Abstract
The obscurin family of polypeptides is essential for normal striated muscle function and contributes to the pathogenesis of fatal diseases, including cardiomyopathies and cancers. The single mammalian obscurin gene, OBSCN, gives rise to giant (∼800 kDa) and smaller (∼40–500 kDa) proteins that are composed of tandem adhesion and signaling motifs. Mammalian obscurin proteins are expressed in a variety of cell types, including striated muscles, and localize to distinct subcellular compartments where they contribute to diverse cellular processes. Obscurin homologs in Caenorhabditis elegans and Drosophila possess a similar domain architecture and are also expressed in striated muscles. The long sought after question, “what does obscurin do?” is complex and cannot be addressed without taking into consideration the subcellular distribution of these proteins and local isoform concentration. Herein, we present an overview of the functions of obscurins and begin to define the intricate relationship between their subcellular distributions and functions in striated muscles.
Keywords: UNC-89, Cardiac muscle, Skeletal muscle, Cardiomyopathy, Molecular scaffold
Introduction
Complexity of the domain architectures of obscurins
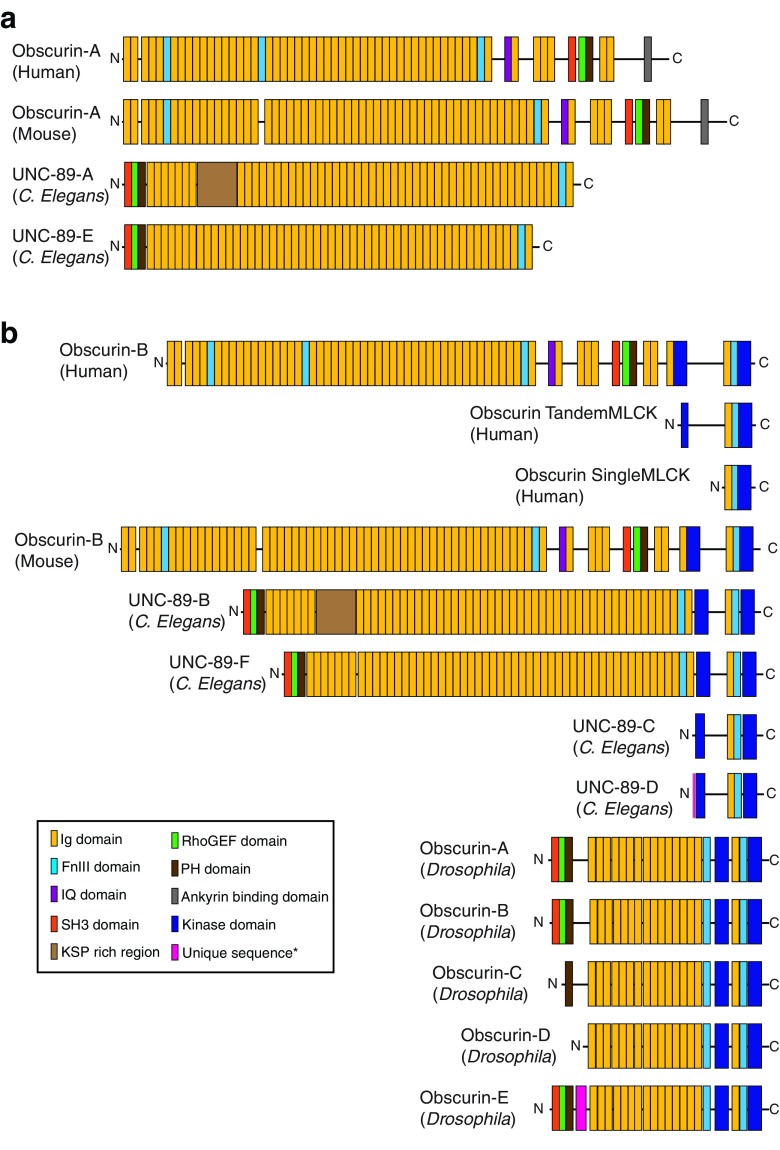
Obscurins have been studied in striated muscle for the past two decades in several organisms, including mammals, Caenorhabditis elegans, Drosophila, and zebrafish. Discovered over 15 years ago, mammalian obscurin was christened for its initial difficulties in detection and characterization (Young et al. 2001). Although detection methods for giant proteins have become more established and characterization of the obscurins more thorough, the name obscurin stuck. However, today, obscurin refers to a family of polypeptides expressed from a single gene. The human obscurin gene (OBSCN) on chromosome 1q42.13 spans 170 kb and is subject to alternative splicing of its 119 exons, which encode for giant (∼800 kDa) proteins and smaller (∼40–500 kDa) isoforms (Fig. 1) (Ackermann et al. 2014; Fukuzawa et al. 2005; Kontrogianni-Konstantopoulos et al. 2009; Perry et al. 2013; Young et al. 2001). The first two-thirds of prototypical human obscurin-A (∼720 kDa) is composed of many immunoglobulin (Ig) and three fibronectin type-III (FnIII) domains, and a single calmodulin-binding IQ motif. These modular domains are followed by a src homology-3 domain (SH3) motif, tandem rho-guanine nucleotide exchange factor (RhoGEF) and pleckstrin homology (PH) domains, and two additional Ig domains. The COOH-terminus of obscurin-A contains a non-modular region with an ankyrin-binding domain (ABD) and putative ERK kinase targets. Alternative splicing at the 3′ end of OBSCN results in the expression of a second giant isoform, obscurin-B. Giant obscurin-B (∼800 kDa) shares most of its domain architecture with obscurin-A, however it lacks the non-modular region and differs in its COOH-terminus. The COOH-terminus of obscurin-B consists of two active Ser/Thr kinase motifs (SK1 and SK2), two additional Ig domains, and another FnIII domain. Alternative transcription and translation initiation sites promote the expression of two smaller kinase-containing isoforms. The tandem kinase isoform encodes a truncated version of the COOH-terminus of obscurin-B, including part of the SK2 motif, the Ig and FnIII domains, and the SK1 motif. The single kinase isoform initiates at the last Ig domain of obscurin-B and includes the FnIII domain and SK1 motif but lacks the SK2 domain. Additionally, alternative splicing of the OBSCN gene, which could give rise to additional giant and smaller polypeptides, has been speculated but not yet experimentally verified.
Fig. 1.
Alternative splicing of the tandem adhesion and signaling motifs encoded by the obscurin gene (OBSCN) creates diverse obscurin proteins. The domain architecture for known human, mouse, and Caenorhabditis elegans isoforms are shown. Predicted domain architecture for Drosophila obscurin isoforms are also shown. Obscurin isoforms are divided into non-kinase isoforms (a) and kinase-containing isoforms (b). See key for domain architecture. Pink box within the NH2-terminus of UNC-89-D and obscurin-E (Drosophila) indicates unique amino acids specific to each isoform and species. FnIII Fibronectin type-III, Ig immunoglobulin, MLCK myosin light-chain kinase, N-, -C NH2- and COOH-terminus, respectively, PH pleckstrin homology, RhoGEF rho-guanine nucleotide exchange factor, SH3 src homology-3
Prior to the identification of mammalian obscurin, UNC-89, the invertebrate homolog of obscurin, was identified in C. elegans (Benian et al. 1996; Sutter et al. 2004). The single UNC-89 gene contains three promoters, is highly alternatively spliced, providing complexity to this family of polypeptides, and generates various isoforms consisting of structural domains and signaling motifs (Fig. 1) (Benian et al. 1996; Ferrara et al. 2005; Qadota et al. 2008a; Small et al. 2004). Similar to mammalian obscurins, there are several UNC-89 isoforms, ranging in size from ∼900 to 156 kDa. UNC-89-E (∼660 kDa) and -F (∼830 kDa) are analogous to mammalian obscurin-A and –B, respectively, where UNC-89-E is the non-kinase-containing isoform and UNC-89-F possesses two pseudo kinases within its COOH-terminus. Notably, the SH3, RhoGEF, and PH triplet of signaling domains is found at the extreme NH2-terminus of UNC-89 isoforms, preceding any Ig domains. UNC-89-A (∼730 kDa) and –B (∼900 kDa) are nearly identical to UNC-89-E and –F, respectively, with the exception of an ∼70 kDa KSP region, rich in lysine/serine/proline residues, found between the NH2-terminal Ig domains. Similar to the mammalian obscurin family of proteins, two small kinase-containing isoforms, UNC-89-C and -D (both ∼156 kDa) are also expressed in C. elegans. Both are truncated versions of the giant kinase-containing isoforms of UNC-89 and contain one Ig domain, one FnIII domain, and two pseudo-kinase domains. They differ only in their unique eight or eleven amino acids prior to the first Ig domain.
More recently, the Drosophila homolog of obscurin has also been identified (Fig. 1) (Katzemich et al. 2012, 2015). Specifically, five obscurin isoforms are expressed in Drosophila in a developmental and tissue-specific manner. In the mature fly, four isoforms (A, B, C, and D) are expressed in the thorax, while two of these isoforms (A and C) are also expressed in the indirect flight muscles (IFM). On the contrary, the larva expresses only obscurin isoform-E (Katzemich et al. 2012, 2015). Unfortunately, their protein composition is not well annotated and lacks experimental confirmation; herein we provide the domain architecture for each predicted isoform (Fig 1). The NH2-terminus of Drosophila obscurin isoforms A, B and E is composed of the SH3, RhoGEF, and PH triplet of signaling motifs, similar to the NH2-terminus of C. elegans UNC-89. Following the signaling motifs there is a non-modular stretch of amino acids, which is preceded by several tandem Ig domains. Notably, obscurin-B differs from obscurin-A in the absence of a short linker region between Ig domains 6 and 7. Obscurin-E, which also lacks the linker region between Ig domains 6 and 7 contains a stretch of amino acids unique to obscurin-E within it NH2-terminus. Obscurin-C lacks the SH3 and RhoGEF domains, while obscurin-D initiates just prior to the stretch of tandem Ig domains. Drosophila obscurin isoforms possess a common COOH-terminal domain, which includes two FnIII domains an Ig domain and two pseudo-kinase domains. Unlike mammalian obscurin-A, there is no analogous ankyrin-binding domain in Drosophila obscurin or C. elegans UNC-89 (Katzemich et al. 2012, 2015).
Zebrafish also express obscurin proteins. However, unlike mammalian, C. elegans, and Drosophila obscurins, which arise from alternative splicing of a single gene, zebrafish obscurins are encoded by two different genes mapping to zebrafish chromosomes 24 and 8 and encoding obscurin and obscurin-MLCK, respectively (Raeker et al. 2006). Zebrafish obscurin contains an ankyrin binding sequence similar to the mammalian obscurin-A, while obscurin-MLCK possesses two kinase domains sharing sequence similarity with mammalian obscurin-B (Raeker et al. 2006).
To date, obscurins have been studied in a piecemeal fashion in terms of isolating the structure, ligands, and function of individual domains (Meyer and Wright 2013; Pernigo et al. 2015; Perry et al. 2013). In this review, we focus on the function of obscurin proteins as they relate to distinct subcellular compartments.
Diversity of the subcellular distribution of obscurins
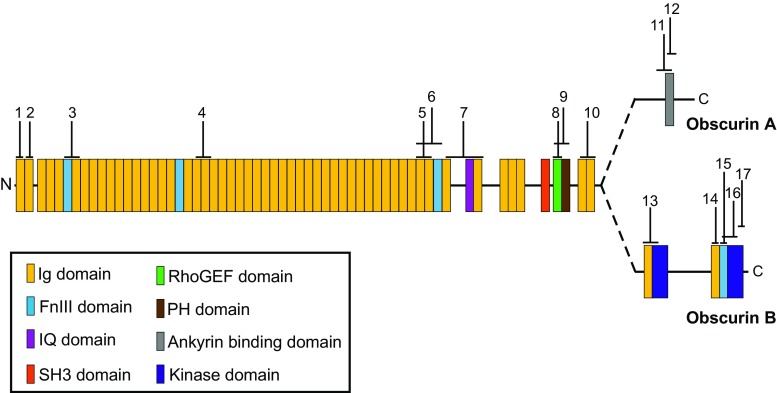
Obscurin isoforms are expressed in both cardiac and skeletal muscles where they concentrate at multiple subcellular locations (Fig. 2) (Bowman et al. 2007; Fukuzawa et al. 2008; Kontrogianni-Konstantopoulos et al. 2006; Shriver et al. 2015). The specific isoforms of obscurin expressed at these locations is not well established. However, the use of antibodies to different epitopes along the length of giant obscurins and contained within multiple obscurin isoforms have facilitated the mapping of obscurin polypeptides to specific subcellular locations (Fig. 3; Table 1). Specifically, obscurin proteins concentrate at distinct regions of the sarcomere. Sarcomeres, the smallest contractile unit of striated muscle, are the foundation for the integration of electrical and mechanical signals across cells that is necessary for muscle contraction. Within the sarcomere, obscurins localize to M-bands, Z-discs, and A/I junctions. In addition, obscurins concentrate at costameres, which are sub-sarcolemmal protein complexes that connect the sarcomere to the sarcolemma and are important for force transmission across myocytes. Obscurins are also enriched at the intercalated discs (ID) of cardiomyocytes. The ID is a specialized membrane structure integrating electrical signals and mechanical communication between neighboring cardiomyocytes to enable the synchronous beating of the heart. Obscurins also localize to sites of network sarcoplasmic reticulum (SR), which is necessary to maintain Ca2+ homeostasis and promote muscle contraction. Other subcellular locations where obscurin proteins concentrate include the nucleus and the muscle side of neuromuscular junctions; however their functions at these sites remain elusive and therefore will not be discussed in this review.
Fig. 2.
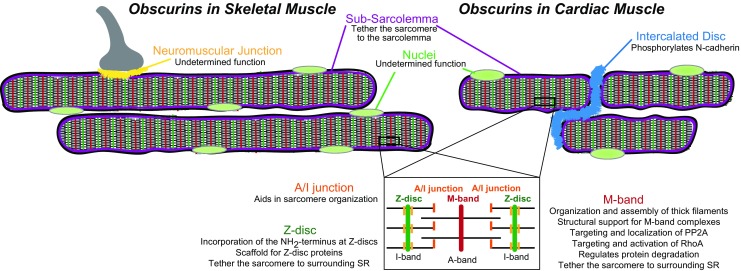
Subcellular distribution of mammalian obscurin proteins in skeletal and cardiac muscle. Known locations for obscurins are highlighted by a distinct color: purple subsarcolemma, light green nulei, blue intercalated disc, yellow neuromuscular junction, orange A/I junction, dark green Z-disc, red M-band. A summary of the functions of obscurin at these locations is also noted. PP2A Protein phosphatase 2, RhoA Ras homolog gene family, member A, SR sarcoplasmic reticulum
Fig. 3.
Epitope mapping of published antibodies of mammalian obscurins. For simplicity, antibodies specific for C. elegans and Drosophila obscurins are not included. See Table 1 for complete description of antibodies, subcellular distributions and references and Fig. 1 for definitions of domains
Table 1.
Subcellular distribution of the various published mammalian obscurin antibodies
| Epitope no.a | Antibody nameb | Subcellular localization | Reference |
|---|---|---|---|
| 1 | 5H10 | M-band (C/S) | Kontrogianni-Konstantopoulos et al. 2006 |
| 2 | 4A8 | M-band (C/S) | Kontrogianni-Konstantopoulos et al. 2006 |
| 3 | F6/I7 | M-band (C/S); Z-disc (C/S) | Bang et al. 2001 |
| 4 | Ob19-20 | M-band (C)c | Young et al. 2001 |
| 5 | Ob48-49 | M-band (Cc | Young et al. 2001 |
| 6 | I48/I49/F50 | Z-disc (C)c | Bang et al. 2001 |
| 7 | Ob51-52 | M-band (C)c | Young et al. 2001 |
| 8 | ObDH | M-band (C)c | Young et al. 2001 |
| 9 | RhoGEF | M-band (C/S) | Bowman et al. 2007 |
| 10 | ObscCOOH | M-band (C/S); Z-disc (C/S) | Kontrogianni-Konstantopoulos et al. 2003 |
| 11 | Ob6215-6353 | M-band (S)c | Bagnato et al. 2003 |
| 12 | ABD | M-band (C/S); Z-disc (C/S) | Ackermann et al. 2014 |
| 13 | SK2 | Nuclei (C); Z-disc (C)c | Borisov et al. 2008 |
| 14 | ObsKin-1 | M-band (C); Z-disc (C); ID (C)c | Hu and Kontrogianni-Konstantopoulos 2013 |
| 15 | ObsKin-2 | M-band (C); Z-disc (C); ID (C)c | Hu and Kontrogianni-Konstantopoulos 2013 |
| 16 | Link7 | Nuclei (C); Z-disc (C)c | Borisov et al. 2008 |
| 17 | ObscKin-3 | M-band (C); Z-disc (C); ID (C)c | Hu and Kontrogianni-Konstantopoulos 2013 |
ABD Ankyrin-binding domain; ID, intercalated disc; Ob, obscurin; RhoGEF, rho-guanine nucleotide exchange factor; SK2, one of two active Ser/Thr kinase motifs in the COOH-terminus of obscurin-B
aEpitope number corresponds to the epitope map found in Fig. 3
bAntibodies along the length of giant obscurins map to distinct epitopes of the proteins
cThose antibodies which have only been tested in either cardiac (C) OR skeletal (S) muscle, as indicated. All others in column (without superscript C or S) have been tested in both types of muscle
In C. elegans, UNC-89 localizes to M-bands within sarcomeres and to dense bodies of body wall muscle cells (Warner et al. 2013). The dense bodies are anchored in the muscle membrane and function to link the actin filaments to the extracellular matrix through connections at Z-discs and costameres. In Drosophila, obscurin concentrates within the sarcomere at the M-bands (Katzemich et al. 2015). The multiple subcellular distributions of this family of proteins gives rise to multiple functions for obscurins.
Varied ligands of obscurins
The subcellular distribution of obscurins at multiple locations within a myocyte can partially explain its diverse functions. However, it is also necessary to consider the various ligands for obscurin proteins and their local concentrations. Within the sarcomere at the level of the M-band, mammalian obscurins interact with titin, myomesin, ankyrin-B, Ras homolog gene family, member A (RhoA), and slow skeletal myosin binding protein C variant 1 (sMyBP-Cv1) (Perry et al. 2013). In C. elegans at the M-band, UNC-89 interacts with copine domain protein atypical-1 (CPNA-1), UNC-15 (paramyosin), small CTD-phosphatase-like-1 (SCPL-1), four and a half LIM domains protein 9 (LIM9/FHL), maternal effect lethal 26 (MEL-26), and Rho1 (Gieseler et al. 2016; Qadota et al. 2008a, b; Warner et al. 2013; Wilson et al. 2012) and Drosophila obscurins interact with Ball and multiple ankyrin repeats single KH domain (MASK) at the M-band (Katzemich et al. 2015). In addition, mammalian obscurins interact with the NH2-terminus of titin and Ran-binding protein 9 (RanBP9) at Z-discs (Bowman et al. 2008), small ankyrin-1 at the SR (Kontrogianni-Konstantopoulos et al. 2003), and ankyrin-B at costameres (Randazzo et al. 2013). At the ID of mammalian cardiac muscle, obscurin polypeptides interact with N-cadherin and Na+/K+ ATPase (Hu and Kontrogianni-Konstantopoulos 2013). The binding partners of obscurin proteins, including their functions, is discussed in detail in the following sections of this review.
Other ligands of mammalian obscurins include calmodulin and calcium/calmodulin-dependent protein kinase I (CaMKII), both of which promiscuously localize throughout myocytes; consequently, the location and function of the interaction between obscurins and these proteins remain elusive. Calmodulin interacts with the IQ motif of obscurins, which localizes to M-bands of developing and adult heart and skeletal muscles, as well as to the sarcolemma and neuromuscular junction of skeletal muscles (Carlsson et al. 2008). Calmodulin is a major calcium sensor that is essential for orchestrating the regulation of cellular proteins of the cytoplasm (Rhoads and Friedberg 1997). Notably, the obscurin IQ motif interacts with calmodulin in a Ca2+-independent manner (Young et al. 2001), suggesting that the interaction is structural in nature, with obscurins providing a docking site for calmodulin at distinct sites within the cytoplasm. In addition, through Ig domain thirty-six obscurin interacts with CaMKII (Hund et al. 2010). CaMKII is involved in many signaling cascades and essential for Ca2+ homeostasis (Balke and Shorofsky 1998). Within cardiomyocytes, CaMKII localizes to the ID and transverse (t)-tubules where it regulates sodium channels to modulate the action potential (Hund et al. 2010). We can speculate that obscurins act as a structural scaffold to dock CaMKII at distinct subcellular regions within the myocyte. Alternatively, obscurins may act to regulate CaMKII as the binding motif within Ig domain thirty-six has a high similarity to the CaMKII auto-regulatory domain (Hund et al. 2010). Additional studies are necessary to investigate the role of obscurins’ interaction with calmodulin and CaMKII.
Defining the location—function relationship of obscurin proteins
“What does obscurin do?” is an intriguing question. To define its physiological function, one must consider the subcellular distribution and local ligand(s) of obscurins. In striated muscle, obscurins can be summarized as essential adaptors in myofibrillogenesis and cytoskeletal organization; in addition they play an important role in Ca2+ homeostasis. In this review we focus on the location–function relationship of obscurin polypeptides in skeletal and cardiac muscles (summarized in Table 2). By defining the specialized functions of obscurins at distinct subcellular locations we may gain a better understanding of obscurins’ role(s) in developing skeletal and cardiac myopathies.
Table 2.
Obscurin/UNC-89 ligands and location-specific functions
| Subcellular localization | Ligand | Function | Reference |
|---|---|---|---|
| Obscurins within the sarcomere | |||
| M-band | Myomesina | Structural support for thick filaments and M-band complexes | Kontrogianni-Konstantopoulos et al. 2004, 2006; Ackermann et al. 2009 |
| sMyBPCa | |||
| Titina | |||
| AnkBa | Target PP2A to M-band | Cunha and Mohler 2008 | |
| RhoAa | Activation of RhoA GTPase activity | Ford-Speelman et al. 2009 | |
| SCPL-1b | Undefined | Qadota et al. 2008b | |
| Rho-1b | Activation of Rho-1 | Qadota et al. 2008a | |
| CPNA-1b | Localization of UNC-89 to M-band | Warner et al. 2013 | |
| UNC-15b | Structural support for M-band complexes | Qadota et al. 2016 | |
| MEL-26b | Regulation of protein degradation | Wilson et al. 2012 | |
| LIM9b | Localization of UNC-89 to M-band | Xiong et al. 2009 | |
| Ballc | Structural support for M-band complexes | Katzemich et al. 2015 | |
| MASKc | |||
| Z-disc | Titina | Scaffold for Z-disc proteins | Young et al. 2001 |
| RanBP9a | Incorporation of titin at Z-discs | Bowman et al. 2008 | |
| Obscurins at cellular membranes | |||
| Sarcoplasmic reticulum | sAnk1a | Tether the sarcomere to the surrounding sarcoplasmic reticulum | Kontrogianni-Konstantopoulos et al. 2003 |
| Costameres | AnkBa | Tether the sarcomere to sarcolemma | Randazzo et al. 2013 |
| Intercalated dscs | N-cadherina | Phosphorylate N-cadherin | Hu and Kontrogianni-Konstantopoulos 2013 |
| Na+/K+ ATPasea | Undefined | ||
AnkB, Ankyrin-B; CPNA-1, copine domain protein atypical-1; LIM9/FHL, four and a half LIM domains protein 9; MASK, multiple ankyrin repeats single KH domain; MEL-26, maternal effect lethal 26; PP2A, protein phosphatase 2; RanBP9, Ran binding protein 9; Rho, Ras homolog gene family; SCPL-1, small CTD-phosphatase-like-1; sMyBP, slow skeletal myosin binding protein; UNC-89, mammalian obscurin
aMammalian ligand
b Caenorhabditis elegans ligand
c Drosophila ligand
Obscurins within the sarcomere
Involvement of obscurins in the formation of the sarcomere
Myofibrillogenesis is the complex process of assembling sarcomeric proteins into complete and functional sarcomeres (Sanger et al. 2002). Obscurin proteins serve multiple roles during myofibrillogenesis, including providing overall structural integrity and myofibril stabilization and, more specifically, providing proper spatial positioning of other contractile proteins, such as myosin (Borisov et al. 2006; Kontrogianni-Konstantopoulos et al. 2009). The specific role played by the obscurin proteins in myofibrillogenesis depends on their spatial/temporal distribution and interactions with key sarcomeric proteins. In support of this, knockdown of obscurins via small, interfering RNA (siRNA) in primary rat adult cardiomyocytes resulted in impaired assembly of new myofibrillar clusters and aberrant contractile apparatus structure (Borisov et al. 2006). In addition, in this same study depletion of obscurin proteins resulted in a loss of rigidity of the myofibrils, modified titin distribution within the sarcomere, and resulted in poorly formed A-bands with a diffuse myosin distribution (Borisov et al. 2006). Notably, during mammalian myocyte development obscurins are first observed at M-bands and later localize to Z-discs after the incorporation of α-actinin, a key Z-disc protein (Borisov et al. 2004; Borisov et al. 2008; Kontrogianni-Konstantopoulos and Bloch 2005). During zebrafish development, obscurin localizes to M-bands and Z-discs nearly simultaneously at approximately 24 hours post-fertilization (Raeker et al. 2006). Interestingly, at 72 h post-fertilization, giant obscurin-A localizes more strongly to M-bands and giant obscurin-B, the kinase-containing isoform, localizes to Z-discs (Raeker et al. 2006). Together, these observations support the involvement of obscurins in multiple stages of myofibrillogenesis and suggest different functions at distinct subcellular distributions throughout development.
Obscurins at the M-band mediate the assembly of thick filaments
Located in the center of sarcomeric A-bands, which house myosin thick filaments, M-bands, devoid of myosin heads but containing several adaptor proteins, serve as a scaffold for thick filaments, thus playing an important role in myosin assembly during myofibrillogenesis (Agarkova and Perriard 2005). In addition to obscurin proteins, other key mammalian M-band proteins of striated muscle include M-protein, myomesin, myosin, titin, obscurin-like 1, and sMyBP-C, which is specific for skeletal muscle (Ackermann et al. 2009; Agarkova and Perriard 2005; Fukuzawa et al. 2008). At M-bands, the NH2-terminal Ig domains of obscurins form a ternary complex with the extreme COOH-terminus of titin and the NH2-terminus of myomesin. In addition, the COOH-terminal domain of titin interacts with the NH2-terminal domain of obscurin like-1, a protein similar to obscurin but transcribed from a separate gene. Notably, the interaction between obscurin like-1 and titin is independent of titin’s interaction with obscurin. Recent structural evidence suggests that these interactions could independently direct the binding of specific additional ligands at the M-band in a special or temporal manner (Pernigo et al. 2015). In skeletal muscle, the NH2-terminal Ig domains of obscurins also interact with sMyBP-Cv1 to provide additional structural support for the M-band lattice (Ackermann et al. 2009).
The necessity of obscurins for the integration of myosin into A-bands of sarcomeres can be demonstrated by several in vitro studies (Borisov et al. 2004, 2006; Kontrogianni-Konstantopoulos et al. 2004, 2006; Randazzo et al. 2013). Overexpression of the COOH-terminal region of obscurin in developing primary skeletal myotubes dramatically reduces A-band myosin organization, but other proteins of M-bands and Z-discs remain unchanged (Kontrogianni-Konstantopoulos et al. 2004). Overexpression of the second Ig domain of obscurin or suppression of giant obscurins in developing primary myotubes inhibits the assembly of the A-bands and thick filament structures (Ackermann et al. 2009). In addition, primary skeletal myotubes with reduced levels of giant obscurins show disruption of M-bands and A-bands (Kontrogianni-Konstantopoulos et al. 2006). These studies support a role for mammalian giant obscurins in the assembly and stability of A- and M-bands of myocytes.
Studies in other organisms, such as C. elegans, zebrafish, and Drosophila, also support a role for giant obscurins in M-band assembly. Notably, the SH3 domain of C. elegans UNC-89 interacts with paramyosin, a major structural component of thick filaments; this interaction is important for thick filament assembly at M-bands (Qadota et al. 2016). UNC-89 mutants lacking the SH3 domain result in paramyosin aggregates, suggesting that the SH3 domain of UNC-89 is essential for paramyosin localization and therefore thick filament assembly (Qadota et al. 2016). In addition, a zebrafish model depleted of obscurins resulted in myofibril disarray and impaired lateral alignment of adjacent myofibrils in both cardiac and skeletal muscle (Raeker et al. 2006). In the fly, decreased expression of Drosophila obscurin in the IFM during pupal development negatively affects myosin assembly (Katzemich et al. 2012), and knockdown of Drosophila obscurins in the adult IFM results in extensive disarray of thick filaments, sarcomeres lacking M-bands and flightless flies (Katzemich et al. 2012). Also, through their pseudo-kinase domains Drosophila obscurins interact with Ball, a Ser/Thr kinase, and MASK, a protein with multiple ankyrin repeats (Katzemich et al. 2015). Ball and MASK control cell proliferation and differentiation in other tissue types, but their function in striated muscle is unclear (Herzig et al. 2014; Smith et al. 2002; Yakulov et al. 2014). Drosophila obscurins function as a scaffold for Ball and MASK, which act downstream to define Z-disc and M-band structures during development (Katzemich et al. 2015). Collectively, these studies indicate that the interactions of obscurins, together with their spatial-temporal expression are key to their role in myofibrillogenesis and assembly and stability of M-band structures across species.
Obscurins facilitate titin’s sarcomeric incorporation at the Z-disc
Obscurins play a predominant role in the assembly of thick filaments through its M-band localization and interactions (discussed above). However, obscurins also mediate the sarcomeric incorporation of titin at the Z-disc (Bowman et al. 2008). Overexpression of the RhoGEF domain of obscurins in developing skeletal myotubes inhibits the incorporation of the NH2-terminus of titin into Z-discs and alters the alignment of A/I junctions but do not affect other sarcomeric structures or Z-disc proteins (Bowman et al. 2008). Notably, the RhoGEF domain of obscurins interacts with RanBP9, a scaffolding protein that binds to RanGTPases to regulate nuclear import and export (Bowman et al. 2008). RanBP9 is initially expressed in the nucleus and begins to translocate to the Z-disc where it colocalizes and interacts with obscurins in differentiating myotubes (Bowman et al. 2008). Similar to overexpression of the obscurin RhoGEF domain, overexpression of the obscurin binding region of RanBP9 also inhibits the incorporation of the NH2-terminus of titin into Z-discs and disrupts A/I junctions (Bowman et al. 2008). Taken together, these results support a role for obscurins, through their interaction with RanBP9 at the Z-disc, in facilitating titin’s sarcomeric incorporation at Z-discs during myocyte development.
Obscurins at the M-band
Obscurins provide structural support for thick filaments and M-band complexes in mature myocytes
The M-band scaffold between the NH2-terminus of obscurin, the COOH-terminus of titin, and the NH2-terminus of myomesin, which is essential for the assembly of thick filaments during myofibrillogenesis (discussed above), is also necessary for the stability of M-bands in mature myocytes. Structural and biochemical studies have shown that this complex is necessary for crosslinking thick filaments at M-bands between adjacent myofibrils (Fukuzawa et al. 2008; Kontrogianni-Konstantopoulos et al. 2004; Kontrogianni-Konstantopoulos et al. 2006; Pernigo et al. 2015).
Similar to mammalian obscurins, C. elegans UNC-89 localizes to M-bands (Benian et al. 1996); this process is mediated via interactions between the NH2-terminus of UNC-89 and CPNA-1, a copine domain protein that provides structural stability for integrin adhesion complexes (Warner et al. 2013). Notably, mutations in the UNC-89 gene results in M-band instability. During embryogenesis, UNC-89 larval mutants show normal assembly of myosin into M-bands (Spooner et al. 2012). However, adult nematodes carrying the same mutations in UNC-89 exhibit disorganized muscle structure, a lack of thick filament incorporation into A-bands, and an absence of M-bands (Spooner et al. 2012). Taken together, these studies across multiple species support an essential and conserved role for obscurins in thick filament stabilization and M-band structural support.
Obscurins function as signaling mediators at M-bands
The scaffolding roles of obscurins throughout the sarcomere are well-established; however the role of obscurins in signal transduction is less developed. Epitopes mapping to the RhoGEF domain of mammalian obscurins localize predominantly at sarcomeric M-bands (Table 1) (Young et al. 2001). RhoGEF domains catalyze the nucleotide exchange on Rho-GTPases (Schiller et al. 2006). Replacement of guanosine-5′-diphosphate (GDP) by guanosine-5′-triphosphate (GTP) activates Rho-GTPases, which function in the rearrangement of cytoskeletal components, trafficking of proteins to the membrane, translational regulation, and cell signaling (Schiller et al. 2006). The RhoGEF domain of obscurin interacts and colocalizes with RhoA, a small GTPase, at the M-band (Ford-Speelman et al. 2009). Exogenous expression of the obscurin RhoGEF domain in adult skeletal muscle activates RhoA, resulting in increased GTP-bound RhoA and a partial redistribution of RhoA to the cytoplasm, the I-band, and the Z-disc (Ford-Speelman et al. 2009). A downstream target of RhoA, Rho associated coiled-coil containing kinase 1 (ROCK1), which functions in a variety of cellular activities, including actin cytoskeletal organization, cell adhesion, and proliferation, also exhibited increased activation in the same system (Ford-Speelman et al. 2009). Similarly, in C. elegans, the RhoGEF domain of giant UNC-89 activates Rho-1 GTPase, the C. elegans homolog of mammalian RhoA, and functions to stabilize cytoskeletal components, specifically myosin filaments at M-bands (Qadota et al. 2008a).
Notably, following contraction-induced muscle injury in mammalian skeletal muscle with the ectopically expressed obscurin RhoGEF domain, there is also a redistribution of RhoA and activation of downstream molecules (Ford-Speelman et al. 2009). In addition, giant obscurins expressing the RhoGEF epitope are significantly increased in murine models of aortic constriction (Borisov et al. 2003), and increased RhoA and ROCK1 activity have been observed in murine models of cardiac hypertrophy (Ford-Speelman et al. 2009). Taken together, these findings support a role of the obscurin RhoGEF domain in regulating RhoA activity and suggest possible signaling functions for obscurins in the response to myocyte injury.
In addition to its direct role in signal transduction, obscurins also act as an adaptor to mediate phosphatase signaling at the M-band. To this end, obscurin interacts with ankyrin-B, a cytoskeletal adaptor protein, which targets protein phosphatase 2 (PP2A) to the M-band in cardiomyocytes (Cunha and Mohler 2008). A muscle-specific exon (exon 43′) of ANK2, the gene for ankyrin-B, encodes a region of the COOH-terminus that contains the obscurin binding site (Cunha and Mohler 2008). Notably, ankyrin binding is unique to obscurin isoforms containing the ankyrin binding domain, present in giant obscurin-A. Cunha and Mohler (2008) designed and used ankyrin-B constructs either missing or containing exon 43′ to define the interaction of obscurin and ankyrin-B at the M-band of cardiac muscle. Ectopic expression of the ankyrin-B construct lacking the obscurin binding site in rat neonatal cardiomyocytes resulted in normal localization of endogenous ankyrin-B at the M-band. To the contrary, exogenous expression of the ankyrin-B exon 43′ construct acted as a dominant negative to reduce M-band localization of endogenous ankyrin-B. Following expression of this dominant negative construct in primary cardiomyocytes, localization of PP2A to the M-band was also reduced (Cunha and Mohler 2008). These findings suggest the obscurin–ankyrin-B complex is necessary for recruitment of PP2A to the M-band. PP2A, a major protein phosphatase in the heart, is responsible for modulating Ca2+ signaling through its regulation of essential ion channels and pumps (Gergs et al. 2004; Lei et al. 2015, 2016). Aberrant expression or mislocalization of PP2A is associated with cardiac pathophysiology, including heart failure and arrhythmias (Gergs et al. 2004; Lei et al. 2015; Li et al. 2016). In addition, in C. elegans the protein phosphatase SCPL-1, which localizes to the M-band, interacts with the pseudo-kinase regions of UNC-89 (Qadota et al. 2008b; Xiong et al. 2009). SCPL-1 acts as a molecular bridge between two obscurin polypeptides (Qadota et al. 2008b). Another protein, LIM-9, which connects integrin proteins, also interacts with the obscurin–SCPL-1 complex (Xiong et al. 2009). The obscurin/SCPL-1/LIM-9 interaction likely functions to modulate the signaling activities of the phosphatase, but further studies are necessary to confirm this.
Also unique to C. elegans, the NH2-terminus and kinase regions of UNC-89 interact with MEL-26 at sarcomeric M-bands (Wilson et al. 2012). MEL-26 is a substrate recognition protein for cullin-3, an E3 ubiquitin ligase that is necessary for the assembly of the ubiquitin protein degradation machinery (Wilson et al. 2012). The interaction of UNC-89 with MEL-26 likely provides a docking site, thereby mediating the regulation of protein turnover. Collectively, these studies demonstrate that obscurins can function as an adaptor protein to mediate signaling activities at the M-band.
Obscurins at the Z-disc
Obscurin is a binding partner of titin at the Z-disc
The Z-disc is a structure composed of numerous proteins, which together function as a scaffold for cytoskeletal and myofilament proteins and also provide support to mediate the transduction of biochemical and mechanical signals (Frank and Frey 2011; Knoll et al. 2011; Pyle and Solaro 2004). Epitopes mapping to the NH2- and COOH-termini of giant obscurins localize to the Z-disc, while an epitope to the SK2 region of obscurin-B maps to the outer periphery of the Z-disc (Bowman et al. 2007), supporting the notion that multiple obscurin isoforms localize to the Z-disc region of the sarcomere (Table 1; Fig. 2). At the Z-disc, COOH-terminal Ig domains of obscurin interact with the NH2-terminus of titin (Young et al. 2001). It is likely that the obscurin–titin interaction at the Z-disc provides the basis for a scaffold of proteins necessary for proper Z-disc function; however further studies are necessary to confirm this.
Obscurins at cellular membranes
Obscurins at sites of internal membranes
Obscurins support normal Ca2+ homeostasis through links with the SR
Obscurins surround myofibrils at the level of Z-discs and M-bands where they interact with small ankyrin-1.5 (sAnk1), an integral protein of the network SR (Kontrogianni-Konstantopoulos et al. 2003). The SR is a striated muscle-specific, specialized form of smooth endoplasmic reticulum which functions to maintain intracellular Ca2+ homeostasis necessary for excitation–contraction coupling (Armani et al. 2006; Barone et al. 2015). The SR is organized into two main functional domains, namely, the junctional SR and the network SR, and surrounds the contractile apparatus at regular intervals throughout the sarcomere (Gyorke 2004). Junctional SR localizes at the level of the A/I junction of skeletal muscle in close proximity to t-tubules and is the main site for storage and release of Ca2+. Ryanodine receptors, the primary Ca2+ release channel of the SR, are located in the junctional SR and coupled to the L-type Ca2+ channels of the t-tubules (Gyorke et al. 2004). The network SR primarily surrounds the M-bands and Z-discs of skeletal muscle and forms a fluid membrane system with the junctional SR (Franzini-Armstrong et al. 2005). The sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA1), is mainly expressed within the network SR and is essential for Ca2+ reuptake mediating myocyte relaxation (Jorgensen et al. 1982). sAnk1, expressed in both cardiac and skeletal muscle, is one of five small isoforms of the ANK1 gene, which also encodes a much larger ankyrin-R protein (Bagnato et al. 2003; Borzok et al. 2007). The NH2−terminus of sAnk1 contains a transmembrane domain which inserts into the membranes of the SR while its COOH-terminus, located on the cytoplasmic side of the SR, interacts with the ankyrin binding domain of the COOH-terminus of obscurin-A (Barone et al. 2015; Kontrogianni-Konstantopoulos et al. 2003). The tight interaction between obscurin and sAnk1 is mediated by electrophilic interactions of positively charged amino acids on sAnk1 and negatively charged residues within the ankyrin binding domain of obscurin (Borzok et al. 2007; Busby et al. 2011). Reduction of sAnk1 in primary myofibers results in loss of network SR integrity, misloclaization of SERCA1, and aberrant Ca2+ handling (Ackermann et al. 2011). Recently, sAnk1 was identified as a novel regulatory protein of SERCA1 activity in skeletal muscle (Desmond et al. 2015).
Notably, the localization and function of sAnk1 is dependent upon the expression and subcellular distribution of obscurin within the sarcomere. In skeletal muscles of mice lacking giant obscurin proteins, network SR architecture is disrupted, and sAnk1 protein expression is decreased due to increased degradation via the potassium (K) channel tetramerization domain containing 6 (KCTD6)/cullin-3 complex (Lange et al. 2009). Cullin-3 binds sAnk1 through the substrate adaptor protein KCTD6 (Lange et al. 2012). The binding of sAnk1 to KCTD is regulated by several posttranslational modifications, including ubiquitination and acetylation of its COOH-terminal lysine residues (Lange et al. 2012). Together this work suggests that obscurin is necessary for the proper localization of sAnk1 and also protects sAnk1 from posttranslational modifications and subsequent degradation by the cullin-3/KCTD6 complex. Myocytes with reduced levels of sAnk1 result in aberrant Ca2+homeostasis; therefore, obscurin is a key mediator of Ca2+homeostasis in the myocyte.
In support of this notion, depletion of zebrafish obscurin containing the ankyirn binding domain, but not of zebrafish obscurin-MLCK and mutant forms of UNC-89, in C. elegans result in impaired organization of the SR and aberrant Ca2+ signaling (Raeker et al. 2006; Spooner et al. 2012). Together these studies highlight the important roles of obscurins in SR stability and normal Ca2+ homeostasis.
Obscurins at costameres
Function of obscurins as adaptors to link sarcomeric complexes to the sarcolemma
Costameres, specialized subsarcolemma structures, connect the contractile apparatus of the sarcomere to the cell membrane (Frank and Frey 2011). Obscurin polypeptides interact with ankyrin-B to provide a molecular bridge between membrane and cytoskeletal proteins, thereby promoting the assembly of essential protein complexes in skeletal muscle (Randazzo et al. 2013). At costameres, ankyrin-B complexes with dynactin-4 and β2-spectrin, both of which are necessary for the proper localization of dystrophin, a key cytoskeletal protein (Ayalon et al. 2008, 2011). In the absence of giant obscurins, ankyrin-B is absent from costameres, which results in mislocalized dystrophin and alterations in the organization of costamere-associated microtubules (Randazzo et al. 2013). Obscurin knockout mice injected with Evans Blue Dye (EBD) prior to exercise exhibited an increased number of EBD-positive muscle fibers, indicative of muscle fibers with increased muscle damage, similar to the mdx mouse model of muscular dystrophy (Randazzo et al. 2013). Therefore, the interaction of obscurin and ankyrin-B is necessary for the proper assembly of the dystrophin complex at costameres and is necessary to maintain sarcolemma integrity in skeletal muscle.
Obscurin at the intercalated disc
Obscurins function to mediate signal transduction at the ID
Kinase-containing isoforms of obscurin localize to the IDs of cardiomyocytes where they interact with N-cadherin and the Na+/K+ ATPase (Hu and Kontrogianni-Konstantopoulos 2013). Specifically, the SK2 domain of obscurins directly binds and phosphorylates N-cadherin (Hu and Kontrogianni-Konstantopoulos 2013). In addition, the SK2 kinase domain of obscurins autophosphorylates residues within the SK2 domain (Hu and Kontrogianni-Konstantopoulos 2013), suggesting an internal regulatory mechanism for the more 5′ kinase domain of obscurin. Through its SK2 domain, kinase-containing obscurins may play a role in modulating cardiomyocyte adhesion by its ability to phosphorylate N-cadherin. Further studies are necessary to confirm this. Additionally, the SK1 domain of obscurin interacts directly with the Na+/K+ ATPase (Hu and Kontrogianni-Konstantopoulos 2013); however the functional implications of that interaction are still undefined. Notably, targeted deletion of RhoGEF containing obscurin isoforms in zebrafish embryos resulted in a loss of organized IDs and absent myocyte–myocyte connections, rescuable by the introduction of a mini-obscurin containing many of the signaling motifs, including the SH3, RhoGEF and PH domains, but lacking the obscurin kinase domains (Fukuzawa et al. 2005; Raeker et al. 2010). Together these studies suggest a role for the many signaling domains of obscurin in ID stability and potentially cellular adhesion.
Role of obscurins in the development of myopathies
Variants of OBSCN are linked to human heart failure and cardiomyopathies
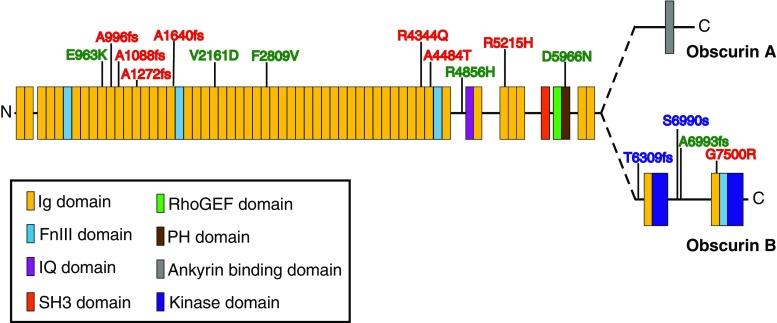
In support of the essential roles of obscurins in diverse cellular processes within striated muscle, obscurins have been implicated in fatal myopathies. Specifically, genomic linkage analysis has identified several variants of the OBSCN gene that are directly linked with the development of hypertrophic (HCM) and dilated (DCM) cardiomyopathy as well as left ventricular noncompaction (Fig. 4) (Arimura et al. 2007; Marston et al. 2015; Rowland et al. 2016; Xu et al. 2015). While the impact of these mutations on obscurin protein function is unknown, plausible consequences on the protein function can be speculated.
Fig. 4.
A cartoon depiction of the variants of the obscurin gene (OBSCN) and where they map to on the obscurin protein. Dilated and hypertrophic cardiomyopathies and left ventricular non-compaction are highlighted in green, red, and blue, respectively. fs Frameshift mutations
About a decade ago, the first two variants of OBSCN were identified in HCM patients (Arimura et al. 2007). To date eight variants of OBSCN are linked to HCM. Specifically, R4344Q and A4484T, which were identified in a single HCM patient, map to the COOH-terminal Ig domains of obscurin that interact with the Z-disc NH2-terminal domains of titin (Arimura et al. 2007). Molecular modeling of the obscurin Ig domains carrying the mutations predicted a structural change with the R4344Q variant but not the A4484T variant (Arimura et al. 2007). The R4344Q variant also shows decreased binding to titin, and ectopic expression of the Ig domains carrying this mutation result in impaired localization to the Z-disc compared to overexpression of wild-type Ig domains (Arimura et al. 2007). A possible pathogenic mechanism for the R4344Q variant of obscurin is decreased stability and integrity of sarcomeric structure at the Z-disc through disruption of the obscurin–titin complex; however, it cannot be denied that the R4344Q mutation of obscurin is involved in the development of HCM through an alternative mechanism. More recently, six additional variants of OBSCN were identified in HCM patients (Xu et al. 2015). Specifically, missense variants R5215H and G7500R map to one of the Ig domains between the IQ motif and SH3 domain and to the Ig domain just upstream of SK2, respectively. The functional significance of these regions and therefore of these variants is as yet undefined. In addition, four frameshift variants, namely, A966fs, A1640fs, A1088fs, and A1272fs, occurring prior to Ig domain 20 result in truncated proteins devoid of the signaling motifs and are classified as loss-of-function mutations (Xu et al. 2015).
Sequencing of OBSCN in patients with DCM identified four novel potentially pathogenic OBSCN variants (Marston et al. 2015). Notably, two of the four variants, E963K and R4856H, which map to regions of obscurins with unknown function, are found in conjunction with variants of DSP, the gene encoding desmoplakin, and SCN5A, which encodes the predominant voltage gated sodium channel of cardiomyocytes, Nav1.5, respectively (Marston et al. 2015). Variants in desmoplakin and Nav1.5 have been individually linked to DCM and arrhythmogenic cardiomyopathies (Hershberger et al. 2013), and therefore the pathogenic significance of these two obscurin variants remains unclear. Two other variants in OBSCN, V2161D and D5966N, which map to Ig domain 21, with unknown function, and the PH domain of obscurins, respectively, were also identified in separate DCM patients (Marston et al. 2015). In breast epithelial cells, the obscurin PH domain interacts with PI3K to mediate the activation of the PI3K pathway (Shriver et al. 2016). It is likely that the D5966N variant within the PH domain of obscurin impacts the activation of the PI3K pathway, leading to the pathogenesis of DCM. Notably, patients carrying OBSCN variants have lower levels of giant obscurin proteins than control DCM heart samples lacking OBSCN variants (Marston et al. 2015). In addition, patients with DCM of unknown origin and therefore lacking OBSCN variants also exhibit lower amounts of full-length giant obscurins compared to control non-failing hearts (Makarenko et al. 2004).
More recently, four novel variants of the OBSCN gene have been identified in patients with left ventricular noncompaction, a rare form of cardiomyopathy (Rowland et al. 2016). All four variants were localized to the COOH-terminus of obscurin-B upstream of the second protein kinase domain, SK1. Obscurin variants S7947fs and A7950fs cause frameshifts resulting in truncated obscurin proteins lacking SK1, while the T7266fs variant, located further upstream, lacks both SK2 and SK1. The fourth variant identified is a splicing variant, which also occurs within the 3′ end of OBSCN. The placement of these variants at the COOH-terminus suggests that the function of the NH2-terminal domains would not be affected; however these variants would be present in smaller kinase-containing isoforms of obscurin and likely greatly impact their functions.
Animal models of heart failure support the role of obscurins in the development of human cardiomyopathies. In a murine model of myocardial hypertrophy induced by pressure overload, the levels of specific regions of obscurin transcripts, in part encoding the RhoGEF and SK2 domains, have been shown to increase in response to cardiac stress (Borisov et al. 2003). The upregulation of transcripts encoding these signaling domains suggests their importance in processes occurring during cardiac hypertrophy, such as increased sarcomere assembly and myocyte growth. Although the pathogenic mechanism of the involvement of obscurins in the development of severe and fatal myopathies is undetermined, collectively these studies provide strong evidence to support the role of obscurins in the pathogenesis of cardiomyopathies.
Support for the involvement of obscurins in skeletal myopathies
Obscurins have also been implicated in the development of severe skeletal myopathies, such as limb-girdle muscular dystrophy 2 J and tibial muscular dystrophy. Specifically, variants of the extreme COOH-terminus of titin that are linked to these myopathies affect binding to the NH2-terminus of obscurin (Fukuzawa et al. 2008). Biopsies from a subset of patients with these variants revealed a distinct loss of obscurin at the M-band of sarcomeres (Fukuzawa et al. 2008). One of the hallmarks of muscular dystrophy is an increase in centrally nucleated fibers in skeletal muscle. Mice deficient in giant obscurins show an increase in central nucleated skeletal muscle fibers after 12 months of age compared to wild-type littermates (Lange et al. 2009). Together these studies suggest that within skeletal muscle obscurins contribute to the pathophysiology of select muscular dystrophies.
Concluding remarks
Obscurins are multi-domain proteins localized throughout myocytes as illustrated in Fig. 2. The diverse subcellular distributions and distinct domains of obscurin proteins allow not only the interaction of this family of proteins with numerous binding partners but also facilitate multiple functions for this protein family. Interestingly, obscurins can interact with a single ligand at multiple subcellular regions to elicit different functions. An example of this is the interaction of obscurins with ankyrin-B. At the M-band, obscurin acts as an adaptor to tether ankyrin-B and target PP2A to the M-band. However, its localization and interaction with ankyrin-B at costameres is necessary to provide a scaffold for essential costameric proteins, including dystrophin. Therefore, it is paramount that we consider the subcellular distribution of obscurins when defining their function.
Overall, the obscurin family of proteins is important for proper myocyte development, normal muscle function, and Ca2+ homeostasis. These proteins provide molecular scaffolds at distinct regions throughout the cell, serve as adaptor proteins to elicit signaling responses at specific locations, and play a direct role in signal transduction at specific sites throughout the myocyte. As detailed in this review, the function of obscurin is intimately linked to its localization within the myocyte as well as its local binding partners. To properly define the function of obscurin, one must first address its subcellular distribution and its local ligands. These important facts must be considered as we move forward in defining novel functions of obscurins and the role they play in the development of myopathies.
Acknowledgments
MAA acknowledges funding from the National Institutes of Health (HL116778).
Compliance with ethical standards
Conflict of interest
Maegen A. Ackermann declares that none of the authors have any conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
Footnotes
This article is part of a Special Issue on ‘Titin and its Binding Proteins in Striated Muscles’ edited by Amy Li and Cristobal G. dos Remedios.
References
- Ackermann MA, Hu LY, Bowman AL, Bloch RJ, Kontrogianni-Konstantopoulos A. Obscurin interacts with a novel isoform of MyBP-C slow at the periphery of the sarcomeric M-band and regulates thick filament assembly. Mol Biol Cell. 2009;20:2963–2978. doi: 10.1091/mbc.E08-12-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann MA, Shriver M, Perry NA, Hu LY, Kontrogianni-Konstantopoulos A. Obscurins: Goliaths and Davids take over non-muscle tissues. PLoS One. 2014;9:e88162. doi: 10.1371/journal.pone.0088162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann MA, Ziman AP, Strong J et al (2011) Integrity of the network sarcoplasmic reticulum in skeletal muscle requires small ankyrin 1. J Cell Sci 124:3619–3630. doi:10.1242/jcs.085159 [DOI] [PMC free article] [PubMed]
- Agarkova I, Perriard JC (2005) The M-band: an elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol 15:477–485. doi:10.1016/j.tcb.2005.07.001 [DOI] [PubMed]
- Arimura T, Matsumoto Y, Okazaki O et al (2007) Structural analysis of obscurin gene in hypertrophic cardiomyopathy. Biochem Biophys Res Commun 362:281–287. doi:10.1016/j.bbrc.2007.07.183 [DOI] [PubMed]
- Armani A, Galli S, Giacomello E et al (2006) Molecular interactions with obscurin are involved in the localization of muscle-specific small ankyrin1 isoforms to subcompartments of the sarcoplasmic reticulum. Exp Cell Res 312:3546–3558. doi:10.1016/j.yexcr.2006.07.027 [DOI] [PubMed]
- Ayalon G, Davis JQ, Scotland PB, Bennett V. An ankyrin-based mechanism for functional organization of dystrophin and dystroglycan. Cell. 2008;135:1189–1200. doi: 10.1016/j.cell.2008.10.018. [DOI] [PubMed] [Google Scholar]
- Ayalon G, Hostettler JD, Hoffman J, Kizhatil K, Davis JQ, Bennett V. Ankyrin-B interactions with spectrin and dynactin-4 are required for dystrophin-based protection of skeletal muscle from exercise injury. J Biol Chem. 2011;286:7370–7378. doi: 10.1074/jbc.M110.187831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang ML, Centner T, Fornoff F et al (2001) The complete gene sequence of titin, expression of an unusual ≈700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ Res 89:1065–72. doi:10.1161/hh2301.100981 [DOI] [PubMed]
- Bagnato P, Barone V, Giacomello E, Rossi D, Sorrentino V. Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J Cell Biol. 2003;160:245–253. doi: 10.1083/jcb.200208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balke CW, Shorofsky SR. Alterations in calcium handling in cardiac hypertrophy and heart failure. Cardiovasc Res. 1998;37:290–299. doi: 10.1016/S0008-6363(97)00272-1. [DOI] [PubMed] [Google Scholar]
- Barone V, Randazzo D, Del Re V, Sorrentino V, Rossi D. Organization of junctional sarcoplasmic reticulum proteins in skeletal muscle fibers. J Muscle Res Cell Motil. 2015;36:501–515. doi: 10.1007/s10974-015-9421-5. [DOI] [PubMed] [Google Scholar]
- Benian GM, Tinley TL, Tang X, Borodovsky M (1996) The Caenorhabditis elegans gene unc-89, required fpr muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J Cell Biol 132:835–848 [DOI] [PMC free article] [PubMed]
- Borisov AB, Kontrogianni-Konstantopoulos A, Bloch RJ, Westfall MV, Russell MW. Dynamics of obscurin localization during differentiation and remodeling of cardiac myocytes: obscurin as an integrator of myofibrillar structure. J Histochem Cytochem. 2004;52:1117–1127. doi: 10.1369/jhc.3A6183.2004. [DOI] [PubMed] [Google Scholar]
- Borisov AB, Raeker MO, Kontrogianni-Konstantopoulos A et al (2003) Rapid response of cardiac obscurin gene cluster to aortic stenosis: differential activation of Rho-GEF and MLCK and involvement in hypertrophic growth. Biochem Biophys Res Commun 310:910–918 [DOI] [PubMed]
- Borisov AB, Raeker MO, Russell MW. Developmental expression and differential cellular localization of obscurin and obscurin-associated kinase in cardiac muscle cells. J Cell Biochem. 2008;103:1621–1635. doi: 10.1002/jcb.21551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov AB, Sutter SB, Kontrogianni-Konstantopoulos A, Bloch RJ, Westfall MV, Russell MW. Essential role of obscurin in cardiac myofibrillogenesis and hypertrophic response: evidence from small interfering RNA-mediated gene silencing. Histochem Cell Biol. 2006;125:227–238. doi: 10.1007/s00418-005-0069-x. [DOI] [PubMed] [Google Scholar]
- Borzok MA, Catino DH, Nicholson JD, Kontrogianni-Konstantopoulos A, Bloch RJ. Mapping the binding site on small ankyrin 1 for obscurin. J Biol Chem. 2007;282:32384–32396. doi: 10.1074/jbc.M704089200. [DOI] [PubMed] [Google Scholar]
- Bowman AL, Catino DH, Strong JC, Randall WR, Kontrogianni-Konstantopoulos A, Bloch RJ. The rho-guanine nucleotide exchange factor domain of obscurin regulates assembly of titin at the Z-disk through interactions with Ran binding protein 9. Mol Biol Cell. 2008;19:3782–3792. doi: 10.1091/mbc.E08-03-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman AL, Kontrogianni-Konstantopoulos A, Hirsch SS, Geisler SB, Gonzalez-Serratos H, Russell MW, Bloch RJ. Different obscurin isoforms localize to distinct sites at sarcomeres. FEBS Lett. 2007;581:1549–1554. doi: 10.1016/j.febslet.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby B, Oashi T, Willis CD et al (2011) Electrostatic interactions mediate binding of obscurin to small ankyrin 1: biochemical and molecular modeling studies. J Mol Biol 408:321–334. doi:10.1016/j.jmb.2011.01.053 [DOI] [PMC free article] [PubMed]
- Carlsson L, Yu JG, Thornell LE. New aspects of obscurin in human striated muscles. Histochem Cell Biol. 2008;130:91–103. doi: 10.1007/s00418-008-0413-z. [DOI] [PubMed] [Google Scholar]
- Cunha SR, Mohler PJ. Obscurin targets ankyrin-B and protein phosphatase 2A to the cardiac M-line. J Biol Chem. 2008;283:31968–31980. doi: 10.1074/jbc.M806050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmond PF, Muriel J, Markwardt ML, Rizzo MA, Bloch RJ. Identification of small ankyrin 1 as a novel sarco(endo)plasmic reticulum Ca2 + −ATPase 1 (SERCA1) regulatory protein in skeletal muscle. J Biol Chem. 2015;290:27854–27867. doi: 10.1074/jbc.M115.676585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara TM, Flaherty DB, Benian GM (2005) Titin/connectin-related proteins in C. elegans: a review and new findings. J Muscle Res Cell Motil 26:435–447. doi:10.1007/s10974-005-9027-4 [DOI] [PubMed]
- Ford-Speelman DL, Roche JA, Bowman AL, Bloch RJ. The rho-guanine nucleotide exchange factor domain of obscurin activates rhoA signaling in skeletal muscle. Mol Biol Cell. 2009;20:3905–3917. doi: 10.1091/mbc.E08-10-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D, Frey N. Cardiac Z-disc signaling network. J Biol Chem. 2011;286:9897–9904. doi: 10.1074/jbc.R110.174268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzini-Armstrong C, Protasi F, Tijskens P. The assembly of calcium release units in cardiac muscle. Ann N Y Acad Sci. 2005;1047:76–85. doi: 10.1196/annals.1341.007. [DOI] [PubMed] [Google Scholar]
- Fukuzawa A, Idowu S, Gautel M. Complete human gene structure of obscurin: implications for isoform generation by differential splicing. J Muscle Res Cell Motil. 2005;26:427–434. doi: 10.1007/s10974-005-9025-6. [DOI] [PubMed] [Google Scholar]
- Fukuzawa A, Lange S, Holt M et al (2008) Interactions with titin and myomesin target obscurin and obscurin-like 1 to the M-band: implications for hereditary myopathies. J Cell Sci 121:1841–1851. doi:10.1242/jcs.028019 [DOI] [PubMed]
- Gergs U, Boknik P, Buchwalow I et al (2004) Overexpression of the catalytic subunit of protein phosphatase 2A impairs cardiac function. J Biol Chem 279:40827–40834. doi:10.1074/jbc.M405770200 [DOI] [PubMed]
- Gieseler K, Qadota H, Benian GM (2016) Development, structure, and maintenance of C. elegans body wall muscle. In: The C.elegans Research Community (ed) WormBook, pp 1–59. doi:10.1895/wormbook.1.81.2 [DOI] [PMC free article] [PubMed]
- Gyorke S, Gyorke I, Terentyev D, Viatchenko-Karpinski S, Williams SC. Modulation of sarcoplasmic reticulum calcium release by calsequestrin in cardiac myocytes. Biol Res. 2004;37:603–607. doi: 10.4067/S0716-97602004000400014. [DOI] [PubMed] [Google Scholar]
- Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10:531–547. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- Herzig B, Yakulov TA, Klinge K, Gunesdogan U, Jackle H, Herzig A (2014) Ballchen is required for self-renewal of germline stem cells in Drosophila melanogaster. Biol Open 3:510–521. doi:10.1242/bio.20147690 [DOI] [PMC free article] [PubMed]
- Hu LY, Kontrogianni-Konstantopoulos A. The kinase domains of obscurin interact with intercellular adhesion proteins. FASEB J. 2013;27:2001–2012. doi: 10.1096/fj.12-221317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hund TJ, Koval OM, Li J et al (2010) A beta(IV)-spectrin/CaMKII signaling complex is essential for membrane excitability in mice. J Clin Invest 120:3508–3519. doi:10.1172/JCI43621 [DOI] [PMC free article] [PubMed]
- Jorgensen AO, Shen AC, MacLennan DH, Tokuyasu KT (1982) Ultrastructural localization of the Ca2+ + Mg2+ −dependent ATPase of sarcoplasmic reticulum in rat skeletal muscle by immunoferritin labeling of ultrathin frozen sections. J Cell Biol 92:409–416 [DOI] [PMC free article] [PubMed]
- Katzemich A, Kreiskӧther N, Alexandrovich A et al (2012) The function of the M-line protein obscurin in controlling the symmetry of the sarcomere in the flight muscle of Drosophila. J Cell Sci 125:3367–3379. doi:10.1242/jcs.097345 [DOI] [PMC free article] [PubMed]
- Katzemich A, West RJ, Fukuzawa A et al (2015) Binding partners of the kinase domains in Drosophila obscurin and their effect on the structure of the flight muscle. J Cell Sci 128:3386–3397. doi:10.1242/jcs.170639 [DOI] [PMC free article] [PubMed]
- Knoll R, Buyandelger B, Lab M (2011) The sarcomeric Z-disc and Z-discopathies J Biomed Biotechnol 2011:569628. doi:10.1155/2011/569628 [DOI] [PMC free article] [PubMed]
- Kontrogianni-Konstantopoulos A, Ackermann MA, Bowman AL, Yap SV, Bloch RJ. Muscle giants: molecular scaffolds in sarcomerogenesis. Physiol Rev. 2009;89:1217–1267. doi: 10.1152/physrev.00017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A, Bloch RJ. Obscurin: a multitasking muscle giant. J Muscle Res Cell Motil. 2005;26:419–426. doi: 10.1007/s10974-005-9024-7. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A, Catino DH, Strong JC, Randall WR, Bloch RJ. Obscurin regulates the organization of myosin into A bands. Am J Physiol Cell Physiol. 2004;287:C209–217. doi: 10.1152/ajpcell.00497.2003. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A, Catino DH, Strong JC et al (2006) Obscurin modulates the assembly and organization of sarcomeres and the sarcoplasmic reticulum. FASEB J 20:2102–2111. doi:10.1096/fj.06-5761com [DOI] [PubMed]
- Kontrogianni-Konstantopoulos A, Jones EM, Van Rossum DB, Bloch RJ. Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol Biol Cell. 2003;14:1138–1148. doi: 10.1091/mbc.E02-07-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S, Ouyang K, Meyer G et al (2009) Obscurin determines the architecture of the longitudinal sarcoplasmic reticulum. J Cell Sci 122:2640–2650. doi:10.1242/jcs.046193 [DOI] [PMC free article] [PubMed]
- Lange S, Perera S, Teh P, Chen J. Obscurin and KCTD6 regulate cullin-dependent small ankyrin-1 (sAnk1.5) protein turnover. Mol Biol Cell. 2012;23:2490–2504. doi: 10.1091/mbc.E12-01-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Wang X, Ke Y, Solaro RJ. Regulation of Ca(2+) transient by PP2A in normal and failing heart. Front Physiol. 2015;6:13. doi: 10.3389/fphys.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Fang C, Xu D, Xu Y, Fu H, Li J. Cardiomyocyte specific deletion of PP2A causes cardiac hypertrophy Am J. Transl Res. 2016;8:1769–1779. [PMC free article] [PubMed] [Google Scholar]
- Makarenko I, Opitz CA, Leake MC et al (2004) Passive stiffness changes caused by upregulation of compliant titin isoforms in human dilated cardiomyopathy hearts. Circ Res 95:708–716. doi:10.1161/01.RES.0000143901.37063.2f [DOI] [PubMed]
- Marston S, Montgiraud C, Munster AB et al (2015) OBSCN Mutations associated with dilated cardiomyopathy and haploinsufficiency. PLoS One 10:e0138568. doi:10.1371/journal.pone.0138568 [DOI] [PMC free article] [PubMed]
- Meyer LC, Wright NT. Structure of giant muscle proteins. Front Physiol. 2013;4:368. doi: 10.3389/fphys.2013.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernigo S, Fukuzawa A, Pandini A et al (2015) The crystal structure of the human titin:obscurin complex reveals a conserved yet specific muscle M-band zipper module. J Mol Biol 427:718–736. doi:10.1016/j.jmb.2014.11.019 [DOI] [PubMed]
- Perry NA, Ackermann MA, Shriver M, Hu LY, Kontrogianni-Konstantopoulos A. Obscurins: unassuming giants enter the spotlight. IUBMB Life. 2013;65:479–486. doi: 10.1002/iub.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle WG, Solaro RJ. At the crossroads of myocardial signaling: the role of Z-discs in intracellular signaling and cardiac function. Circ Res. 2004;94:296–305. doi: 10.1161/01.RES.0000116143.74830.A9. [DOI] [PubMed] [Google Scholar]
- Qadota H, Blangy A, Xiong G, Benian GM (2008a) The DH-PH region of the giant protein UNC-89 activates RHO-1 GTPase in Caenorhabditis elegans body wall muscle. J Mol Biol 383:747–752. doi:10.1016/j.jmb.2008.08.083 [DOI] [PMC free article] [PubMed]
- Qadota H, Mayans O, Matsunaga Y et al (2016) The SH3 domain of UNC-89 (obscurin) interacts with paramyosin, a coiled-coil protein, in Caenorhabditis elegans muscle. Mol Biol Cell 27:1606–1620. doi:10.1091/mbc.E15-09-0675 [DOI] [PMC free article] [PubMed]
- Qadota H, McGaha LA, Mercer KB, Stark TJ, Ferrara TM, Benian GM (2008b) A novel protein phosphatase is a binding partner for the protein kinase domains of UNC-89 (Obscurin) in Caenorhabditis elegans. Mol Biol Cell 19:2424–2432. doi:10.1091/mbc.E08-01-0053 [DOI] [PMC free article] [PubMed]
- Raeker MO, Bieniek AN, Ryan AS, Tsai HJ, Zahn KM, Russell MW. Targeted deletion of the zebrafish obscurin A RhoGEF domain affects heart, skeletal muscle and brain development. Dev Biol. 2010;337:432–443. doi: 10.1016/j.ydbio.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeker MO, Su F, Geisler SB et al (2006) Obscurin is required for the lateral alignment of striated myofibrils in zebrafish. Dev Dyn 235:2018–2029. doi:10.1002/dvdy.20812 [DOI] [PubMed]
- Randazzo D, Giacomello E, Lorzenzini S et al (2013) Obscurin is required for ankyrinB-dependent dystrophin localization and sarcolemma integrity. J Cell Biol 200:523–536. doi:10.1083/jcb.201205118 [DOI] [PMC free article] [PubMed]
- Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. FASEB J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- Rowland TJ, Graw SL, Sweet ME, Gigli M, Taylor MR, Mestroni L. Obscurin Variants in Patients With Left Ventricular Noncompaction. J Am Coll Cardiol. 2016;68:2237–2238. doi: 10.1016/j.jacc.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger JW, Chowrashi P, Shaner NC et al (2002) Myofibrillogenesis in skeletal muscle cells. Clin Orthop Relat Res:S153-162 [DOI] [PubMed]
- Schiller MR, Chakrabarti K, King GF, Schiller NI, Eipper BA, Maciejewski MW. Regulation of RhoGEF activity by intramolecular and intermolecular SH3 domain interactions. J Biol Chem. 2006;281:18774–18786. doi: 10.1074/jbc.M512482200. [DOI] [PubMed] [Google Scholar]
- Shriver M, Marimuthu S, Paul C et la (2016) Giant obscurins regulate the PI3K cascade in breast epithelial cells via direct binding to the PI3K/p85 regulatory subunit. Oncotarget. doi:10.18632/oncotarget.9985 [DOI] [PMC free article] [PubMed]
- Shriver M, Stroka KM, Vitolo MI et al (2015) Loss of giant obscurins from breast epithelium promotes epithelialto- mesenchymal transition, tumorigenicity and metastasis. Oncogene 34:4248–4259. doi:10.1038/onc.2014.358 [DOI] [PMC free article] [PubMed]
- Small TM, Gernert KM, Flaherty DB, Mercer KB, Borodovsky M, Benian GM (2004) Three new isoforms of Caenorhabditis elegans UNC-89 containing MLCK-like protein kinase domains. J Mol Biol 342:91–108. doi:10.1016/j.jmb.2004.07.006 [DOI] [PubMed]
- Smith RK, Carroll PM, Allard JD, Simon MA (2002) MASK, a large ankyrin repeat and KH domain-containing protein involved in Drosophila receptor tyrosine kinase signaling. Development 129:71–82 [DOI] [PubMed]
- Spooner PM, Bonner J, Maricq AV, Benian GM, Norman KR (2012) Large isoforms of UNC-89 (obscurin) are required for muscle cell architecture and optimal calcium release in Caenorhabditis elegans. PLoS One 7:e40182. doi:10.1371/journal.pone.0040182 [DOI] [PMC free article] [PubMed]
- Sutter SB, Raeker MO, Borisov AB, Russell MW. Orthologous relationship of obscurin and Unc-89: phylogeny of a novel family of tandem myosin light chain kinases. Dev Genes Evol. 2004;214:352–359. doi: 10.1007/s00427-004-0413-5. [DOI] [PubMed] [Google Scholar]
- Warner A, Xiong G, Qadota H et al (2013) CPNA-1, a copine domain protein, is located at integrin adhesion sites and is required for myofilament stability in Caenorhabditis elegans. Mol Biol Cell 24:601–616. doi:10.1091/mbc.E12-06-0478 [DOI] [PMC free article] [PubMed]
- Wilson KJ, Qadota H, Mains PE, Benian GM (2012) UNC-89 (obscurin) binds to MEL-26, a BTB-domain protein, and affects the function of MEI-1 (katanin) in striated muscle of Caenorhabditis elegans. Mol Biol Cell 23:2623–2634. doi:10.1091/mbc.E12-01-0055 [DOI] [PMC free article] [PubMed]
- Xiong G, Qadota H, Mercer KB, McGaha LA, Oberhauser AF, Benian GM (2009) A LIM-9 (FHL)/SCPL-1 (SCP) complex interacts with the C-terminal protein kinase regions of UNC-89 (obscurin) in Caenorhabditis elegans muscle. J Mol Biol 386:976–988 [DOI] [PMC free article] [PubMed]
- Xu J, Li Z, Ren X et al (2015) Investigation of pathogenic genes in Chinese sporadic hypertrophic cardiomyopathy patients by whole exome sequencing. Sci Rep 5:16609. doi:10.1038/srep16609 [DOI] [PMC free article] [PubMed]
- Yakulov T, Gunesdogan U, Jackle H, Herzig A (2014) Ballchen participates in proliferation control and prevents the differentiation of Drosophila melanogaster neuronal stem cells. Biol Open 3:881–886. doi:10.1242/bio.20148631 [DOI] [PMC free article] [PubMed]
- Young P, Ehler E, Gautel M. Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly. J Cell Biol. 2001;154:123–136. doi: 10.1083/jcb.200102110. [DOI] [PMC free article] [PubMed] [Google Scholar]