMI) Working Group Report to the Advisory Committee to the Director N. 2015 http://www.nih.gov/sites/default/files/research-training/initiatives/pmi/pmi-working-group-report-20150917-2.pdf.
- 3.Bielinski SJ, Olson JE, Pathak J, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc. 2014;89(1):25–33. doi: 10.1016/j.mayocp.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Owen RP, Sangkuhl K, Klein TE, Altman RB. Cytochrome P450 2D6. Pharmacogenet Genomics. 2009;19(7):559–562. doi: 10.1097/FPC.0b013e32832e0e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olson JE, Ryu E, Johnson KJ, et al. The Mayo Clinic Biobank: a building block for individualized medicine. Mayo Clin Proc. 2013;88(9):952–962. doi: 10.1016/j.mayocp.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji Y, Skierka JM, Blommel JH, et al. Preemptive Pharmacogenomic Testing for Precision Medicine: A Comprehensive Analysis of Five Actionable Pharmacogenomic Genes Using Next-Generation DNA Sequencing and a Customized CYP2D6 Genotyping Cascade. J Mol Diagn. 2016;18(3):438–445. doi: 10.1016/j.jmoldx.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krueger KP, Berger BA, Felkey B. Medication adherence and persistence: a comprehensive review. Adv Ther. 2005;22(4):313–356. doi: 10.1007/BF02850081. [DOI] [PubMed] [Google Scholar]
- 8.Iuga AO, McGuire MJ. Adherence and health care costs. Risk Manag Healthc Policy. 2014;7:35–44. doi: 10.2147/RMHP.S19801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.St Sauver JL, Bielinski SJ, Olson JE, et al. Integrating Pharmacogenomics into Clinical Practice: Promise vs Reality. Am J Med. 2016;129(10):1093–1099 e1091. doi: 10.1016/j.amjmed.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.