Abstract
The inherited cardiomyopathies, hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM) and left ventricular non-compaction (LVNC), have been frequently associated with mutations in sarcomeric proteins. In recent years, advances in DNA sequencing technology has allowed the study of the giant proteins of the sarcomere, such as titin and nebulin. Obscurin has been somewhat neglected in these studies, largely because its functional role is far from clear, although there was an isolated report in 2007 of obscurin mutations associated with HCM. Recently, whole exome sequencing methodology (WES) has been used to address mutations in OBSCN, the gene for obscurin, and OBSCN variants were found to be relatively common in inherited cardiomyopathies. In different studies, 5 OBSCN unique variants have been found in a group of 30 end-stage failing hearts, 6 OBSCN unique variants in 74 HCM cases and 3 OBSCN unique variants in 10 LVNC patients. As yet, the number of known potentially disease-causing OBSCN variants is quite small. The reason for this is that mutations in the OBSCN gene have not been recognised as potentially disease-causing until recently, and were not included in large-scale genetic surveys. OBSCN mutations may be causative of HCM, DCM and LVNC and other cardiomyopathies, or they may work in concert with other variants in the same or other genes to initiate the pathology. Currently, the function of obscurin is not well understood, but we anticipate that many more OBSCN variants linked to cardiomyopathy will be found when the large cohorts of patient sequences available are tested. It is to be hoped that the establishment of the importance of obscurin in pathology will stimulate a thorough investigation of obscurin function.
Keywords: Cardiac muscle, Obscurin, Mutation, Dilated carriomyopathy, Hypertrophic cardiomyopathy
The inherited cardiomyopathies, particularly hypertrophic cardiomyopathy (HCM) dilated cardiomyopathy (DCM) and left ventricular non-compaction (LVNC), have been frequently associated with mutations in the sarcomeric proteins. In recent years, advances in DNA sequencing technology have allowed the study of the giant proteins of the sarcomere, notably truncating mutations in the titin gene (TTNtv), which have been found to be responsible for up to 25% of familial DCM cases (Roberts et al. 2015), and mutations in nebulin (NEB), the most common mutations causing skeletal muscle myopathies such as nemaline myopathy (Lehtokari et al. 2014).
Obscurin has been somewhat neglected in these studies, largely because its functional role is far from clear. There was an isolated report in 2007 of obscurin mutations associated with HCM but this was not followed up for several years. Recently, whole exome sequencing methodology (WES) has been used to address mutations in OBSCN, the gene for obscurin, and OBSCN variants were found to be relatively common in inherited cardiomyopathies. Marston et al.’s (2015) study found 5 OBSCN unique variants in 4 individual hearts from a group of 30 end-stage failing hearts, Xu et al. (2015) found 6 OBSCN unique variants in 74 HCM cases and Rowland et al. (2016) found 4 OBSCN unique variants in 335 DCM and LVNC patients, of which OBSCN variants were associated with 3 out of 11 LVNC cases (Table 1).
Table 1.
Cardiomyopathy-linked Obscurin mutations based on the Obscurin B sequence NP_001092093 (Fukuzawa et al. 2005)
| Mutation | Pathology | Domain | |
|---|---|---|---|
| Arimura et al. | R4344Q | HCM | Ob58 |
| A4484T | HCM | Ob59 | |
| Marston et al. | E963K | DCM | Ob 9 |
| V2161D | DCM | Ob 21 | |
| F2809 V | DCM | Ob 27 | |
| R4856H | DCM | Ob 47 | |
| D5966N | DCM | PH | |
| Rowland et al. | T6309R | LVNC | Between Ob66 and Ob67 |
| S6990P | LVNC | Between kinase I and Ob69 | |
| A6993P | DCM | Between kinase I and Ob69 | |
| c25367-1G > C | LVNC | ||
| Xu et al. | A996fs | HCM | Ob 10 |
| A1088fs | HCM | Ob 11 | |
| A1272fs | HCM | Ob13 | |
| A1640fs | HCM | Ob 17 | |
| G7500R | HCM | Ob 69 |
The locations of the mutations are shown in the obscurin B sequence in Fig. 1. Note that the published amino acid numbers in Rowland et al. 2016 were based on NP_001258152 and have been converted to NP_001092093 numbering for comparison
On this basis, potentially disease-causing OBSCN variants seem to be fairly common in all three diseases. As yet, the number of known potentially disease-causing OBSCN variants is quite small. The reason for this is that mutations in the OBSCN gene have not been recognised as potentially disease-causing until recently, and are not included in current large-scale genetic surveys, For instance, Alfares et al. (2015) studied 2912 HCM cases, Ware et al. (2016) studied 172 peripartum cardiomyopathy cases and Haas et al. (2014) studied 639 DCM cases. It is noteworthy that the largest study to date, in which 46 genes in 7855 cardiomyopathy cases were compared with 60,706 reference samples, did not analyse variants in the OBSCN gene (Walsh et al. 2017). We anticipate that, when these large cohorts are re-examined for OBSCN disease-related variants, a considerable number will be found.
All the recent studies on OBSCN variants have involved whole exon sequencing with the unique variants being identified by comparison with large reference databases, such as Exacs and EVS, and their pathogenicity predicted using algorithms such as SIFT. This approach, based on small populations, may be biased in patient ethnicity and other factors. It is recognised that such conclusions may give a false positive association of disease with variants especially in a large protein (Walsh et al. 2017). Variants that cause chain termination, as found in the Rowland et al. (2016) and Xu et al. (2015) studies, are more likely to be pathogenic than missense mutations; however, none of the current studies on OBSCN variants have any family history showing the variant segregating with the disease, which is required to confirm causality. In the absence of additional data, we cannot exclude the possibility that the observed variants are simply bystander effects and that the true disease-causing mutation is elsewhere.
The only study that has directly investigated the obscurin molecule in heart muscle with identified mutations was Marston et al.’s (2015) study of familial DCM. where obscurin haploinsufficiency was demonstrated for all the OBSCN variant samples compared with fDCM with mutations identified in other genes or in donor heart samples. This, therefore, increases the confidence that OBSCN variants and familial DCM are linked in this particular group of samples from patients who had transplants for end-stage heart failure. In their studies on TTNtv mutations, Roberts et al. (2015) found a strong correlation of end-stage failure with mutation; however, the other studies on OBSCN used larger groups with less severe disease and the association is less certain.
OBSCN variants may be monogenic causes of cardiomyopathy or they may contribute to the disease phenotype in concert with other variants. It is therefore relevant to note that a significant proportion of the reported OBSCN variant samples also had a second disease-related variant. The HCM patient studied by Arimura et al. (2007) had two OBSCN mutations: R4344Q and A4484T. Arimura suggested that only the R4344Q variant was pathogenic, and it has since been shown that the A4484T variant is, in fact, common (15%) in black Americans. The A4484T variant has recently been downgraded to “disease-causing?” in a recent analysis of pathogenicity of putative HCM mutations (Manrai et al. 2016); however, there is no direct evidence for the relative roles of these variants and both may be required for pathogenicity. Similarly, Marston et al. (2015) found a DCM sample with two OBSCN mutations: V2161D and F2809V. This paper also lists two other double variants: OBSCN E963K + DSP R1537C and OBSCN R4856H + SCN5A S216L; both the DSP and the SCN5A variant have been previously noted as being associated with cardiomyopathies but not causative (Marangoni et al. 2011; Xu et al. 2010). On balance, it seems likely that most OBSCN variants contribute to cardiomyopathy rather than being a monogenic cause.
With other disease-causing mutations such as MyBP-C and nebulin, the discovery of disease-related mutations has stimulated investigation of the structure and function of the protein in muscle, but this has not happened for obscurin. The structure of obscurin is well established based on the amino acid sequence derived from cDNA sequencing (Fig. 1). The canonical sequence of obscurin B, the largest isoform, has 7968 amino acids. It is a typical modular protein being largely made up from multiple Ig domains plus three Fn3 domains, one IQ domain, one DH domain and one PH domain (Fukuzawa et al. 2005). In muscle, two large alternatively spliced isoforms are found, obscurin A and B. Obscurin A contains a C-terminal interaction site with sAnk1 and other ankryn isoforms, whilst obscurin B has two C-terminal kinase domains. Three of the OBSCN variants were found to be in the two-kinase domain sequence specific to obscurin B and none have been found in the obscurin A C-terminal sequence (Table 1). Western blots of obscurin in human heart muscle show a single band with an apparent molecular mass of 960 kD, suggesting that obscurin B is the main isoform in human heart (Marston et al. 2015), although both isoforms are present in rat and mouse hearts. It should be recognised that there are further isoforms of obscurin A and B expressed in many non-muscle tissues with shorter N-terminal sequences and a common C-terminus (Ackermann et al. 2014). OBSCN variants have been associated with pathologies including Wilm’s tumour and with aspirin sensitivity in asthmatics which could involve these smaller isoforms (Perry et al. 2013).
Fig. 1.
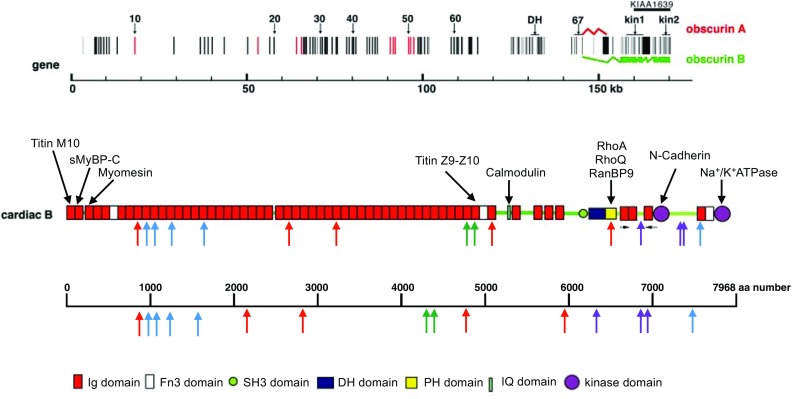
Obscurin structure and cardiomyopathy-associated mutations, based on the model of Fukuzawa et al. (2005). Top gene structure, showing the exons and alternative splicing that yields obscurin A or B. Middle schematic domain structure of obscurin B. The proposed interaction sites are shown above the model and the location of the mutations are shown below (refer to Table 1). Note that this model is not to scale. Bottom location of the mutations in the 7688 amino acid sequence of obscurin B (NP_001092093)
Obscurin has been found to interact with numerous other proteins in vitro. Best characterised are the N-terminal domains that interact with M-line titin and myomesin which anchor the N-terminus of obscurin to the M-line complex and contribute to its stability. Titin M10 Ig domain interacts with the N-terminal Ig domain of obscurin and the myomesin Ig domain 4 interacts with the third Ig domain of obscurin (Gautel 2011; Pernigo et al. 2010, 2017). In addition, obscurin domains Ob58/59 interact with titin Ig domains Z9/Z10 in vitro, potentially anchoring obscurin to the Z-disk (Bang et al. 2001).
The sAnk1 binding site in the C-terminus of obscurinA has also been extensively studied. This interaction is particularly significant because it represents a direct molecular link between the contractile sarcomere and the SR membrane, illuminating a possible mechanism for the alignment and anchoring of the SR around the developing myofibril and during contraction (Bagnato et al. 2003; Kontrogianni-Konstantopoulos et al. 2003; Perry et al. 2013). The PH domain interacts with Rho proteins. It can specifically induce exchange of GDP for GTP in the small GTPases, RhoA and RhoQ (Ford-Speelman et al. 2009; Perry et al. 2013; Russell et al. 2002), leading to activation of their downstream effectors. Since RhoA activity plays an important role in normal myofibril growth and during pathologic hypertrophy, this interaction could be physiologically significant. The kinase domains of obscurin appear to be active and have been shown to phosphorylate N-cadherin and Na+/K+ ATPase (Hu and Kontrogianni-Konstantopoulos 2013; Russell et al. 2002). In striated muscles, obscurin has been shown to interact with a novel isoform of sleletal MyBP-C (Ackermann et al. 2009) . The functional relevance of these domain interactions is still not well understood, but the current consensus is that obscurin is involved in the development of the myofibril and sarcoplasmic reticulum. Obscurin also forms a physical link between the M-line and the sarcoplasmic reticulum or sarcolemma which could include a mechanosensing function.
The OBSCN variants found so far seem to be distributed throughout the molecule (Fig. 1), mostly with no apparent connection to any functional domain. Arimura et al. (2007) have proposed that the R4344Q mutation affects binding of obscurin to Z-line titin (domains Z9-Z10), and Marston et al. (2015) noted obscurin haploinsufficiency but did not measure any functional properties. The best clues to the physiological function of obscurin are provided by the obscurin knockout mouse model (Lange et al. 2009); however, the effects of knockout have only been fully studied in skeletal muscle rather than in cardiac muscle and no cardiac defect was reported. In skeletal muscle, absence of obscurin results in disorganisation of microtubules at the sarcolemma and delocalisation of dystrophin at costameres. More striking abnormalities appear with intense exercise, including fibre damage, atrophy and degeneration which revert when exercise is stopped (Randazzo et al. 2017). It was proposed that the absence of obscurin causes the failure of the muscle sarcolemma to support the mechanical stress of intensive chronic contraction. In the heart, this may be manifested as development of a hypo-contractile heart under stress which could trigger DCM.
In conclusion, despite limited investigations, mutations in the OBSCN gene are emerging as significant in a range of cardiomyopathies. OBSCN mutations may be causative of HCM, DCM and LVNC and other cardiomyopathies, or they may work in concert with other variants in the same or other genes to initiate the pathology. Currently, the function of obscurin is not well understood other than the suggestion that it is not an essential gene and that its functions involve stabilisation of sarcomere to sarcoplasmic-reticulum and sarcomlemma links that may only become important during high levels of contraction. We anticipate that many more OBSCN variants linked to cardiomyopathy will be found when the large cohorts of patient sequences available are tested. It is to be hoped that the establishment of the importance of obscurin in pathology will stimulate a thorough investigation of obscurin function.
Acknowledgement
This work was supported by a grant from the British Heart Foundation (RG/11/20/29266).
Compliance with ethical standards
Conflict of interest
Steven Marston declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on ‘Titin and its Binding Proteins in Striated Muscles’ edited by Amy Li and Cristobal G. dos Remedios.
References
- Ackermann MA, Hu L-YR, Bowman AL, Bloch RJ, Kontrogianni-Konstantopoulos A. Obscurin interacts with a novel isoform of MyBP-C slow at the periphery of the sarcomeric M-band and regulates thick filament assembly. Mol Biol Cell. 2009;20:2963–2978. doi: 10.1091/mbc.E08-12-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann MA, Shriver M, Perry NA, Hu L-YR, Kontrogianni-Konstantopoulos A. Obscurins: goliaths and Davids take over non-muscle tissues. PLoS ONE. 2014;9:e88162. doi: 10.1371/journal.pone.0088162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfares AA, et al. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med. 2015;17:880–888. doi: 10.1038/gim.2014.205. [DOI] [PubMed] [Google Scholar]
- Arimura T, et al. Structural analysis of obscurin gene in hypertrophic cardiomyopathy. Biochem Biophys Res Commun. 2007;362:281–287. doi: 10.1016/j.bbrc.2007.07.183. [DOI] [PubMed] [Google Scholar]
- Bagnato P, Barone V, Giacomello E, Rossi D, Sorrentino V. Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J Cell Biol. 2003;160:245–253. doi: 10.1083/jcb.200208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang ML, et al. The complete Gene sequence of titin, expression of an unusual 700-kDa titin isoform, and its interaction with Obscurin identify a novel Z-line to I-band linking system. Circ Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- Ford-Speelman DL, Roche JA, Bowman AL, Bloch RJ. The rho-guanine nucleotide exchange factor domain of obscurin activates rhoA signaling in skeletal muscle. Mol Biol Cell. 2009;20:3905–3917. doi: 10.1091/mbc.E08-10-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa A, Idowu S, Gautel M (2005) Complete human gene structure of obscurin: implications for isoform generation by differential splicing. J Musc Res Cell Motil 26(6–8):427–434 [DOI] [PubMed]
- Gautel M. The sarcomeric cytoskeleton: who picks up the strain? Curr Opin Cell Biol. 2011;23:39–46. doi: 10.1016/j.ceb.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Haas J, et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. 2014;36:1123–1135. doi: 10.1093/eurheartj/ehu301. [DOI] [PubMed] [Google Scholar]
- Hu LYR, Kontrogianni-Konstantopoulos A. The kinase domains of obscurin interact with intercellular adhesion proteins. FASEB J. 2013;27:2001–2012. doi: 10.1096/fj.12-221317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A, Jones EM, Van Rossum DB, Bloch RJ. Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol Biol Cell. 2003;14:1138–1148. doi: 10.1091/mbc.E02-07-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S, Ouyang K, Meyer G, Cui L, Cheng H, Lieber RL, Chen J. Obscurin determines the architecture of the longitudinal sarcoplasmic reticulum. J Cell Sci. 2009;122:2640–2650. doi: 10.1242/jcs.046193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtokari V-L, et al. Mutation update: the spectra of nebulin variants and associated myopathies. Hum Mutat. 2014;35:1418–1426. doi: 10.1002/humu.22693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrai AK, et al. Genetic misdiagnoses and the potential for health disparities. New Eng J Med. 2016;375:655–665. doi: 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marangoni S, et al. A Brugada syndrome mutation (p.S216L) and its modulation by p.H558R polymorphism: standard and dynamic characterization. Cardiovasc Res. 2011;91:606–616. doi: 10.1093/cvr/cvr142. [DOI] [PubMed] [Google Scholar]
- Marston S, et al. OBSCN mutations associated with dilated cardiomyopathy and Haploinsufficiency. PLoS ONE. 2015;10:e0138568. doi: 10.1371/journal.pone.0138568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernigo S, Fukuzawa A, Bertz M, Holt M, Rief M, Steiner RA, Gautel M. Structural insight into M-band assembly and mechanics from the titin-obscurin-like-1 complex. Proc Natl Acad Sci U S A. 2010;107:2908–2913. doi: 10.1073/pnas.0913736107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernigo S, et al. Binding of Myomesin to Obscurin-like-1 at the muscle M-band provides a strategy for isoform-specific mechanical protection. Structure. 2017;25:107–120. doi: 10.1016/j.str.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry NA, Ackermann MA, Shriver M, Hu L-YR, Kontrogianni-Konstantopoulos A. Obscurins: unassuming giants enter the spotlight. IUBMB Life. 2013;65:479–486. doi: 10.1002/iub.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo D, et al. Exercise-induced alterations and loss of sarcomeric M-line organization in the diaphragm muscle of obscurin knockout mice. Am J Physiol Cell physiol. 2017;312:C16–C28. doi: 10.1152/ajpcell.00098.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AM, et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci Transl Med. 2015;7:270ra276. doi: 10.1126/scitranslmed.3010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland TJ, Graw SL, Sweet ME, Gigli M, Taylor MRG, Mestroni L. Obscurin variants in patients with Left ventricular Noncompaction. J Am Coll Cardiol. 2016;68:2237–2238. doi: 10.1016/j.jacc.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell MW, Raeker MO, Korytkowski KA, Sonneman KJ. Identification, tissue expression and chromosomal localization of human Obscurin-MLCK, a member of the titin and Dbl families of myosin light chain kinases. Gene. 2002;282:237–246. doi: 10.1016/S0378-1119(01)00795-8. [DOI] [PubMed] [Google Scholar]
- Walsh R, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19:192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware JS, et al. Shared genetic predisposition in Peripartum and dilated cardiomyopathies. N Engl J Med. 2016;374:233–241. doi: 10.1056/NEJMoa1505517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, et al. Compound and digenic heterozygosity contributes to arrhythmogenic right ventricular cardiomyopathy. J Am Coll Cardiol. 2010;55:587–597. doi: 10.1016/j.jacc.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, et al. Investigation of pathogenic genes in Chinese sporadic hypertrophic cardiomyopathy patients by whole exome sequencing. Sci Rep. 2015;5:16609. doi: 10.1038/srep16609. [DOI] [PMC free article] [PubMed] [Google Scholar]


