Abstract
Circadian rhythms orchestrate biochemical and physiological processes in living organisms to respond the day/night cycle. In mammals, nearly all cells hold self-sustained circadian clocks meanwhile couple the intrinsic rhythms to systemic changes in a hierarchical manner. The suprachiasmatic nucleus (SCN) of the hypothalamus functions as the master pacemaker to initiate daily synchronization according to the photoperiod, in turn determines the phase of peripheral cellular clocks through a variety of signaling relays, including endocrine rhythms and metabolic cycles. With aging, circadian desynchrony occurs at the expense of peripheral metabolic pathologies and central neurodegenerative disorders with sleep symptoms, and genetic ablation of circadian genes in model organisms resembled the aging-related features. Notably, a number of studies have linked longevity nutrient sensing pathways in modulating circadian clocks. Therapeutic strategies that bridge the nutrient sensing pathways and circadian clock might be rational designs to defy aging.
Keywords: circadian rhythms, SCN, longevity
INTRODUCTION
With seminal successes in biomedical researches, the improved medical conditions markedly lengthened human lifespan however also led to emerging threats as known the age-associated complexities (Kaeberlein et al., 2015). The wide range of age-associated diseases, including neurodegenerative diseases, cardiovascular disorder, type-2 diabetes, and higher cancer incidences, are driven by the causes of time, genetic and environmental situations that remain difficult to dissect for major effector(s) in individuals. Over decades, researches on aging have revealed the retardation of physiological decline and lifespan extension are conceivable by genetic perturbations in model organisms, the results now offered potential therapeutic strategies to prolong both healthspan and lifespan (Lopez-Otin et al., 2016). Dietary restriction (DR), a chronic reduction of dietary intake regime was proven as a major link of connecting these genetic longevity studies. DR increases lifespan in many model organisms, including budding yeast Saccharomyces cerevisiae, nematode Caenorhabditis elegans, and fruitfly Drosophila melanogaster. These relatively simplified models rendered further analyses of longevity genes and pathways that are activated upon low-energy challenges thus mimicked DR effects (Fontana and Partridge, 2015; Guarente, 2013). Importantly, salutary effects of DR is evolutionarily conserved as also observed in primates (Colman et al., 2009; Colman et al., 2014). The significance of DR emphases the idea that energy homeostasis is centered in longevity, while aging is largely caused by aberrant energy condition and metabolic inflexibility (Riera and Dillin, 2015). Thus interactions among calorie intake, meal frequency and timing, as organized by the daily circadian rhythm program, are likely key to maintain the cellular and organ fitness.
Circadian rhythms govern a wide range of physiological and behavioral systems, such as energy metabolism, sleep-wake cycles, body temperature and locomotor activity (Panda et al., 2002; Reppert and Weaver, 2002). Declined circadian rhythmicity in endocrine rhythm, phase alignment and sleep are commonly seen with aging (Mattis and Sehgal, 2016). Consistently, experimental disruptions of circadian rhythms seriously impede functional physiology, lifespan and endorse cancer incident (Filipski et al., 2003; Froy, 2013; Fu et al., 2002; Kondratova and Kondratov, 2012; Penev et al., 1998). Even a milder circadian challenge, chronic jet-lag, imposes on aged wild-type mice can markedly increase mortality (Davidson et al., 2006). On the other hand, implant functional circadian clock with fetal suprachiasmatic nucleus in aged rodents allowed higher amplitude rhythm behavior and longer surviorship (Hurd and Ralph, 1998; Li and Satinoff, 1998). The evidence pictured the pivotal contributions of robust circadian rhythms in upholding the healthy physiology and likely the extension of lifespan.
This review includes an overview of the molecular mechanism of circadian control, and molecular deficiencies implicated in age-related malfunctions. It discusses the central circadian clock system and the pathologies with aging, including the impacts to neurodegenerative diseases and sleep. Finally, advises the links of circadian components to energy-sensing pathways that modulate mammalian lifespan, furthermore their potential as therapeutic targets to treat age-associated loss in physiological homeostasis.
MOLECULAR OSCILLATORS IN THE CIRCADIAN CLOCK
Circadian oscillations are generated via transcriptional–translational feedback loops in a cell autonomous manner in mammals (Bass and Takahashi, 2010; Dibner et al., 2010). The core transcription factors CLOCK and BMAL1 heterodimerize and bind to E-box motif-containing clock-controlled genes (CCGs) in a time-dependent manner. There are at least two interconnected feedback loops involved in the transcriptional regulation (Fig. 1). In the primary feedback loop, CLOCK:BMAL1 initiates the transcription of Period (Per) and Cryptochrome (Cry) through the binding of E-box elements. The transcriptional control is also facilitated by recruiting various coactivators including CBP/p300 (Hosoda et al., 2009; Li et al., 2010), TRAP150 (Lande-Diner et al., 2013) and SRC-2 (Stashi et al., 2014). When CRYs and PERs proteins accumulate to critical levels, they assemble into hetero-complexes and function as corepressors via direct binding to CLOCK:BMAL1 thus repress their own expression. The repression is facilitated by posttranslational modifications, for instance phosphorylation of PERs for nuclear translocation hence binding to CLOCK:BMAL1 (Lee et al., 2001). The repression is later relieved by the degradation of CRYs and PERs over time, allows another circadian cycle of CRYs and PERs expressions taking place. In the secondary loop, the nuclear orphan receptors REV-ERBα, REV-ERBβ, RORα, RORβ and RORγ are involved in controlling the temporal expression of BMAL1 and CLOCK. Of note, these nuclear orphan receptors are also CCGs under CLOCK:BMAL1 regulation. By recognizing RORE elements within the promoters of Bmal1 and Clock genes, ROR collaborates with PGC-1α to transcriptionally activate Bmal1 and Clock. REV-ERB competes for the RORE binding at circadian times with concentration advantage over ROR, and executes as BMAL1 and CLOCK repressor (Cho et al., 2012; Preitner et al., 2002; Sato et al., 2004). The repressing activity requires the recruitment of a NCoR1-HDAC3 corepressor complex (Everett and Lazar, 2014). Recent findings of hypoxia-inducible factor 1α (HIF1α) in additional regulatory route indicated oxygen level is an auxiliary cue in clock. HIF1α itself is a CCG and at hypoxia, activates per expression via binding to the hypoxia-responsive element (HRE) in a complex with ARNT. The results are of clinical interests to cardioprotection and phase conditions such as jetlag (Adamovich et al., 2017; Peek et al., 2017; Wu et al., 2017). Other transcriptional regulations in particular cell types, for instance ZBTB20 in rhythmic expression of prokineticin receptor-2 (Prokr2) in the suprachiasmatic nucleus neurons, is critical for the bimodal activity behavior in mice (Qu et al., 2016). It will be important to decipher known, or identify new regulatory mechanisms in cell types that are responsible for unique circadian behaviors.
Figure 1.
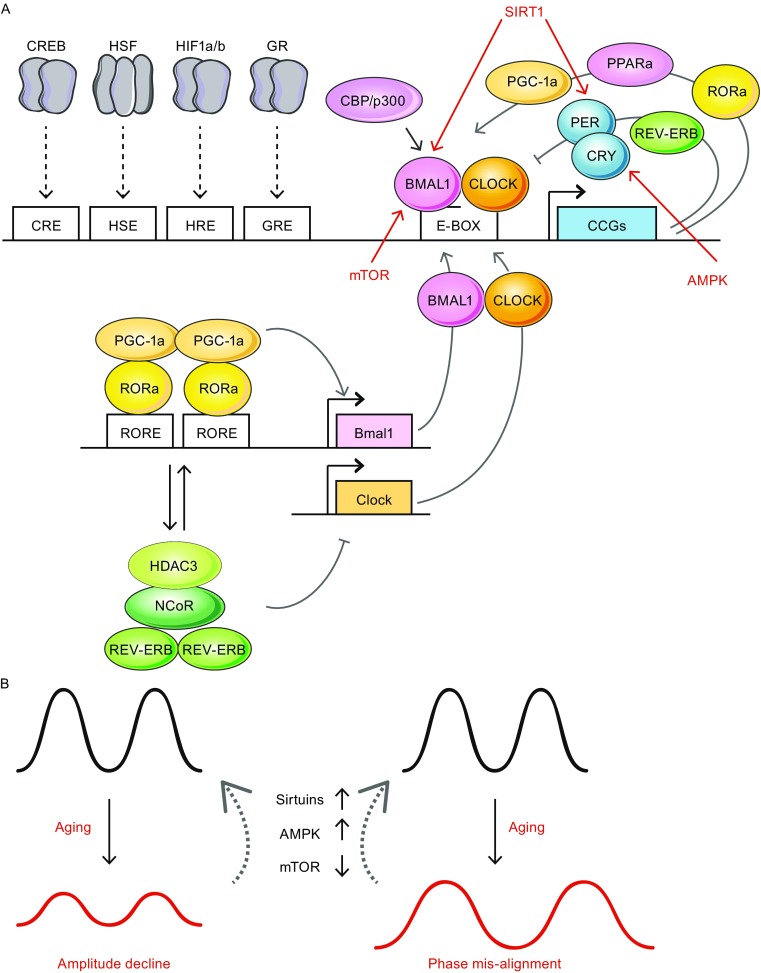
Molecular oscillators in circadian control. (A) Transcription factor complex CLOCK:BMAL1 binds to E-box containing motifs, allows the transcriptional activation of clock-controlled genes (CCGs) such as Pers, Crys, Ror and Rev-Erb. The activation is facilitated by recruiting coactivators such as CBP/p300. CCG transcriptions are as well regulated by transcription factors relaying the external cues. Examples include cAMP responsive element binding protein (CREB), heat shock factor 1 (HSF1), hypoxia-inducible factor 1α (HIF1α) and glucocorticoid receptor (GR) that bind to their respective regulatory elements (Bollinger and Schibler, 2014; Wu et al., 2016). Two interconnected feedback loops involved in the circadian transcriptional regulation. In the primary feedback loop, PER and CRY assemble into repressor complexes next attenuate the activity of CLOCK:BMAL1. In the second feedback loop, ROR (also a CCG protein) can complex with coactivator PGC-1α and bind to RORE element for Bmal1 (and likely Clock) activation(s). REV-ERB works as a repressor in Bmal1 transcription by concentration-dependent competition at the same RORE sequence. The repression involved the recruitment of NCoR/HDAC3 corepressor complexes. (B) Energy sensors such as Sirtuins, AMPK and mTOR participate in circadian modulations via post-translational modification of circadian components, as depicted in (A). Interventions target the pathways are of potential to treat age-associated circadian amplitude decline and phase mis-alignment
Notably, both the primary and secondary feedback loops are modulated by post-translational modifications in versatile ways, e.g, protein ubiquitination, phosphorylation/dephosphorylation, acetylation/deacetylation, poly ADP-ribosylation and O-GlcNAcylation (Reddy and Rey, 2014). These modifications indicate evident basis linking circadian and metabolic cycles at timely manner. Identify new post-translation modifications and classify the modifications in the central and peripheral tissues will be of great value to understand circadian physiology.
CLOCK GENES AND AGE-RELATED DISORDERS
Aging is a major risk factor for many human pathologies, including cancer, diabetes, cardiovascular disorders and neurodegenerative diseases (Lopez-Otin et al., 2013). Genetic models of circadian disruption pheno-copied aging and metabolic disorders frequently. A prominent case is the loss of BMAL1. Mice deficient for Bmal1 are suffered from a series of conditions related to aging. e.g., sarcopenia (with both reduction in muscle fiber size and quantity), cataracts, cornea inflammation, osteoporosis, premature hair loss, and failed to form adequate visceral and subcutaneous adipose storage (Kondratov et al., 2006). The strain is severely short-lived with average lifespan of 37.0 ± 12.1 weeks, compared to longer than 110 weeks of lifespan in same background wild type animals (Nadon, 2006). The findings coordinate well with the roles of BMAL1 in homeostatic maintenance of the glucose level (Rudic et al., 2004), and in adipogenesis regulation (Shimba et al., 2005). Consistently, it has been noticed that Bmal1 mRNA amplitude declined with altered peak phase in natural aging in rodents (Kolker et al., 2003).
As a reciprocal component of BMAL1, CLOCK deficiency also results in shorter average lifespan to approximately 15% reduction compared to wild type, and premature pathologies including cataracts and dermatitis (Dubrovsky et al., 2010). CLOCK appears to be crucial in glucose homeostasis as well, as both whole body and conditional disruptions of CLOCK caused hypoinsulinaemia hence diabetes mellitus in rodents. Same study demonstrated BMAL1 is also participated in sustaining the pancreatic clock (Marcheva et al., 2010). Of note, Clock∆19 strain, the CLOCK truncated line that was originally identified for its significant period change from a random mutagenesis screen, is with milder aging phenotypes such as diurnal activity/feeding rhythms and obesity in normal housing conditions compared to the knockout strain (Turek et al., 2005). Additional challenges such as post ionizing irradiation triggered an accelerated aging program in the strain (Antoch et al., 2008). The results suggested that the particular CLOCK truncation might be partially functional in protecting from premature aging, at a condition that the intrinsic period is far from optimal. Loss of PER2, a core circadian component, is linked to cancer predisposing. The animals are sensitive to γ irradiation later developed salivary gland hyperplasia, teratoma and malignant lymphomas (Fu et al., 2002). Further, genetic ablation of both Per1 and Per2 caused an arrhythmic phenotype together with premature aging conditions, e.g., early decline in fertility, kyphosis and predisposed tumor incidences (Lee, 2005). The DNA damage response and p53-mediated apoptosis are defective in these animals. The studies demonstrated that circadian clock components are also important regulators in cell cycle and proliferation likely specific in adulthood, as the double knockouts seem developmentally normal at birth. Another component CRY1 is shown to modulate hepatic gluconeogenesis by regulating the cAMP signaling. Rhythmic expression of CRY1 directly adjusts intracellular cAMP concentrations and the phosphorylation level of cAMP response element-binding protein (CREB) by protein kinase A (Zhang et al., 2010). Lipid metabolism is linked to circadian clock in the cases of REV-ERB and ROR families. They are important for regulating lipogenesis, lipid storage and adipocyte differentiation in a rhythmic manner (Bray and Young, 2007; Chawla and Lazar, 1993; Torra et al., 2000). REV-ERBs act as decent targets in treating obesity. The agonists work against fat mass accumulation in high fat fed mice, consequently improve dyslipidemia and hyperglycemia (Cho et al., 2012; Solt et al., 2012).
The role of circadian genes in Cancer Biology remains to be a complicated conundrum. As contrast to the tumor suppressing effect of PER2, deletion of Cry1/2 in p53 null mice protected the early onset of cancer incidence, and sensitized the p53 deficient cells to apoptosis upon genotoxic stress (Ozturk et al., 2009). A recent finding of targeting BMAL1 and CLOCK for acute myeloid leukemia (AML) therapy indicates further the pro-cancerous option of clock components (Puram et al., 2016). Many core circadian proteins are involved in the cell cycle and the DNA damage response (Sahar and Sassone-Corsi, 2009), thus may facilitate the proliferation of transformed malignant cancer cells while normal post-mitotic cells should be exempted from the risk. Careful analyses of cancer types and the associations to circadian gene alterations are essential to address the paradox.
THE CENTRAL CIRCADIAN CLOCK SYSTEM
To organize physiology and behavior for proper functioning according to the 24-hour environmental light/dark cycle, mammals rely on a central pacemaker known as the suprachiasmatic nucleus (SCN) for systemic synchronization. SCN resides at the anterior hypothalamus and directly contacts optic chiasm for sensing the external photic input. It is composed by paired nuclei lateral to either side of the third ventricle (Colwell, 2011). Though with limited neurons (~20,000 in mouse), SCN contains considerable neuron heterogeneity. There includes calretinin, neurotensin (NT), gastrin releasing peptide (GRP), angiotensin II, prokineticin 2 (PK2), neuromedin S (NMS), vasoactive intestinal peptide (VIP) and arginine vasopressin (AVP) expressing neurons (Welsh et al., 2010). Among them VIP and AVP neurons are key neuron types that mark the ventral core and dorsal shell subdivisions of SCN, respectively (Golombek and Rosenstein, 2010). Most SCN neurons are GABAergic (Moore and Speh, 1993; Morin et al., 2006).
Neurons in the core are considered to incorporate external inputs, such as photic light cue from the retinohypothalamic tract (RHT), and likely also the projections from the raphe nuclei (Morin and Allen, 2006). The environmental information is then coupled and communicated to the rest parts of the SCN (Fig. 2). Among the core neurons, VIP neuron is essential in the SCN oscillation coupling; likewise GRP, NT neurons and neurotransmitter GABA contribute significantly to the process as well (Aida et al., 2002; Choi et al., 2008; De Jeu and Pennartz, 2002; Meyer-Spasche et al., 2002; Shinohara et al., 2000). The sensory core neurons display lesser clock gene expression amplitudes, perhaps is suitable for faster resetting when respond to environmental changes, as has been predicted in mathematical modeling work (Pulivarthy et al., 2007). This is by contrast to AVP, PK2, and even GABAergic neurons in the dorsal shell SCN that circadian genes including Per1 and Per2 are robustly oscillated in the subdivision (Hamada et al., 2004; Nakamura et al., 2005; Yan and Okamura, 2002).
Figure 2.
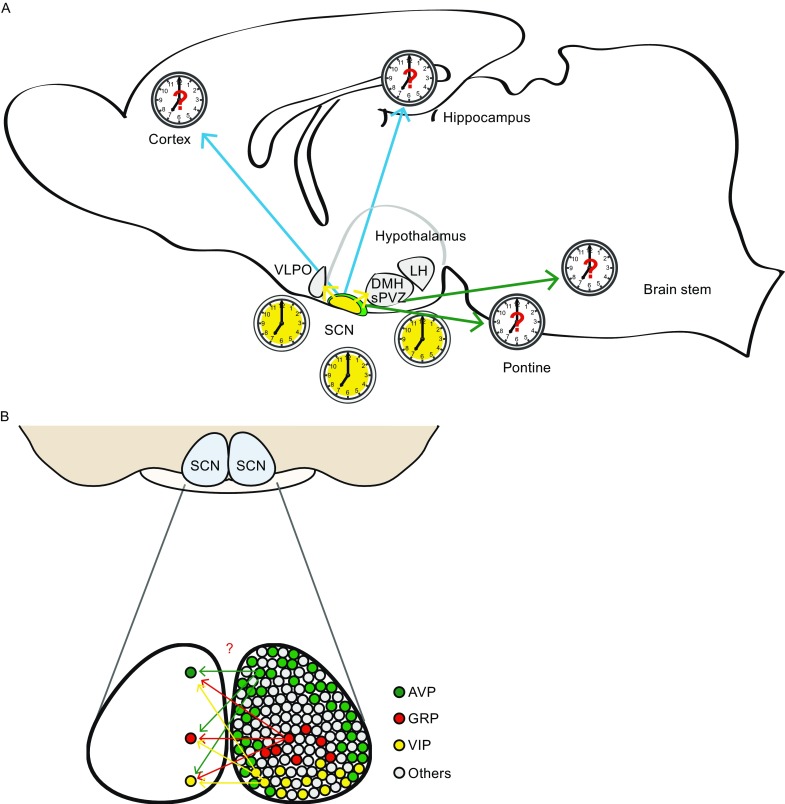
Schematic functional map of circadian control. Studies have demonstrated the SCN efferent primarily travel to other hypothalamic nuclei incuding dorsomedial hypothalamus (DMH), subparaventricular zone (sPVZ) and more. A map of direct SCN neuron projections to sleep-awake or cognitive centers in the brain (A) and the intra-SCN (bilateral) connectomes (B), with the details in connection density and neuron types, remains elusive at the moment
It has been observed that most of the projections from the core terminate within the shell, stresses the point that the interplay between the two regions is most important among all circadian outputs from the SCN (Antle and Silver, 2005). Several studies have demonstrated the SCN efferent from both core and shell travel chiefly to other hypothalamic nuclei, e.g., dorsomedial hypothalamus (DMH), paraventricular hypothalamic nucleus (PVN), arcuate hypothalamic nucleus (ARC), subparaventricular zone (sPVZ), and more (Abrahamson and Moore, 2001; Kalsbeek et al., 2006; Yan et al., 2007). The projections cover the nervous and endocrine systems for temporal control of the daily oscillation in the body (Dibner et al., 2010). Notably, the projections to central sleep system, for example the ventrolateral preoptic area (VLPO), locus coeruleus (LC) and lateral hypothalamus (LH), are suggested as indirect or sparse (Abrahamson et al., 2001; Aston-Jones et al., 2001; Chou et al., 2002; Novak and Nunez, 2000). This postulates either the sparse connections are sufficient for temporal cues to structure the sleep program, or there exists a central hub, such as DMH, for the communications in between (Chou et al., 2003; Mattis and Sehgal, 2016). A thorough SCN map with neuron type accuracy via connectome works will be of great help to elucidate the functional circadian circuitry (Fig. 2).
AGE-ASSOCIATED DECLINE IN CENTRAL CIRCADIAN SYSTEM
Important features of functional circadian rhythms include, e.g., sustaining at a sufficient oscillation amplitude through out the daily cycle; composing a phase that is properly aligned with the light/dark condition and can be entrained by light; and maintaining in a near 24 h period to reflect the Earth day (Bass and Takahashi, 2010; Welsh et al., 2010). Aging hampers amplitude both in circadian gene expressions (Hofman and Swaab, 2006; Yamazaki et al., 2002), and several physical indexes including melatonin level, sleep-wake disruptions, lowered locomotor activity (Duffy and Czeisler, 2002; Valentinuzzi et al., 1997; Weinert, 2000; Yoon et al., 2003). Further, phase shifts and re-entrainment difficulty are also common drawbacks with aging (Gibson et al., 2009; Scarbrough et al., 1997; Valentinuzzi et al., 1997). While many factors account for these physical changes, the central clock SCN is likely to stand as a key element responsible for this age-related decline.
The central clock SCN decay, considering the direct or indirect contacts to variable brain regions, would reveal degeneration in at least two aspects: the SCN projections, and secreted signals from the SCN. While projection details of young versus old await careful investigations, SCN secreting outputs have been studied. For example, aging affects SCN prominently in the AVP neuron population. In human, diurnal oscillation of the neuropeptide in young is evident, but in elderly people (over 50 years of age) the change becomes subtle. Further, the peaking time in the early morning in young is reversed to low-amplitude night peaking in the elderly (Hofman and Swaab, 1994). Interestingly, the annual cycle of AVP expression is also lost with aging. Young subjects are normally with lowest AVP-immunoreactive values during the summer and highest in autumn (Hofman and Swaab, 1995). These results suggest that the activities of human SCN, both for the diurnal and the seasonal rhythms, become disturbed later in life. VIP neuron is another, perhaps more sensitive, example to reflect human SCN aging. In young male subjects (10–40 years), the number of VIP neurons in the SCN is highest. However in the age of 40–65 years old, the VIP neuron number dramatically decreased by about 60% further does not show significant decline in later ages (Hofman et al., 1996; Zhou et al., 1995).
Studies in rodents offered further evidences for SCN activity change. First, the AVP neuron is significantly reduced in aged rats while the total SCN neuron number stayed similar (Roozendaal et al., 1987), a change reminiscent to the human case. SCN neurons become desynchronized and are with decreased phase coherence with aging (Farajnia et al., 2012). In vivo multiunit neural activity (MUA) recordings in the SCN in young (3–5 months) versus aged (13–18 months) mice revealed that the day and night amplitude differences in the older are significantly reduced. Similar decline in neural activity rhythms was also observed in the subparaventricular zone, one of the major SCN output regions (Nakamura et al., 2011). Whether molecular clock components are good indicators for SCN aging remain unclear. Nakamura et al. applied PER2 as the molecular marker and only revealed subtle change in the two groups, suggested the electrical activity rhythm is the more sensitive circadian output measurement compared to molecular components of the clock. Of note, other experiments showed significant age-related decline of BMAL1 and CLOCK in the SCN as well in few other brain areas, when compared 4 versus 16 month-old mice (Wyse and Coogan, 2010). Further characteristic analyses on SCN aging are crucial to reason the basis of circadian dampens.
CIRCADIAN DISRUPTION AND NEURODEGENERATIVE DISEASES
Circadian disorders with sleep symptoms are commonly seen in patients with neurodegenerative diseases (Kondratova and Kondratov, 2012). For instance, Parkinson’s disease (PD) patients are disrupted for the cortisol and melatonin rhythms (Breen et al., 2014; Videnovic et al., 2014), and displayed Bmal1 reduce in blood samples from PD patients (Cai et al., 2010), these all point toward the deteriorated situations in circadian control. A PD transgenic model via Thy-1 promoter mediated α-synuclein over-expression exhibited several circadian phenotypes in aging: a clear reduced wheel-running activity with altered period, altered temporal distribution of sleep, and decayed spontaneous neural activity in the SCN, suggested circadian rhythm is severely disrupted in the PD model (Kudo et al., 2011).
Another PD model also links phenotypes to circadian impairments. The strategy is to selectively inactivate mitochondrial transcription factor A (Tfam) in dopamine neurons thus mimics PD progression particularly for dopamine neuron degeneration. The conditional knockouts have reduced physical activity as early as by 5 months of age, and then dampened for both circadian amplitude and stability. The animals also showed abolished rhythmic locomotion in constant dark or constant light conditions (Fifel and Cooper, 2014). Besides circadian phenotypes, sleep disturbance is another hallmark during PD progression. Several sleep disorders are discovered in PD patients, including insomnia, restless leg syndrome and REM behavior disorder (RBD), a symptom that permits motor activity during REM sleep (Barone et al., 2009; Iranzo, 2013). RBD is potentially useful for the prediction of PD onset, as PD pathology occurs in the brainstem earlier than the substantia nigra (Braak et al., 2004). Loss of hypocretin neurons in the lateral hypothalamus could also explain the malfunction of sleep/arousal program in PD patients (Fronczek et al., 2007).
Alzheimer’s disease (AD) patients are long recognized for SCN neuronal loss, of that VIP neuron is a prominent case (Swaab et al., 1985; Zhou et al., 1995). A recent analysis of actogram associated to post-mortem brain tissue demonstrated that besides locomotor activity phenotype, AD patients are also diagnosed with false rhythmic control of core-body temperature and rest-activity (Satlin et al., 1995; van Someren et al., 1996). Sleep–wake cycle dysfunction and increased daytime sleepiness are regard as risk factors for AD related dementia (Lee et al., 2007). Consistently, the transgenic 3xTg-AD mouse strain that exhibits both Aβ and tau pathology (as in human AD) were scored for similar circadian phenotypes. The results indicated that prior to AD pathology, activity during daytime and period change were observed in the transgenic, interesting more in the males. The number of VIP neuron is decreased in the SCN, suggested again that circadian dysfunction is predictive in early AD onset (Sterniczuk et al., 2010a; Sterniczuk et al., 2010b).
Patients with Huntington’s disease (HD) have sleep symptoms including advanced sleep phase, insomnia and reduced REM sleep (Arnulf et al., 2008; Goodman and Barker, 2010). Neuropathological analyses demonstrated that HD patients are depleted for many hypothalamic neuropeptides, i.e., AVP, oxytocin and hypocretin, hence disturbed regular sleep/awake daily cycle (Aziz et al., 2008; Gabery et al., 2010). The HD model, R6/2 transgenic strain displayed reduced expression of Per2 and blunted oscillation of Bmal1 in the SCN, as well reduced in motor cortex and striatum. The increased daytime activity is likely associated with reduced prokineticin 2 expression that is critical for suppressing daytime activity in nocturnal animals (Morton et al., 2005). VIP and VPAC2 receptor are down regulated in R6/2 animals in the SCN (Fahrenkrug et al., 2007).
Together, the findings indicate circadian parameters can serve as the basis for prognostic purposes. Sustaining efficient circadian activities are likely key to prevent age-related disorders, including neurodegenerative diseases.
LONGEVITY MEDIATORS IN CIRCADIAN REGULATION
Vast amount of evidences have pointed out the importance of circadian rhythm in functional physiology, however data that are suggestive to longevity remain unclear. Our current understandings mostly based on loss-of-function studies (Eckel-Mahan et al., 2013), while genetic manipulation of circadian gene has yet been reported with lifespan extension outcome. Interestingly, many longevity mediators and pathways exert the beneficial effects via cooperating with multiple circadian components. One evident example is Sirt1, a longevity gene that mediates calorie restriction (CR) benefits (Guarente, 2013), is involved in circadian regulation (Chang and Guarente, 2014; Jung-Hynes et al., 2010). SIRT1 is a NAD+-dependent deacetylase that is involved in regulating circadian gene transcriptions via deacetylating histone H3 K9/K14 at the promoters regions (Nakahata et al., 2008). It had been suggested that CLOCK works as a histone acetyltransferase (HAT) in BMAL1 acetylation to facilitate rhythmic circadian gene transcriptions (Hirayama et al., 2007). SIRT1 appears to counterbalance the BMAL1 acetylation status in both fibroblast culture and the liver (Nakahata et al., 2008). Alternatively, SIRT1 deacetylates PER2 thus regulates PER2 stability further adjusts the circadian feedback inhibition (Asher et al., 2008). Notably, the synthesis of SIRT1 cofactor NAD+ also follows a circadian expression pattern. Nicotinamide phosphoribosyltransferase (NAMPT), the rate-limiting enzyme for NAD+ salvage pathway, is a rhythmically expressed protein that under E-box transcriptional control (Nakahata et al., 2009; Ramsey et al., 2009). Together with facts that SIRT1 oscillates in a circadian manner, and the level of NAD+ declines with aging (Gomes et al., 2013), SIRT1 in the interconnected loops revealed a strong correlation between energy and circadian rhythms.
SIRT1 has been demonstrated for numerous vital roles in upholding neuronal health, including neuronal development, memory formation and neurodegenerative disease preventions (Herskovits and Guarente, 2014). Owing to the critical function of hypothalamus in metabolic regulations, many SIRT1 related studies have been carried out in different hypothalamic nuclei. For instance, SIRT1 activities are important for POMC neuron in the ARC, and for SF1 neuron in the VMH to maintain systemic glucose homeostasis hence prevent obesity (Ramadori et al., 2011; Ramadori et al., 2010). A transgenic strain overexpressing SIRT1 in DMH and LH showed improved sleep quality with extended lifespan (Satoh et al., 2013). In the SCN, SIRT1 prevents age-associated circadian phenotypes via supporting molecular oscillation of clock genes (Chang and Guarente, 2013). Of note, SIRT6 (a SIRT1 homolog) also participates in circadian regulation of fatty acid and cholesterol metabolism (Masri et al., 2014). Whether new post-translational modifications take place in circadian modulation, perhaps link to the versatile activities found in other sirtuins (Choudhary et al., 2014), is of great interest to pursue in the future.
Adenosine monophosphate-activated protein kinase (AMPK), another longevity mediator that is important for sensing low energy state, contribute to relieve the PER/CRY mediated circadian feedback repression. AMPK phosphorylates and activates casein kinase I epsilon (CKIε) for the subsequent phosphorylation of PER, therefore promote PER degradation (Um et al., 2007). In a similar action, AMPK phosphorylates CRY directly and facilitates CRY degradation (Lamia et al., 2009). The stimulation of AMPK activity leads to a phase advance effect. AMPK has been studied in many hypothalamic nuclei such as ARC, VMH and DMH, for energy balance and metabolism control (Lopez et al., 2016), yet the role in central circadian regulation in the SCN remains unclear.
The mammalian target of rapamycin (mTOR), an important sensor of insulin, growth factor, and mitogen inputs, has been revealed in circadian control through many effector proteins. For instance ribosomal S6 protein kinase 1 (S6K1), an important regulator of translation acting downstream of mTOR activation, can rhythmically phosphorylate BMAL1. The particular modification allows BMAL1 to work as a translation factor in a timely manner with response to the mTOR signaling, in addition to the canonical role in circadian transcription (Lipton et al., 2015). Of note, BMAL1 deficiency caused elevated activity of mTORC1 both in cell culture and in vivo. In vivo administration of the mTORC1 inhibitor Rapatar increased Bmal1 null mice lifespan by 50% (Khapre et al., 2014), the results suggested complex, bi-directional regulations may exist between BMAL1 and mTOR. In the SCN, the photic signal activates mTOR signaling and promotes the translation of VIP by repressing 4E-BP1. Accordingly, the 4E-BP1 deficient mice exhibit accelerated re-entrainment upon light/dark shift and are more resilient to constant light mediated circadian disruption (Cao et al., 2013). Together, the findings reveal strong energy links among aging, metabolism and circadian physiology.
CONCLUDING REMARKS
It is clear that age-related diseases such as cancer, type-2 diabetes, obesity and neurodegenerative disorders are profoundly metabolic-associated (Lopez-Otin et al., 2013), and functional circadian activities maybe key to preclude the abnormalities (Asher and Sassone-Corsi, 2015). Numerous data indicate nutrient-sensing/longevity pathways such as SIRT1 and others assist circadian control, and impose regulatory loops in coordinating photic and non-photic feeding stimuli. The beneficial effects offered viewpoints that interventions for promoting healthy aging and longevity may as well treat circadian disorders. Initiated trials on sirtuin-activating compounds (STACs) such as resveratrol, SRT2104 or NAD+ precursors (Bonkowski and Sinclair, 2016; Wood et al., 2004); metformin for AMPK activation (Barzilai et al., 2016); and rapamycin for mTOR inhibition (Harrison et al., 2009; Li et al., 2014) are suitable candidates for such interventions. Effects of these on central versus peripheral clocks, and the underlying mechanisms await careful analyses in the future.
ACKNOWLEDGEMENTS
We thank Y.-C. Tang and the members in Chang laboratory for discussions. We are grateful to members of the Chang laboratory for their critical reading of the manuscript. H.-C. Chang is supported by General Program of National Natural Science Foundation of China (Grant No. 31671221) and Chinese Academy of Sciences (2013OHTP03).
ABBREVIATIONS
AD, Alzheimer’s disease; AMPK, adenosine monophosphate-activated protein kinase; ARC, arcuate hypothalamic nucleus; AVP, arginine vasopressin; CKIε, casein kinase I epsilon; CR, calorie restriction; CREB, cAMP response element-binding protein; DMH, dorsomedial hypothalamus; DR, dietary restriction; GRP, gastrin releasing peptide; HAT, histone acetyltransferase; HD, Huntington’s disease; HIF1α, hypoxia-inducible factor 1α; HRE, hypoxia-responsive element; LC, locus coeruleus; LH, lateral hypothalamus; mTOR, mammalian target of rapamycin; MUA, multiunit neural activity; NAMPT, Nicotinamide phosphoribosyltransferase; NMS, neuromedin S; NT, neurotensin; PD, Parkinson’s disease; PK2, prokineticin 2; PVN, paraventricular hypothalamic nucleus; RBD, REM behavior disorder; RHT, retinohypothalamic tract; SCN, suprachiasmatic nucleus; sPVZ, subparaventricular zone; VIP, vasoactive intestinal peptide; VLPO, ventrolateral preoptic area
COMPLIANCE WITH ETHICAL STANDARDS
Fang Liu and Hung-Chun Chang declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172–191. doi: 10.1016/S0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- Abrahamson EE, Leak RK, Moore RY. The suprachiasmatic nucleus projects to posterior hypothalamic arousal systems. NeuroReport. 2001;12:435–440. doi: 10.1097/00001756-200102120-00048. [DOI] [PubMed] [Google Scholar]
- Adamovich Y, Ladeuix B, Golik M, Koeners MP, Asher G. Rhythmic oxygen levels reset circadian clocks through HIF1alpha. Cell Metab. 2017 doi: 10.1016/j.cmet.2016.09.014. [DOI] [PubMed] [Google Scholar]
- Aida R, Moriya T, Araki M, Akiyama M, Wada K, Wada E, Shibata S. Gastrin-releasing peptide mediates photic entrainable signals to dorsal subsets of suprachiasmatic nucleus via induction of period gene in mice. Mol Pharmacol. 2002;61:26–34. doi: 10.1124/mol.61.1.26. [DOI] [PubMed] [Google Scholar]
- Antle MC, Silver R. Orchestrating time: arrangements of the brain circadian clock. Trends Neurosci. 2005;28:145–151. doi: 10.1016/j.tins.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Antoch MP, Gorbacheva VY, Vykhovanets O, Toshkov IA, Kondratov RV, Kondratova AA, Lee C, Nikitin AY. Disruption of the circadian clock due to the clock mutation has discrete effects on aging and carcinogenesis. Cell Cycle. 2008;7:1197–1204. doi: 10.4161/cc.7.9.5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnulf I, Nielsen J, Lohmann E, Schiefer J, Wild E, Jennum P, Konofal E, Walker M, Oudiette D, Tabrizi S, et al. Rapid eye movement sleep disturbances in Huntington disease. Arch Neurol. 2008;65:482–488. doi: 10.1001/archneur.65.4.482. [DOI] [PubMed] [Google Scholar]
- Asher G, Sassone-Corsi P. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015;161:84–92. doi: 10.1016/j.cell.2015.03.015. [DOI] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Aston-Jones G, Chen S, Zhu Y, Oshinsky ML. A neural circuit for circadian regulation of arousal. Nat Neurosci. 2001;4:732–738. doi: 10.1038/89522. [DOI] [PubMed] [Google Scholar]
- Aziz A, Fronczek R, Maat-Schieman M, Unmehopa U, Roelandse F, Overeem S, van Duinen S, Lammers GJ, Swaab D, Roos R. Hypocretin and melanin-concentrating hormone in patients with Huntington disease. Brain Pathol. 2008;18:474–483. doi: 10.1111/j.1750-3639.2008.00135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone P, Antonini A, Colosimo C, Marconi R, Morgante L, Avarello TP, Bottacchi E, Cannas A, Ceravolo G, Ceravolo R, et al. The PRIAMO study: a multicenter assessment of nonmotor symptoms and their impact on quality of life in Parkinson’s disease. Mov Dis. 2009;24:1641–1649. doi: 10.1002/mds.22643. [DOI] [PubMed] [Google Scholar]
- Barzilai N, Crandall JP, Kritchevsky SB, Espeland MA. Metformin as a tool to target aging. Cell Metab. 2016;23:1060–1065. doi: 10.1016/j.cmet.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger T, Schibler U. Circadian rhythms—from genes to physiology and disease. Swiss Med Wkly. 2014;144:w13984. doi: 10.4414/smw.2014.13984. [DOI] [PubMed] [Google Scholar]
- Bonkowski MS, Sinclair DA. Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nat Rev Mol Cell Biol. 2016;17:679–690. doi: 10.1038/nrm.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Ghebremedhin E, Rub U, Bratzke H, Del Tredici K. Stages in the development of Parkinson’s disease-related pathology. Cell Tissue Res. 2004;318:121–134. doi: 10.1007/s00441-004-0956-9. [DOI] [PubMed] [Google Scholar]
- Bray MS, Young ME. Circadian rhythms in the development of obesity: potential role for the circadian clock within the adipocyte. Obes Rev. 2007;8:169–181. doi: 10.1111/j.1467-789X.2006.00277.x. [DOI] [PubMed] [Google Scholar]
- Breen DP, Vuono R, Nawarathna U, Fisher K, Shneerson JM, Reddy AB, Barker RA. Sleep and circadian rhythm regulation in early Parkinson disease. JAMA Neurol. 2014;71:589–595. doi: 10.1001/jamaneurol.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Liu S, Sothern RB, Xu S, Chan P. Expression of clock genes Per1 and Bmal1 in total leukocytes in health and Parkinson’s disease. Eur J Neurol. 2010;17:550–554. doi: 10.1111/j.1468-1331.2009.02848.x. [DOI] [PubMed] [Google Scholar]
- Cao R, Robinson B, Xu H, Gkogkas C, Khoutorsky A, Alain T, Yanagiya A, Nevarko T, Liu AC, Amir S, et al. Translational control of entrainment and synchrony of the suprachiasmatic circadian clock by mTOR/4E-BP1 signaling. Neuron. 2013;79:712–724. doi: 10.1016/j.neuron.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Guarente L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell. 2013;153:1448–1460. doi: 10.1016/j.cell.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab. 2014;25:138–145. doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Lazar MA. Induction of Rev-ErbA alpha, an orphan receptor encoded on the opposite strand of the alpha-thyroid hormone receptor gene, during adipocyte differentiation. J Biol Chem. 1993;268:16265–16269. [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HJ, Lee CJ, Schroeder A, Kim YS, Jung SH, Kim JS, Kim DY, Son EJ, Han HC, Hong SK, et al. Excitatory actions of GABA in the suprachiasmatic nucleus. J Neurosci. 2008;28:5450–5459. doi: 10.1523/JNEUROSCI.5750-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC, Bjorkum AA, Gaus SE, Lu J, Scammell TE, Saper CB. Afferents to the ventrolateral preoptic nucleus. J Neurosci. 2002;22:977–990. doi: 10.1523/JNEUROSCI.22-03-00977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC, Scammell TE, Gooley JJ, Gaus SE, Saper CB, Lu J. Critical role of dorsomedial hypothalamic nucleus in a wide range of behavioral circadian rhythms. J Neurosci. 2003;23:10691–10702. doi: 10.1523/JNEUROSCI.23-33-10691.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Beasley TM, Kemnitz JW, Johnson SC, Weindruch R, Anderson RM. Caloric restriction reduces age-related and all-cause mortality in rhesus monkeys. Nat Commun. 2014;5:3557. doi: 10.1038/ncomms4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS. Linking neural activity and molecular oscillations in the SCN. Nat Rev Neurosci. 2011;12:553–569. doi: 10.1038/nrn3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AJ, Straume M, Block GD, Menaker M. Daily timed meals dissociate circadian rhythms in hepatoma and healthy host liver. Int J Cancer. 2006;118:1623–1627. doi: 10.1002/ijc.21591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jeu M, Pennartz C. Circadian modulation of GABA function in the rat suprachiasmatic nucleus: excitatory effects during the night phase. J Neurophysiol. 2002;87:834–844. doi: 10.1152/jn.00241.2001. [DOI] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Dubrovsky YV, Samsa WE, Kondratov RV. Deficiency of circadian protein CLOCK reduces lifespan and increases age-related cataract development in mice. Aging. 2010;2:936–944. doi: 10.18632/aging.100241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JF, Czeisler CA. Age-related change in the relationship between circadian period, circadian phase, and diurnal preference in humans. Neurosci Lett. 2002;318:117–120. doi: 10.1016/S0304-3940(01)02427-2. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155:1464–1478. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett LJ, Lazar MA. Nuclear receptor Rev-erbalpha: up, down, and all around. Trends Endocrinol Metab. 2014;25:586–592. doi: 10.1016/j.tem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrug J, Popovic N, Georg B, Brundin P, Hannibal J. Decreased VIP and VPAC2 receptor expression in the biological clock of the R6/2 Huntington’s disease mouse. J Mol Neurosci. 2007;31:139–148. doi: 10.1385/jmn/31:02:139. [DOI] [PubMed] [Google Scholar]
- Farajnia S, Michel S, Deboer T, vanderLeest HT, Houben T, Rohling JH, Ramkisoensing A, Yasenkov R, Meijer JH. Evidence for neuronal desynchrony in the aged suprachiasmatic nucleus clock. J Neurosci. 2012;32:5891–5899. doi: 10.1523/JNEUROSCI.0469-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fifel K, Cooper HM. Loss of dopamine disrupts circadian rhythms in a mouse model of Parkinson’s disease. Neurobiol Dis. 2014;71:359–369. doi: 10.1016/j.nbd.2014.08.024. [DOI] [PubMed] [Google Scholar]
- Filipski E, King VM, Li X, Granda TG, Mormont MC, Claustrat B, Hastings MH, Levi F. Disruption of circadian coordination accelerates malignant growth in mice. Pathol Biol. 2003;51:216–219. doi: 10.1016/S0369-8114(03)00034-8. [DOI] [PubMed] [Google Scholar]
- Fontana L, Partridge L. Promoting health and longevity through diet: from model organisms to humans. Cell. 2015;161:106–118. doi: 10.1016/j.cell.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronczek R, Overeem S, Lee SY, Hegeman IM, van Pelt J, van Duinen SG, Lammers GJ, Swaab DF. Hypocretin (orexin) loss in Parkinson’s disease. Brain. 2007;130:1577–1585. doi: 10.1093/brain/awm090. [DOI] [PubMed] [Google Scholar]
- Froy O. Circadian aspects of energy metabolism and aging. Ageing Res Rev. 2013;12:931–940. doi: 10.1016/j.arr.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/S0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- Gabery S, Murphy K, Schultz K, Loy CT, McCusker E, Kirik D, Halliday G, Petersen A. Changes in key hypothalamic neuropeptide populations in Huntington disease revealed by neuropathological analyses. Acta Neuropathol. 2010;120:777–788. doi: 10.1007/s00401-010-0742-6. [DOI] [PubMed] [Google Scholar]
- Gibson EM, Williams WP, 3rd, Kriegsfeld LJ. Aging in the circadian system: considerations for health, disease prevention and longevity. Exp Gerontol. 2009;44:51–56. doi: 10.1016/j.exger.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golombek DA, Rosenstein RE. Physiology of circadian entrainment. Physiol Rev. 2010;90:1063–1102. doi: 10.1152/physrev.00009.2009. [DOI] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AO, Barker RA. How vital is sleep in Huntington’s disease? J Neurol. 2010;257:882–897. doi: 10.1007/s00415-010-5517-4. [DOI] [PubMed] [Google Scholar]
- Guarente L. Calorie restriction and sirtuins revisited. Genes Dev. 2013;27:2072–2085. doi: 10.1101/gad.227439.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T, Antle MC, Silver R. Temporal and spatial expression patterns of canonical clock genes and clock-controlled genes in the suprachiasmatic nucleus. Eur J Neurosci. 2004;19:1741–1748. doi: 10.1111/j.1460-9568.2004.03275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskovits AZ, Guarente L. SIRT1 in neurodevelopment and brain senescence. Neuron. 2014;81:471–483. doi: 10.1016/j.neuron.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- Hofman MA, Swaab DF. Alterations in circadian rhythmicity of the vasopressin-producing neurons of the human suprachiasmatic nucleus (SCN) with aging. Brain Res. 1994;651:134–142. doi: 10.1016/0006-8993(94)90689-0. [DOI] [PubMed] [Google Scholar]
- Hofman MA, Swaab DF. Influence of aging on the seasonal rhythm of the vasopressin-expressing neurons in the human suprachiasmatic nucleus. Neurobiol Aging. 1995;16:965–971. doi: 10.1016/0197-4580(95)02016-0. [DOI] [PubMed] [Google Scholar]
- Hofman MA, Swaab DF. Living by the clock: the circadian pacemaker in older people. Ageing Res Rev. 2006;5:33–51. doi: 10.1016/j.arr.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Hofman MA, Zhou JN, Swaab DF. No evidence for a diurnal vasoactive intestinal polypeptide (VIP) rhythm in the human suprachiasmatic nucleus. Brain Res. 1996;722:78–82. doi: 10.1016/0006-8993(96)00180-1. [DOI] [PubMed] [Google Scholar]
- Hosoda H, Kato K, Asano H, Ito M, Kato H, Iwamoto T, Suzuki A, Masushige S, Kida S. CBP/p300 is a cell type-specific modulator of CLOCK/BMAL1-mediated transcription. Mol Brain. 2009;2:34. doi: 10.1186/1756-6606-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd MW, Ralph MR. The significance of circadian organization for longevity in the golden hamster. J Biol Rhythms. 1998;13:430–436. doi: 10.1177/074873098129000255. [DOI] [PubMed] [Google Scholar]
- Iranzo A. Parkinson disease and sleep: sleep-wake changes in the premotor stage of Parkinson disease; impaired olfaction and other prodromal features. Curr Neurol Neurosci Rep. 2013;13:373. doi: 10.1007/s11910-013-0373-0. [DOI] [PubMed] [Google Scholar]
- Jung-Hynes B, Reiter RJ, Ahmad N. Sirtuins, melatonin and circadian rhythms: building a bridge between aging and cancer. J Pineal Res. 2010;48:9–19. doi: 10.1111/j.1600-079X.2009.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Rabinovitch PS, Martin GM. Healthy aging: the ultimate preventative medicine. Science. 2015;350:1191–1193. doi: 10.1126/science.aad3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsbeek A, Palm IF, La Fleur SE, Scheer FA, Perreau-Lenz S, Ruiter M, Kreier F, Cailotto C, Buijs RM. SCN outputs and the hypothalamic balance of life. J Biol Rhythms. 2006;21:458–469. doi: 10.1177/0748730406293854. [DOI] [PubMed] [Google Scholar]
- Khapre RV, Kondratova AA, Patel S, Dubrovsky Y, Wrobel M, Antoch MP, Kondratov RV. BMAL1-dependent regulation of the mTOR signaling pathway delays aging. Aging. 2014;6:48–57. doi: 10.18632/aging.100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolker DE, Fukuyama H, Huang DS, Takahashi JS, Horton TH, Turek FW. Aging alters circadian and light-induced expression of clock genes in golden hamsters. J Biol Rhythms. 2003;18:159–169. doi: 10.1177/0748730403251802. [DOI] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratova AA, Kondratov RV. The circadian clock and pathology of the ageing brain. Nat Rev Neurosci. 2012;13:325–335. doi: 10.1038/nrn3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo T, Loh DH, Truong D, Wu Y, Colwell CS. Circadian dysfunction in a mouse model of Parkinson’s disease. Exp Neurol. 2011;232:66–75. doi: 10.1016/j.expneurol.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande-Diner L, Boyault C, Kim JY, Weitz CJ. A positive feedback loop links circadian clock factor CLOCK-BMAL1 to the basic transcriptional machinery. Proc Natl Acad Sci USA. 2013;110:16021–16026. doi: 10.1073/pnas.1305980110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC. The circadian clock and tumor suppression by mammalian period genes. Methods Enzymol. 2005;393:852–861. doi: 10.1016/S0076-6879(05)93045-0. [DOI] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/S0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Lee JH, Bliwise DL, Ansari FP, Goldstein FC, Cellar JS, Lah JJ, Levey AI. Daytime sleepiness and functional impairment in Alzheimer disease. Am J Geriatr Psychiatry. 2007;15:620–626. doi: 10.1097/JGP.0b013e3180381521. [DOI] [PubMed] [Google Scholar]
- Li H, Satinoff E. Fetal tissue containing the suprachiasmatic nucleus restores multiple circadian rhythms in old rats. Am J Physiol. 1998;275:R1735–R1744. doi: 10.1152/ajpregu.1998.275.6.R1735. [DOI] [PubMed] [Google Scholar]
- Li XM, Delaunay F, Dulong S, Claustrat B, Zampera S, Fujii Y, Teboul M, Beau J, Levi F. Cancer inhibition through circadian reprogramming of tumor transcriptome with meal timing. Cancer Res. 2010;70:3351–3360. doi: 10.1158/0008-5472.CAN-09-4235. [DOI] [PubMed] [Google Scholar]
- Li J, Kim SG, Blenis J. Rapamycin: one drug, many effects. Cell Metab. 2014;19:373–379. doi: 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, Guttler T, Davis F, Asara JM, Sahin M. The circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation. Cell. 2015;161:1138–1151. doi: 10.1016/j.cell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez M, Nogueiras R, Tena-Sempere M, Dieguez C. Hypothalamic AMPK: a canonical regulator of whole-body energy balance. Nat Rev Endocrinol. 2016;12:421–432. doi: 10.1038/nrendo.2016.67. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Galluzzi L, Freije JM, Madeo F, Kroemer G. Metabolic control of longevity. Cell. 2016;166:802–821. doi: 10.1016/j.cell.2016.07.031. [DOI] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao CY, Roqueta-Rivera M, Deng CX, Osborne TF, Mostoslavsky R, et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell. 2014;158:659–672. doi: 10.1016/j.cell.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattis J, Sehgal A. Circadian rhythms, sleep, and disorders of aging. Trends Endocrinol Metab. 2016;27:192–203. doi: 10.1016/j.tem.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Spasche A, Reed HE, Piggins HD. Neurotensin phase-shifts the firing rate rhythm of neurons in the rat suprachiasmatic nuclei in vitro. Eur J Neurosci. 2002;16:339–344. doi: 10.1046/j.1460-9568.2002.02067.x. [DOI] [PubMed] [Google Scholar]
- Moore RY, Speh JC. GABA is the principal neurotransmitter of the circadian system. Neurosci Lett. 1993;150:112–116. doi: 10.1016/0304-3940(93)90120-A. [DOI] [PubMed] [Google Scholar]
- Morin LP, Allen CN. The circadian visual system, 2005. Brain Res Rev. 2006;51:1–60. doi: 10.1016/j.brainresrev.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Morin LP, Shivers KY, Blanchard JH, Muscat L. Complex organization of mouse and rat suprachiasmatic nucleus. Neuroscience. 2006;137:1285–1297. doi: 10.1016/j.neuroscience.2005.10.030. [DOI] [PubMed] [Google Scholar]
- Morton AJ, Wood NI, Hastings MH, Hurelbrink C, Barker RA, Maywood ES. Disintegration of the sleep-wake cycle and circadian timing in Huntington’s disease. J Neurosci. 2005;25:157–163. doi: 10.1523/JNEUROSCI.3842-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadon NL. Exploiting the rodent model for studies on the pharmacology of lifespan extension. Aging Cell. 2006;5:9–15. doi: 10.1111/j.1474-9726.2006.00185.x. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura W, Yamazaki S, Takasu NN, Mishima K, Block GD. Differential response of Period 1 expression within the suprachiasmatic nucleus. J Neurosci. 2005;25:5481–5487. doi: 10.1523/JNEUROSCI.0889-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TJ, Nakamura W, Yamazaki S, Kudo T, Cutler T, Colwell CS, Block GD. Age-related decline in circadian output. J Neurosci. 2011;31:10201–10205. doi: 10.1523/JNEUROSCI.0451-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak CM, Nunez AA. A sparse projection from the suprachiasmatic nucleus to the sleep active ventrolateral preoptic area in the rat. NeuroReport. 2000;11:93–96. doi: 10.1097/00001756-200001170-00019. [DOI] [PubMed] [Google Scholar]
- Ozturk N, Lee JH, Gaddameedhi S, Sancar A. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proc Natl Acad Sci USA. 2009;106:2841–2846. doi: 10.1073/pnas.0813028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- Peek CB, Levine DC, Cedernaes J, Taguchi A, Kobayashi Y, Tsai SJ, Bonar NA, McNulty MR, Ramsey KM, Bass J. Circadian clock interaction with HIF1alpha mediates oxygenic metabolism and anaerobic glycolysis in skeletal muscle. Cell Metab. 2017 doi: 10.1016/j.cmet.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penev PD, Kolker DE, Zee PC, Turek FW. Chronic circadian desynchronization decreases the survival of animals with cardiomyopathic heart disease. Am J Physiol. 1998;275:H2334–H2337. doi: 10.1152/ajpheart.1998.275.6.H2334. [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/S0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Pulivarthy SR, Tanaka N, Welsh DK, De Haro L, Verma IM, Panda S. Reciprocity between phase shifts and amplitude changes in the mammalian circadian clock. Proc Natl Acad Sci USA. 2007;104:20356–20361. doi: 10.1073/pnas.0708877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram RV, Kowalczyk MS, de Boer CG, Schneider RK, Miller PG, McConkey M, Tothova Z, Tejero H, Heckl D, Jaras M, et al. Core circadian clock genes regulate leukemia stem cells in AML. Cell. 2016;165:303–316. doi: 10.1016/j.cell.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z, Zhang H, Huang M, Shi G, Liu Z, Xie P, Li H, Wang W, Xu G, Zhang Y, et al. Loss of ZBTB20 impairs circadian output and leads to unimodal behavioral rhythms. eLife. 2016;5:e17171. doi: 10.7554/eLife.17171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadori G, Fujikawa T, Fukuda M, Anderson J, Morgan DA, Mostoslavsky R, Stuart RC, Perello M, Vianna CR, Nillni EA, et al. SIRT1 deacetylase in POMC neurons is required for homeostatic defenses against diet-induced obesity. Cell Metab. 2010;12:78–87. doi: 10.1016/j.cmet.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadori G, Fujikawa T, Anderson J, Berglund ED, Frazao R, Michan S, Vianna CR, Sinclair DA, Elias CF, Coppari R. SIRT1 deacetylase in SF1 neurons protects against metabolic imbalance. Cell Metab. 2011;14:301–312. doi: 10.1016/j.cmet.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, Rey G. Metabolic and nontranscriptional circadian clocks: eukaryotes. Annu Rev Biochem. 2014;83:165–189. doi: 10.1146/annurev-biochem-060713-035623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Riera CE, Dillin A. Tipping the metabolic scales towards increased longevity in mammals. Nat Cell Biol. 2015;17:196–203. doi: 10.1038/ncb3107. [DOI] [PubMed] [Google Scholar]
- Roozendaal B, van Gool WA, Swaab DF, Hoogendijk JE, Mirmiran M. Changes in vasopressin cells of the rat suprachiasmatic nucleus with aging. Brain Res. 1987;409:259–264. doi: 10.1016/0006-8993(87)90710-4. [DOI] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- Satlin A, Volicer L, Stopa EG, Harper D. Circadian locomotor activity and core-body temperature rhythms in Alzheimer’s disease. Neurobiol Aging. 1995;16:765–771. doi: 10.1016/0197-4580(95)00059-N. [DOI] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, Yamada KA, Imai S. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013;18:416–430. doi: 10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarbrough K, Losee-Olson S, Wallen EP, Turek FW. Aging and photoperiod affect entrainment and quantitative aspects of locomotor behavior in Syrian hamsters. Am J Physiol. 1997;272:R1219–R1225. doi: 10.1152/ajpregu.1997.272.4.R1219. [DOI] [PubMed] [Google Scholar]
- Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M, Wada T, Aoyagi T, Tezuka M. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci USA. 2005;102:12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara K, Hiruma H, Funabashi T, Kimura F. GABAergic modulation of gap junction communication in slice cultures of the rat suprachiasmatic nucleus. Neuroscience. 2000;96:591–596. doi: 10.1016/S0306-4522(99)00556-4. [DOI] [PubMed] [Google Scholar]
- Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485:62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stashi E, Lanz RB, Mao J, Michailidis G, Zhu B, Kettner NM, Putluri N, Reineke EL, Reineke LC, Dasgupta S, et al. SRC-2 is an essential coactivator for orchestrating metabolism and circadian rhythm. Cell reports. 2014;6:633–645. doi: 10.1016/j.celrep.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterniczuk R, Antle MC, Laferla FM, Dyck RH. Characterization of the 3xTg-AD mouse model of Alzheimer’s disease: part 2. Behavioral and cognitive changes. Brain Res. 2010;1348:149–155. doi: 10.1016/j.brainres.2010.06.011. [DOI] [PubMed] [Google Scholar]
- Sterniczuk R, Dyck RH, Laferla FM, Antle MC. Characterization of the 3xTg-AD mouse model of Alzheimer’s disease: part 1. Circadian changes. Brain Res. 2010;1348:139–148. doi: 10.1016/j.brainres.2010.05.013. [DOI] [PubMed] [Google Scholar]
- Swaab DF, Fliers E, Partiman TS. The suprachiasmatic nucleus of the human brain in relation to sex, age and senile dementia. Brain Res. 1985;342:37–44. doi: 10.1016/0006-8993(85)91350-2. [DOI] [PubMed] [Google Scholar]
- Torra IP, Tsibulsky V, Delaunay F, Saladin R, Laudet V, Fruchart JC, Kosykh V, Staels B. Circadian and glucocorticoid regulation of Rev-erbalpha expression in liver. Endocrinology. 2000;141:3799–3806. doi: 10.1210/endo.141.10.7708. [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, Chung JH. Activation of 5’-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J Biol Chem. 2007;282:20794–20798. doi: 10.1074/jbc.C700070200. [DOI] [PubMed] [Google Scholar]
- Valentinuzzi VS, Scarbrough K, Takahashi JS, Turek FW. Effects of aging on the circadian rhythm of wheel-running activity in C57BL/6 mice. Am J Physiol. 1997;273:R1957–R1964. doi: 10.1152/ajpregu.1997.273.6.R1957. [DOI] [PubMed] [Google Scholar]
- van Someren EJ, Hagebeuk EE, Lijzenga C, Scheltens P, de Rooij SE, Jonker C, Pot AM, Mirmiran M, Swaab DF. Circadian rest-activity rhythm disturbances in Alzheimer’s disease. Biol Psychiatry. 1996;40:259–270. doi: 10.1016/0006-3223(95)00370-3. [DOI] [PubMed] [Google Scholar]
- Videnovic A, Lazar AS, Barker RA, Overeem S. ‘The clocks that time us’–circadian rhythms in neurodegenerative disorders. Nat Rev Neurol. 2014;10:683–693. doi: 10.1038/nrneurol.2014.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert D. Age-dependent changes of the circadian system. Chronobiol Int. 2000;17:261–283. doi: 10.1081/CBI-100101048. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Wu Y, Tang D, Liu N, Xiong W, Huang H, Li Y, Ma Z, Zhao H, Chen P, Qi X, et al. Reciprocal regulation between the circadian clock and hypoxia signaling at the genome level in mammals. Cell Metab. 2017 doi: 10.1016/j.cmet.2016.09.009. [DOI] [PubMed] [Google Scholar]
- Wyse CA, Coogan AN. Impact of aging on diurnal expression patterns of CLOCK and BMAL1 in the mouse brain. Brain Res. 2010;1337:21–31. doi: 10.1016/j.brainres.2010.03.113. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Effects of aging on central and peripheral mammalian clocks. Proc Natl Acad Sci USA. 2002;99:10801–10806. doi: 10.1073/pnas.152318499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Okamura H. Gradients in the circadian expression of Per1 and Per2 genes in the rat suprachiasmatic nucleus. Eur J Neurosci. 2002;15:1153–1162. doi: 10.1046/j.1460-9568.2002.01955.x. [DOI] [PubMed] [Google Scholar]
- Yan L, Karatsoreos I, Lesauter J, Welsh DK, Kay S, Foley D, Silver R. Exploring spatiotemporal organization of SCN circuits. Cold Spring Harb Symp Quant Biol. 2007;72:527–541. doi: 10.1101/sqb.2007.72.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon IY, Kripke DF, Elliott JA, Youngstedt SD, Rex KM, Hauger RL. Age-related changes of circadian rhythms and sleep-wake cycles. J Am Geriatr Soc. 2003;51:1085–1091. doi: 10.1046/j.1532-5415.2003.51356.x. [DOI] [PubMed] [Google Scholar]
- Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JN, Hofman MA, Swaab DF. VIP neurons in the human SCN in relation to sex, age, and Alzheimer’s disease. Neurobiol Aging. 1995;16:571–576. doi: 10.1016/0197-4580(95)00043-E. [DOI] [PubMed] [Google Scholar]


