Abstract
The Didymellaceae is one of the most species-rich families in the fungal kingdom, and includes species that inhabit a wide range of ecosystems. The taxonomy of Didymellaceae has recently been revised on the basis of multi-locus DNA sequence data. In the present study, we investigated 108 Didymellaceae isolates newly obtained from 40 host plant species in 27 plant families, and various substrates from caves, including air, water and carbonatite, originating from Argentina, Australia, Canada, China, Hungary, Israel, Italy, Japan, South Africa, the Netherlands, the USA and former Yugoslavia. Among these, 68 isolates representing 32 new taxa are recognised based on the multi-locus phylogeny using sequences of LSU, ITS, rpb2 and tub2, and morphological differences. Within the Didymellaceae, five genera appeared to be limited to specific host families, with other genera having broader host ranges. In total 19 genera are recognised in the family, with Heracleicola being reduced to synonymy under Ascochyta. This study has significantly improved our understanding on the distribution and biodiversity of Didymellaceae, although the placement of several genera still need to be clarified.
Key words: Host-associated, Karst caves, Multi-locus phylogeny, Phoma, Taxonomy
Taxonomic novelties: New species: Allophomaoligotrophica Q. Chen, Crous & L. Cai; Ascochytaboeremae L.W. Hou, Crous & L. Cai; Calophomarosae Q. Chen, Crous & L. Cai; Didymellaaeria Q. Chen, Crous & L. Cai, D. aquatica Q. Chen, Crous & L. Cai, D. chloroguttulata Q. Chen, Crous & L. Cai, D. ellipsoidea Q. Chen, Crous & L. Cai, D. ilicicola Q. Chen, Crous & L. Cai, D. infuscatispora Q. Chen, Crous & L. Cai, D. macrophylla Q. Chen, Crous & L. Cai, D. ocimicola Q. Chen, Crous & L. Cai, D. pteridis L.W. Hou, Crous & L. Cai, D. sinensis Q. Chen, Crous & L. Cai, D. suiyangensis Q. Chen, Crous & L. Cai; Epicoccumcamelliae Q. Chen, Crous & L. Cai, E. dendrobii Q. Chen, Crous & L. Cai, E. duchesneae Q. Chen, Crous & L. Cai, E. hordei Q. Chen, Crous & L. Cai, E. italicum Q. Chen, Crous & L. Cai, E. latusicollum Q. Chen, Crous & L. Cai, E. layuense Q. Chen, Crous & L. Cai, E. poae Q. Chen, Crous & L. Cai, E. viticis Q. Chen, Crous & L. Cai; Heterophomaverbascicola Q. Chen, Crous & L. Cai; Neoascochytaargentina L.W. Hou, Crous & L. Cai, Neoa. soli Q. Chen, Crous & L. Cai, Neoa. triticicola L.W. Hou, Crous & L. Cai; Neodidymelliopsisachlydis L.W. Hou, Crous & L. Cai, Neod. longicolla L.W. Hou, Crous & L. Cai; Stagonosporopsisbomiensis Q. Chen, Crous & L. Cai, S. papillata Q. Chen, Crous & L. Cai
New variety: Boeremiaexigua var. opuli Q. Chen, Crous & L. Cai
New combinations: Ascochytapremilcurensis (Tibpromma et al.) Q. Chen, Crous & L. Cai; Didymellasegeticola (Q. Chen) Q. Chen, Crous & L. Cai
Introduction
The Didymellaceae is the largest family in the Pleosporales (Ascomycota, Pezizomycotina, Dothideomycetes), with more than 5 400 taxon names listed in MycoBank (Crous et al. 2004). The family Didymellaceae was established by de Gruyter et al. (2009) to encompass three main genera, viz. Ascochyta, Didymella and Phoma, and other allied phoma-like genera which grouped in the Didymellaceae. Aveskamp et al. (2010) circumscribed the boundaries of Didymellaceae, redefined the genera Epicoccum, Peyronellaea and Stagonosporopsis, and established the genus Boeremia. He also acknowledged two sexual genera in the family, namely Leptosphaerulina and Macroventuria. In spite of these studies, the polyphyly of Ascochyta, Didymella and Phoma remained unresolved. A revision of the Didymellaceae has recently been published, comprising 17 well-supported monophyletic clades which were treated as individual genera (Chen et al. 2015a). Moreover, the generic delimitations of Ascochyta, Didymella, Epicoccum and Phoma were further emended to reveal more natural evolutionary relationships (Chen et al. 2015a). Subsequent to this revision, several additional genera were added, namely Briansuttonomyces (Crous & Groenewald 2016), Neomicrosphaeropsis (Thambugala et al. 2017), Didymellocamarosporium (Wijayawardene et al. 2016), Heracleicola and Neodidymella (Ariyawansa et al. 2015).
Species of Didymellaceae are cosmopolitan and distributed throughout a broad range of environments. Most of the members in this family are plant pathogens of a wide range of hosts, mainly causing leaf and stem lesions; some are of quarantine significance (Aveskamp et al., 2008, Aveskamp et al., 2010, Chen et al., 2015a, Chen et al., 2015b). Several species belonging to Ascochyta and Nothophoma have been reported to be host-specific to a single plant genus or family (Aveskamp et al., 2010, Chen et al., 2015a). Nevertheless, host specificity in genera of Didymellaceae has not been specifically addressed.
Correct species identification in this family has always proven difficult, chiefly relying on morphology and plant host association (Aveskamp et al., 2010, Chen et al., 2015a). However, a robust backbone tree based on internal transcribed spacer regions and intervening 5.8S nrDNA (ITS), partial 28S large subunit nrDNA (LSU) sequences, and partial regions of RNA polymerase II second largest subunit (rpb2) and β-tubulin (tub2) genes provide a relatively robust phylogenetic backbone for taxon determination (Chen et al. 2015a).
The present study reports on a collection of 108 Didymellaceae isolates obtained from 40 host plant species in 27 plant families in China, as well as several other countries. Of these, 68 isolates representing 32 new taxa are described by employing a polyphasic approach using morphological characteristics and multi-locus phylogenetics.
Materials and methods
Sampling and isolation
The majority of Didymellaceae strains were isolated from diseased plants in seven provinces of China (Gansu, Guizhou, Inner Mongolia, Jiangxi, Qianghai, Shandong and Tibet), as well as Australia, Italy, Japan and the USA. Some strains isolated from air, soil, water and faeces were collected from the Mingyong Glacier in Yunnan Province and inside the Karst caves in Guizhou Province in China. The air, soil and water samples were collected from inside the cave following the methods used by Zhang et al. (2017). Several strains were obtained from the Herbarium BRIP (Dutton Park, Queensland, Australia), the International Collection of Microorganisms from Plants (ICMP, Landcare Research, Auckland, New Zealand), and the Westerdijk Fungal Biodiversity Institute (CBS, Utrecht, the Netherlands), as listed in Table 1.
Table 1.
Isolates used in this study and their GenBank accession numbers. New taxa and new combinations introduced in the present study and newly generated sequences are indicated in bold.
| Species | Strain number1 | Status2 | Host, substrate | Host family | Country | GenBank accession numbers3 |
|||
|---|---|---|---|---|---|---|---|---|---|
| LSU | ITS | RPB2 | TUB | ||||||
| Allophoma labilis | CBS 124.93; PD 87/269 | Lycopersicon esculentum | Solanaceae | Netherlands | GU238091 | GU237765 | KT389552 | GU237619 | |
| Al. minor | CBS 325.82 | T | Syzygium aromaticum | Myrtaceae | Indonesia | GU238107 | GU237831 | KT389553 | GU237632 |
| Al. nicaraguensis | CBS 506.91; PD 91/876; IMI 215229 | T | Coffea arabica | Rubiaceae | Nicaragua | GU238058 | GU237876 | KT389551 | GU237596 |
| Al. oligotrophica | CGMCC 3.18114; LC 6245 | T | Air | China | KY742194 | KY742040 | KY742128 | KY742282 | |
| CGMCC 3.18115; LC 6246 | Air | China | KY742195 | KY742041 | KY742129 | KY742283 | |||
| CGMCC 3.18116; LC 6247 | Air | China | KY742196 | KY742042 | KY742130 | KY742284 | |||
| Al. piperis | CBS 268.93; CBS 108.93; PD 88/720 | T | Peperomia pereskiifolia | Piperaceae | Netherlands | GU238129 | GU237816 | KT389554 | GU237644 |
| CBS 108.93; PD 90/2011 | Peperomia sp. | Piperaceae | Netherlands | GU238130 | GU237921 | KT389555 | GU237645 | ||
| Al. tropica | CBS 436.75; DSM 63365 | T | Saintpaulia ionantha | Gesneriaceae | Germany | GU238149 | GU237864 | KT389556 | GU237663 |
| Al. zantedeschiae | CBS 131.93; PD 69/140 | Calla sp. | Araceae | Netherlands | GU238159 | FJ427084 | KT389557 | FJ427188 | |
| CBS 229.32 | Cicer arietinum | Fabeceae | Romania | KT389690 | KT389473 | KT389558 | KT389767 | ||
| ICMP 16850 | Lycopersicon esculentum | Solanaceae | Hungary | KY742197 | KY742043 | KY742131 | KY742285 | ||
| Ascochyta boeremae | CBS 372.84; PD 80/1246 | T | Pisum sativum | Fabeceae | Australia | KT389697 | KT389480 | — | KT389774 |
| As. boeremae | CBS 373.84; PD 80/1247 | Pisum sativum | Fabeceae | Australia | KT389698 | KT389481 | KT389560 | KT389775 | |
| As. fabae | CBS 524.77 | Phaseolus vulgaris | Fabeceae | Belgium | GU237963 | GU237880 | — | GU237526 | |
| CBS 649.71 | Vicia faba | Fabeceae | Netherlands | GU237964 | GU237902 | — | GU237527 | ||
| PD 83/492 | Phaseolus vulgaris | Fabeceae | Netherlands | GU237965 | GU237917 | — | GU237528 | ||
| As. herbicola | CBS 629.97; PD 76/1017 | R | Water | USA | GU238083 | GU237898 | KP330421 | GU237614 | |
| As. lentis | CBS 370.84; PD 81/783 | Lens culinaris | Fabeceae | — | KT389691 | KT389474 | — | KT389768 | |
| As. medicaginicola var. macrospora | BRIP 45051; LC 5258 | Medicago sativa | Fabeceae | Australia | KY742198 | KY742044 | KY742132 | KY742286 | |
| CBS 112.53 | T | Medicago sativa | Fabeceae | USA | GU238101 | GU237749 | — | GU237628 | |
| CBS 404.65; IMI 116999 | R | Medicago sativa | Fabeceae | Canada | GU238102 | GU237859 | KP330423 | GU237629 | |
| As. medicaginicola var. medicaginicola | CBS 316.90 | Medicago sativa | Fabeceae | Czech Republic | GU238103 | GU237828 | — | GU237630 | |
| As. nigripycnidia | CBS 116.96; PD 95/7930 | T | Vicia cracca | Fabeceae | Russia | GU238118 | GU237756 | — | GU237637 |
| As. phacae | CBS 184.55 | T | Phaca alpina | Fabeceae | Switzerland | KT389692 | KT389475 | — | KT389769 |
| As. pisi | CBS 122750; ATCC 201619 | Pisum sativum | Fabeceae | USA | KT389694 | KT389477 | — | KT389771 | |
| CBS 122751; ATCC 201620 | Pisum sativum | Fabeceae | Canada | KP330444 | KP330432 | EU874867 | KP330388 | ||
| CBS 122785; PD 78/517 | T | Pisum sativum | Fabeceae | Netherlands | GU237969 | GU237763 | — | GU237532 | |
| CBS 126.54 | Pisum sativum | Fabeceae | Netherlands | EU754137 | GU237772 | DQ677967 | GU237531 | ||
| CBS 108.49 | Juglans regia | Juglandaceae | Netherlands | KT389693 | KT389476 | — | KT389770 | ||
| As. premilcurensis | MFLUCC 14-0518 | T | Heracleum sphondylium | Apiaceae | Italy | KT326695 | KT326694 | — | — |
| As. rabiei | CBS 206.30 | — | — | — | KT389695 | KT389478 | KT389559 | KT389772 | |
| CBS 237.37 | T | Cicer arietinum | Fabeceae | Bulgaria | KT389696 | KT389479 | — | KT389773 | |
| CBS 534.65 | Cicer arietinum | Fabeceae | India | GU237970 | GU237886 | KP330405 | GU237533 | ||
| As. syringae | CBS 545.72 | Syringa vulgaris | Oleaceae | Netherlands | KT389700 | KT389483 | — | KT389777 | |
| As. versabilis | CBS 876.97; PD 82/1008 | R | Silene sp. | Caryophyllaceae | Netherlands | GU238152 | GU237909 | KT389561 | GU237664 |
| As. viciae | CBS 451.68 | Vicia sepium | Fabeceae | Netherlands | KT389701 | KT389484 | KT389562 | KT389778 | |
| As. viciae-pannonicae | CBS 254.92 | Vicia pannonica | Fabeceae | Czech Republic | KT389702 | KT389485 | — | KT389779 | |
| Boeremia crinicola | CBS 109.79; PD 77/747 | R | Crinum powellii | Amaryllidaceae | Netherlands | GU237927 | GU237737 | KT389563 | GU237489 |
| B. diversispora | CBS 102.80; IMI 331907; PD 79/61 | Phaseolus vulgaris | Fabeceae | Kenya | GU237930 | GU237725 | KT389565 | GU237492 | |
| CBS 101194; PD 79/687; IMI 373349 | Phaseolus vulgaris | Fabeceae | Netherlands | GU237929 | GU237716 | KT389564 | GU237491 | ||
| B. exigua var. coffeae | CBS 119730 | Coffea arabica | Rubiaceae | Brazil | GU237942 | GU237759 | KT389567 | GU237504 | |
| CBS 109183; PD 2000/10506; IMI 300060 | R | Coffea arabica | Rubiaceae | Cameroon | GU237943 | GU237748 | KT389566 | GU237505 | |
| CBS 431.74; PD 74/2447 | R | Solanum tuberosum | Solanaceae | Netherlands | EU754183 | FJ427001 | KT389569 | FJ427112 | |
| B. exigua var. forsythiae | CBS 101197; PD 95/721 | Forsythia sp. | Oleaceae | Netherlands | GU237931 | GU237718 | KT389570 | GU237493 | |
| CBS 101213; PD 92/959 | R | Forsythia sp. | Oleaceae | Netherlands | GU237932 | GU237723 | KT389571 | GU237494 | |
| B. exigua var. gilvescens | CBS 101150; PD 79/118 | Cichorium intybus | Asteraceae | Netherlands | EU754182 | GU237715 | KT389568 | GU237495 | |
| B. exigua var. heteromorpha | CBS 443.94 | T | Nerium oleander | Apocynaceae | Italy | GU237935 | GU237866 | KT389573 | GU237497 |
| CBS 101196; PD 79/176 | Nerium oleander | Apocynaceae | France | GU237934 | GU237717 | KT389572 | GU237496 | ||
| B. exigua var. linicola | CBS 114.28 | Linum usitatissimum | Linaceae | Netherlands | GU237937 | GU237752 | — | GU237499 | |
| CBS 116.76; ATCC 32332; IMI 197074; PD 75/544 | R | Linum usitatissimum | Linaceae | Netherlands | GU237938 | GU237754 | KT389574 | GU237500 | |
| CBS 248.38 | Nemophila insignis | Hydrophyllaceae | Netherlands | KT389703 | KT389486 | KT389575 | KT389780 | ||
| B. exigua var. opuli | CGMCC 3.18354; LC 8117 | T | Viburnum opulus | Caprifoliaceae | USA | KY742199 | KY742045 | KY742133 | KY742287 |
| LC 8118 | Viburnum opulus | Caprifoliaceae | USA | KY742200 | KY742046 | KY742134 | KY742288 | ||
| B. exigua var. populi | CBS 100167; PD 93/217 | T | Populus (×) euramericana | Salicaceae | Netherlands | GU237939 | GU237707 | — | GU237501 |
| B. exigua var. pseudolilacis | CBS 423.67 | Lathyrus sp. | Fabeceae | Netherlands | KT389704 | KT389487 | KT389576 | KT389781 | |
| CBS 462.67 | Lamium maculatum | Lamiaceae | Netherlands | KT389705 | KT389488 | — | KT389782 | ||
| CBS 101207; PD 94/614 | T | Syringa vulgaris | Oleaceae | Netherlands | GU237941 | GU237721 | — | GU237503 | |
| B. exigua var. viburni | CBS 100354; PD 83/448 | R | Viburnum opulus | Caprifoliaceae | Netherlands | GU237944 | GU237711 | KT389577 | GU237506 |
| B. foveata | CBS 109176; PD 94/1394 | R | Solanum tuberosum | Solanaceae | Bulgaria | GU237946 | GU237742 | KT389578 | GU237508 |
| B. hedericola | CBS 367.91; PD 87/229 | R | Hedera helix | Araliaceae | Netherlands | GU237949 | GU237842 | KT389579 | GU237511 |
| B. lilacis | CBS 569.79; PD 72/741; IMI 331909 | R | Syringa vulgaris | Oleaceae | Netherlands | GU237936 | GU237892 | — | GU237498 |
| CBS 588.67 | Philadelphus sp. | Saxifragaceae | Netherlands | KT389709 | KT389492 | — | KT389786 | ||
| LC 5178 | Lonicera japonica | Caprifoliaceae | China | KY742201 | KY742047 | — | KY742289 | ||
| LC 8116 | Ocimum sp. | Lamiaceae | China | KY742202 | KY742048 | — | KY742290 | ||
| B. lycopersici | CBS 378.67; PD 67/276 | R | Lycopersicon esculentum | Solanaceae | Netherlands | GU237950 | GU237848 | KT389580 | GU237512 |
| B. noackiana | CBS 100353; PD 87/718 | R | Phaseolus vulgaris | Fabeceae | Guatemala | GU237952 | GU237710 | — | GU237514 |
| CBS 101203; PD 79/1114 | Phaseolus vulgaris | Fabeceae | Colombia | GU237953 | GU237720 | KT389581 | GU237515 | ||
| B. sambuci-nigrae | CBS 629.68; CECT 20048; IMI 331913; PD 67/753 | T | Sambucus nigra | Caprifoliaceae | Netherlands | GU237955 | GU237897 | — | GU237517 |
| B. strasseri | CBS 126.93; PD 73/642 | Mentha sp. | Lamiaceae | Netherlands | GU237956 | GU237773 | KT389584 | GU237518 | |
| B. telephii | CBS 760.73; PD 71/1616 | R | Sedum telephium | Crassulaceae | Netherlands | GU237959 | GU237905 | — | GU237521 |
| CBS 109175; PD 79/524 | R | Sedum telephium | Crassulaceae | Netherlands | GU237958 | GU237741 | KT389585 | GU237520 | |
| B. trachelospermi | CGMCC 3.18222; LC 8105 | T | Trachelospermum jasminoides | Apocynaceae | USA | KY064032 | KY064028 | KY064033 | KY064051 |
| Briansuttonomyces eucalypti | CBS 114879; CPC 362 | T | Eucalyptus sp. | Myrtaceae | South Africa | KU728519 | KU728479 | — | KU728595 |
| CBS 114887; CPC 363 | Eucalyptus sp. | Myrtaceae | South Africa | KU728520 | KU728480 | — | KU728596 | ||
| Calophoma aquilegiicola | CBS 107.31 | Aquilegia sp. | Ranunculaceae | — | KT389710 | KT389493 | — | KT389787 | |
| C. aquilegiicola | CBS 107.96; PD 73/598 | R | Aconitum pyramidale | Ranunculaceae | Netherlands | GU238041 | GU237735 | KT389586 | GU237581 |
| CBS 108.96; PD 79/611 | R | Aquilegia sp. | Ranunculaceae | Netherlands | GU238042 | GU237736 | — | GU237582 | |
| CBS 109.96; PD 83/832 | Aquilegia sp. | Ranunculaceae | Netherlands | KT389711 | KT389494 | — | KT389788 | ||
| CBS 116402 | Thalictrum dipterocarpum | Ranunculaceae | New Zealand | KT389712 | KT389495 | — | KT389789 | ||
| C. clematidina | CBS 102.66 | Clematis sp. | Ranunculaceae | UK | FJ515630 | FJ426988 | KT389587 | FJ427099 | |
| CBS 108.79; PD 78/522 | T | Clematis sp. | Ranunculaceae | Netherlands | FJ515632 | FJ426989 | KT389588 | FJ427100 | |
| C. clematidis-rectae | CBS 507.63; PD 07/03486747; MUCL 9574 | Clematis sp. | Ranunculaceae | Netherlands | FJ515647 | FJ515606 | KT389589 | FJ515624 | |
| C. complanata | CBS 268.92; PD 75/3 | Angelica sylvestris | Umbelliferae | Netherlands | EU754180 | FJ515608 | GU371778 | FJ515626 | |
| CBS 100311 | Heracleum sphondylium | Umbelliferae | Netherlands | EU754181 | GU237709 | KT389590 | GU237594 | ||
| C. glaucii | CBS 112.96; PD 79/765 | Dicentra sp. | Papaveraceae | Netherlands | GU238077 | GU237750 | — | GU237610 | |
| CBS 114.96; PD 94/888 | Chelidonium majus | Papaveraceae | Netherlands | FJ515649 | FJ515609 | — | FJ515627 | ||
| C. rosae | CGMCC 3.18347; LC 5169 | T | Rosa sp. | Rosaceae | China | KY742203 | KY742049 | KY742135 | KY742291 |
| LC 8119 | Rosa sp. | Rosaceae | China | KY742204 | KY742050 | KY742136 | KY742292 | ||
| C. vodakii | CBS 173.53 | T | Hepatica triloba | Ranunculaceae | Switzerland | KT389714 | KT389497 | — | KT389791 |
| Didymella acetosellae | CBS 179.97 | Rumex hydrolapathum | Polygonaceae | Netherlands | GU238034 | GU237793 | KP330415 | GU237575 | |
| D. aeria | CGMCC 3.18353; LC 7441 | T | Air | China | KY742205 | KY742051 | KY742137 | KY742293 | |
| LC 8120 | Air | China | KY742206 | KY742052 | KY742138 | KY742294 | |||
| D. aliena | CBS 379.93; PD 82/945 | Berberis sp. | Berberidaceae | Netherlands | GU238037 | GU237851 | KP330416 | GU237578 | |
| LC 8121 | Pyrus calleryana | Rosaceae | Italy | KY742207 | KY742053 | — | KY742295 | ||
| D. americana | CBS 185.85; PD 80/1191 | R | Zea mays | Poaceae | USA | GU237990 | FJ426972 | KT389594 | FJ427088 |
| CBS 568.97; ATCC 44494; PD 94/1544 | Glycine max | Fabeceae | USA | GU237991 | FJ426974 | — | FJ427090 | ||
| LC 5157 | Sorghum bicolor | Poaceae | China | KY742208 | KY742054 | KY742139 | KY742296 | ||
| D. anserina | CBS 253.80 | — | — | Germany | KT389715 | KT389498 | KT389595 | KT389795 | |
| CBS 285.29 | Calluna sp. | Ericaceae | UK | KT389716 | KT389499 | — | KT389796 | ||
| CBS 360.84 | R | Potato flour | Netherlands | GU237993 | GU237839 | KT389596 | GU237551 | ||
| CBS 397.65 | Plastic | Germany | KT389717 | KT389500 | KT389597 | KT389797 | |||
| D. aquatica | CGMCC 3.18349; LC 5556 | T | Water | China | KY742209 | KY742055 | KY742140 | KY742297 | |
| LC 5555 | Water | China | KY742210 | KY742056 | KY742141 | KY742298 | |||
| D. arachidicola | CBS 333.75; ATCC 28333; IMI 386092; PREM 44889 | T | Arachis hypogaea | Fabeceae | South Africa | GU237996 | GU237833 | KT389598 | GU237554 |
| D. aurea | CBS 269.93; PD 78/1087 | T | Medicago polymorpha | Fabeceae | New Zealand | GU237999 | GU237818 | KT389599 | GU237557 |
| D. bellidis | CBS 714.85; PD 74/265 | R | Bellis perennis | Asteraceae | Netherlands | GU238046 | GU237904 | KP330417 | GU237586 |
| PD 94/886 | Bellis sp. | Asteraceae | Netherlands | GU238047 | GU237923 | — | GU237587 | ||
| D. boeremae | CBS 109942; PD 84/402 | T | Medicago littoralis cv. Harbinger | Fabeceae | Australia | GU238048 | FJ426982 | KT389600 | FJ427097 |
| D. calidophila | CBS 448.83 | T | Soil | Egypt | GU238052 | FJ427059 | — | FJ427168 | |
| PD 84/109 | Cucumis sativus | Cucurbitaceae | Netherlands | GU238053 | FJ427060 | — | FJ427169 | ||
| D. chenopodii | CBS 128.93; PD 79/140 | R | Chenopodium quinoa cv. Sajana | Chenopodiaceae | Peru | GU238055 | GU237775 | KT389602 | GU237591 |
| D. chloroguttulata | CGMCC 3.18351; LC 7435 | T | Air | China | KY742211 | KY742057 | KY742142 | KY742299 | |
| LC 8122 | Air | China | KY742212 | KY742058 | KY742143 | KY742300 | |||
| D. coffeae-arabicae | CBS 123380; PD 84/1013 | T | Coffea arabica | Rubiaceae | Ethiopia | GU238005 | FJ426993 | KT389603 | FJ427104 |
| LC 8975 | Lagerstroemia indica | Lythraceae | Italy | KY742213 | KY742059 | KY742144 | KY742301 | ||
| D. curtisii | CBS 251.92; PD 86/1145 | R | Nerine sp. | Amaryllidaceae | Netherlands | GU238013 | FJ427038 | — | FJ427148 |
| PD 92/1460 | Sprekelia sp. | Amaryllidaceae | Netherlands | GU238012 | FJ427041 | KT389604 | FJ427151 | ||
| D. dactylidis | CBS 124513; PD 73/1414 | T | Dactylis glomerata | Poaceae | USA | GU238061 | GU237766 | — | GU237599 |
| D. dimorpha | CBS 346.82 | T | Opuntia sp | Cactaceae | Spain | GU238068 | GU237835 | — | GU237606 |
| D. ellipsoidea | CGMCC 3.18350; LC 7434 | T | Air | China | KY742214 | KY742060 | KY742145 | KY742302 | |
| LC 8123 | Air | China | KY742215 | KY742061 | KY742146 | KY742303 | |||
| D. eucalyptica | CBS 377.91; PD 79/210 | R | Eucalyptus sp. | Myrtaceae | Australia | GU238007 | GU237846 | KT389605 | GU237562 |
| D. exigua | CBS 183.55 | T | Rumex arifolius | Polygonaceae | France | EU754155 | GU237794 | EU874850 | GU237525 |
| D. gardeniae | CBS 626.68; IMI 108771 | T | Gardenia jasminoides | Rubiaceae | India | GQ387595 | FJ427003 | KT389606 | FJ427114 |
| D. glomerata | CBS 133.72 | Fresco in church | Romania | KT389718 | FJ427004 | — | FJ427115 | ||
| CBS 528.66; PD 63/590 | R | Chrysanthemum sp. | Asteraceae | Netherlands | EU754184 | FJ427013 | GU371781 | FJ427124 | |
| LC 4963 | Leymus chinensis | Poaceae | China | KY742216 | KY742062 | KY742147 | KY742304 | ||
| LC 8124 | Faeces | China | KY742217 | KY742063 | KY742148 | KY742305 | |||
| D. heteroderae | CBS 109.92; PD 73/1405 | T | Undefined food material | Netherlands | GU238002 | FJ426983 | KT389601 | FJ427098 | |
| LC 8125 | Hydrangea macrophylla | Saxifragaceae | China | KY742218 | KY742064 | KY742149 | KY742306 | ||
| D. ilicicola | CGMCC 3.18355; LC 8126; LC 8127 | T | Ilex chinensis | Aquifoliaceae | Italy | KY742219 | KY742065 | KY742150 | KY742307 |
| LC 8127 | Ilex chinensis | Aquifoliaceae | Italy | KY742220 | KY742066 | KY742151 | KY742308 | ||
| D. infuscatispora | CGMCC 3.18356; LC 8128 | T | Chrysanthemum indicum | Asteraceae | China | KY742221 | KY742067 | KY742152 | KY742309 |
| LC 8129 | Chrysanthemum indicum | Asteraceae | China | KY742222 | KY742068 | — | KY742310 | ||
| D. lethalis | CBS 103.25 | — | — | — | GU238010 | GU237729 | KT389607 | GU237564 | |
| LC 8130 | Liquidambar styraciflua | Hamamelidaceae | Italy | KY742223 | KY742069 | KY742153 | KY742311 | ||
| D. longicolla | CBS 124514; PD 80/1189 | T | Opuntia sp. | Cactaceae | Spain | GU238095 | GU237767 | — | GU237622 |
| D. macrophylla | CGMCC 3.18357; LC 8131 | T | Hydrangea macrophylla | Saxifragaceae | Italy | KY742224 | KY742070 | KY742154 | KY742312 |
| LC 8132 | Hydrangea macrophylla | Saxifragaceae | Italy | KY742225 | KY742071 | KY742155 | KY742313 | ||
| D. mascrostoma | CBS 223.69 | R | Acer pseudoplatanus | Aceraceae | Switzerland | GU238096 | GU237801 | KT389608 | GU237623 |
| CBS 247.38 | Pinus nigra var. astriaca | Pinaceae | — | KT389719 | KT389501 | — | KT389798 | ||
| CBS 482.95 | Larix decidua | Pinaceae | Germany | GU238099 | GU237869 | KT389609 | GU237626 | ||
| CBS 529.66; PD 66/521 | R | Malus sylvestris | Rosaceae | Netherlands | GU238098 | GU237885 | — | GU237625 | |
| LC 5203 | Soil | China | KY742226 | KY742072 | KY742156 | KY742314 | |||
| D. maydis | CBS 588.69 | T | Zea mays | Poaceae | USA | EU754192 | FJ427086 | GU371782 | FJ427190 |
| D. microchlamydospora | CBS 105.95 | T | Eucalyptus sp. | Myrtaceae | UK | GU238104 | FJ427028 | KP330424 | FJ427138 |
| D. molleriana | CBS 229.79; LEV 7660 | R | Digitalis purpurea | Scrophulariaceae | New Zealand | GU238067 | GU237802 | KP330418 | GU237605 |
| CBS 109179; PD 90/835-1 | Digitalis sp. | Scrophulariaceae | Netherlands | GU238066 | GU237744 | — | GU237604 | ||
| D. musae | CBS 463.69 | R | Mangifera indica | Anacardiaceae | India | GU238011 | FJ427026 | — | FJ427136 |
| D. negriana | CBS 358.71 | R | Vitis vinifera | Vitaceae | Germany | GU238116 | GU237838 | KT389610 | GU237635 |
| ICMP 10845; LC 5249 | Vitis vinifera | Vitaceae | former Yugoslavia | KY742227 | KY742073 | — | KY742315 | ||
| D. nigricans | CBS 444.81; PDDCC 6546 | T | Actinidia chinensis | Actinidiaceae | New Zealand | GU238000 | GU237867 | — | GU237558 |
| LC 8133 | Robinia pseudoacacia f. decaisneana | Fabeceae | Italy | KY742228 | KY742074 | KY742157 | KY742316 | ||
| LC 8134 | Acer palmatum | Aceraceae | Japan | KY742229 | KY742075 | KY742158 | KY742317 | ||
| LC 8135 | Acer palmatum | Aceraceae | Japan | KY742230 | KY742076 | KY742159 | KY742318 | ||
| LC 8136 | Acer palmatum | Aceraceae | Japan | KY742231 | KY742077 | KY742160 | KY742319 | ||
| PD 77/919 | Actinidia chinensis | Actinidiaceae | New Zealand | GU238001 | GU237915 | KT389611 | GU237559 | ||
| D. ocimicola | CGMCC 3.18358; LC 8137 | T | Ocimum sp. | Lamiaceae | China | KY742232 | KY742078 | — | KY742320 |
| LC 8138 | Ocimum sp. | Lamiaceae | China | KY742233 | KY742079 | — | KY742321 | ||
| D. pedeiae | CBS 124517; PD 92/612A | T | Schefflera elegantissima | Araliaceae | Netherlands | GU238127 | GU237770 | KT389612 | GU237642 |
| D. pinodella | CBS 318.90; PD 81/729 | Pisum sativum | Fabeceae | Netherlands | GU238016 | FJ427051 | — | FJ427161 | |
| CBS 531.66 | Trifolium pretense | Fabeceae | USA | GU238017 | FJ427052 | KT389613 | FJ427162 | ||
| LC 8139 | Acer palmatum | Aceraceae | Japan | KY742234 | KY742080 | KY742161 | KY742322 | ||
| D. pinodes | CBS 525.77 | T | Pisum sativum | Fabeceae | Belgium | GU238023 | GU237883 | KT389614 | GU237572 |
| D. pomorum | CBS 285.76; ATCC 26241; IMI 176742; VKM F-1843 | Heracleum dissectum | Umbelliferae | Russia | GU238025 | FJ427053 | KT389615 | FJ427163 | |
| CBS 354.52 | Triticum spelta | Poaceae | Switzerland | KT389720 | KT389502 | KT389616 | KT389799 | ||
| CBS 388.80 | Triticum sp. | Poaceae | South Africa | GU238027 | FJ427055 | KT389617 | FJ427165 | ||
| CBS 539.66; ATCC 16791; IMI 122266; PD 64/914 | R | Polygonum tataricum | Polygonaceae | Netherlands | GU238028 | FJ427056 | KT389618 | FJ427166 | |
| LC 5185 | Gentiana straminea | Gentianaceae | China | KY742235 | KY742081 | KY742162 | KY742323 | ||
| LC 8140 | Dendrobium fimbriatum | Orchidaceae | China | KY742236 | KY742082 | — | KY742324 | ||
| D. protuberans | CBS 132.96; PD 93/853 | Rhinanthus major | Scrophulariaceae | Netherlands | GU237989 | GU237778 | — | GU237550 | |
| CBS 377.93; PD 80/976 | Daucus carota | Umbelliferae | Netherlands | GU238014 | GU237847 | KT389619 | GU237565 | ||
| CBS 381.96; PD 71/706 | T | Lycium halifolium | Solanaceae | Netherlands | GU238029 | GU237853 | KT389620 | GU237574 | |
| CBS 391.93; PD 80/87 | Spinacia oleracea | Chenopodiaceae | Netherlands | GU238015 | GU237858 | KT389621 | GU237566 | ||
| D. pteridis | CBS 379.96 | T | Pteris sp. | Pteridaceae | Netherlands | KT389722 | KT389504 | KT389624 | KT389801 |
| D. rhei | BRIP 5562; LC 5251 | Rheum rhaponticum | Polygonaceae | Australia | KY742237 | KY742083 | KY742163 | KY742325 | |
| CBS 109177; LEV 15165; PD 2000/9941 | R | Rheum rhaponticum | Polygonaceae | New Zealand | GU238139 | GU237743 | KP330428 | GU237653 | |
| D. rumicicola | CBS 683.79; LEV 15094 | T | Rumex obtusifolius | Polygonaceae | New Zealand | KT389721 | KT389503 | KT389622 | KT389800 |
| D. sancta | CBS 281.83 | T | Ailanthus altissima | Simaroubaceae | South Africa | GU238030 | FJ427063 | KT389623 | FJ427170 |
| D. segeticola | CGMCC 3.17489; LC 1636 | T | Cirsium segetum | Asteraceae | China | KP330455 | KP330443 | KP330414 | KP330399 |
| CGMCC 3.17498; LC 1635 | Cirsium segetum | Asteraceae | China | KP330454 | KP330442 | KP330413 | KP330398 | ||
| LC 1633 | Cirsium segetum | Asteraceae | China | KP330452 | KP330440 | KP330411 | KP330396 | ||
| LC 1634 | Cirsium segetum | Asteraceae | China | KP330453 | KP330441 | KP330412 | KP330397 | ||
| LC 8141 | Camellia sasanqua | Theaceae | Japan | KY742238 | KY742084 | KY742164 | KY742326 | ||
| D. senecionicola | CBS 160.78; LEV 11451 | R | Senecio jacobaea | Asteraceae | New Zealand | GU238143 | GU237787 | — | GU237657 |
| D. sinensis | CGMCC 3.18348; LC 5210 | T | Cerasus pseudocerasus | Rosaceae | China | KY742239 | KY742085 | — | KY742327 |
| LC 5246 | Urticaceae | Urticaceae | China | KY742240 | KY742086 | KY742165 | KY742328 | ||
| LC 8142 | Dendrobium officinale | Orchidaceae | China | KY742241 | KY742087 | KY742166 | KY742329 | ||
| LC 8143 | Dendrobium officinale | Orchidaceae | China | KY742242 | KY742088 | KY742167 | KY742330 | ||
| D. subglomerata | CBS 110.92; PD 76/1010 | R | Triticum sp. | Poaceae | USA | GU238032 | FJ427080 | KT389626 | FJ427186 |
| D. subherbarum | CBS 249.92; PD 78/1088 | Solanum sp. | Solanaceae | Peru | GU238144 | GU237808 | — | GU237658 | |
| CBS 250.92; DAOM 171914; PD 92/371 | T | Zea mays | Poaceae | Canada | GU238145 | GU237809 | — | GU237659 | |
| D. suiyangensis | CGMCC 3.18352; LC 7439 | T | Air | China | KY742243 | KY742089 | KY742168 | KY742330 | |
| LC 8144 | Air | China | KY742244 | KY742090 | KY742169 | KY742332 | |||
| D. viburnicola | CBS 523.73; PD 69/800 | R | Viburnum cassioides | Caprifoliaceae | Netherlands | GU238155 | GU237879 | KP330430 | GU237667 |
| Didymellocamarosporium tamaricis | MFLUCC 14-0241 | T | Tamarix sp. | Tamaricaceae | Italy | KU848183 | — | — | — |
| Endocoryneum festucae | MFLUCC 14-0461 | T | Festuca sp. | Poaceae | Italy | KU848203 | — | — | — |
| Epicoccum brasiliense | CBS 120105 | T | Amaranthus sp. | Amaranthaceae | Brazil | GU238049 | GU237760 | KT389627 | GU237588 |
| E. camelliae | CGMCC 3.18343; LC 4858 | T | Camellia sinensis | Theaceae | China | KY742245 | KY742091 | KY742170 | KY742333 |
| LC 4862 | Camellia sinensis | Theaceae | China | KY742246 | KY742092 | KY742171 | KY742334 | ||
| E. dendrobii | CGMCC 3.18359; LC 8145 | T | Dendrobium fimbriatum | Orchidaceae | China | KY742247 | KY742093 | — | KY742335 |
| LC 8146 | Dendrobium fimbriatum | Orchidaceae | China | KY742248 | KY74209 | — | KY742336 | ||
| E. draconis | CBS 186.83; PD 82/47 | R | Dracaena sp. | Agavaceae | Rwanda | GU238070 | GU237795 | KT389628 | GU237607 |
| E. duchesneae | CGMCC 3.18345; LC 5139 | T | Duchesnea indica | Rosaceae | China | KY742249 | KY742095 | — | KY742337 |
| LC 8147 | Duchesnea indica | Rosaceae | China | KY742250 | KY742096 | — | KY742338 | ||
| E. henningsii | CBS 104.80; PD 74/1017 | R | Acacia mearnsii | Fabeceae | Kenya | GU238081 | GU237731 | KT389629 | GU237612 |
| E. hordei | CGMCC 3.18360; LC 8148 | T | Hordeum vulgare | Poaceae | Australia | KY742251 | KY742097 | — | KY742339 |
| LC 8149 | Hordeum vulgare | Poaceae | Australia | KY742252 | KY742098 | — | KY742340 | ||
| E. huancayense | CBS 105.80; PD 75/908 | T | Solanum sp. | Solanaceae | Peru | GU238084 | GU237732 | KT389630 | GU237615 |
| E. italicum | CGMCC 3.18361; LC 8150 | T | Acca sellowiana | Myrtaceae | Italy | KY742253 | KY742099 | KY742172 | KY742341 |
| LC 8151 | Acca sellowiana | Myrtaceae | Italy | KY74225 | KY742100 | KY742173 | KY742342 | ||
| E. latusicollum | CGMCC 3.18346; LC 5158 | T | Sorghum bicolor | Poaceae | China | KY742255 | KY742101 | KY742174 | KY742343 |
| LC 4859 | Camellia sinensis | Theaceae | China | KY742256 | KY742102 | KY742175 | KY742344 | ||
| LC 5124 | Vitex negundo | Verbenaceae | China | KY742257 | KY742103 | — | KY742345 | ||
| LC 8152 | Podocarpus macrophyllus | Podocarpaceae | Japan | KY742258 | KY742104 | KY742176 | KY742346 | ||
| LC 8153 | Podocarpus macrophyllus | Podocarpaceae | Japan | KY742259 | KY742105 | KY742177 | KY742347 | ||
| LC 8154 | Acer palmatum | Aceraceae | Japan | KY742260 | KY742106 | — | KY742348 | ||
| E. layuense | CGMCC 3.18362; LC 8155 | T | Perilla sp. | Lamiaceae | China | KY742261 | KY742107 | — | KY742349 |
| LC 8156 | Perilla sp. | Lamiaceae | China | KY742262 | KY742108 | — | KY742350 | ||
| E. nigrum | CBS 125.82; IMI 331914; CECT 20044 | Human toenail | Netherlands | GU237974 | FJ426995 | KT389631 | FJ427106 | ||
| CBS 173.73; ATCC 24428; IMI 164070 | T | Dactylis glomerata | Poaceae | USA | GU237975 | FJ426996 | KT389632 | FJ427107 | |
| LC 5180 | Lonicera japonica | Caprifoliaceae | China | KY742263 | KY742109 | KY742178 | KY742351 | ||
| LC 8157 | Ocimum sp. | Lamiaceae | China | KY742264 | KY742110 | KY742179 | KY742352 | ||
| LC 8158 | Poa annua | Poaceae | USA | KY742265 | KY742111 | KY742180 | KY742353 | ||
| LC 8159 | Poa annua | Poaceae | USA | KY742266 | KY742112 | KY742181 | KY742354 | ||
| E. pimprinum | CBS 246.60; ATCC 22237; ATCC 16652; IMI 81601 | T | Soil | India | GU237976 | FJ427049 | — | FJ427159 | |
| PD 77/1028 | Soil | India | GU237977 | FJ427050 | KT389633 | FJ427160 | |||
| E. plurivorum | CBS 558.81; PDDCC 6873 | T | Setaria sp. | Poaceae | New Zealand | GU238132 | GU237888 | KT389634 | GU237647 |
| E. poae | CGMCC 3.18363; LC 8160 | T | Poa annua | Poaceae | USA | KY742267 | KY742113 | KY742182 | KY742355 |
| LC 8161 | Poa annua | Poaceae | USA | KY742268 | KY742114 | KY742183 | KY742356 | ||
| LC 8162 | Poa annua | Poaceae | USA | KY742269 | KY742115 | KY742184 | KY742357 | ||
| E. sorghinum | CBS 179.80; PD 76/1018 | Sorghum vulgare | Poaceae | Puerto Rico | GU237978 | FJ427067 | KT389635 | FJ427173 | |
| CBS 627.68; PD 66/926 | Citrus sp. | Rutaceae | France | GU237979 | FJ427072 | KT389636 | FJ427178 | ||
| LC 4860 | Camellia sinensis | Theaceae | China | KY742270 | KY742116 | KY742185 | KY742358 | ||
| E. viticis | BRIP 29294; LC 5257 | Andropogon gayanus | Poaceae | Australia | KY742271 | KY742117 | — | KY742359 | |
| CGMCC 3.18344; LC 5126 | T | Vitex negundo | Verbenaceae | China | KY742272 | KY742118 | KY742186 | KY742360 | |
| Heterophoma adonidis | CBS 114309; UPSC 2982 | Adonis vernalis | Ranunculaceae | Sweden | KT389724 | KT389506 | KT389637 | KT389803 | |
| H. dictamnicola | CBS 507.91; PD 74/148 | Dictamnus albus | Rutaceae | Netherlands | GU238065 | GU237877 | KT389638 | GU237603 | |
| H. novae-verbascicola | CBS 127.93; PD 92/347 | Verbascum densiflorum | Scrophulariaceae | Netherlands | GU238120 | GU237774 | — | GU237639 | |
| H. poolensis | CBS 113.20; PD 92/774 | — | — | — | GU238119 | GU237751 | — | GU237638 | |
| CBS 116.93; PD 71/884 | Antirrhinum majus | Scrophulariaceae | Netherlands | GU238134 | GU237755 | — | GU237649 | ||
| H. sylvatica | CBS 874.97; PD 93/764 | Melampyrum pratense | Scrophulariaceae | Netherlands | GU238148 | GU237907 | — | GU237662 | |
| H. verbascicola | CGMCC 3.18364; LC 8163 | T | Verbascum thapsus | Scrophulariaceae | China | KY742273 | KY742119 | KY742187 | KY742361 |
| LC 8164 | Verbascum thapsus | Scrophulariaceae | China | KY742274 | KY742120 | KY742188 | KY742362 | ||
| Leptosphaeria conoidea | CBS 616.75; ATCC 32813; IMI 199777; PD 74/56 | Lunaria annua | Cruciferae | Netherlands | JF740279 | JF740201 | KT389639 | KT389804 | |
| Leptosphaeria doliolum | CBS 505.75 | T | Urtica dioica | Urticaceae | Netherlands | GQ387576 | JF740205 | KT389640 | JF740144 |
| Leptosphaerulina americana | CBS 213.55 | Trifolium pratense | Fabeceae | USA | GU237981 | GU237799 | KT389641 | GU237539 | |
| L. arachidicola | CBS 275.59; ATCC 13446 | Arachis hypogaea | Fabeceae | Taiwan, China | GU237983 | GU237820 | — | GU237543 | |
| L. australis | CBS 317.83 | Eugenia aromatica | Myrtaceae | Indonesia | EU754166 | GU237829 | GU371790 | GU237540 | |
| L. trifolii | CBS 235.58 | Trifolium sp. | Fabeceae | Netherlands | GU237982 | GU237806 | — | GU237542 | |
| Macroventuria anomochaeta | CBS 502.72 | Medicago sativa | Fabeceae | South Africa | GU237985 | GU237873 | — | GU237545 | |
| CBS 525.71 | T | Decayed canvas | South Africa | GU237984 | GU237881 | GU456346 | GU237544 | ||
| M. wentii | CBS 526.71 | T | Plant litter | USA | GU237986 | GU237884 | KT389642 | GU237546 | |
| Neoascochyta argentina | CBS 112524 | T | Triticum aestivum | Poaceae | Argentina | KT389742 | KT389524 | — | KT389822 |
| Neoa. desmazieri | CBS 247.79 | Poaceae | Poaceae | Austria | KT389725 | KT389507 | — | KT389805 | |
| CBS 297.69 | T | Lolium perenne | Poaceae | Germany | KT389726 | KT389508 | KT389644 | KT389806 | |
| CBS 758.97 | Hay | Norway | KT389727 | KT389509 | — | KT389807 | |||
| Neoa. europaea | CBS 819.84 | Hordeum vulgare | Poaceae | Germany | KT389728 | KT389510 | KT389645 | KT389808 | |
| CBS 820.84 | T | Hordeum vulgare | Poaceae | Germany | KT389729 | KT389511 | KT389646 | KT389809 | |
| Neoa. exitialis | CBS 118.40 | — | — | — | KT389732 | KT389514 | KT389647 | KT389812 | |
| CBS 389.86 | Triticum aestivum | Poaceae | Switzerland | KT389733 | KT389515 | KT389648 | KT389813 | ||
| CBS 811.84 | Secale cereale | Poaceae | Germany | KT389734 | KT389516 | — | KT389814 | ||
| CBS 812.84 | Hordeum vulgare | Poaceae | Germany | KT389735 | KT389517 | — | KT389815 | ||
| CBS 110124 | Triticum sp. | Poaceae | Netherlands | KT389730 | KT389512 | — | KT389810 | ||
| CBS 113693; UPSC 1929 | Allium sp. | Liliaceae | Sweden | KT389731 | KT389513 | — | KT389811 | ||
| Neoa. graminicola | CBS 301.69 | Lolium multiflorum | Poaceae | Germany | KT389737 | KT389519 | KT389650 | KT389817 | |
| CBS 447.82 | Triticum aestivum | Poaceae | Germany | KT389738 | KT389520 | — | KT389818 | ||
| CBS 586.79 | Hordeum vulgare | Poaceae | Belgium | KT389739 | KT389521 | — | KT389819 | ||
| CBS 815.84 | Hordeum vulgare | Poaceae | Germany | KT389740 | KT389522 | — | KT389820 | ||
| CBS 816.84 | Hordeum vulgare | Poaceae | Germany | KT389741 | KT389523 | KT389651 | KT389821 | ||
| CBS 102789 | R | Lolium perenne | Poaceae | New Zealand | KT389736 | KT389518 | KT389649 | KT389816 | |
| Neoa. paspali | CBS 560.81; PD 92/1569 | T | Paspalum dilatatum | Poaceae | New Zealand | GU238124 | FJ427048 | KP330426 | FJ427158 |
| Neoa. soli | CGMCC 3.18365; LC 8165 | T | Soil | China | KY742275 | KY742121 | — | KY742363 | |
| LC 8166 | Soil | China | KY742276 | KY742122 | — | KY742364 | |||
| Neoa. triticicola | CBS 544.74 | T | Triticum aestivum | Poaceae | South Africa | EU754134 | GU237887 | KT389652 | GU237488 |
| Neodidymelliopsis achlydis | CBS 256.77 | T | Achlys triphylla | Berberidaceae | Canada | KT389749 | KT389531 | — | KT389829 |
| Neod. cannabis | CBS 121.75; ATCC 32164; IMI 194767; PD 73/584 | T | Urtica dioica | Urticaceae | Netherlands | GU237972 | GU237761 | — | GU237535 |
| CBS 234.37 | Cannabis sativa | Moraceae | — | GU237961 | GU237804 | KP330403 | GU237523 | ||
| CBS 591.67 | Urtica dioica | Urticaceae | Netherlands | KT389746 | KT389528 | — | KT389826 | ||
| CBS 629.76 | Packing material | Netherlands | KT389747 | KT389529 | — | KT389827 | |||
| Neod. longicolla | CBS 382.96 | T | Soil in desert | Israel | KT389750 | KT389532 | — | KT389830 | |
| Neod. polemonii | CBS 375.67 | Polemonium caeruleum | Polemoniaceae | Netherlands | KT389748 | KT389530 | — | KT389828 | |
| CBS 109181; PD 83/757 | T | Polemonium caeruleum | Polemoniaceae | Netherlands | GU238133 | GU237746 | KP330427 | GU237648 | |
| Neod. xanthina | CBS 168.70 | Delphinium sp. | Ranunculaceae | Netherlands | KT389751 | KT389533 | — | KT389831 | |
| CBS 383.68 | T | Delphinium sp. | Ranunculaceae | Netherlands | GU238157 | GU237855 | KP330431 | GU237668 | |
| Neomicrosphaeropsis italica | MFLUCC 15-0485; ICMP 21253 | T | Tamarix sp. | Tamaricaceae | Italy | KU729854 | KU900318 | KU674820 | — |
| MFLUCC 15-0484 | Tamarix sp. | Tamaricaceae | Italy | KU729853 | KU900319 | KU695539 | KX453298 | ||
| MFLUCC 16-0284 | Tamarix sp. | Tamaricaceae | Italy | KU900296 | KU900321 | KU714604 | KX453299 | ||
| Neom. novorossica | MFLUCC 14-0578; ICMP 20751 | T | Tamarix ramosissima | Tamaricaceae | Russia | KX198710 | KX198709 | — | — |
| Neom. rossica | MFLUCC 14-0586; ICMP 20753 | T | Tamarix ramosissima | Tamaricaceae | Russia | KU729855 | KU752192 | — | — |
| Neom. tamaricicola | MFLUCC 14-0443; ICMP 20708 | Tamarix gallica | Tamaricaceae | Italy | KU729851 | KU900322 | — | — | |
| MFLUCC 14-0439; ICMP 20743 | Tamarix gallica | Tamaricaceae | Italy | KU729858 | KU900323 | — | — | ||
| Nothophoma anigozanthi | CBS 381.91; PD 79/1110 | T | Anigozanthus maugleisii | Haemodoraceae | Netherlands | GU238039 | GU237852 | KT389655 | GU237580 |
| No. arachidis-hypogaeae | CBS 125.93; PD 77/1029 | R | Arachis hypogaea | Fabeceae | India | GU238043 | GU237771 | KT389656 | GU237583 |
| No. gossypiicola | CBS 377.67 | Gossypium sp. | Malvaceae | USA | GU238079 | GU237845 | KT389658 | GU237611 | |
| No. infossa | CBS 123395 | T | Fraxinus pennsylvanica | Oleaceae | Argentina | GU238089 | FJ427025 | KT389659 | FJ427135 |
| No. quercina | CBS 633.92; ATCC 36786; VKM MF-325 | Microsphaera alphitoides from Quercus sp. | Ukraine | EU754127 | GU237900 | KT389657 | GU237609 | ||
| Paraboeremia adianticola | CBS 187.83; PD 82/128 | Polystichum adiantiforme | Dryopteridaceae | USA | GU238035 | GU237796 | KP330401 | GU237576 | |
| CBS 260.92; PD 86/1103 | Pteris ensiformis | Pteridaceae | — | KT389752 | KT389534 | — | KT389832 | ||
| Pa. camellae | CGMCC 3.18106; LC 4852 | T | Camellia sp. | Theaceae | China | KX829042 | KX829034 | KX829050 | KX829058 |
| CGMCC 3.18107; LC 6253 | Camellia sp. | Theaceae | China | KX829043 | KX829035 | KX829051 | KX829059 | ||
| CGMCC 3.18108; LC 6254 | Camellia sp. | Theaceae | China | KX829044 | KX829036 | KX829052 | KX829060 | ||
| Pa. litseae | CGMCC 3.18109; LC 5028 | T | Litsea sp. | Lauraceae | China | KX829037 | KX829029 | KX829045 | KX829053 |
| CGMCC 3.18110; LC 5030 | Litsea sp. | Lauraceae | China | KX829038 | KX829030 | KX829046 | KX829054 | ||
| Pa. oligotrophica | CGMCC 3.18111; LC 6250 | T | Carbonatite | China | KX829039 | KX829031 | KX829047 | KX829055 | |
| CGMCC 3.18112; LC 6251 | Carbonatite | China | KX829040 | KX829032 | KX829048 | KX829056 | |||
| CGMCC 3.18113; LC 6252 | Carbonatite | China | KX829041 | KX829033 | KX829049 | KX829057 | |||
| Pa. putaminum | CBS 130.69; CECT 20054; IMI 331916 | R | Malus sylvestris | Rosaceae | Denmark | GU238138 | GU237777 | — | GU237652 |
| CBS 372.91; PD 75/960 | R | Ulmus sp. | Ulmaceae | Netherlands | GU238137 | GU237843 | — | GU237651 | |
| Pa. selaginellae | CBS 122.93; PD 77/1049 | T | Selaginella sp. | Selaginellaceae | Netherlands | GU238142 | GU237762 | — | GU237656 |
| Phoma herbarum | CBS 134.96; PD 84/676 | Delphinium sp. | Ranunculaceae | Netherlands | KT389753 | KT389535 | KT389661 | KT389834 | |
| CBS 274.37 | Picea excelsa | Pinaceae | UK | KT389754 | KT389537 | KT389662 | KT389835 | ||
| CBS 304.51 | Achillea millefolium | Asteraceae | Switzerland | KT389755 | KT389538 | — | KT389836 | ||
| CBS 377.92; IMI 213845 | Human leg | Netherlands | KT389756 | KT389536 | KT389663 | KT389837 | |||
| CBS 502.91; PD 82/276 | Nerium sp. | Apocynaceae | Netherlands | GU238082 | GU237874 | KP330419 | GU237613 | ||
| CBS 615.75; PD 73/665; IMI 199779 | R | Rosa multiflora cv. Cathayensis | Rosaceae | Netherlands | EU754186 | FJ427022 | KP330420 | FJ427133 | |
| CBS 127589; UAMH 10909 | Polytrichum juniperinum | Polytrichaceae | USA | KT389757 | KT389539 | KT389664 | KT389838 | ||
| Phomatodes aubrietiae | CBS 383.67; PD 65/223 | R | Aubrietia hybrida cv. Superbissima | Cruciferae | Netherlands | GU238044 | GU237854 | — | GU237584 |
| Phomat. aubrietiae | CBS 627.97; PD 70/714 | T | Aubrietia sp. | Cruciferae | Netherlands | GU238045 | GU237895 | KT389665 | GU237585 |
| Phomat. nebulosa | CBS 117.93; PD 83/90 | Mercurialis perennis | Euphorbiaceae | Netherlands | GU238114 | GU237757 | KP330425 | GU237633 | |
| CBS 740.96 | Armoracia rusticana | Cruciferae | Netherlands | KT389758 | KT389540 | KT389667 | KT389839 | ||
| CBS 100191 | Thlaspi arvense | Cruciferae | Poland | KP330446 | KP330434 | KT389666 | KP330390 | ||
| Pseudohendersonia galiorum | MFLUCC 14–0452 | T | Galium sp. | Rubiaceae | Italy | KU848207 | — | — | — |
| Stagonosporopsis actaeae | CBS 106.96; PD 94/1318 | T | Actaea spicata | Ranunculaceae | Netherlands | GU238166 | GU237734 | KT389672 | GU237671 |
| CBS 114303; UPSC 2962 | Actaea spicata | Ranunculaceae | Sweden | KT389760 | KT389544 | — | KT389847 | ||
| S. ajacis | CBS 177.93; PD 90/115 | T | Delphinium sp. | Ranunculaceae | Kenya | GU238168 | GU237791 | KT389673 | GU237673 |
| S. andigena | CBS 101.80; PD 75/909; IMI 386090 | R | Solanum sp. | Solanaceae | Peru | GU238169 | GU237714 | — | GU237674 |
| CBS 269.80; PD 75/914 | Solanum sp. | Solanaceae | Peru | GU238170 | GU237817 | — | GU237675 | ||
| S. artemisiicola | CBS 102636; PD 73/1409 | R | Artemisia dracunculus | Asteraceae | France | GU238171 | GU237728 | KT389674 | GU237676 |
| S. astragali | CBS 178.25; MUCL 9915 | R | Astragalus sp. | Fabeceae | — | GU238172 | GU237792 | — | GU237677 |
| S. bomiensis | CGMCC 3.18366; LC 8167 | T | Boraginaceae | Boraginaceae | China | KY742277 | KY742123 | KY742189 | KY742365 |
| LC 8168 | Boraginaceae | Boraginaceae | China | KY742278 | KY742124 | KY742190 | KY742366 | ||
| S. caricae | CBS 248.90 | Carica papaya | Caricaceae | Chile | GU238175 | GU237807 | — | GU237680 | |
| CBS 282.76 | Brassica sp. | Cruciferae | Indonesia | GU238177 | GU237821 | — | GU237682 | ||
| S. chrysanthemi | CBS 500.63; MUCL 8090 | R | Chrysanthemum indicum | Asteraceae | Germany | GU238190 | GU237871 | — | GU237695 |
| CBS 137.96; PD 84/75 | R | Chrysanthemum indicum | Asteraceae | Netherlands | GU238191 | GU237783 | — | GU237696 | |
| S. crystalliniformis | CBS 713.85; ATCC 76027; PD 83/826 | T | Lycopersicon esculentum | Solanaceae | Colombia | GU238178 | GU237903 | KT389675 | GU237683 |
| S. cucurbitacearum | CBS 133.96; PD 79/127 | Cucumis sp. | Cucurbitaceae | New Zealand | GU238181 | GU237780 | KT389676 | GU237686 | |
| S. dennisii | CBS 631.68; PD 68/147 | T | Solidago floribunda | Asteraceae | Netherlands | GU238182 | GU237899 | KT389677 | GU237687 |
| S. dorenboschii | CBS 426.90; IMI 386093; PD 86/551 | T | Physostegia virginiana | Lamiaceaee | Netherlands | GU238185 | GU237862 | KT389678 | GU237690 |
| S. helianthi | CBS 200.87 | T | Helianthus annuus | Asteraceae | Italy | KT389761 | KT389545 | KT389683 | KT389848 |
| S. heliopsidis | CBS 109182; PD 74/231 | R | Heliopsis patula | Asteraceae | Netherlands | GU238186 | GU237747 | KT389679 | GU237691 |
| S. hortensis | CBS 104.42 | R | — | Netherlands | GU238198 | GU237730 | KT389680 | GU237703 | |
| CBS 572.85; PD 79/269 | R | Phaseolus vulgaris | Fabeceae | Netherlands | GU238199 | GU237893 | KT389681 | GU237704 | |
| S. inoxydabilis | CBS 425.90; PD 81/520 | T | Chrysanthemum parthenii | Asteraceae | Netherlands | GU238188 | GU237861 | KT389682 | GU237693 |
| S. loticola | CBS 562.81; PDDCC 6884 | T | Lotus pedunculatus | Fabeceae | New Zealand | GU238192 | GU237890 | KT389684 | GU237697 |
| S. lupini | CBS 101494; PD 98/5247 | T | Lupinus albus | Fabeceae | UK | GU238194 | GU237724 | KT389685 | GU237699 |
| S. oculo-hominis | CBS 634.92; IMI 193307 | T | Human corneal ulcer | USA | GU238196 | GU237901 | KT389686 | GU237701 | |
| S. papillatus | CGMCC 3.18367; LC 8169 | T | Rumex nepalensis | Polygonaceae | China | KY742279 | KY742125 | KY742191 | KY742367 |
| LC 8170 | Rumex nepalensis | Polygonaceae | China | KY742280 | KY742126 | KY742192 | KY742368 | ||
| LC 8171 | Boraginaceae | Boraginaceae | China | KY742281 | KY742127 | KY742193 | KY742369 | ||
| S. rudbeckiae | CBS 109180; PD 79/175 | R | Rudbeckia bicolor | Asteraceae | Netherlands | GU238197 | GU237745 | — | GU237702 |
| S. tanaceti | CBS 131484 | T | Tanacetum cinerariifolium | Asteraceae | Australia | JQ897461 | NR_111724 | — | JQ897496 |
| S. trachelii | CBS 379.91; PD 77/675 | R | Campanula isophylla | Campanulaceae | Netherlands | GU238173 | GU237850 | KT389687 | GU237678 |
| CBS 384.68 | R | Campanula isophylla | Campanulaceae | Sweden | GU238174 | GU237856 | — | GU237679 | |
| S. valerianellae | CBS 273.92; PD 82/43 | Valerianella locusta | Caprifoliaceae | Netherlands | GU238200 | GU237819 | — | GU237705 | |
| CBS 329.67; PD 66/302 | T | Valerianella locusta var. oleracea | Caprifoliaceae | Netherlands | GU238201 | GU237832 | — | GU237706 | |
| Xenodidymella applanata | CBS 195.36 | T | Rubus idaeus | Rosaceae | Netherlands | KT389764 | KT389548 | — | KT389852 |
| X. applanata | CBS 205.63 | Rubus idaeus | Rosaceae | Netherlands | GU237998 | GU237798 | KP330402 | GU237556 | |
| CBS 115577 | Rubus idaeus | Rosaceae | Sweden | KT389762 | KT389546 | KT389688 | KT389850 | ||
| CBS 115578 | Rubus arcticus nothossp. stellarcticus | Rosaceae | Sweden | KT389763 | KT389547 | — | KT389851 | ||
| X. asphodeli | CBS 375.62 | T | Asphodelus albus | Asphodelaceae | France | KT389765 | KT389549 | KT389689 | — |
| CBS 499.72 | Asphodelus ramosus | Asphodelaceae | Italy | KT389766 | KT389550 | — | KT389853 | ||
| X. catariae | CBS 102635; PD 77/1131 | Nepeta cataria | Lamiaceaee | Netherlands | GU237962 | GU237727 | KP330404 | GU237524 | |
| X. humicola | CBS 220.85; PD 71/1030 | R | Franseria sp. | Asteraceae | USA | GU238086 | GU237800 | KP330422 | GU237617 |
ATCC: American Type Culture Collection, Virginia, U.S.A.; BRIP: Plant Pathology Herbarium, Department of Employment, Economic, Development and Innovation, Queensland, Australia; CBS: Westerdijk Fungal Biodiversity Institute (formerly CBS-KNAW), Utrecht, The Netherlands; CECT: Colección Española de Cultivos Tipo, Valencia University, Spain; CGMCC: China General Microbiological Culture Collection, Beijing, China; CPC: Culture collection of Pedro Crous, housed at CBS; DAOM: Canadian Collection of Fungal Cultures, Ottawa, Canada; DSM: Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany; FMR, Facultat de Medicina, Universitat Rovira i Virgili, Reus, Spain; ICMP: International Collection of Microorganisms from Plants, Auckland, New Zealand; IMI: International Mycological Institute, CABI-Bioscience, Egham, Bakeham Lane, U.K.; LC: Corresponding author's personal collection deposited in laboratory, housed at CAS, China; LEV: Plant Health and Diagnostic Station, Auckland, New Zealand; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; MUCL: Mycotheque de l'Universite catholique de Louvain, Louvain-la-Neuve, Belgium; PD: Plant Protection Service, Wageningen, the Netherlands; PDDCC: Plant Diseases Division Culture Collection, Auckland, New Zealand; PREM: National Collection of Fungi: Culture Collection, Pretoria, South Africa; UAMH: University of Albert Microfungus Colletion and Herbarium, Canada; UPSC: Uppsala University Culture Collection, Sweden; UTHSC, Fungus Testing Laboratory at the University of Texas Health Science Center, San Antonio, Texas, USA; VKM: All-Russian Collection of Microorganisms, Pushchino, Russia.
T: ex-type strain; R: representative strain.
ITS: internal transcibed spacer regions 1 & 2 including 5.8S nrDNA gene; LSU: 28S large subunit of the nrRNA gene; RPB2: RNA polymerase II second subunit; TUB: ß-tubulin.
Plant-associated isolates were obtained from symptomatic tissue with sporocarps using the single spore isolation protocols of Choi et al. (1999) and Zhang et al. (2013), and from tissue according to the techniques outlined by Cai et al. (2009). Isolates from other substrates were obtained following the methods described by Zhang et al. (2017) and further screened with carbon-free silica gel medium to select the oligotrophic strains (Wainwright & Al-Talhi 1999). All the Didymellaceae isolates were primarily identified based on morphology and ITS sequence data, which distinguished them from other groups of fungi. Type specimens of new species in this study were deposited in the Mycological Herbarium of Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS), with the ex-type living cultures deposited in China General Microbiological Culture Collection Center (CGMCC), or the other Biological Resource Centres cited above.
Morphology
Isolates were incubated on oatmeal agar (OA), malt extract agar (MEA) and potato dextrose agar (PDA) (Crous et al. 2009) at 25 °C, and under near-ultraviolet (UV) light (12 h light/12 h dark) or on pine needle agar (PNA) (Smith et al. 1996) to induce sporulation. Colony diameters were measured after 7 d of incubation, and the culture characters were determined after 14 d (Boerema et al. 2004). Colony colours were rated according to the colour charts of Rayner (1970). Preparations were mounted in distilled water to study the micromorphological structures of mature ascomata/conidiomata, ascospores/conidia and conidiogenous cells from OA cultures (Aveskamp et al., 2010, Chen et al., 2015a, Chen et al., 2015b). Observations were conducted with a Leica M125 dissecting microscope and a Nikon Eclipse 80i compound microscope under differential interference contrast (DIC) illumination. To study the pseudothecial/pycnidial wall, sections of mature pseudothecia/pycnidia were made by a Leica CM1950 freezing microtome (Aveskamp et al., 2010, Chen et al., 2015a, Chen et al., 2015b). The NaOH spot test was carried out by a drop of 1N NaOH to determine the secretion of metabolite E on MEA cultures (Boerema et al. 2004).
DNA isolation, amplification and phylogenetic analyses
Total genomic DNA was extracted from fresh mycelia using the MP Fastprep-24 sample preparation system, according to the protocol described by Cubero et al. (1999). The primers V9G (de Hoog & Gerrits van den Ende 1998) and ITS4 (White et al. 1990) were used to amplify part of the nuclear rDNA operon (ITS) spanning the 3′ end of the 18S rRNA gene, the first internal transcribed spacer (ITS1), the 5.8S rRNA gene, the second ITS region (ITS2), and the first 100 bp of the 5′ end of the 28S rRNA gene (LSU); the primers LR0R (Rehner & Samuels 1994), LR7 and LR5 (Vilgalys & Hester 1990) were used for LSU amplification; Btub2Fd and Btub4Rd (Woudenberg et al. 2009) for the partial β-tubulin (tub2) gene region, and RPB2-5F2 (Sung et al. 2007) and fRPB2-7cR (Liu et al. 1999) for the RNA polymerase II second largest subunit (rpb2). Amplicons for each locus were generated following the protocols listed in Chen et al. (2015a).
Sequencing was conducted in both directions with the same primer pair used for amplification at the Omega Genetics Company (Beijing, China). Consensus sequences were assembled in MEGA v. 6.0 (Tamura et al. 2013) and additional reference sequences were obtained from GenBank (Table 1). Subsequent alignments for each locus were generated with MAFFT v. 7 (http://mafft.cbrc.jp/alignment/server/index.html; Katoh & Standley 2013), and manually corrected when necessary. The concatenated aligned dataset and each locus were analysed separately using Maximum Likelihood (ML) and Bayesian Inference (BI). The best-fit models of evolution for the four loci tested (SYM+I+G for ITS and GTR+I+G for LSU, rpb2 and tub2) were estimated by MrModeltest v. 2.3 (Nylander 2004).
The ML analyses were conducted with RAxML v. 7.2.6 (Stamatakis & Alachiotis 2010) using a GTRGAMMA substitution model with 1 000 bootstrap replicates. The robustness of the analyses was evaluated by bootstrap support (MLBS). Bayesian (BI) analyses were performed on MrBayes v. 3.2.1 (Ronquist et al. 2012) based on the models selected by the MrModeltest. The Markov Chain Monte Carlo (MCMC) algorithm of four chains was initiated in parallel from a random tree topology. The analyses lasted until the average standard deviation of split frequencies was below 0.01 with trees saved each 1 000 generations. The first 25 % of trees were removed as burn-in phase and the remaining trees were used to calculate posterior probabilities. Posterior probabilities values of the BI analyses (BPP) over 0.95 were considered significant. Leptosphaeria conoidea (CBS 616.75) and L. doliolum (CBS 505.75) were selected as outgroup. Sequences generated in this study were deposited in GenBank (Table 1), the final matrices and trees in TreeBASE (www.treebase.org; accession number: S20724), and novel taxonomic descriptions and nomenclature in MycoBank (www.MycoBank.org; Crous et al. 2004).
Unique fixed nucleotide positions are used to describe a sterile species (see Taxonomy below), and the closest phylogenetic neighbour was selected and subjected to single nucleotide polymorphism (SNP) analyses using MEGA v. 6.0 (Tamura et al. 2013).
Statistical analysis
A heatmap showing the host distribution of each genus of Didymellaceae was generated with R v. 3.3.1 heatmap.2 (https://www.r-project.org/).
Results
Phylogeny
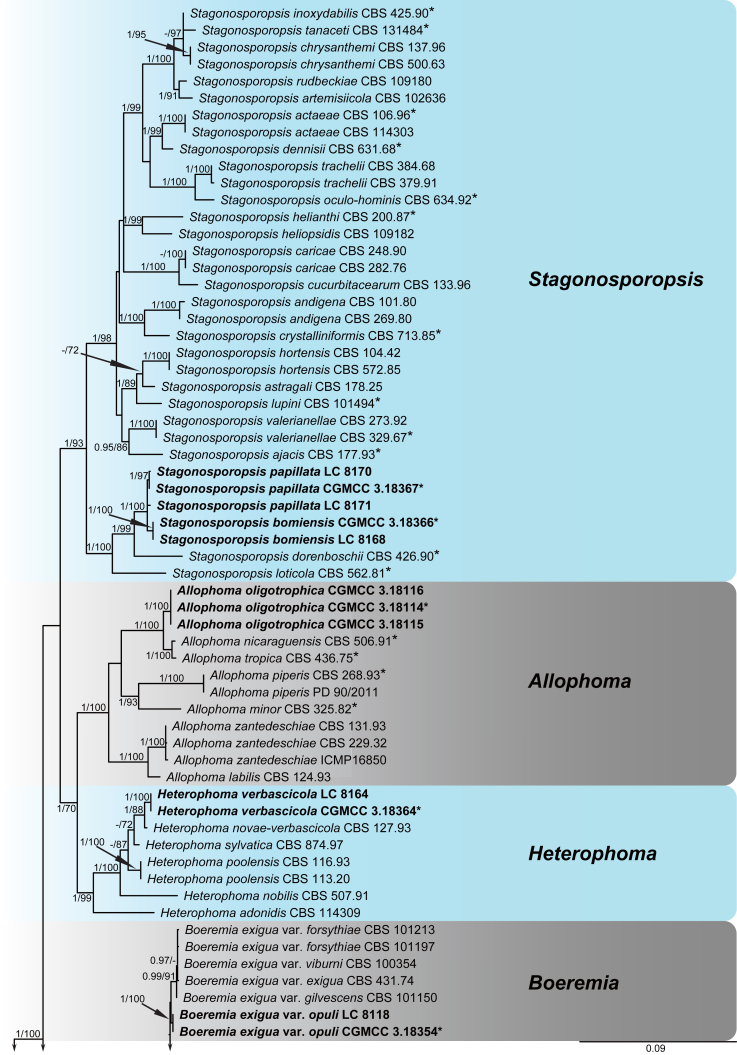
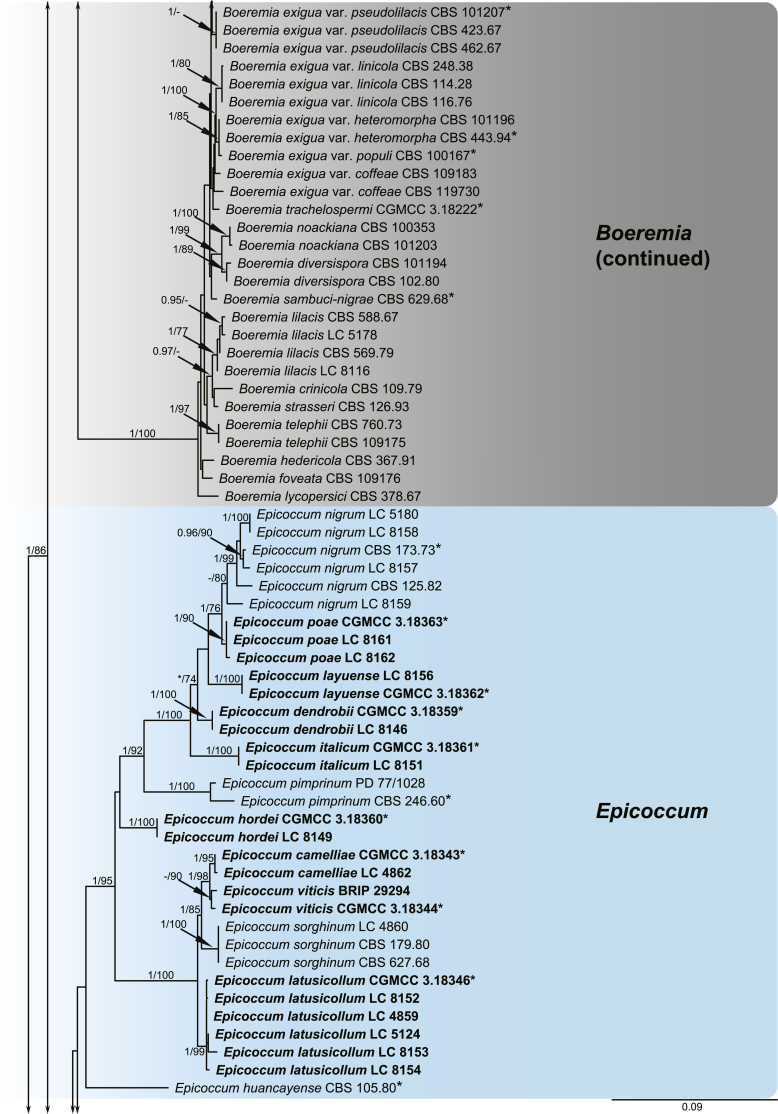
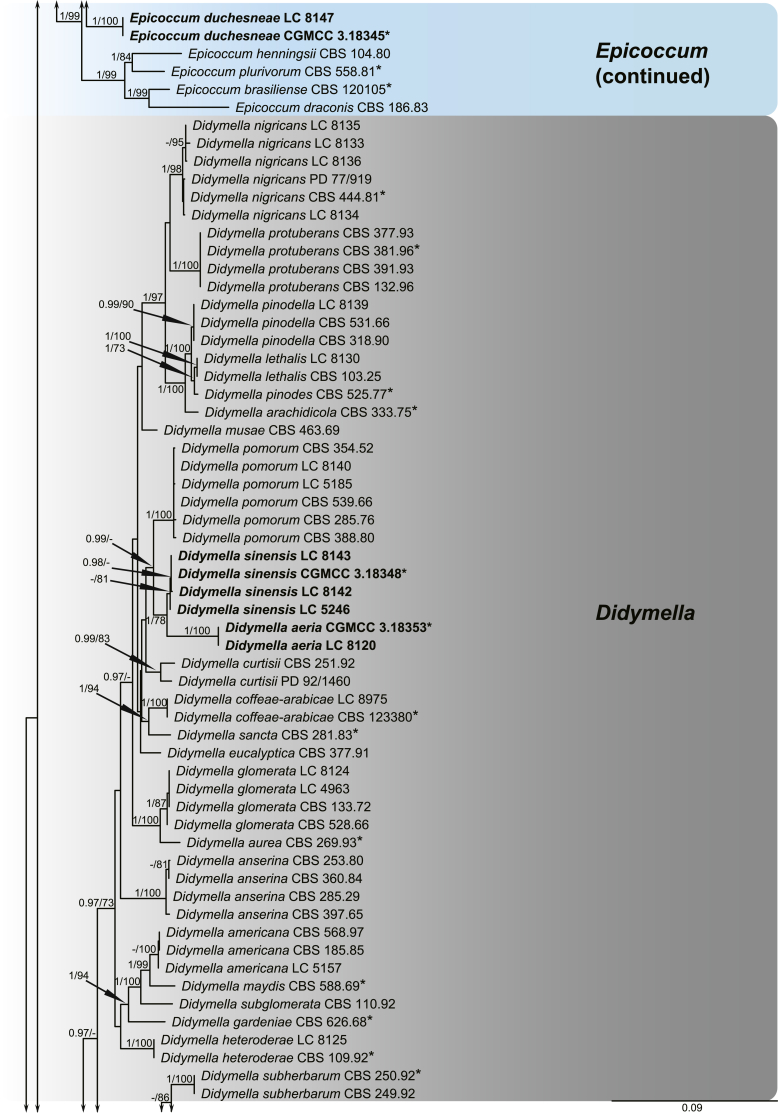
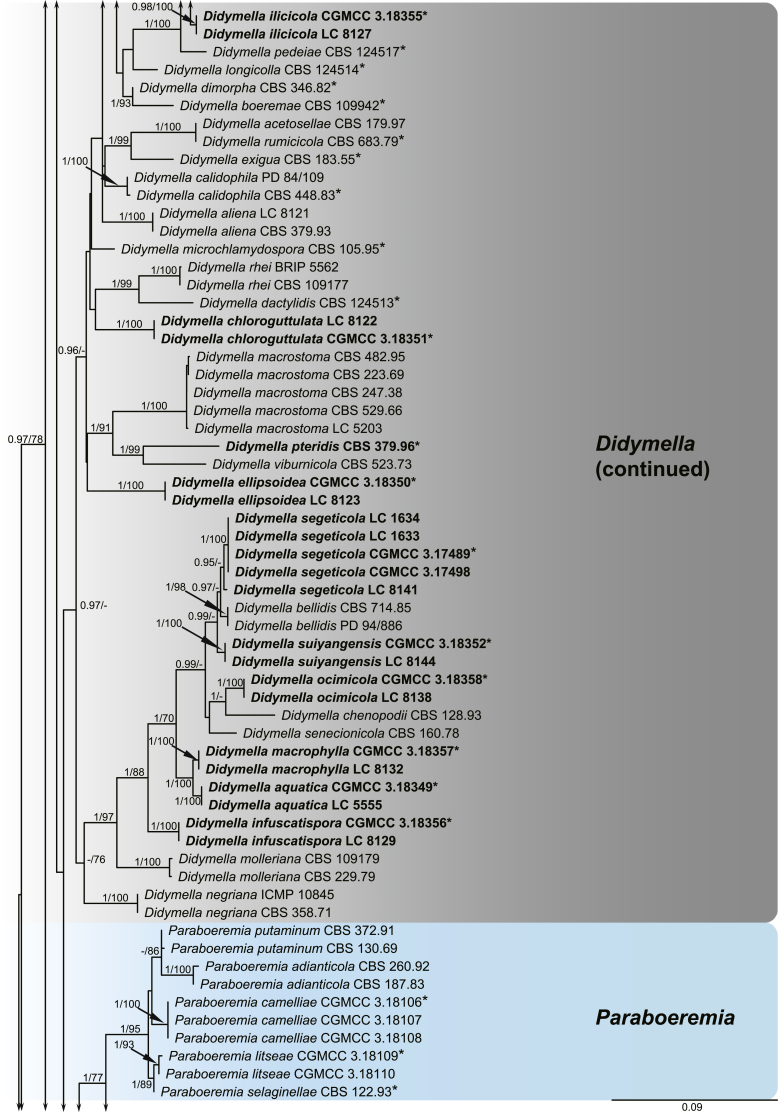
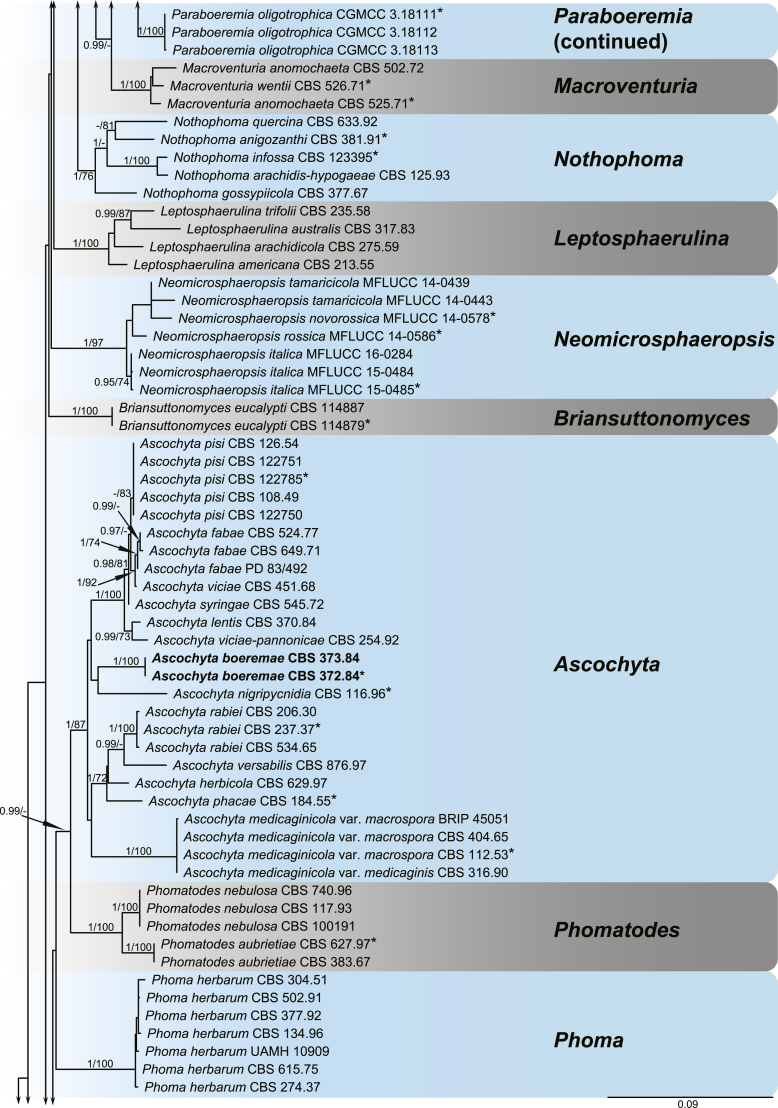
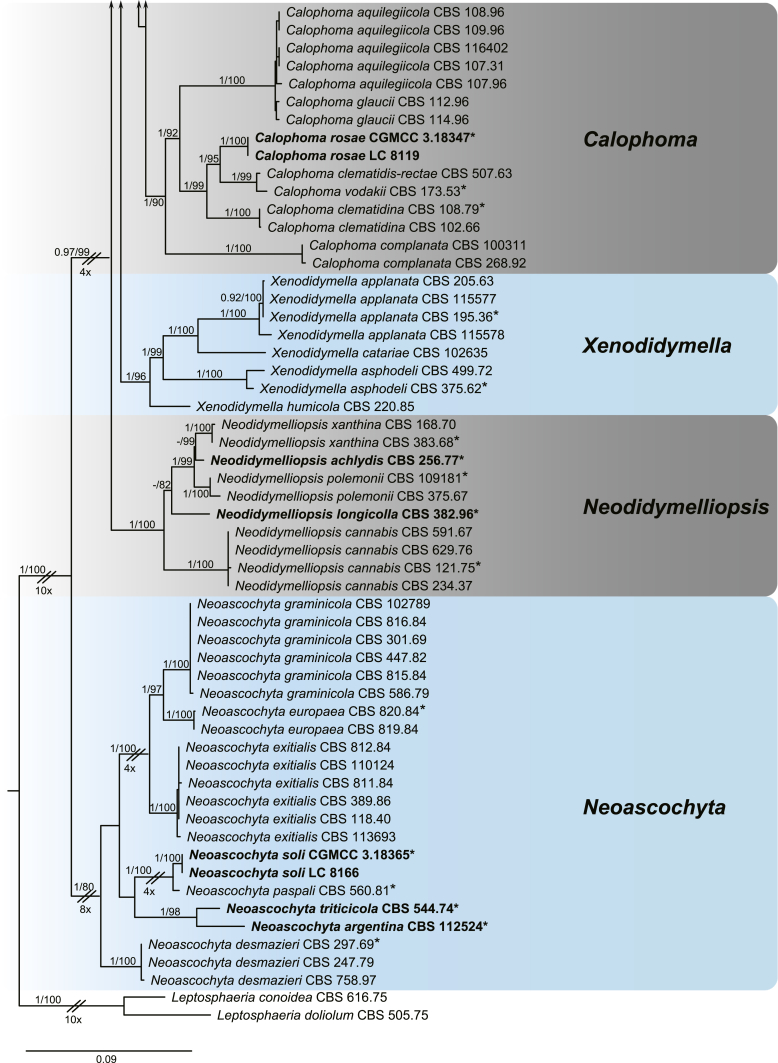
A multi-locus phylogeny, based on four loci, was used to infer the relationships among species in Didymellaceae (Fig. 1). The resulting concatenated aligned dataset comprised 360 ingroup isolates belonging to 194 taxa and consisted of 2 460 characters (964 for LSU, 531 for ITS, 599 for rpb2 and 354 for tub2, including alignment gaps), of which 265 are conserved and 901 are phylogenetically informative (173 for LSU, 230 for ITS, 310 for rpb2 and 188 for tub2). The trees generated from ML and Bayesian analyses of the individual loci (data not shown) and the combined dataset showed essentially congruent topologies. The ML tree based on the combined dataset was presented, with bootstrap support values (MLBS) and Bayesian posterior probabilities (BPP) indicated for well-supported clades in Fig. 1. The LSU sequences were the least successful in resolving species with only 59 out of 194 taxa resolved (30 %), followed by ITS with 104 out of 194 taxa (54 %), and tub2 (90 %) and rpb2 (92 %) which proved to be more suitable for the resolution of species.
Fig. 1.
Phylogenetic tree inferred from a Maximum likelihood analysis based on a concatenated alignment of LSU, ITS, rpb2 and tub2 sequences of 360 strains representing species in Didymellaceae. The RAxML bootstrap support values (MLBS) and Bayesian posterior probabilities (BPP) are given at the nodes (BPP/MLBS). Some branches were shortened to fit them to the page – these are indicated by two diagonal lines with the number of times a branch was shortened indicated next to the lines. New taxa and new combination introduced in this study are formatted in bold. Ex-type strains are marked by an asterisk (*). The tree was rooted to Leptosphaeria conoidea (CBS 616.75) and L. doliolum (CBS 505.75).
A total of 194 ingroup taxa formed a clade (BPP = 1; MLBS = 100 %) representing the Didymellaceae, which include 19 monophyletic generic clades. Seventeen genera previously recognised, namely Allophoma (BPP = 1; MLBS = 100 %), Ascochyta (BPP = 1; MLBS = 87 %), Boeremia (BPP = 1; MLBS = 100 %), Calophoma (BPP = 1; MLBS = 90 %), Didymella (BPP = 0.97; MLBS = 60 %), Epicoccum (BPP = 1; MLBS = 99 %), Heterophoma (BPP = 1; MLBS = 99 %), Leptosphaerulina (BPP = 1; MLBS = 100 %), Macroventuria (BPP = 1; MLBS = 100 %), Neoascochyta (BPP = 1; MLBS = 80 %), Neodidymelliopsis (BPP = 1; MLBS = 100 %), Nothophoma (BPP = 1; MLBS = 76 %), Paraboeremia (BPP = 1; MLBS = 77 %), Phoma (BPP = 1; MLBS = 100 %), Phomatodes (BPP = 1; MLBS = 100 %), Stagonosporopsis (BPP = 1; MLBS = 93 %) and Xenodidymella (BPP = 1; MLBS = 96 %), and two genera recently added in this family, namely Briansuttonomyces (BPP = 1; MLBS = 100 %) and Neomicrosphaeropsis (BPP = 1; MLBS = 97 %) were highly supported as independent groups.
Host specificity analysis
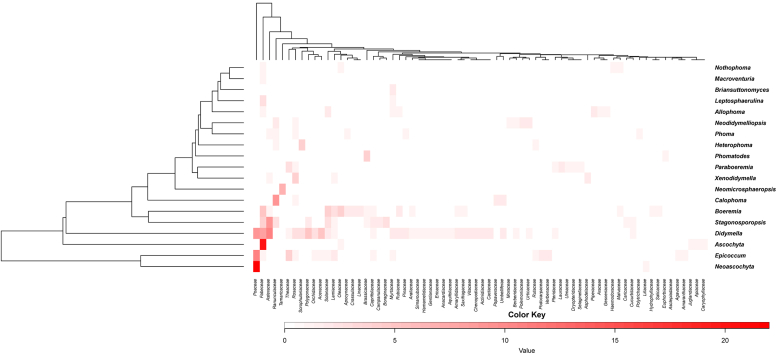
The heatmap was plotted to reveal the distribution of Didymellaceae species in various host families. The colour-coding columns indicate the number of species in each fungal genus that are associated with a particular host family. A darker colour indicates more fungal species related to the host family. In the present study, all the plant-associated species are linked to 70 different host families in total, of which Asteraceae, Fabaceae, Poaceae, Ranunculaceae, Rosaceae and Solanaceae are the most common hosts for Didymellaceae. Most of the Didymellaceae genera have a wide host range, while Ascochyta, Neoascochyta and Neomicrosphaeropsis showed relatively high host specificity within Fabaceae, Poaceae and Tamaricaceae, respectively (Fig. 2).
Fig. 2.
Heatmap of relative abundances of different host plant families in each genus of Didymellaceae. The colour-coding for columns indicate the number of species in each fungal genus that are associated with a particular host family.
Taxonomy
As a result of morphological comparisons and multi-locus sequence analysis of 360 strains, including 108 strains studied in the present paper and 252 reference strains, 194 taxa are recognised in 19 different genera of Didymellaceae. Recognised clades of novel taxa are described and illustrated, and two new combinations are proposed below. One species proved to be sterile in culture, and therefore is described based on DNA sequence data, following the approach of Gomes et al. (2013) and Lombard et al. (2016). Novel taxa are arranged in alphabetical order by genus and species.
Allophoma Q. Chen & L. Cai, Stud. Mycol. 82: 162. 2015.
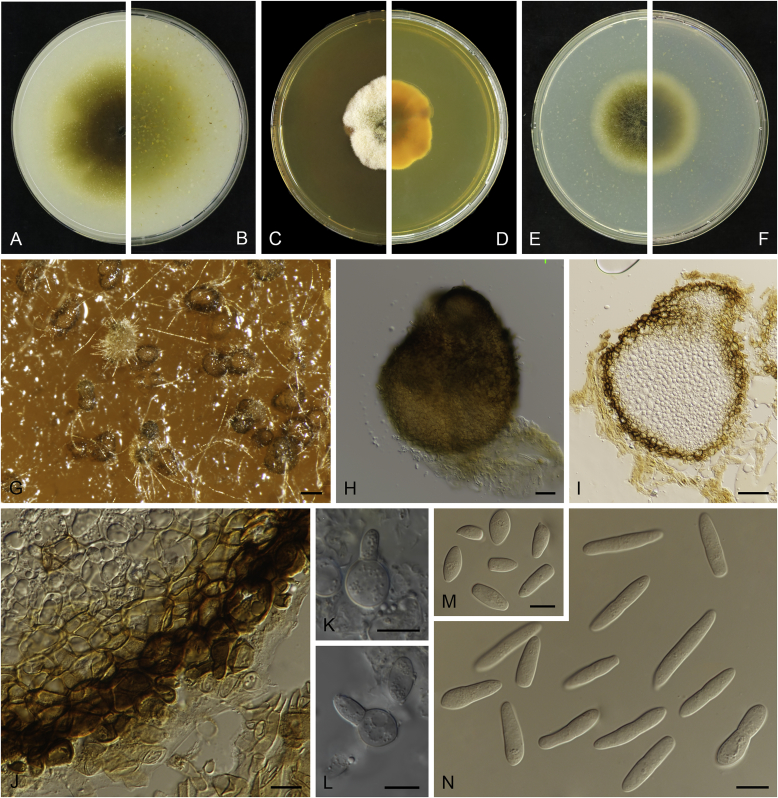
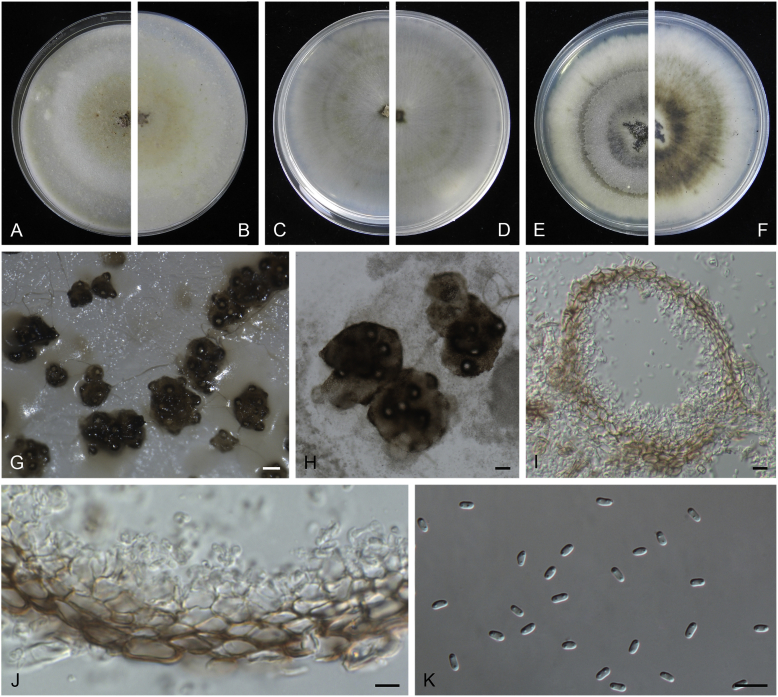
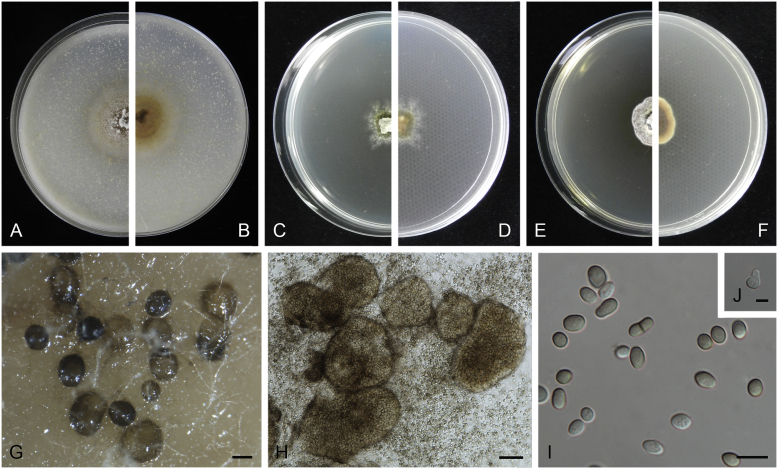
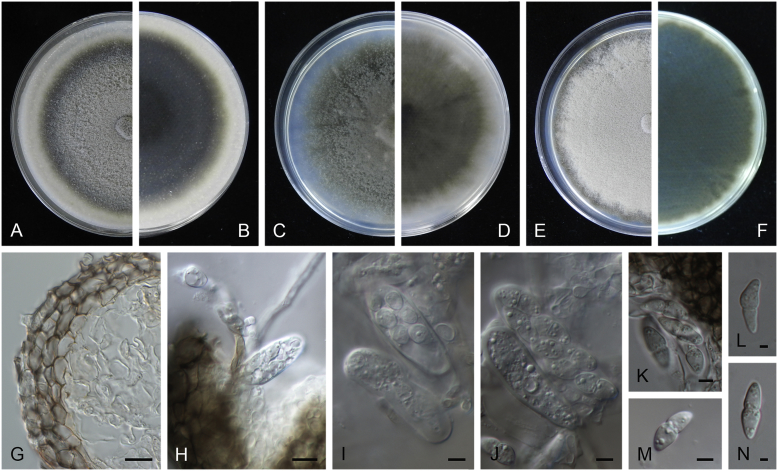
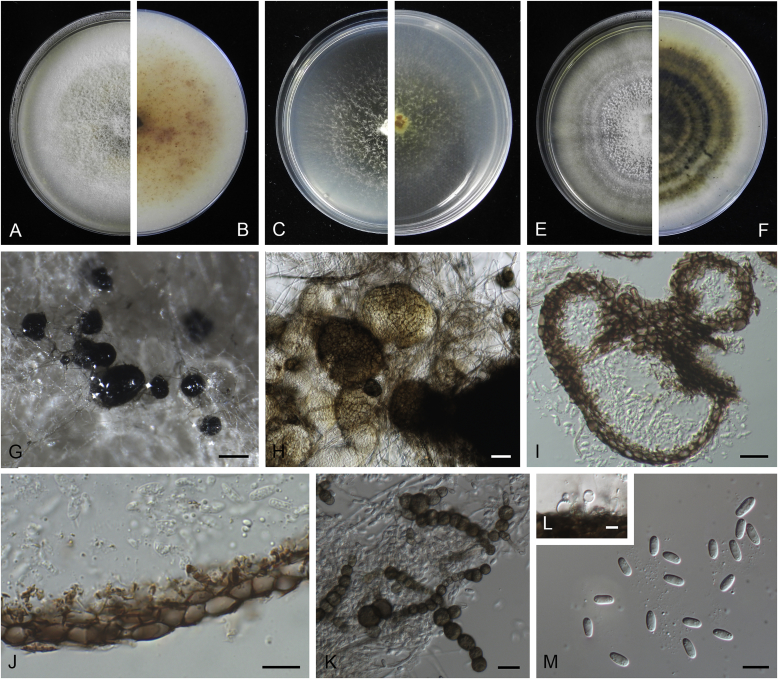
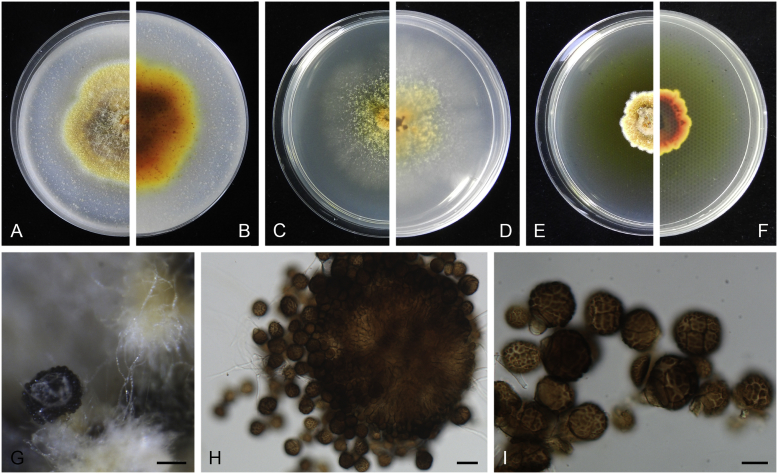
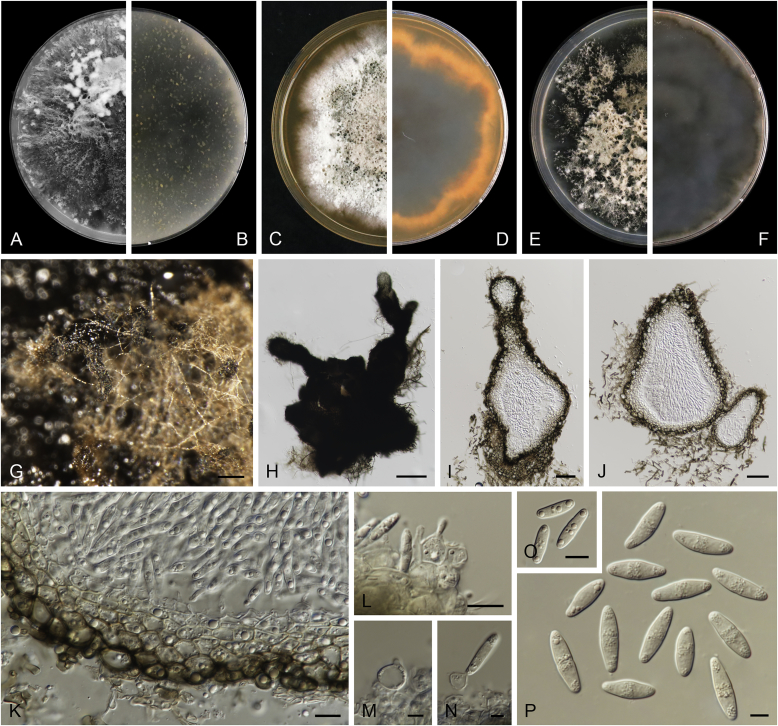
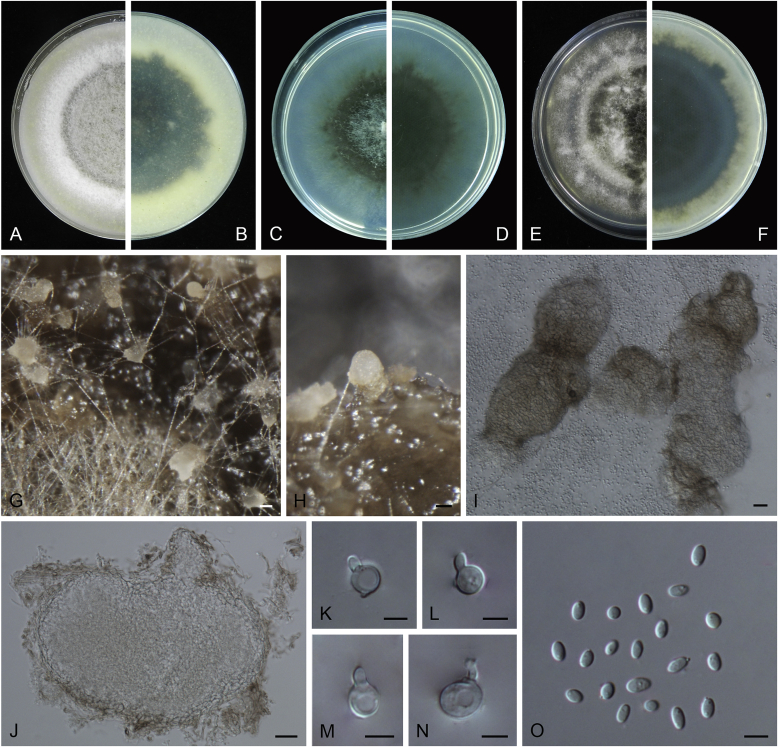
Allophoma oligotrophica Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818956. Fig. 3.
Fig. 3.
Allophoma oligotrophica (CGMCC 3.18114). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia producing on OA. H. Pycnidium. I. Section of pycnidium. J. Section of pycnidial wall. K. Conidiogenous cells. L. Conidia. Scale bars: G = 100 μm; H–I = 50 μm; J, L = 10 μm; K = 5 μm.
Etymology: Oligotrophica, referring to the oligotrophic substrate of the fungus.
Conidiomata pycnidial, solitary, globose to subglobose, brown, glabrous, semi-immersed or immersed, 150–440(–590) × 145–420 μm. Ostioles single, slightly papillate. Pycnidial wall pseudoparenchymatous, composed of oblong to isodiametric cells, 3–5 layers, 11–19.5 μm thick. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 4.5–7 × 3.5–6.5 μm. Conidia oblong to cylindrical, smooth- and thin-walled, hyaline, aseptate, 3–4.5 × 1.5–2.5 μm, with 2 distinct pale green polar guttules. Conidia matrix whitish.
Culture characteristics: Colonies on OA, 45–50 mm diam after 7 d, margin regular, covered by white floccose aerial mycelia, white to pale olivaceous; reverse buff, with pale olivaceous concentric rings near the centre. Colonies on MEA 50–55 mm diam after 7 d, margin regular, aerial mycelia sparse, olivaceous, white near the centre; reverse olivaceous. Colonies on PDA, 50–55 mm diam after 7 d, margin regular, covered by dense white felty aerial mycelia, white, olivaceous near the centre; reverse buff, olivaceous near the centre. NaOH test negative.
Specimens examined: China, Guizhou, Shuanghe Cave National Geopark, from air, 8 May 2015, Z.F. Zhang (holotype HMAS 247035, dried culture, ex-holotype living culture CGMCC 3.18114 = LC 6245); ibid. CGMCC 3.18115 = LC 6246; ibid. CGMCC 3.18116 = LC 6247.
Notes: Species of Allophoma were hitherto all known as plant pathogens, while Al. oligotrophica is the first species which was isolated from air using carbon-free silica gel medium (Jiang et al. 2017). Allophoma oligotrophica is closely related to Al. nicaraguensis (1 bp difference in ITS, 14 in rpb2 and 3 in tub2) and Al. tropica (1 bp difference in ITS, 15 in rpb2 and 2 in tub2) (Fig. 1). Morphologically, Al. oligotrophica produces larger pycnidia (150–440 × 145–420 μm vs. 30–150 × 28–120 μm) and longer conidiogenous cells (4.5–7 × 3.5–6.5 μm vs. 3–4.5 × 3.5–4.5 μm) than Al. nicaraguensis (Chen et al. 2015a), and differs from Al. tropica in its slightly larger conidiogenous cells (4.5–7 × 3.5–6.5 μm vs. 2–6 × 3–6 μm) and oblong to cylindrical conidia (de Gruyter & Noordeloos 1992).
Ascochyta Lib. emend. Q. Chen & L. Cai. Stud. Mycol. 82: 185. 2015.
Synonym: Heracleicola Tibpromma et al., Fungal Divers. 75: 58. 2015.
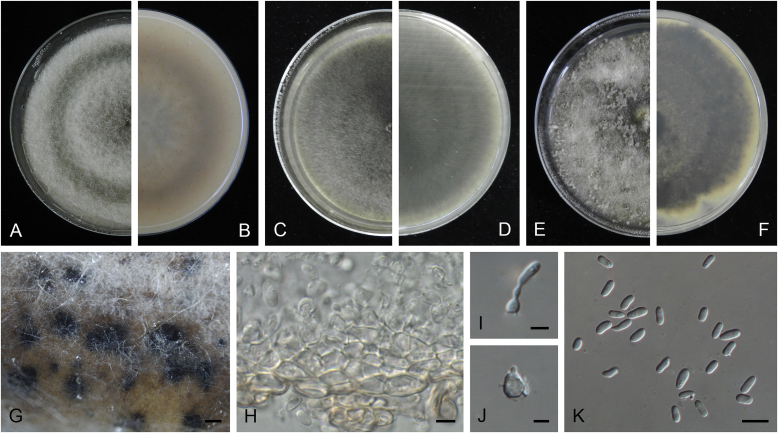
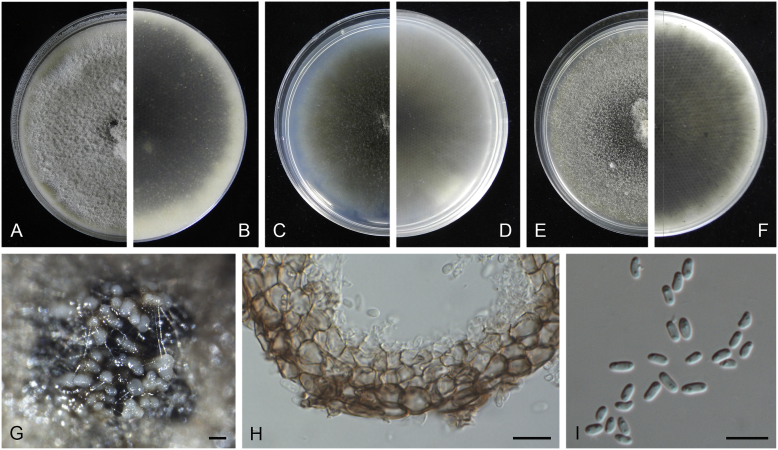
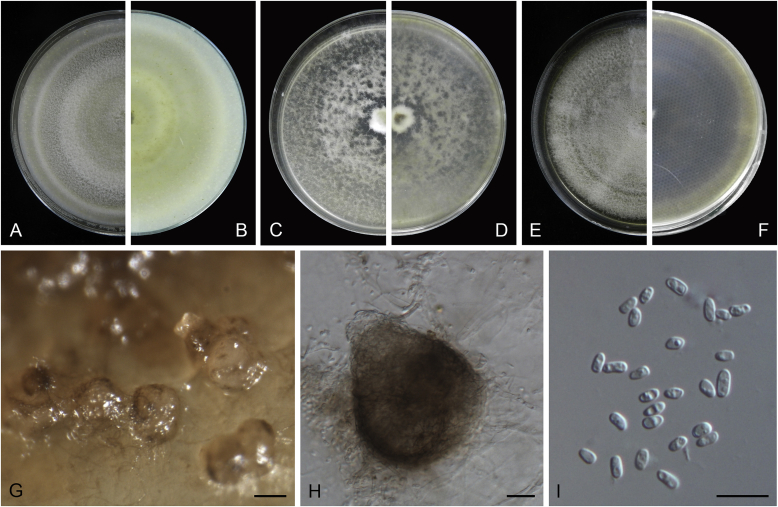
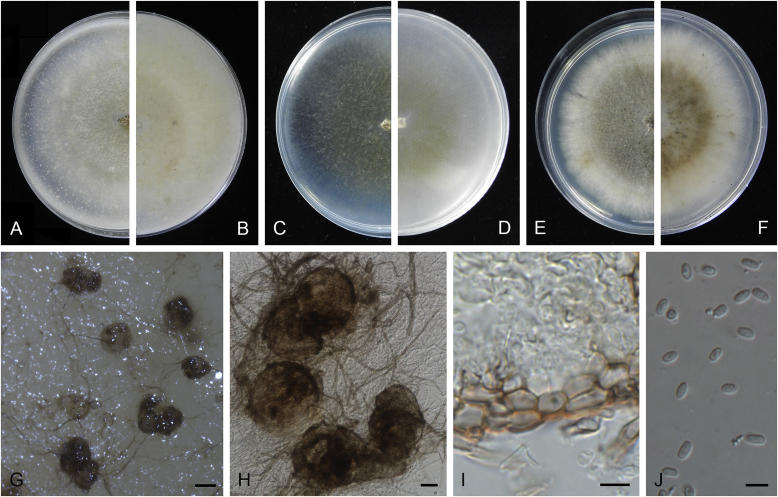
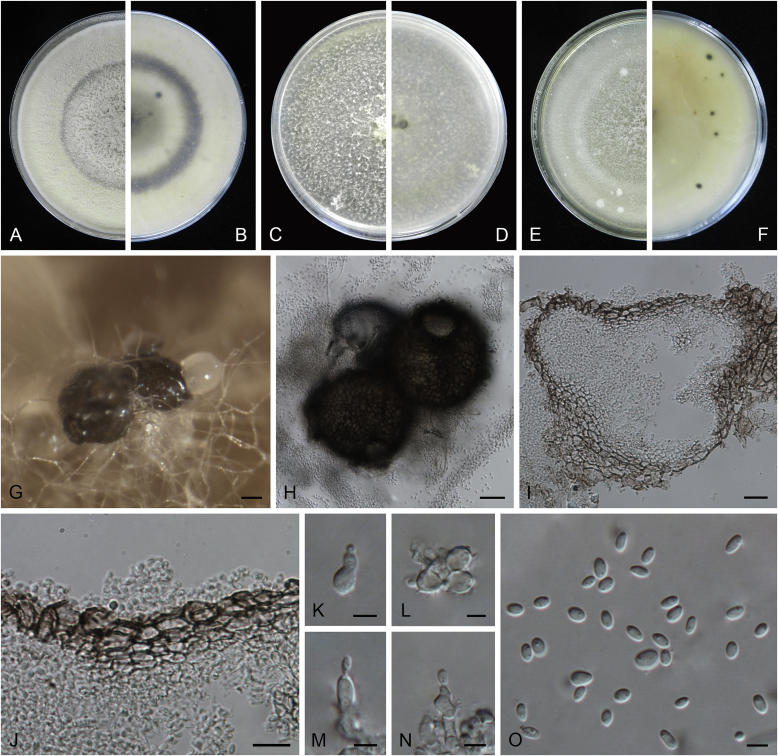
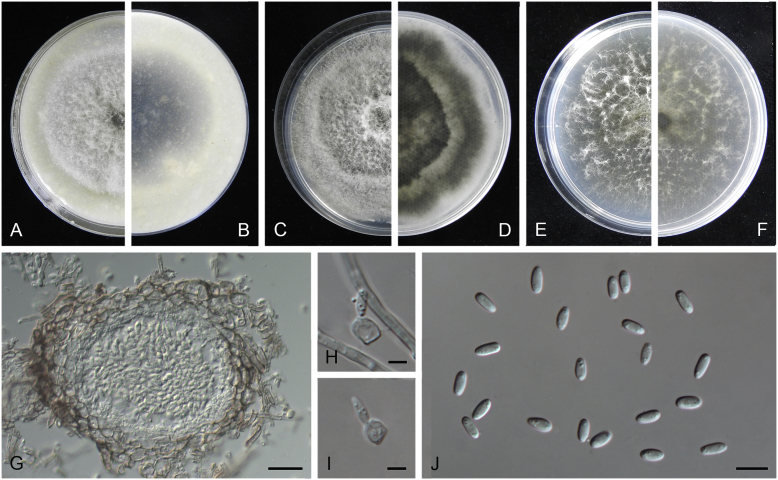
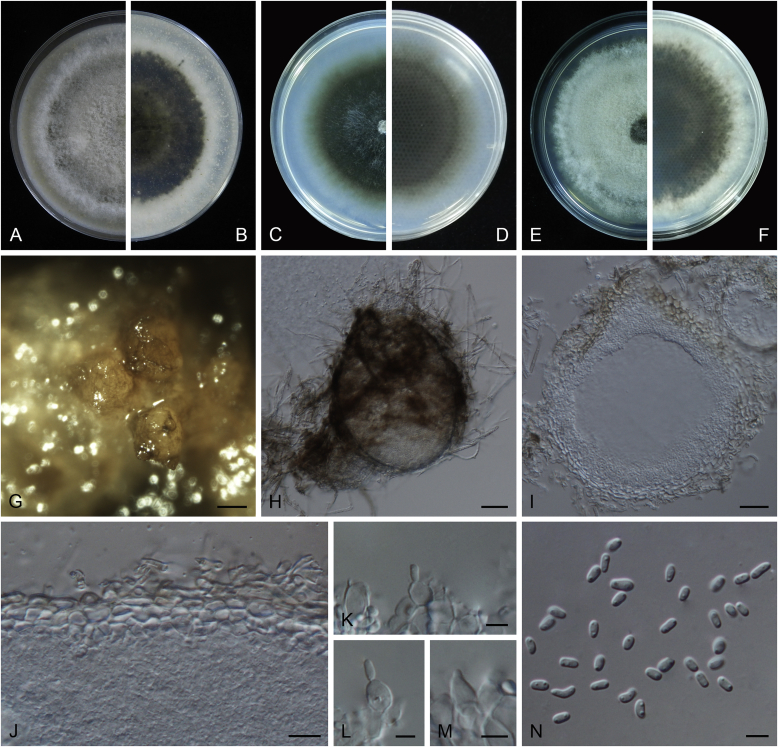
Ascochyta boeremae L.W. Hou, Crous & L. Cai, sp. nov. MycoBank MB820000. Fig. 4.
Fig. 4.
Ascochyta boeremae (CBS 372.84). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Pycnidium. I. Section of pycnidium. J. Section of pycnidial wall. K–L. Conidiogenous cells. M–N. Conidia. Scale bars: G = 300 μm; H–I = 50 μm; J–N = 10 μm.
Etymology: Named after Gerhard H. Boerema, who collected the holotype of this species.
Conidiomata pycnidial, mostly solitary, sometimes confluent, (sub-)globose or flask-shaped, glabrous, semi-immersed in or superficial on the agar, ostiolate, 170–550(–650) × 140–400(–650) μm. Ostiole single, slightly papillate, sometimes elongated as a short neck. Pycnidial wall pseudoparenchymatous, composed of oblong to isodiametric cells, 3–6-layers, with outer 2–3-layers pigmented, 25–50 μm thick. Conidiogenous cells phialidic, hyaline, smooth, ampulliform or doliiform, 9.5–14.5 × 8.5–13 μm. Conidia greatly variable in shape and size, large conidia mostly oblong to bacilliform, or fusiform, mainly aseptate but sometimes uniseptate; small conidia ellipsoidal to oval, broadly ovoid, smooth- and thin-walled, hyaline, aseptate, (14–)16.5–26(–32) × 4.5–7.5(–8.5) μm, eguttulate or sometimes with 1–2 guttules per cell. Conidial matrix whitish cream.
Culture characteristics: Colonies on OA, 25–30 mm diam after 7 d, margin regular, covered by sparsely flat aerial mycelia, yellowish olivaceous; reverse concolourous. Colonies on MEA 20–25 mm diam after 7 d, margin regular, covered with floccose aerial mycelia, white, grey near the centre; reverse sienna to pale brown. Colonies on PDA, 15–20 mm diam after 7 d, margin regular, covered by woolly aerial mycelia, greenish olivaceous, buff near the margin; reverse concolourous. NaOH spot test: a dark reddish brown discolouration on MEA.
Specimens examined: Australia, from a leaf of Pisum sativum, deposited in CBS Sep. 1984, G.H. Boerema (holotype CBS H-23017, dried culture, ex-holotype living culture CBS 372.84 = PD 80/1246); from a leaf of Pisum sativum, deposited in CBS Sep. 1984, G.H. Boerema, CBS H-9078, culture CBS 373.84 = PD 80/1247.
Notes: CBS 372.84 and CBS 373.84 were originally deposited as “Ascochyta fabae”, but are distinct from the authentic cultures of As. fabae (CBS 524.77, CBS 649.71 and PD 83/492) in the phylogenetic tree. Morphologically, these two strains produce aseptate conidia differing from the uniseptate conidia of As. fabae (Saccardo 1902). Therefore, we describe it as a new species, As. boeremae. Ascochyta boeremae is genetically closely related to As. nigripycnidia (Fig. 1), but differs morphologically from the latter by producing larger conidia (14–32 × 4.5–8.5 μm vs. 5.5–15 × 1.5–4 μm; Boerema et al. 2004).
Ascochyta premilcurensis (Tibpromma et al.) Q. Chen, Crous & L. Cai, comb. nov. MycoBank MB820001.
Basionym: Heracleicola premilcurensis Tibpromma et al., Fungal Divers. 75: 59. 2015.
Description: Ariyawansa et al. (2015).
Specimen examined: Italy, Premilcuore, Province of Forli-Cesena, Valbura, on dead stem of Heracleum sphondylium, 6 Jun. 2014, E. Camporesi (holotype MFLU 14-0725, ex-holotype living culture MFLUCC 14-0518).
Notes: The genus Heracleicola was introduced by Ariyawansa et al. (2015) to accommodate a single species Heracleicola premilcurensis, which is located in the genus Ascochyta based on combined LSU and ITS analysis (Supplementary Fig. S1) in the present study. Heracleicola is therefore synonymised under Ascochyta, and a new combination in Ascochyta proposed.
Boeremia Aveskamp et al., Stud. Mycol. 65: 36. 2010.
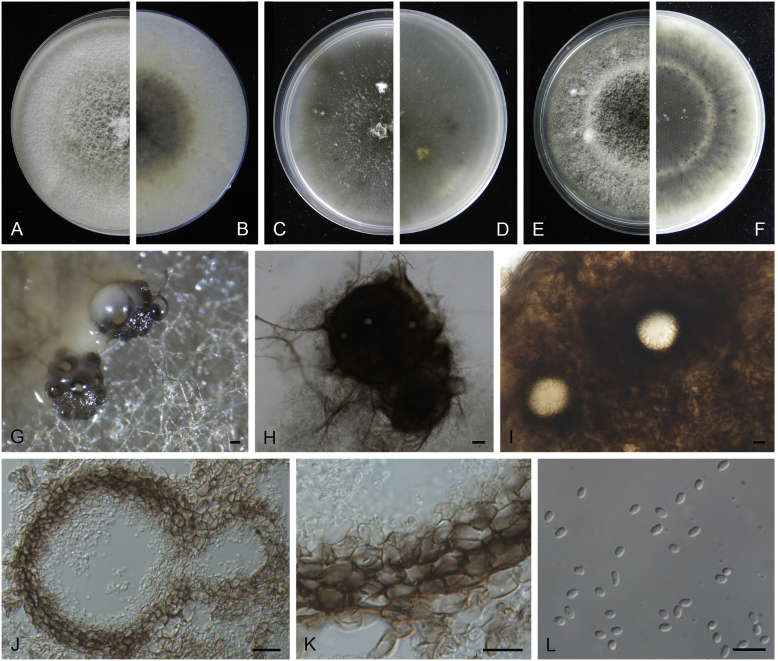
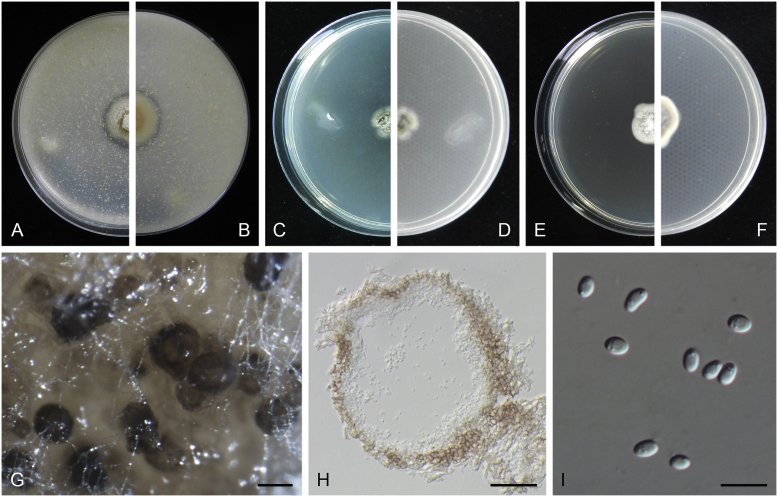
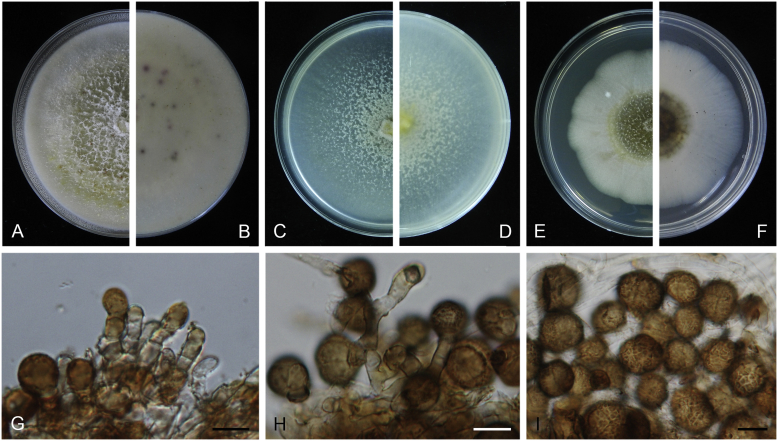
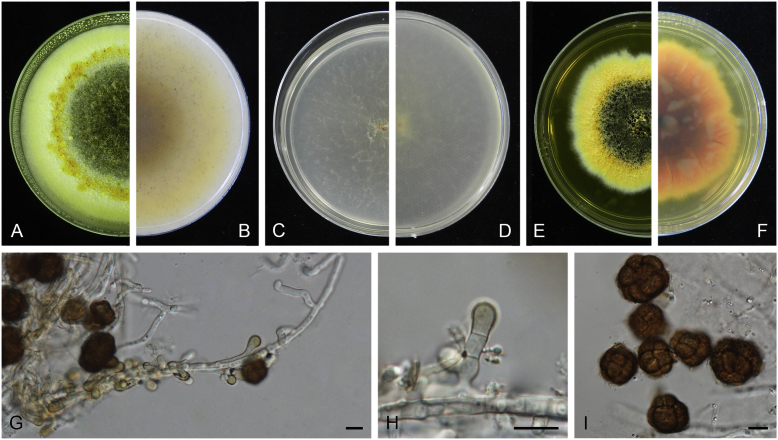
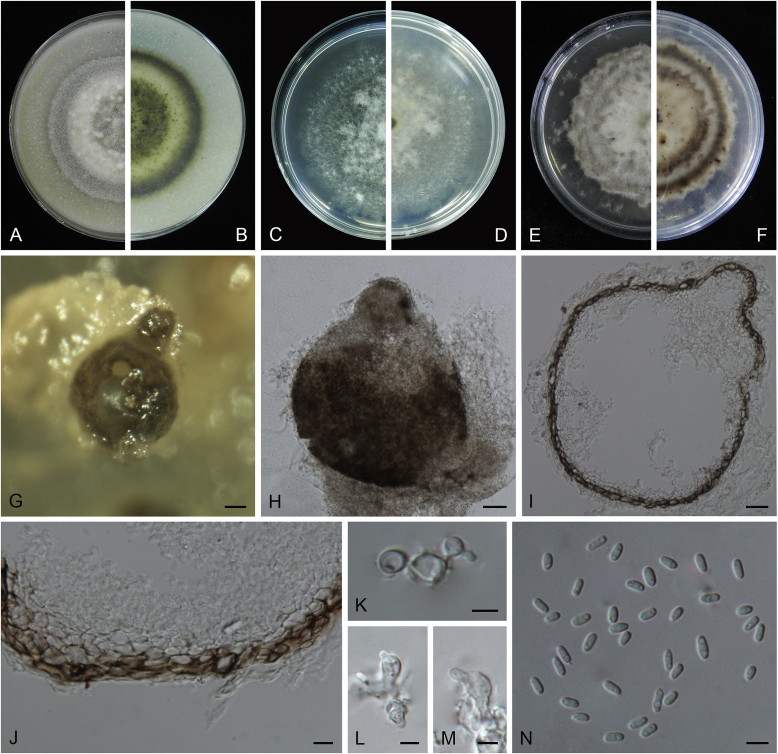
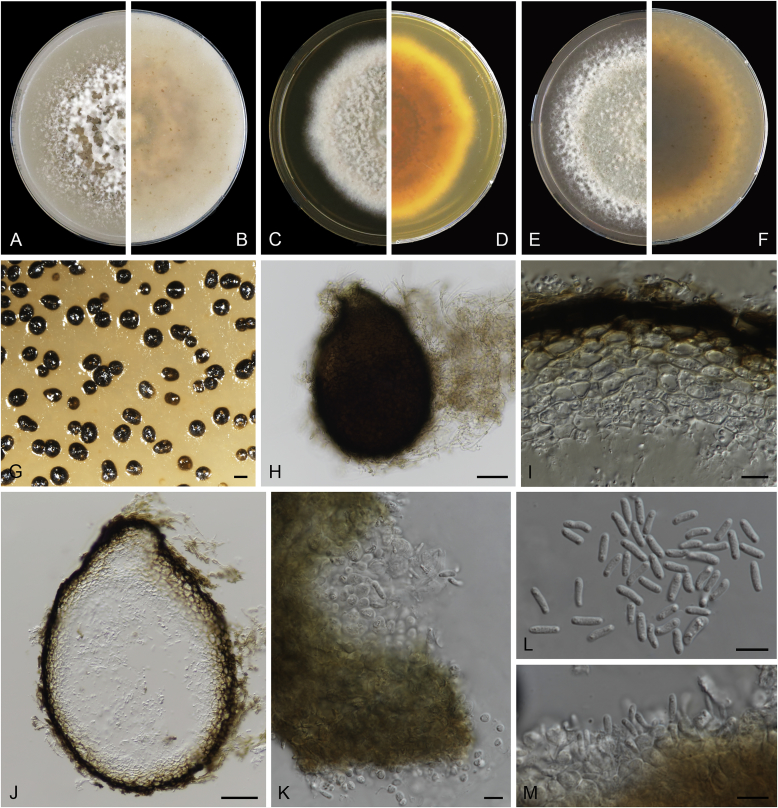
Boeremia exigua var. opuli Q. Chen, Crous & L. Cai, var. nov. MycoBank MB818957. Fig. 5.
Fig. 5.
Boeremia exigua var. opuli (CGMCC 3.18354). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Section of pycnidial wall. I–J. Conidiogenous cells. K. Conidia. Scale bars: G = 400 μm; H–J = 5 μm; K = 10 μm.
Etymology: Named after the host species from which the holotype was collected, Viburmum opulus.
Conidiomata pycnidial, solitary or aggregated, globose to subglobose, brown, covered with hyphae, produced on the agar surface or (semi-)immersed, 245–360 × 200–305 μm. Ostiole single, slightly papillate. Pycnidial wall pseudoparenchymatous, composed of isodiametric cells, 4–5 layers, 20–37.5 μm thick. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 4–9(–10) × 4–7.5 μm. Conidia oblong to cylindrical, obovoid, incidentally slightly curved, or reniform, smooth- and thin-walled, hyaline, aseptate, 5.5–9.5 × 2.5–4 μm, with 2 or several minute guttules. Conidia matrix cream.
Culture characteristics: Colonies on OA, 70–76 mm diam after 7 d, margin regular, covered by white floccose aerial mycelia, white with a pale green concentric ring, pale olivaceous near the centre; reverse reddish brown, grey near the centre. Colonies on MEA 70–75 mm diam after 7 d, margin regular, aerial mycelia white, velvety, olivaceous; reverse concolourous. Colonies on PDA, 65–80 mm diam after 7 d, margin regular, aerial mycelia white, felty, in some sectors covered by a low mat of floccose white to grey aerial mycelia, olivaceous near the centre; reverse olivaceous, with a buff margin. Application of NaOH results in a pale green discolouration of the agar.
Specimens examined: USA, from seedlings of Viburmum opulus, 2014, W.J. Duan (holotype HMAS 247147, dried culture, ex-holotype living culture CGMCC 3.18354 = LC 8117); ibid. LC 8118.
Notes: Boeremia exigua var. opuli is phylogenetically closely related to B. exigua var. exigua, B. exigua var. forsythiae, B. exigua var. glivescens and B. exigua var. viburni (Fig. 1). Although similar in conidial dimensions, pycnidia of B. exigua var. opuli (245–360 × 200–305 μm) are much larger than those of the other four varieties (75–200 μm; van der Aa et al. 2000). Boeremia exigua var. opuli also differs from those four varieties in seven positions in the rpb2 locus. Varieties of B. exigua are morphologically very similar and phylogenetically closely related to each other. Boeremia exigua var. exigua and var. forsythia have a wide host range, while other varieties appear host specific to a certain group of plants, such as var. coffeae to Coffea arabica (Rubiaceae), var. forsythia to Forsythia hybrids (Oleaceae), var. heteromorpha to Nerium oleander and Vinca spp. (Apocynaceae), var. linicola to Linum usitatissimum (Linaceae), var. populi to Populus and Salix (Salicaceae), and var. viburni to Viburnum spp. and occasionally Lonicera sp. (Caprifoliaceae) (Boerema et al. 2004). Besides, B. exigua var. pseudolilacis has been found only on Syringa vulgaris (Oleaceae; Aveskamp et al. 2010) and var. opuli only on Viburnum opulus. A host-range determination of B. exigua var. rhapontica indicates that this variety also has a very narrow host range (Berner et al. 2015). Thus, the plant generic inter-relatedness is presumed to be the basis for susceptibility to Boeremia exigua varieties (Berner et al. 2015).
Calophoma Q. Chen & L. Cai, Stud. Mycol. 82: 191. 2015.
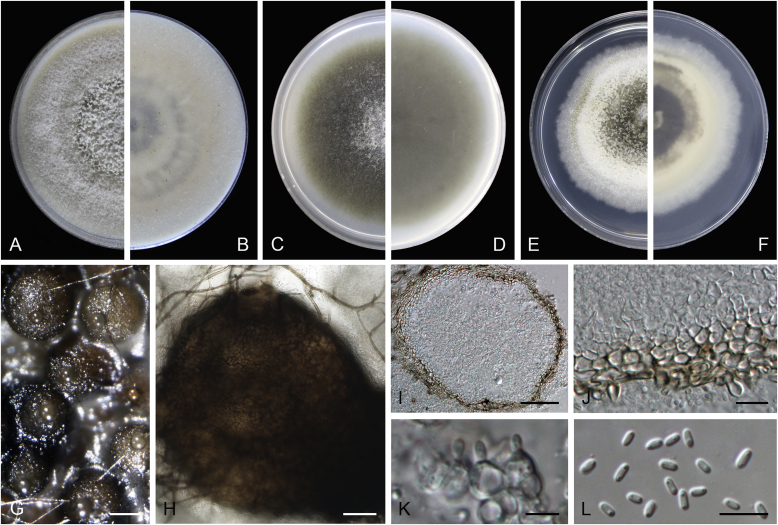
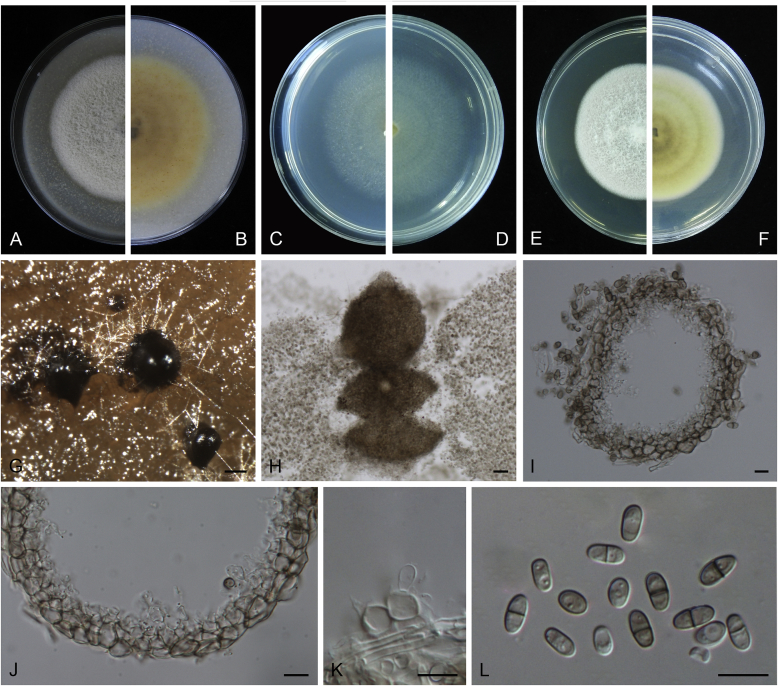
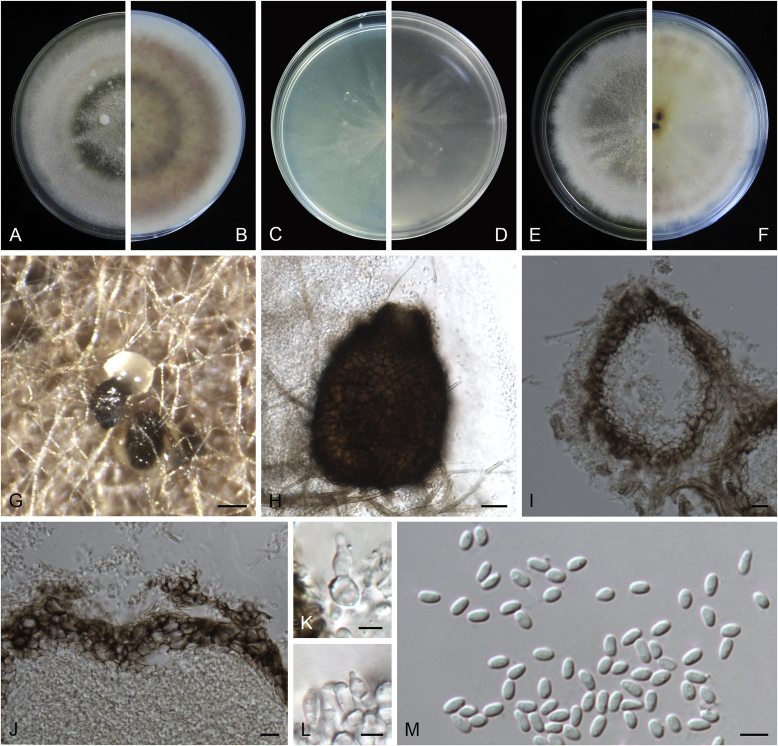
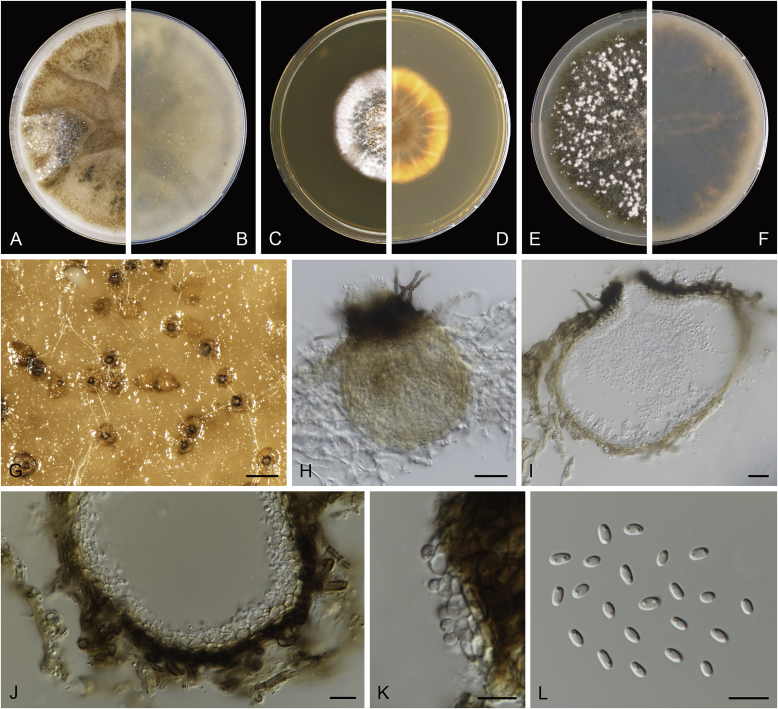
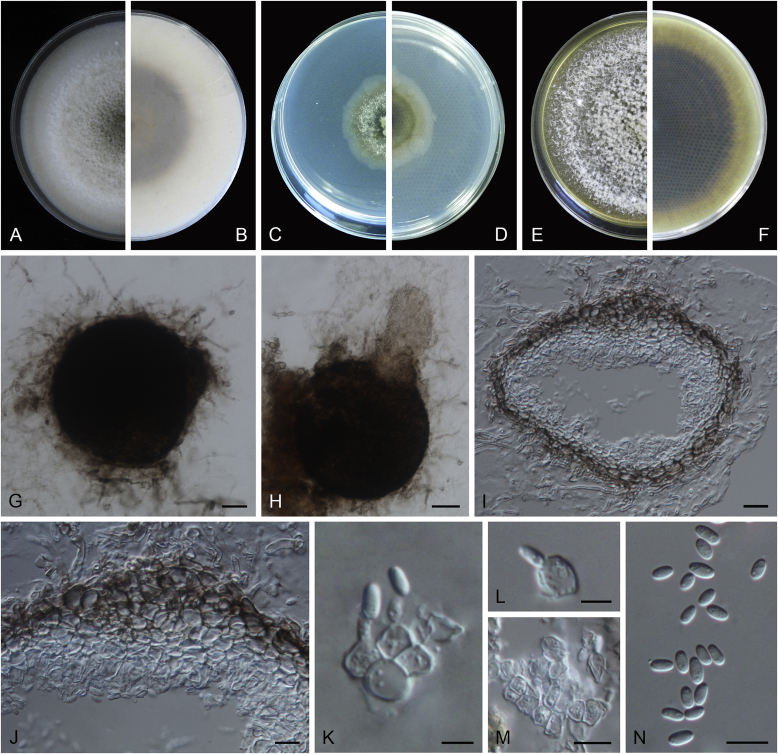
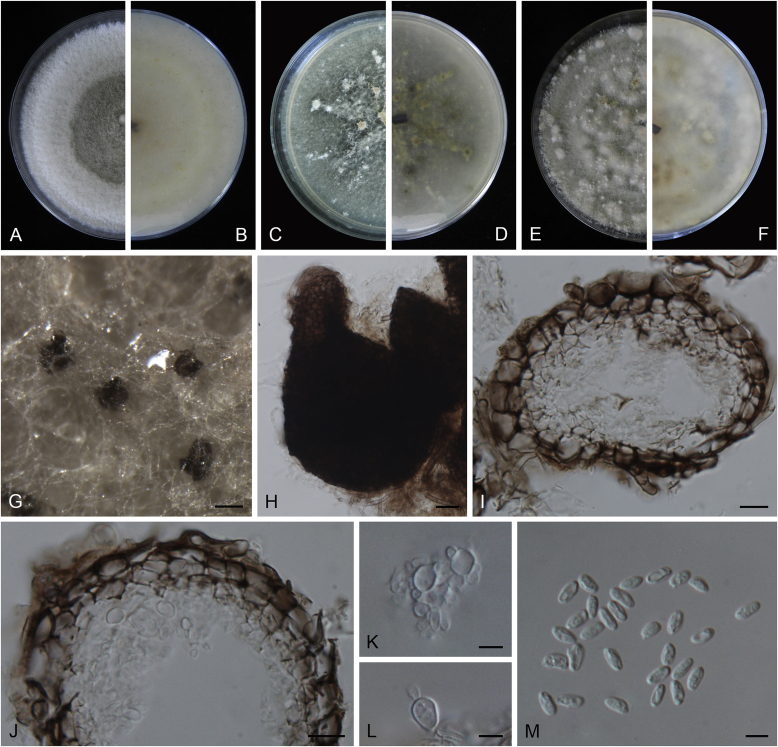
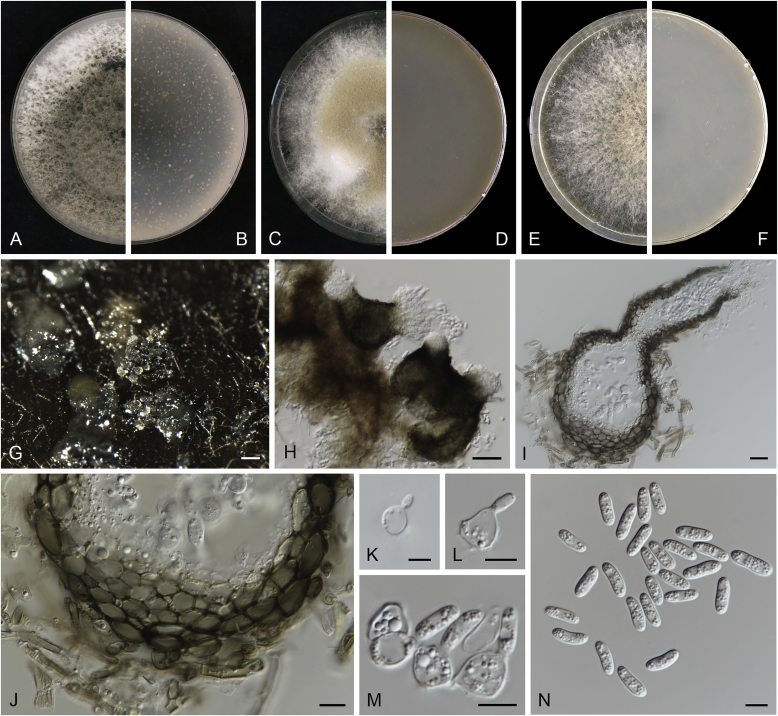
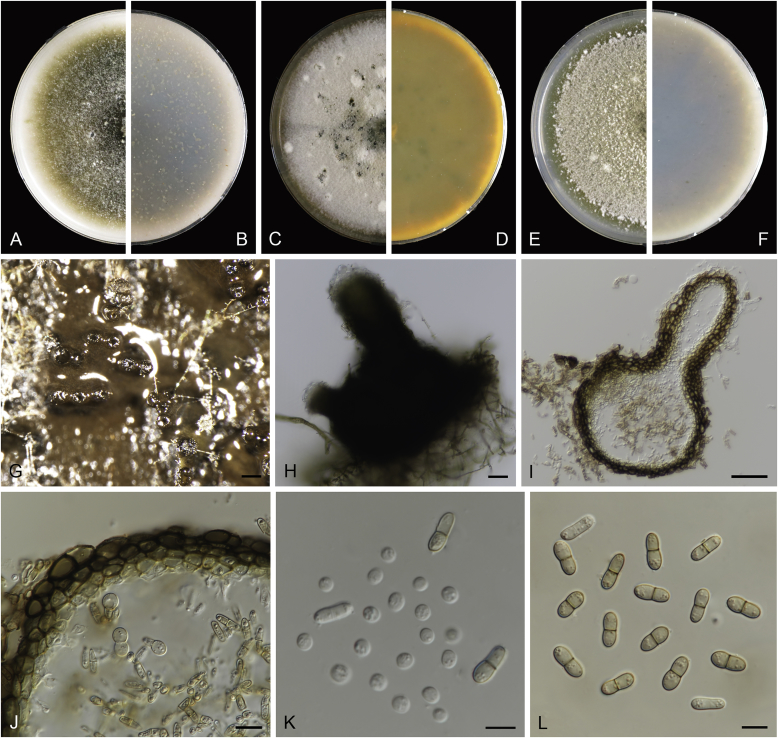
Calophoma rosae Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818976. Fig. 6A.
Fig. 6.
Calophoma rosae (CGMCC 3.18347). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Pycnidia. I. Section of pycnidium. J. Section of pycnidial wall. K. Conidiogenous cells. L. Conidia. Scale bars: G = 100 μm; H = 40 μm; I–L = 10 μm.
Etymology: Named after the host genus Rosa, from which the holotype was isolated.
Leaf spots amphigenous, circular to irregular, up to 15 mm diam, occurring on or close to the tip of the leaf, brown, surrounded by a dark purple border (Fig. 7). Conidiomata pycnidial, mostly aggregated but sometimes solitary, globose to subglobose, brown, glabrous or covered with some hyphal outgrowths, semi-immersed in or superficial on the agar, ostiolate, (110–)130–210 × (110–)130–180 μm. Ostiole single, sometimes with short necks, slightly papillate. Pycnidial wall pseudoparenchymatous, composed of isodiametric cells, 3–4 layers, 11–20 μm thick, pigmented. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 6.5–7 × 7–8.5 μm. Conidia ellipsoidal to oblong, smooth- and thin-walled, 0–1-septate, hyaline, later becoming pale brown with ageing, 6–10 × 3–4.5 μm, eguttulate or sometimes with several guttules. Conidial matrix initially buff, gradually becoming dark brown.
Fig. 7.
Symptoms on diseased leaves. A.Calophoma rosae on Rosa sp. B.Didymella infuscatispora on Chrysanthemum indicum. C.Didymella ocimicola on Ocimum sp. D.Didymella sinensis on Cerasus pseudocerasus. E.Epicoccum dendrobii on Dendrobium fimbriatum. F.Epicoccum duchesneae on Duchesnea indica. G.Stagonosporopsis bomiensis on Boraginaceae. H.Epicoccum viticis on Vitex negundo. I.Epicoccum layuense on Perilla sp. J.Heterophoma verbascicola on Verbascum thapsus. K.Stagonosporopsis papillata on Rumex nepalensis.
Culture characteristics: Colonies on OA, 35–40 mm diam after 7 d, margin regular, covered by floccose aerial mycelia, dense, white; reverse buff. Colonies on MEA 33–35 mm diam after 7 d, margin regular, aerial mycelia sparse, flattened, white; reverse concolourous. Colonies on PDA, 30–36 mm diam after 7 d, margin regular, aerial mycelia covering the whole colony, floccose, dense, white; reverse yellowish green, with concentric rings. NaOH test negative.
Specimens examined: China, Qinghai, Xunhua, from leaves of Rosa sp., 2 Sep. 2013, Q. Chen (holotype HMAS 247148, dried culture, ex-holotype living culture CGMCC 3.18347 = LC 5169); ibid. LC 8119.
Notes: Calophoma rosae is phylogenetically closely related to C. clematidis-rectae and C. vodakii (Fig. 1). Morphologically C. rosae differs from C. clematidis-rectae in having larger conidiogenous cells (6.5–7 × 7–8.5 μm vs. 3–5 × 2.5–4.5 μm), larger conidia (6–10 × 3–4.5 μm vs. 3–8 × 2–3.5 μm) (Aveskamp et al. 2010), and from C. vodakii in having shorter and wider conidia (6.5–7 × 7–8.5 μm vs. 14–22 × 4–4.5 μm; Saccardo and Trotter, 1913, Müller, 1953).
Calophoma rosae is the first and only record thus far from the Rosaceae, while most species in this genus are associated with species of Ranunculaceae.
Didymella Sacc. ex Sacc., Syll. Fung. 1: 545. 1882, emend. Q. Chen & L. Cai, Stud. Mycol. 82: 173. 2015.
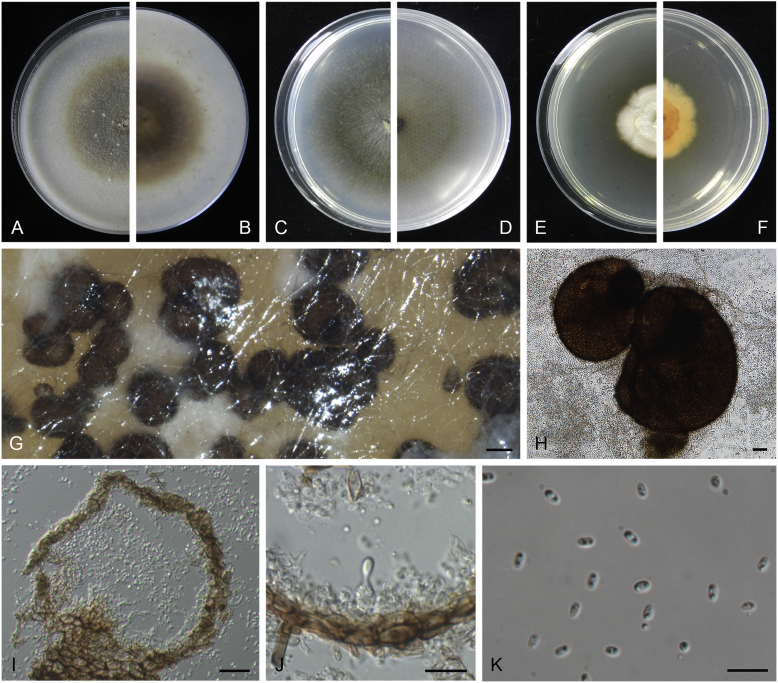
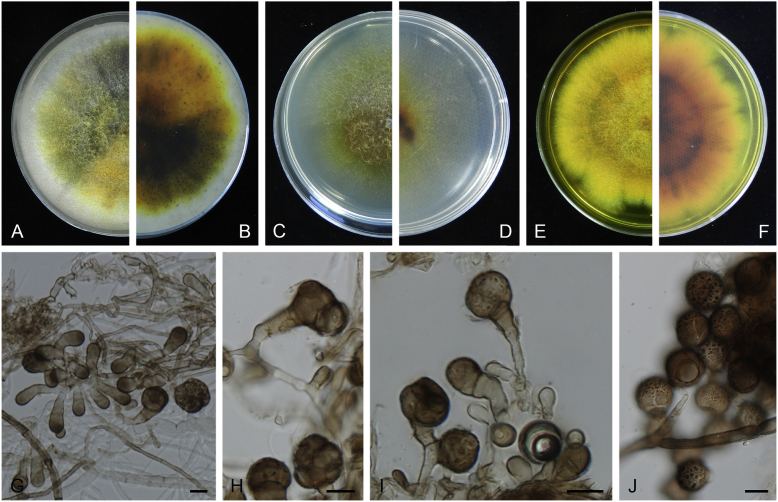
Didymella aeria Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818968. Fig. 8.
Fig. 8.
Didymella aeria (CGMCC 3.18353). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Pycnidia. I. Section of pycnidium. J. Section of pycnidial wall. K. Conidia. Scale bars: G = 200 μm; H = 30 μm; I = 20 μm; J–K = 10 μm.
Etymology: Name linked to the fact that this species was collected from air.
Conidiomata pycnidial, solitary or aggregated, globose to subglobose, later becoming irregular, brown, glabrous, superficial or semi-immersed, 155–375(–460) × 130–340(–460) μm. Ostiole single, with a short neck, slightly papillate or non-papillate. Pycnidial wall pseudoparenchymatous, composed of oblong to isodiametric cells, 2–3 layers, 8.5–25 μm thick, brown-pigmented. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 5–7 × 4.5–6 μm. Conidia ellipsoidal, smooth- and thin-walled, hyaline, aseptate, 3–5 × 2–3 μm, with 2 large dull green polar guttules. Conidial matrix salmon.
Culture characteristics: Colonies on OA, 55–60 mm diam after 7 d, margin regular, white aerial mycelia sparse, brownish olivaceous; reverse white to reddish brown. Colonies on MEA 44–48 mm diam after 7 d, margin regular, white to olivaceous, with sparse white aerial mycelia spreading over the colony; reverse concolourous. Colonies on PDA, 15–20 mm diam after 7 d, margin irregular, fluffy to felty, white; reverse amber to saffron. NaOH spot test: a brown discolouration on MEA.
Specimens examined: China, Guizhou, Zunyi, Shuanghe Cave National Geopark, from air, 8 May 2015, Z.F. Zhang (holotype HMAS 247149, dried culture, ex-holotype living culture CGMCC 3.18353 = LC 7441); ibid. LC 8120.
Notes: The most closely related species to Didymella aeria are D. sinensis and D. pomorum (Fig. 1), but with respectively 33 bp and 55 bp differences in four sequenced loci. Didymella aeria produces hyaline conidia measuring 3–5 × 2–3 μm, while D. pomorum produces longer, brown conidia (4–8 × 1.5–3 μm; Boerema 1993). The asexual morph of D. sinensis was unfortunately not observed. Didymella aeria was trapped from air in a Karst cave in China.
Didymella aquatica Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818973. Fig. 9.
Fig. 9.
Didymella aquatica (CGMCC 3.18349). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia sporulating on OA. H. Pycnidia. I. Section of pycnidium. J. Section of pycnidial wall. K. Conidia. Scale bars: G = 100 μm; H = 50 μm; I, K = 10 μm; J = 5 μm.
Etymology: Name derived from the substrate where the holotype was collected, water.
Conidiomata pycnidial, solitary, sometimes aggregated, globose to subglobose, brown, glabrous or covered with some hyphal outgrowths, superficial, ostiolate, 105–355 × 95–315 μm. Ostioles 2–13, sometimes elongated as a short neck, up to 50.5 μm long, papillate. Pycnidial wall pseudoparenchymatous, composed of oblong to isodiametric cells, 2–5 layers, 14–35 μm thick, outer wall 2–3-layers pigmented. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 4–5 × 3.5–5 μm. Conidia ellipsoidal to oblong, smooth- and thin-walled, hyaline, aseptate, 4–5.5 × 2–3 μm, with 2 distinct polar guttules. Conidial matrix cream.
Culture characteristics: Colonies on OA, 15–40 mm diam after 7 d, margin regular, covered by velvety aerial mycelia, flat, white to amber; reverse concolourous. Colonies on MEA 46–53 mm diam after 7 d, margin regular, white to pale green, with sparse aerial mycelia near the centre; reverse concolourous. Colonies on PDA, 54–56 mm diam after 7 d, margin regular, floccose to felty, white to grey, iron grey near the centre; reverse white, hazel to brown. NaOH test negative.
Specimens examined: China, Guizhou, Kuankuoshui National Geopark, water, 23 Jul. 2014, Z.F. Zhang (holotype HMAS 247150, dried culture, ex-holotype living culture CGMCC 3.18349 = LC 5556); ibid. LC 5555.
Notes: Didymella aquatica formed a distinct lineage sister to D. macrophylla, with 6 bp differences in both rpb2 and tub2 loci. Morphologically, D. aquatica is clearly differentiated from D. macrophylla in producing smaller conidiogenous cells (4–5 × 3.5–5 μm vs. 6–8 × 4.5–8 μm), longer and narrower conidia (4–5.5 × 2–3 μm vs. 1.5–2.5 × 3.5–5.5 μm), and in the number of conidiomatal ostioles (2–13 vs. 1). This is the first Didymella species known from water.
Didymella chloroguttulata Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818970. Fig. 10.
Fig. 10.
Didymella chloroguttulata (CGMCC 3.18351). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Section of pycnidial wall. I. Conidia. Scale bars: G = 200 μm; H–I = 10 μm.
Etymology: Latin, chloro- = green, referring to the two green guttules of the conidia.
Conidiomata pycnidial, confluent, globose to subglobose, brown, glabrous, superficial, 145–260(–410) × 130–230(–365) μm. Ostiole single, sometimes with a short neck, slightly papillate or non-papillate. Pycnidial wall pseudoparenchymatous, composed of isodiametric cells, 3–4 layers, 14.5–22 μm thick, pigmented. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 5.5–8 × 4–6.5 μm. Conidia oblong to cylindrical, incidentally slightly curved, smooth- and thin-walled, hyaline, aseptate, 4–6 × 2–3 μm, with 2–3 dull green polar guttules. Conidial exudates not recorded.
Culture characteristics: Colonies on OA, 54–57 mm diam after 7 d, margin regular, covered by floccose aerial mycelia, dense, grey; reverse black. Colonies on MEA 44–47 mm diam after 7 d, margin regular, white aerial mycelia sparse, fluffy, greenish brown; reverse concolourous. Colonies on PDA, 57–62 mm diam after 7 d, margin regular, floccose, grey to leaden-black; reverse leaden-black. NaOH spot test: a pale reddish brown discolouration on MEA.
Specimens examined: China, Guizhou, Zunyi, Shuanghe Cave National Geopark, air, 8 May 2015, Z.F. Zhang (holotype HMAS 247151, dried culture, ex-holotype living culture CGMCC 3.18351 = LC 7435); ibid. LC 8122.
Notes: Didymella chloroguttulata is characterised by having two to three dull green polar guttules in its oblong to cylindrical conidia and sometimes having conidiomata with a short neck. In the phylogenetic tree, it formed a distinct clade sister to D. dactylidis and D. rhei (Fig. 1). Didymella chloroguttulata is well distinguished from these two species in the NaOH reactions (pale reddish brown discolouration on D. chloroguttulata, slight greenish discolouration on D. dactylidis, and no effect on D. rhei) (de Gruyter et al. 2002, Aveskamp et al. 2010).
Didymella ellipsoidea Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818971. Fig. 11.
Fig. 11.
Didymella ellipsoidea (CGMCC 3.18350). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia sporulating on OA. H. Pycnidia. I. Ostioles on pycnidium. J. Section of pycnidium. K. Section of pycnidial wall. L. Conidia. Scale bars: G = 100 μm; H = 50 μm; I, K–L = 10 μm; J = 20 μm.
Etymology: Name refers to its ellipsoidal conidia.
Conidiomata pycnidial, solitary, globose to subglobose, brown, glabrous or covered with some hyphal outgrowths, superficial, ostiolate, 335–400(–460) × 290–340(–440) μm. Ostioles 1–8, often developing to elongated short necks, up to 80 μm long, papillate. Pycnidial wall pseudoparenchymatous, composed of isodiametric cells, 3–5 layers, 23.5–50 μm thick, pigmented. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 5.5–7.5 × 4.5–6.5 μm. Conidia ellipsoidal, smooth- and thin-walled, hyaline, aseptate, 3–4.5 × 2–3 μm, with 2 pale green guttules. Conidial matrix cream.
Culture characteristics: Colonies on OA, 56–62 mm diam after 7 d, margin regular, covered by floccose aerial mycelia, dense, white to pale brown; reverse white to greenish brown. Colonies on MEA 61–64 mm diam after 7 d, margin regular, white aerial mycelia sparse, fluffy, grey to olivaceous; reverse concolourous. Colonies on PDA, 52–54 mm diam after 7 d, margin regular, floccose, grey to leaden-black, with a white concentric ring near the centre; reverse concolourous. NaOH test negative.
Specimen examined: China, Guizhou, Zunyi, Shuanghe Cave National Geopark, air, 8 May 2015, Z.F. Zhang (holotype HMAS 247152, dried culture, ex-holotype living culture CGMCC 3.18350 = LC 7434); ibid. LC 8123.
Notes: This species is represented by two isolates trapped from air in a Karst cave which cluster in a distinct lineage clearly differentiated from other species in Didymella (Fig. 1). Morphologically, Didymella ellipsoidea is distinguishable from its closest neighbours, D. viburnicola, in producing wider conidia (3–4.5 × 2–3 μm vs. 3.5–5.5 × 1.5–2 μm; de Gruyter & Noordeloos 1992), from D. macrostoma in having shorter conidia (3–4.5 × 2–3 μm vs. 4–11 × 2–4 μm; de Gruyter et al. 2002), and from D. pteridis in producing larger conidiogenous cells (5.5–7.5 × 4.5–6.5 μm vs. 4–5 × 3.5–4.5 μm).
Didymella ilicicola Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818969. Fig. 12.
Fig. 12.
Didymella ilicicola (CGMCC 3.18355). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Pycnidium. I. Section of pycnidium. J. Section of pycnidial wall. K–L. Conidiogenous cells. M. Conidia. Scale bars: G = 100 μm; H = 20 μm; I–J = 10 μm; K–M = 5 μm.
Etymology: Name derived from Ilex, the plant from which the holotype was collected.
Conidiomata pycnidial, solitary or aggregated, (sub-)globose to flask-shaped, or obpyriform, brown, later becoming irregular when matured, covered with hyphal outgrowths, mostly erumpent, sometimes semi-immersed, ostiolate, (80–)150–200 × (70–)150–180 μm. Ostioles 2–3, elongated as short papillate necks. Pycnidial wall pseudoparenchymatous, composed of oblong to isodiametric cells, 2–5 layers, 15–20 μm thick, outer wall 2–3-layers pigmented. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 4.5–8 × 3.5–5 μm. Conidia ellipsoidal to oblong, smooth- and thin-walled, hyaline, aseptate, 3–4 × 1.5–2.5 μm, with two minute guttules. Conidial matrix cream to buff.
Culture characteristics: Colonies on OA, 43–50 mm diam after 7 d, margin regular, covered by floccose aerial mycelia, white to pale buff, with a dull green concentric ring near the centre; reverse reddish brown to buff, with a brown concentric ring. Colonies on MEA 56–65 mm diam after 7 d, margin irregular, white, aerial mycelia sparse; reverse concolourous. Colonies on PDA, 62–65 mm diam after 7 d, margin regular, felty to floccose, dense, white to pale yellow; reverse white to buff with some pale reddish brown tings in concentric rings. NaOH test negative.
Specimens examined: Italy, from seedlings of Ilex chinensis, 2013, W.J. Duan (holotype HMAS 247153, dried culture, ex-holotype living culture CGMCC 3.18355 = LC 8126); ibid. LC 8127.
Notes: Didymella ilicicola clustered in a clade together with D. subherbarum and D. pedeiae (Fig. 1), but with 1 bp and 9 bp differences in ITS and tub2 respectively from D. subherbarum (lack of rpb2 sequence), and 26 bp and 12 bp differences in rpb2 and tub2 respectively from D. pedeiae. Morphologically, D. ilicicola differs from D. subherbarum in producing shorter conidia (3–4 × 1.5–2.5 μm vs. 4–6.5 × 1.5–2 μm; de Gruyter et al. 1993), and from D. pedeiae in producing larger conidiogenous cells (4.5–8 × 3.5–5 μm vs. 3.5–4.5 × 3–4 μm; Aveskamp et al. 2010).
Didymella infuscatispora Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818974. Fig. 13.
Fig. 13.
Didymella infuscatispora (CGMCC 3.18356). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Pycnidia. I. Conidia. J. Conidiogenous cell. Scale bars: G = 200 μm; H = 50 μm; I = 10 μm; J = 2.5 μm.
Etymology: Latin, infuscat- = brownish, referring to the colour of its conidia.
Leaf spots amphigenous, irregular, 3–11 mm diam, extending along leaf margin to the whole leaf, dark grey to dark brown (Fig. 7B). Conidiomata pycnidial, solitary, globose to subglobose, brown, later becoming irregular when matured, covered with some hyphal outgrowths, superficial, ostiolate, (50–)95–265 × (20–)75–165 μm. Ostiole single, sometimes elongated as short necks, slightly papillate. Pycnidial wall pseudoparenchymatous, composed of isodiametric cells, 2–3 layers, 13.5–23 μm thick. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 6–8.5 × 5.5–8 μm. Conidia globose to broadly ellipsoidal, oblong, smooth- and thin-walled, hyaline, later becoming pale brown, mostly aseptate, occasionally 1-septate, 5–8.5 × 3.5–5.5 μm, with several indistinct minute guttules. Conidial matrix dark brown.
Culture characteristics: Colonies on OA, 15–20 mm diam after 7 d, margin regular, covered by felty aerial mycelia, white to buff, pale brown near the centre; reverse white to amber, hazel near the centre. Colonies on MEA 10–15 mm diam after 7 d, margin irregular, aerial mycelia sparse, white to pale green; reverse white to pale green, yellowish brown near the centre. Colonies on PDA, 14–16 mm diam after 7 d, margin regular, aerial mycelia felty, flat, white to pale brown; reverse buff to brown. NaOH test negative.
Specimens examined: China, Tibet, Lulang, on leaves of Chrysanthemum indicum, 15 Jun. 2015, Q. Chen (holotype HMAS 247154, dried culture, ex-holotype living culture CGMCC 3.18356 = LC 8128); ibid. LC 8129.
Note: This species clustered in a distinct lineage separated from other species in this genus, and is characterised by pale brown and broadly ellipsoidal conidia and a dark brown conidial matrix when mature.
Didymella macrophylla Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB819189. Fig. 14.
Fig. 14.
Didymella macrophylla (CGMCC 3.18357). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Pycnidia. I. Conidia. Scale bars: G = 200 μm; H–I = 10 μm.
Etymology: Named after the host species Hydrangea macrophylla, from which the holotype was collected.
Conidiomata pycnidial, mostly solitary, sometime aggregated, globose to subglobose, pale brown, glabrous, semi-immersed or immersed in agar, ostiolate, (80–)120–200 × (60–)100–150 μm. Ostiole single, slightly papillate. Pycnidial wall pseudoparenchymatous, composed of oblong to isodiametric cells, 1–2 layers, 7–15 μm thick, pigmented. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 6–8 × 4.5–8 μm. Conidia obovoid, ellipsoidal to oblong, smooth- and thin-walled, hyaline, aseptate, 3.5–5.5 × 1.5–2.5 μm, with two polar guttules. Conidial matrix buff.
Culture characteristics: Colonies on OA, 44–46 mm diam after 7 d, margin regular, covered by floccose aerial mycelia, white to grey, yellowish grey near the centre; reverse grey to greyish yellow. Colonies on MEA 54–60 mm diam after 7 d, margin regular, covered by white aerial mycelia, fluffy; reverse concolourous. Colonies on PDA, 59–68 mm diam after 7 d, margin regular, aerial mycelia floccose, dense, white to yellowish grey; reverse dark brown with pale olivaceous margin. NaOH test negative.
Specimens examined: Italy, Hydrangea macrophylla, 2013, W.J. Duan (holotype HMAS 247155, dried culture, ex-holotype living culture CGMCC 3.18357 = LC8131); ibid. LC 8132.
Note: Didymella macrophylla is phylogenetically most closely related to D. aquatica (Fig. 1), which is discussed under the notes of the latter species.
Didymella ocimicola Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB819127. Fig. 15.
Fig. 15.
Didymella ocimicola (CGMCC 3.18358). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Section of pycnidium. I. Conidia. Scale bars: G = 100 μm; H = 50 μm; I = 10 μm.
Etymology: Name derived from Ocimum, the plant host from which the holotype was collected.
Leaf spots amphigenous, irregular, 8–15 mm diam, next to or close to the leaf margin, pale brown (Fig. 7C). Conidiomata pycnidial, solitary, sometimes aggregated, globose to flask-shaped, brownish olivaceous, covered by some hyphal outgrowths, superficial or semi-immersed, ostiolate, 100–235 × 95–180 μm. Ostiole single, with an elongated neck, slightly papillate or non-papillate. Pycnidial wall pseudoparenchymatous, composed of isodiametric cells, 2–5 layers, 12–35.5 μm thick, brown-pigmented. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 5–5.5 × 3.5–5 μm. Conidia globose to broadly ellipsoidal, smooth- and thin-walled, hyaline, aseptate, 4–6.5 × 3–4.5 μm, with one to several distinct guttules. Conidial exudates not recorded.
Culture characteristics: Colonies on OA, 10–15 mm diam after 7 d, margin regular, aerial mycelia floccose, flat, white to buff; reverse concolourous. Colonies on MEA 9–12 mm diam after 7 d, margin irregular, aerial mycelia floccose, white, dull green; reverse white to dull green. Colonies on PDA, 10–15 mm diam after 7 d, margin regular, aerial mycelia floccose, white; reverse olivaceous with white to pale brown patches. NaOH test negative.
Specimens examined: China, Tibet, Lulang, on leaves of Ocimum sp., 15 Jun. 2015, Q. Chen (holotype HMAS 247156, dried culture, ex-holotype living culture CGMCC 3.18358 = LC 8137); ibid. LC 8138.
Notes: Didymella ocimicola grouped closely with D. chenopodii and D. senecionicola (Fig. 1), but differs from D. chenopodii in smaller conidiogenous cells (5–5.5 × 3.5–5 μm vs. 4–8 × 4–6 μm) and wider conidia (4–6.5 × 3–4.5 μm vs. 5–5.5 × 2–2.2 μm) and from D. senecionicola in wider conidia (4–6.5 × 3–4.5 μm vs. 4–6.5 × 1.5–2.5 μm) (de Gruyter et al. 1993). Didymella ocimicola has 44 bp and 30 bp differences in three loci (lack of rpb2 sequence) from D. chenopodii and D. senecionicola respectively.
Didymella pteridis L.W. Hou, Crous & L. Cai, sp. nov. MycoBank MB820002. Fig. 16.
Fig. 16.
Didymella pteridis (CBS 379.96). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Pycnidium. I. Section of pycnidium. J. Section of pycnidial wall. K. Conidiogenous cells. L. Conidia. Scale bars: G = 300 μm; H = 40 μm; I = 20 μm; J–L = 10 μm.
Etymology: Named after the host genus Pteris, from which the holotype was collected.
Conidiomata pycnidial, mainly solitary, sometimes aggregated, (sub-)globose or flask-shaped, glabrous or with some mycelial outgrowths, superficial or semi-immersed, ostiolate, 170–350(–430) × 150–330 μm. Ostiole single, papillate, sometimes elongated as a short neck, with dark colour near the ostioles. Pycnidial wall pseduoparenchymatous, composed of oblong to isodiametric cells, 3–6 layers, 9–28 μm thick, with outer 1–2-layers pigmented. Conidiogenous cell phialidic, hyaline, smooth, ampulliform to doliiform, 4–5 × 3.5–4.5 μm. Conidia ovoid to broadly oval, smooth- and thin-walled, hyaline, aseptate, (3–)4–6 × 2.5–3.5 μm, with two polar guttules. Conidial matrix pale salmon.
Culture characteristics: Colonies on OA, 58–60 mm diam after 7 d, margin regular, aerial mycelia flat, cinnamon to hazel, mycelia sparse in some furrowed zone, pycnidia abundant near the margin; reverse buff to pale olivaceous. Colonies on MEA 20–25 mm diam after 7 d, margin regular, aerial mycelia floccose, white, grey near the centre, pale salmon conidial matrix appeared near the centre; reverse yellow in outer ring, changing towards the centre from saffron, hazel, greyish brown to brown. Colonies on PDA, 65–68 mm diam after 7 d, margin regular, densely covered by floccose aerial mycelia, greenish brown, with some white mycelial pellets scattering over the colony; reverse dark brown, buff near the margin. NaOH spot test: a pale reddish brown discolouration on MEA.
Specimen examined: The Netherlands, Wageningen, Alphen aan de Rijn, from a leaf of Pteris sp., deposited in CBS Apr. 1996 (holotype CBS H-23013, dried culture, ex-holotype living culture CBS 379.96).
Notes: CBS 379.96 was originally identified as “Didymella adianticola”, which is currently a synonym of Paraboeremia adianticola. CBS 379.96 is well distinguished from Pa. adianticola both in morphology and phylogeny. Didymella pteridis produces pycnidia with a single ostiole and shorter conidiogenous cells (4–5 × 3.5–4.5 μm), different from Pa. adianticola (pycnidia with 1–3 ostioles, conidiogenous cells 5.5–7 × 3–6.5 μm; Chen et al. 2015a). Thus, we introduce CBS 379.96 as a new species, D. pteridis. Didymella pteridis is closely related to D. viburnicola in the multi-locus phylogenetic analyses (Fig. 1), but D. pteridis is differentiated from the latter by wider conidia (3–6 × 2.5–3.5 μm vs. 3.6–5.6 × 1.6–2.2 μm) and the colour of its conidial matrix (pale salmon vs. whitish) (de Gruyter & Noordeloos 1992).
Didymella segeticola (Q. Chen) Q. Chen, Crous & L. Cai, comb. nov. MycoBank MB819327.
Basionym: Phoma segeticola Q. Chen, Phytotaxa 197: 274. 2015.
Description: Chen et al. (2015b).
Specimens examined: China, Hubei, Shennongjia Forest Region, on diseased leaves of Cirsium segetum, 1 Aug. 2011, K. Zhang (holotype HMAS 245746, ex-holotype living culture CGMCC 3.17489); ibid. CGMCC 3.17498 = LC 1635; ibid. LC 1633; ibid. LC 1634.
Notes: This species was introduced as Phoma segeticola, before the comprehensive revision of Didymellaceae (Chen et al. 2015b). Under current circumstance of Didymellaceae, it belongs to Didymella. Didymella segeticola is closely related to D. bellidis, and has 12 bp differences in four loci from the latter. Morphologically, D. segeticola could be distinguished from the latter in producing wider conidia (4.5–7 × 2.5–4 μm vs. 4–6.5 × 2–2.5 μm; Chen et al. 2015b).
Didymella sinensis Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818967. Fig. 17.
Fig. 17.
Didymella sinensis (CGMCC 3.18348). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Section of pseudothecial wall. H. Asci forming in ascomata. I–J. Asci. K–N. Ascospores. Scale bar: G–H = 10 μm; I–K, M = 5 μm; L, N = 2.5 μm.
Etymology: Epithet derived from the country of origin, China.
Leaf spots amphigenous, angular to irregular, 3–5 mm diam, scatter over the leaf, dark brown to black (Fig. 7D). Ascomata aggregated, globose to irregular, brown, small, up to 170 μm diam, papillate. Pseudothecial wall 18–29.5 μm thick, outer wall consisting of 2–5 layers of cells of textura angularis. Pseudoparaphyses hyaline, 1.5–2 μm diam, septate. Asci bitunicate, clavate to short cylindrical, 32–52 × 8.5–16 μm. Ascospores biseriate, ellipsoidal, straight to slightly curved, 12–18 × 4.5–7.5 μm, hyaline, smooth, apex obtuse, base broadly obtuse to subobtuse, medianly 1-septate, upper cell often wider than lower cell, slightly constricted at the septum.
Culture characteristics: Colonies on OA, 49–52 mm diam after 7 d, margin regular, covered by floccose aerial mycelia, grey to black; reverse black. Colonies on MEA 57–60 mm diam after 7 d, margin regular, greyish brown; reverse concolourous. Colonies on PDA, 56–60 mm diam after 7 d, margin regular, pale grey, with brownish olivaceous margin; reverse dark brown. NaOH spot test: a hazel discolouration on MEA.
Specimens examined: China, Guizhou, Huangguoshu waterfall, on leaves of Cerasus pseudocerasus, 21 Jul. 2014, Q. Chen (holotype HMAS 247157, dried culture, ex-holotype living culture CGMCC 3.18348 = LC 5210); Guizhou, Kuankuoshui National Geopark, Urticaceae, 20 Jul. 2014, Q. Chen, LC 5246; Guizhou, Xingyi, on leaves of Dendrobium officinale, 4 Jul. 2015, Q. Chen, LC 8142; ibid. LC 8143.
Notes: Didymella sinensis has only been observed as a sexual morph, which is not common among species of Didymellaceae. Four isolates from diseased leaves of three host plants in different families were collected, i.e. Cerasus pseudocerasus (Rosaceae), Dendrobium officinale (Orchidaceae) and Urticaceae, indicating an opportunistic pathogen with very broad host range.
Didymella suiyangensis Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818972. Fig. 18.
Fig. 18.
Didymella suiyangensis (CGMCC 3.18352). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Pycnidia. I. Section of pycnidial wall. J. Conidia. Scale bars: G = 300 μm; H = 30 μm; I–J = 5 μm.
Etymology: Epithet derived from the location of origin, Suiyang County in Guizhou, China.
Conidiomata pycnidial, solitary, sometimes aggregated, globose to irregular, brown, covered by some hyphal outgrowths, superficial or semi-immersed, ostiolate, (65–)90–240 × 55–180 μm. Ostiole single, slightly papillate or non-papillate. Pycnidial wall pseudoparenchymatous, composed of oblong to isodiametric cells, 2–4 layers, 15–36.5 μm thick, outer wall 2-layers pigmented. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 4–4.5 × 3–4 μm. Conidia ellipsoidal to oblong, smooth- and thin-walled, hyaline, aseptate, 3.5–7 × 2–3 μm, with indistinct guttules. Conidial matrix cream.
Culture characteristics: Colonies on OA, 52–55 mm diam after 7 d, margin regular, covered by floccose aerial mycelia, sparsely, white to buff; reverse concolourous. Colonies on MEA 59–64 mm diam after 7 d, margin regular, floccose, pale grey to greenish olivaceous; reverse white to yellowish green. Colonies on PDA, 57–61 mm diam after 7 d, margin regular, floccose, white to pale greyish brown; reverse white to hazel. NaOH spot test: a reddish brown discolouration on MEA.
Specimens examined: China, Guizhou, Zunyi, Shuanghe Cave National Geopark, air, 8 May 2015, Z.F. Zhang (holotype HMAS 247158, dried culture, ex-holotype living culture CGMCC 3.18352 = LC 7439); ibid. LC 8144.
Notes: Didymella suiyangensis formed a distinct clade sister to D. bellidis and D. segeticola (Fig. 1), with respectively 18 bp and 19 bp differences in four loci from the latter two species. However, D. suiyangensis is differentiated from D. bellidis and D. segeticola in producing narrower conidiogenous cells (4–4.5 × 3–4 μm vs. 3–6 × 4–8 μm and 5–6.5 × 4–5.5 μm), and the number of ostioles (1 vs. 1–5 and 1–2, respectively). Moreover, the NaOH reactions on MEA showed a reddish brown discolouration on D. suiyangensis, but green to red on D. bellidis and negative on D. segeticola (de Gruyter et al. 1993, Chen et al. 2015b).
Epicoccum Link, Mag. Neuesten Entdeck. Gesammten Naturk. Ges. Naturf. Freunde Berlin 7: 32. 1815, emend. Q. Chen & L. Cai, Stud. Mycol. 82: 171. 2015.
Epicoccum camelliae Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818958.
Etymology: Name refers to the host genus from which the holotype was collected, Camellia.
Cultures sterile. Epicoccum camelliae differs from its closest phylogenetic neighbour E. viticis by unique fixed alleles in three loci based on alignments of the separate loci deposited in TreeBASE (S20724): LSU positions: 66(T), 398(T); tub2 positions: 30(T), 258(C); rpb2 positions: 47(C), 95(C), 197(A), 419(C), 554(A).
Specimens examined: China, Jiangxi, Ganzhou, leaves of Camellia sinensis, 7 Sep. 2013, Y. Zhang (holotype HMAS 247159, dried culture, culture ex-holotype CGMCC 3.18343 = LC 4858); ibid. LC4862.
Notes: Epicoccum camelliae is closely related to E. viticis with a high support value in the phylogenetic tree (Fig. 1), and has 10 bp differences in four loci from the latter. Two isolates of this species are both from Camellia sinensis, one as endophyte in healthy leaves and the other as pathogenic fungus from diseased leaves. Both isolates proved to be sterile on the defined media used in this study.
Epicoccum dendrobii Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818964. Fig. 19.
Fig. 19.
Epicoccum dendrobii (CGMCC 3.18359). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Sporodochia. H–I. Conidia. Scale bars: G–I = 10 μm.
Etymology: Named after the host plant, Dendrobium.
Leaf spots amphigenous, subcircular, up to 10 mm diam, black (Fig. 7E). Conidiomata sporodochial, aggregated, semi-immersed or superficial, clavate, pale brown. Hyphae septate, frequently branched, 2.5–4.5 μm. Conidia globose, aseptate and smooth when young, later becoming multicellular-phragmosporous, verrucose, subglobose-pyriform, brown, with a basal cell, 11–19 μm diam.
Culture characteristics: Colonies on OA, 58–64 mm diam after 7 d, margin regular, covered by floccose aerial mycelia, dense, white to greyish yellow; reverse white to pale grey, with some purple dots scattered over the colony. Colonies on MEA 65–68 mm diam after 7 d, margin regular, grey, with sparse white aerial mycelia; reverse white to yellow. Colonies on PDA, 34–38 mm diam after 7 d, margin regular, aerial mycelia felty to floccose, flat, white to buff, olivaceous near the centre; reverse pale salmon, hazel to brown near the centre. NaOH test negative.
Specimens examined: China, Guizhou, Xingyi, on leaves of Dendrobium fimbriatum, 4 Jul. 2015, Q. Chen (holotype HMAS 247160, dried culture, ex-holotype living culture CGMCC 3.18359 = LC 8145); ibid. LC 8146.
Notes: Epicoccum dendrobii formed a distinct clade basal to E. nigrum, E. poae and E. layuense (Fig. 1). These species all produce typical epicoccoid conidia (multicellular-phragmosporous, verrucose), with phoma-like conidia only observed in E. nigrum. Epicoccum dendrobii differs in the length of its epicoccoid conidia (11–19 μm) from E. nigrum (15–35 μm; Punithalingam et al. 1972) and E. poae (10–23 μm), and in its NaOH reaction (negative) from E. layuense (a pale reddish brown discolouration on MEA, with a yellowish brown margin).
Epicoccum duchesneae Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818966. Fig. 20.
Fig. 20.
Epicoccum duchesneae (CGMCC 3.18345). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G–H. Pycnidia. I. Section of pycnidium. J. Section of pycnidial wall. K–M. Conidiogenous cells. N. Conidia. Scale bars: G–I = 40 μm; J, M–N = 10 μm; K–L = 5 μm.
Etymology: Name derived from Duchesnea, the plant genus from which the holotype was collected.
Leaf spots amphigenous, circular to irregular, 2–5 mm diam, yellowish brown, surrounded by a purple border (Fig. 7F). Conidiomata pycnidial, solitary, globose to subglobose, covered with hyphal outgrowths, immersed in agar, ostiolate, (150–)170–270 × (100–)150–230 μm. Ostiole single, sometimes with an elongated, pale brown neck, slightly papillate. Pycnidial wall pseudoparenchymatous, composed of isodiametric cells, 3–5 layers, 13–30 μm thick, outer wall of 2–3 pigmented layers. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 4.5–9.5 × 3.5–7 μm. Conidia ellipsoidal to oblong, smooth- and thin-walled, hyaline, aseptate, 2.5–3.5 × 1.5–2 μm, egutullate or sometimes with 1(–3) small guttules. Conidial matrix whitish to salmon.
Culture characteristics: Colonies on OA, 62–65 mm diam after 7 d, margin regular, covered by floccose aerial mycelia, white to grey, greyish brown near the centre; reverse white to dark brown. Colonies on MEA 24–27 mm diam after 7 d, margin regular, covered by white, sparse floccose aerial mycelia, grey to pale olivaceous; reverse concolourous. Colonies on PDA, 55–60 mm diam after 7 d, margin regular, aerial mycelia covering the whole colony, floccose, white to grey; reverse greenish olivaceous to dark brown. Application of NaOH results in a pale olivaceous discolouration of the agar.
Specimens examined: China, Jiangxi, Ganzhou, on leaves of Duchesnea indica, 12 May 2013, Q. Chen (holotype HMAS 247161, dried culture, ex-holotype living culture CGMCC 3.18345 = LC 5139); ibid. LC 8147.
Notes: Epicoccum duchesneae formed a distinct lineage close to E. huancayense (Fig. 1). Epicoccum duchesneae differs in producing smaller conidia from E. huancayense, 2.5–3.5 × 1.5–2 μm vs. (4–)5–8(–12) × 2.5–4.5 μm (de Gruyter et al. 1998).
Epicoccum hordei Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818961. Fig. 21.
Fig. 21.
Epicoccum hordei (CGMCC 3.18360). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Pycnidia. I. Section of pycnidium. J. Section of pycnidial wall. K. Chlamydospores. L. Conidiogenous cells. M. Conidia. Scale bars: G = 300 μm; H = 30 μm; I, K = 20 μm; J, M = 10 μm; L = 5 μm.
Etymology: Named after the host genus Hordeum, from which the holotype was isolated.
Conidiomata pycnidial, solitary or aggregated, globose to subglobose, glabrous, semi-immersed or on the surface of agar, ostiolate, (85–)115–190(–260) × (70–)95–180 μm. Ostiole single, non-papillate. Pycnidial wall pseudoparenchymatous, composed of oblong to isodiametric cells, 2–3 layers, 11–18.5 μm thick, pigmented. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 7–8.5 × 5.5–7.5 μm. Conidia obovoid, ellipsoidal to oblong, cylindrical, smooth- and thin-walled, hyaline, aseptate, 6.5–9 × 3–4 μm, with several minute guttules. Conidial matrix pale brown. Chlamydospores unicellular, produced on the agar, yellowish brown to dark brown, intercalary, in chains, globose to subglobose, 6–21.5 μm diam, thick-walled.
Culture characteristics: Colonies on OA, 58–62 mm diam after 7 d, margin regular, covered by floccose aerial mycelia, white to grey, pale olivaceous near the centre; reverse white to amber. Colonies on MEA 43–46 mm diam after 7 d, margin regular, aerial mycelia white, fluffy to floccose, grey to greenish yellow; reverse concolourous. Colonies on PDA, 54–56 mm diam after 7 d, margin regular, aerial mycelia floccose, white to grey, with pale olivaceous concentric rings; reverse pale greenish brown to olivaceous, with concentric rings. Application of NaOH results in a pale brown discolouration of the agar.
Specimens examined: Australia, on seeds of Hordeum vulgare, 2014, W.J. Duan (holotype HMAS 247162, dried culture, ex-holotype living culture CGMCC 3.18360 = LC 8148); ibid. LC 8149.
Notes: Isolates of this species clustered in a lineage closely related to E. pimprinum (49 bp differences in four sequenced loci) (Fig. 1). Morphologically, E. hordei differs in the colour of its conidial matrix (pale brown) from E. pimprinum (salmon) and the absence of elongated necks of pycnidia (with pronounced necks in E. pimprinum) (Boerema 1993).
Epicoccum italicum Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818965. Fig. 22.
Fig. 22.
Epicoccum italicum (CGMCC 3.18361). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G–H. Sporodochia. I. Conidia. Scale bars: G–I = 10 μm.
Etymology: Named after the country where the holotype was collected, Italy.
Conidiomata sporodochial, aggregated, semi-immersed or superficial, clavate, yellowish brown. Hyphae septate, branched, 3.5–5 μm. Conidia multicellular-phragmosporous, verrucose, subglobose-pyriform, brown, with a basal cell, 12.5–28 μm diam.
Culture characteristics: Colonies on OA, 48–50 mm diam after 7 d, margin regular, covered by floccose aerial mycelia, dense, white to yellow, dark iron-grey near the centre, with a pale yellow halo near the margin, and a yellow concentre ring; reverse buff to yellowish brown. Colonies on MEA 50–55 mm diam after 7 d, margin regular, grey to pale yellowish green, with sparse white aerial mycelia; reverse concolourous. Colonies on PDA, 35–39 mm diam after 7 d, margin irregular, aerial mycelia floccose, yellow with a white margin, black near the centre; reverse salmon to saffron, with a yellow margin. Application of NaOH results in a yellow discolouration of the agar.
Specimens examined: Italy, on seedlings of Acca sellowiana, 2013, W.J. Duan (holotype HMAS 247163, dried culture, ex-holotype living culture CGMCC 3.18361 = LC 8150); ibid. LC 8151.
Notes: Phylogenetically, Epicoccum italicum formed a distinct lineage closely related to E. dendrobii. Morphologically, the two species could be distinguished in the length of epicoccoid conidia (12.5–28 μm in E. italicum vs. 11–19 μm in E. dendrobii), and the results of NaOH test (a yellow discolouration in E. italicum vs. negative in E. dendrobii).
Epicoccum latusicollum Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818960. Fig. 23.
Fig. 23.
Epicoccum latusicollum (CGMCC 3.18346). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Pycnidia. I. Section of pycnidium. J. Section of pycnidial wall. K–L. Conidiogenous cells. M. Conidia. Scale bars: G = 100 μm; H = 20 μm; I–J = 10 μm; K–M = 5 μm.
Etymology: Name refers to the wide neck of pycnidia, latus = wide, collum = neck.
Conidiomata pycnidial, mostly solitary, sometime aggregated, globose to subglobose or pyriform, glabrous, produced on the agar surface, ostiolate, 110–155 × 90–130 μm. Ostioles 1–2, sometimes elongated as a short, slightly papillate neck. Pycnidial wall pseudoparenchymatous, composed of oblong to isodiametric cells, 3–4 cell layers of which outer 2–3 are brown pigmented, 15–20 μm thick. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 5–8 × 4–5.5 μm. Conidia ellipsoidal to oblong, smooth- and thin-walled, hyaline, aseptate, 4–6.5 × 2–3 μm, guttulate. Conidial matrix buff.
Culture characteristics: Colonies on OA, 70–72 mm diam after 7 d, margin regular, flattened, whole colony covered by floccose aerial mycelia, white, grey to smoke grey near the centre; reverse white to buff. Colonies on MEA 75–80 mm diam after 7 d, margin regular, aerial mycelia floccose, greyish dull green, forming several mycelial pellets, white or pale salmon; reverse grey, with some yellow sections. Colonies on PDA, 80–85 mm diam after 7 d, margin regular, floccose aerial mycelia covering the whole colony, dense, white to grey, forming several white mycelial pellets; reverse white to hazel. NaOH spot test: a green discolouration on MEA, later changing to three colour layers, via dark green, pale red to purple, from the centre to the outer ring.
Specimens examined: China, Jiangxi, Ganzhou, on leaves of Vitex negundo, 25 Apr. 2013, Q. Chen, LC 5124; Jiangxi, Ganzhou, endophyte of Camellia sinensis, 7 Sep. 2013, Y. Zhang, LC 4859; Shandong, Jining, on leaves of Sorghum bicolor, 3 Aug. 2013, N. Zhou (holotype HMAS 247164, dried culture, ex-holotype living culture CGMCC 3.18346 = LC 5158). Japan, Podocarpus macrophyllus, 2013, W.J. Duan, LC 8152; ibid. LC 8153; on stem of Acer palmatum, LC 8154.
Notes: Isolates of Epicoccum latusicollum clustered in a sister clade to E. camelliae, E. sorghinum and E. viticis (Fig. 1). Although the conidial dimensions are similar in these species, E. latusicollum differs in 1 bp in ITS, 14 bp in rpb2 and 5 bp in tub2 from E. camelliae; 16 bp in rpb2 and 7 bp in tub2 from E. sorghinum; and 1 bp in ITS, 1 bp in LSU, 16 bp in rpb2 and 4 bp in tub2 from E. viticis.
This is the first report of an Epicoccum species from Acer palmatum (Aceraceae), Podocarpus macrophyllus (Podocarpaceae) and Vitex negundo (Verbenaceae).
Epicoccum layuense Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818963. Fig. 24.
Fig. 24.
Epicoccum layuense (CGMCC 3.18362). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Sporodochia. H–I. Conidia producing on sporodochia. J. Conidia. Scale bars: G–I = 10 μm.
Etymology: Epithet derived from the location of origin, Layue Village in Tibet, China.
Leaf spots distinct, angular to irregular, up to 12 mm diam, dark brown. Conidiomata sporodochial, aggregated, superficial, clavate, brown (Fig. 7I). Hyphae septate, branched, 2–5.5 μm. Conidia multicellular-phragmosporous, verrucose, subglobose-pyriform, with a basal cell, dark brown, 13–19.5 μm diam.
Culture characteristics: Colonies on OA, 27–37 mm diam after 7 d, margin regular, covered by floccose aerial mycelia, yellow to greenish yellow, olivaceous; reverse yellow to saffron, with dark brown sections. Colonies on MEA 30–43 mm diam after 7 d, margin irregular, aerial mycelia white, floccose, greenish yellow to pale brown; reverse concolourous. Colonies on PDA, 47–55 mm diam after 7 d, margin irregular, aerial mycelia floccose, bright yellow; reverse yellow to pale brown, with a brown concentric ring near the centre. NaOH spot test: a pale reddish brown discolouration on MEA, with a yellowish brown margin.
Specimens examined: China, Tibet, Lulang, on leaves of Perilla sp., 15 Jun. 2015, Q. Chen (holotype HMAS 247165, dried culture, ex-holotype living culture CGMCC 3.18362 = LC 8155); ibid. LC 8156.
Notes: This species is phylogenetically closely related to E. nigrum and E. poae, but E. layuense has differences at 19 positions from E. nigrum and 14 positions from E. poae in the multi-locus sequences of their ex-type strains. Morphologically, E. layuense produces smaller epicoccoid conidia than E. nigrum (13–19.5 μm vs. 15–35 μm; Punithalingam et al. 1972), and they also differ in their NaOH reactions (a pale reddish brown discolouration on E. layuense, but pale brown on E. poae).
Epicoccum poae Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818962. Fig. 25.
Fig. 25.
Epicoccum poae (CGMCC 3.18363). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Conidia producing on OA. H–I. Conidia. Scale bars: G = 100 μm; H = 20 μm; I = 10 μm.
Etymology: Name derived from Poa, the plant genus from which the holotype was collected.
Conidiomata sporodochial, aggregated, superficial, clavate, brown. Hyphae septate, branched, 2–3 μm. Conidia multicellular-phragmosporous, verrucose, subglobose-pyriform, with a basal cell, dark brown, 10–23 μm diam.
Culture characteristics: Colonies on OA, 49–51 mm diam after 7 d, margin regular, covered by floccose aerial mycelia, yellow, reddish brown to brown near the centre, with a white margin; reverse yellow to saffron, with some brown sections. Colonies on MEA 25–27 mm diam after 7 d, margin irregular, aerial mycelia white to greenish yellow, fluffy to floccose, grey to greenish yellow; reverse white to greenish yellow. Colonies on PDA, 20–22 mm diam after 7 d, margin irregular, aerial mycelia flattened, brownish yellow, with a white margin; reverse yellow to saffron, brown towards the centre. NaOH spot test: a pale brown discolouration on MEA.
Specimens examined: USA, on seeds of Poa annua, Oct. 2014, X.M. Wang, strain isolated by Q. Chen (holotype HMAS 247166, dried culture, ex-holotype living culture CGMCC 3.18363 = LC 8160); ibid. LC 8161, LC 8162.
Notes: Epicoccum poae is phylogenetically closely related to E. nigrum (Fig. 1), but differs in producing smaller epicoccoid conidia (10–23 μm vs. 15–35 μm; Punithalingam et al. 1972). Furthermore, E. poae hasn't been observed to have phoma-like conidia, while E. nigrum readily produces short-cylindrical conidia, 3–7(–10) × 1.5–3(–3.5) μm (Punithalingam et al. 1972).
Epicoccum viticis Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818959. Fig. 26.
Fig. 26.
Epicoccum viticis (CGMCC 3.18344). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Pycnidia. I. Section of pycnidium. J. Section of pycnidial wall. K–N. Conidiogenous cells. O. Conidia. Scale bars: G–H = 40 μm; I = 20 μm; J = 10 μm; K–O = 5 μm.
Etymology: Name derived from Vitex, the plant genus from which the holotype was collected.
Leaf spots amphigenous, circular to irregular, 2–8 mm diam, close to the leaf margin, reddish brown, single lesions may coalesce to form larger lesions and become dark brown (Fig. 7H). Conidiomata pycnidial, aggregated or sometimes solitary, (sub-)globose, glabrous, produced on the agar surface, 120–200 × 100–175 μm. Ostioles 1–2, papillate. Pycnidial wall pseudoparenchymatous, composed of oblong to isodiametric cells, 2–3 cell layers, outer 1–2 layers brown pigmented, 8–16 μm thick. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 5.5–9 × 3–6 μm. Conidia ellipsoidal to obovoid, smooth- and thin-walled, hyaline, aseptate, 3.5–6 × 2–3 μm, with two minute polar guttules. Conidial matrix buff to cinnamon.
Culture characteristics: Colonies on OA, 48–67 mm diam after 7 d, margin regular, aerial mycelia floccose, white to grey, with a greyish olivaceous concentric ring; reverse white to pale olivaceous, with a broad greyish olivaceous concentric ring. Colonies on MEA 75–80 mm diam after 7 d, margin regular, aerial mycelia fluffy to floccose, grey to pale yellowish green; reverse concolourous. Colonies on PDA, 70–75 mm diam after 7 d, margin regular, floccose aerial mycelia covering the whole colony, grey; reverse white to buff, with some dull green dots. NaOH test negative.
Specimens examined: Australia, Darwin, Northern Territory University, Greenhouse, from Andropogon gayanus, 2002, A. Hollingsworth, BRIP 29294 = LC 5257. China, Jiangxi, Ganzhou, on leaves of Vitex negundo, 25 Apr. 2013, Q. Chen (holotype HMAS 247167, dried culture, ex-holotype living culture CGMCC 3.18344 = LC 5126).
Note: Epicoccum viticis is phylogenetically closely related to E. camelliae (Fig. 1), with 10 bp differences in four sequenced loci.
Heterophoma Q. Chen & L. Cai, Stud. Mycol. 82: 165. 2015.
Heterophoma verbascicola Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB819128. Fig. 27.
Fig. 27.
Heterophoma verbascicola (CGMCC 3.18364). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidium forming on OA. H. Pycnidium. I. Section of pycnidium. J. Section of pycnidial wall. K–M. Conidiogenous cells. N. Conidia. Scale bars: G–H = 40 μm; I = 20 μm; J–N = 5 μm.
Etymology: Named after the host genus from which the holotype was collected, Verbascum.
Leaf spots amphigenous, angular to irregular, 2–7 mm diam, scattered over the leaf, brown, with a pale yellow diffuse halo (Fig. 7J). Conidiomata pycnidial, aggregated or solitary, globose to subglobose or obpyriform, brown, covered with some hyphal outgrowths, semi-immersed or superficial, ostiolate, 120–300 × (100–)150–300 μm. Ostioles 2–3, elongated as short necks, slightly papillate or non-papillate. Pycnidial wall pseudoparenchymatous, composed of oblong to isodiametric cells, 2–3 layers, 7–20 μm thick, outer wall 1–2-layers pigmented. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 5.5–6 × 3.5–5 μm. Conidia ellipsoidal to oblong, smooth- and thin-walled, hyaline, aseptate, incidentally produce 1-septate large conidia, 3.5–6(–8) × 1.5–3.5 μm, with 1–2 guttules. Conidial matrix cream to buff.
Culture characteristics: Colonies on OA, 40–45 mm diam after 7 d, margin regular, covered by floccose aerial mycelia, white, with grey margin, greyish olivaceous near the centre; reverse white to pale olivaceous with a broad white concentric ring. Colonies on MEA 50–52 mm diam after 7 d, margin regular, aerial mycelia fluffy to floccose, white; reverse concolourous. Colonies on PDA, 50–53 mm diam after 7 d, margin irregular, crenate, dense, felty, white to mouse-grey; reverse white to hazel, with brown concentric rings. NaOH test negative.
Specimens examined: China, Tibet, Lulang, on leaves of Verbascum thapsus, 15 Jun. 2015, Q. Chen (holotype HMAS 247168, dried culture, ex-holotype living culture CGMCC 3.18364 = LC 8163); ibid. LC 8164.
Notes: Heterophoma verbascicola is phylogenetically closely related to H. novae-verbascicola, but is distinguishable from the latter species in its slightly narrower conidiogenous cells (5.5–6 × 3.5–5 μm vs. 2–6 × 4–6 μm; de Gruyter et al. 1993) and larger conidia (3.5–8 × 1.5–3.5 μm vs. 3.5–5.5 × 1.5–2.5 μm). Moreover, the NaOH test showed a yellowish green discolouration that became reddish in H. novae-verbascicola, but remained negative in H. verbascicola.
Neoascochyta Q. Chen & L. Cai, Stud. Mycol. 82: 198. 2015.
Neoascochyta argentina L.W. Hou, Crous & L. Cai, sp. nov. MycoBank MB820003. Fig. 28.
Fig. 28.
Neoascochyta argentina (CBS 112524). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Pycnidium. I. Section of pycnidium. J. Section of pycnidial wall. K–M. Conidiogenous cells. N. Conidia. Scale bars: G = 100 μm; H = 50 μm; I = 20 μm; J–N = 10 μm.
Etymology: Epithet derived from the country of origin, Argentina.
Conidiomata pycnidial, solitary or aggregated, (sub-)globose of flask-shaped, glabrous, semi-immersed or immersed, ostiolate, 210–390 × 140–270 μm. Ostioles 1–3, sometimes elongated as a long neck (up to 350 μm), papillate. Pycnidial wall pseduoparenchymatous, composed of oblong to isodiametric cells, 4–6 layers, with outer 3–4-layers pigmented, 14.5–52 μm thick. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 7.5–14.5 × 6–13.5 μm. Conidia cylindrical, smooth- and thin-walled, hyaline, 0–1-septate, (10.5–)11.5–14.5(–16) × 3–5 μm, guttulate. Conidial matrix whitish cream.
Culture characteristics: Colonies on OA, 50–55 mm diam after 7 d, margin regular, densely covered by floccose aerial mycelia, greyish olivaceous, with some white zones near the margin; reverse greyish black. Colonies on MEA 55–60 mm diam after 7 d, margin regular, densely covered by woolly aerial mycelia, fawn, white near margin; reverse brown. Colonies on PDA, 60–65 mm diam after 7 d, margin regular, covered by pale grey aerial mycelia, floccose, dark olivaceous near the margin; reverse greyish brown. NaOH spot test: a pale reddish brown discolouration on MEA.
Specimen examined: Argentina, Tandil, from a leaf of Triticum aestivum, Oct. 2002 (holotype CBS H-23014, dried culture, ex-holotype living culture CBS 112524).
Notes: CBS 112524 was initially received as “Ascochyta hordei”. However, this isolate clustered in the Neoascochyta clade, and produces much smaller conidia (10.5–16 × 3–5 μm) than As. hordei (15–22 × 3.5–4.5 μm; Punithalingam 1979). Therefore, Neoa. argentina is introduced as a new species, based on isolate CBS 112524. Neoascochyta argentina is well distinguished from its most closely related species Neoa. triticicola by its smaller conidia (10.5–16 × 3–5 μm vs. 16.5–27 × 5–8.5 μm).
Neoascochyta triticicola L.W. Hou, Crous & L. Cai, sp. nov. MycoBank MB820004. Fig. 29.
Fig. 29.
Neoascochyta triticicola (CBS 544.74). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Pycnidia. I–J. Section of pycnidia. K. Section of pycnidial wall. L–N. Conidiogenous cells. O–P. Conidia. Scale bars: G = 500 μm; H = 200 μm; I–J = 50 μm; K–L, O = 10 μm; M–N, P = 5 μm.
Etymology: Name refers to the host genus Triticum, from which the holotype was collected.
Conidiomata pycnidial, mostly confluent, flask-shaped, glabrous or sometimes with hyphal outgrows, superficial or semi-immersed on the agar, (170–)230–420(–620) × 160–430 μm; conidiomata becoming black, irregular with age, and ostiolate. Ostioles 1–3(–5), developing to conspicuously elongated necks (up to 400 μm tall), papillate. Pycnidial wall pseudoparenchymatous, composed of oblong to isodiametric cells, 4–6 layers, with outer 2–3-layers pigmented, 25–40 μm thick. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 8.5–13 × (4.5–)7.5–12(–13) μm. Conidia bacilliform to fusiform, smooth- and thin-walled, hyaline, mainly uniseptate, occasionally aseptate, (16.5–)20–27 × 5–8.5 μm, guttulate. Conidial matrix whitish cream to pale salmon.
Culture characteristics: Colonies on OA, 55–65 mm diam after 7 d, margin regular, aerial mycelia floccose, greyish black, with some greyish mycelia tufts; reverse concolourous. Colonies on MEA 40–55 mm diam after 7 d, margin irregular, slightly lobate, covered by floccose mycelia, white, greyish olivaceous to greyish pink near the centre; reverse dark brown, saffron near the margin. Colonies on PDA, 55–65 mm diam after 7 d, margin irregular, slightly lobate, covered by floccose, greenish black mycelia, with erected tufts of white mycelia; reverse greyish olivaceous. NaOH spot test: a pale reddish brown discolouration on MEA.
Specimen examined: South Africa, Heilbron, from Triticum aestivum, deposited in CBS Sep. 1974, W.J. Jooste (holotype CBS H-9008, ex-holotype living culture CBS 544.74).
Notes: Isolate CBS 544.74 was originally identified as “Ascochyta hordei” but clustered in the Neoascochyta clade. Morphologically, it differs in producing larger conidia (16.5–27 × 5–8.5 μm) from Ascochyta hordei (15–22 × 3.5–4.5 μm; Punithalingam 1979). Therefore, we introduce CBS 544.74 as a new species, Neoa. triticicola. In Neoascochyta, it should be compared with Neoa. argentina, which is discussed under the notes of the latter species.
Neoascochyta soli Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818975. Fig. 30.
Fig. 30.
Neoascochyta soli (CGMCC 3.18365). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Section of pycnidium. H–I. Conidiogenous cells. J. Conidia. Scale bars: G = 20 μm; H–I = 5 μm; J = 10 μm.
Etymology: Name derived from the substrate where the holotype was collected, soil.
Conidiomata pycnidial, aggregated or solitary, globose to subglobose, dark brown, glabrous, superficial, ostiolate, (135–)390–630 × (110–)340–565 μm. Ostiole single, slightly papillate or non-papillate. Pycnidial wall pseudoparenchymatous, composed of isodiametric cells, 3–5 layers, 18–42 μm thick, outer wall of 1–2-pigmented layers. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 6–10.5 × 5.5–9 μm. Conidia ellipsoidal to oblong, smooth- and thin-walled, hyaline, aseptate, 7–10 × 3–4 μm, with 2 to several polar guttules. Conidial exudates not recorded.
Culture characteristics: Colonies on OA, 62–64 mm diam after 7 d, margin regular, covered by floccose aerial mycelia, dense, white, greyish olivaceous near the centre; reverse white to iron grey. Colonies on MEA 45–47 mm diam after 7 d, margin irregular, grey, white near the centre; reverse white to olivaceous, forming concentric rings. Colonies on PDA, 50–53 mm diam after 7 d, margin regular, aerial mycelia fluffy, white to olivaceous; reverse concolourous. NaOH test negative.
Specimens examined: China, Guizhou, Kuankuoshui National Geopark, soil, 23 Jul. 2014, Z.F. Zhang (holotype HMAS 247169, dried culture, ex-holotype living culture CGMCC 3.18365 = LC 8165); ibid. LC 8166.
Notes: Neoascochyta soli clustered with Neoa. paspali in a distinct clade in this genus, but can be differentiated from the latter in producing larger conidiogenous cells (6–10.5 × 5.5–9 μm vs. 4–6 × 4–6 μm). In addition, the test of metabolite E production was negative for Neoa. soli, while a green to bluish discolouration, becoming red, appeared in Neoa. paspali (de Gruyter et al. 1998).
Neodidymelliopsis Q. Chen & L. Cai, Stud. Mycol. 82: 207. 2015.
Neodidymelliopsis achlydis L.W. Hou, Crous & L. Cai, sp. nov. MycoBank MB820005. Fig. 31.
Fig. 31.
Neodidymelliopsis achlydis (CBS 256.77). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Pycnidium. I. Section of pycnidial wall. J. Section of pycnidium. K, M. Conidiogenous cells. L. Conidia. Scale bars: G = 500 μm; H, J = 50 μm; I, K–M = 10 μm.
Etymology: Named after the host genus Achlys, from which the holotype was collected.
Conidiomata pycnidial, solitary or aggregated, (sub-)globose, glabrous, semi-immersed or superficial, ostiolate, (150–)300–550(–630) × (120–)250–500(–630) μm. Ostioles 1–5, slightly papillate. Pycnidial wall pseudoparenchymatous, composed of oblong to isodiametric cells, 4–9 layers, with outer 2–4-layers pigmented, 30–80 μm thick. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, (4–)6.5–10 × (3.5–)4.5–6.5 μm. Conidia oblong to cylindrical, incidentally slightly curved, smooth- and thin-walled, hyaline, aseptate, 7.5–10(–18) × 2–3.5(–5) μm, with two polar guttules. Conidial matrix whitish cream.
Culture characteristics: Colonies on OA, 45–50 mm diam after 7 d, margin regular, aerial mycelia floccose, white to pale brown; reverse pale salmon, with some pale olivaceous tinges near the centre. Colonies on MEA 40–45 mm diam after 7 d, margin regular, aerial mycelia floccose and compact, white to pale grey; reverse saffron to pale yellowish brown, yellow near margin. Colonies on PDA, 50–52 mm diam after 7 d, margin regular, densely covered by floccose, grey aerial mycelia, white near the margin; reverse pale brown to brown. NaOH spot test: a dull green discolouration with a reddish brown margin on MEA.
Specimen examined: Canada, British Columbia, from a leaf of Achlys triphylla, Jun. 1976, J. Gremmen (holotype CBS H-23015, dried culture, ex-holotype living culture CBS 256.77).
Notes: Isolate CBS 256.77 was received as “Ascochyta achlydis”, which was from the same host (Achlys triphylla) and the same location (Canada) as reported for Ascochyta achlydis (Dearness 1916). However, it produces narrower and aseptate conidia compared to the uniseptate conidia of As. achlydis (7.5–18 × 2–5 μm vs. 14–20 × 5–6.5 μm; Dearness 1916), and is obviously a different species. Phylogenetically, CBS 256.77 clustered in the Neodidymelliopsis clade, closely related to Neod. polemonii and Neod. xanthina (Fig. 1), and has differences at six positions from Neod. polemonii and 12 positions from Neod. xanthina in multi-locus sequences of their ex-type strains. We therefore introduce a new species, Neod. achlydis based on CBS 256.77. Morphologically, Neod. achlydis produces pycnidia with 1–5 ostioles, while Neod. xanthina only has pycnidia with a single ostiole (Boerema et al. 2004). Neodidymelliopsis achlydis differs from Neod. polemonii in its whitish cream conidial matrix from whitish/smoke grey of Neod. polemonii (Boerema et al. 2004). Neodidymelliopsis achlydis is also well distinguished from Neod. polemonii and Neod. xanthina in the NaOH reactions (dull green with pale reddish brown margin in Neod. achlydis, pale sienna to rust colour in Neod. polemonii, reddish brown discolouration in Neod. xanthina; Boerema et al. 2004).
Neodidymelliopsis longicolla L.W. Hou, Crous & L. Cai, sp. nov. MycoBank MB820006. Fig. 32.
Fig. 32.
Neodidymelliopsis longicolla (CBS 382.96). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Pycnidium. I. Section of pycnidium. J. Section of pycnidial wall. K. Conidiogenous cells and conidia. L. Conidia. Scale bars: G = 250 μm; H–I = 50 μm; J = 5 μm; K–L = 10 μm.
Etymology: Name refers to the elongated, long ostiolar necks.
Conidiomata pycnidial, solitary or aggregated, globose to flask-shaped, glabrous or with some hyphal outgrows, superficial or semi-immersed, ostiolate, 200–490 × 150–360 μm. Ostioles 1–3, developing into elongated necks, up to 250 μm tall, papillate. Pycnidial wall pseudoparenchymatous, composed of isodiametric cells, 4–7 layers, outer 3–6-layers pigmented, 20–45 μm thick. Conidiogenous cells phialidic, hyaline, smooth, ampulliform, 4.5–6.5 × 4.5–6 μm. Conidia oblong to cylindrical, smooth- and thin-walled, initially aseptate and hyaline, later becoming 1-septated and pale brown, somewhat constricted at the septum, 12–15(–16.5) × 4–7 μm, guttulate. Conidial matrix brown.
Culture characteristics: Colonies on OA, 45–52 mm diam after 7 d, margin regular, aerial mycelia white and woolly, greenish olivaceous; reverse darker brown. Colonies on MEA 55–57 mm diam after 7 d, margin regular, covered by floccose, white aerial mycelia, black pycnidia visible; reverse brown, saffron near the margin. Colonies on PDA, 55–60 mm diam after 7 d, margin regular, densely covered by floccose aerial mycelia, grey, greenish olivaceous near the margin; reverse dark brown, pale brown near the margin. Application of NaOH results in a pale reddish brown discolouration on MEA.
Specimen examined: Israel, En Avdat, Negev desert, from soil in desert, Feb. 1996, A. van Iperen (holotype CBS H-23016, dried culture, ex-holotype living culture CBS 382.96).
Notes: CBS 382.96 was deposited as “Ascochyta scotinospora”, but differs from As. scotinospora by its larger pycnidia (200–490 × 150–360 μm vs. 140 μm diam) and forming elongated long necks (Punithalingam 1979). Phylogenetically, it clustered in the Neodidymelliopsis clade, basal to Neod. achlydis, Neod. polemonii and Neod. xanthina (Fig. 1). Hence, CBS 382.96 was described as a new species, Neod. longicolla. Neodidymelliopsis longicolla differs from Neod. achlydis in its septate conidia (mainly 1-septated vs. aseptate) and colour of its conidial matrix (brown vs. whitish cream); from Neod. polemonii in producing wider conidia (12–16.5 × 4–7 μm vs. 4.5–7.5 × 1.5–4 μm; Chen et al. 2015a); from Neod. xanthina in the number of pycnidial ostioles (1–3 vs. 1; Boerema et al. 2004).
Phoma Sacc. emend. Q. Chen & L. Cai, Stud. Mycol. 82: 194. 2015.
Phoma herbarum Westend., Bull. Acad. R. Sci. Belg., Cl. Sci. 19(3): 118. 1852, emend. Q. Chen & L. Cai, Stud. Mycol. 82: 195. 2015.
Synonyms: Phoma neerlandica Q. Chen & L. Cai, Stud. Mycol. 82: 197. 2015.
Atradidymella muscivora M.L. Davey & Currah, Amer. J. Bot. 96: 1283. 2009.
Phoma muscivora M.L. Davey & Currah, Amer. J. Bot. 96: 1283. 2009.
Phoma cruris-hominis Punith., Nova Hedwigia 31: 135. 1979.
Specimens examined: Canada, Alberta, Wolf Lake, from gametophytes of Polytrichum juniperinum, 2008, M.L. Davey, UAMH 10909 = CBS 127589. Switzerland, Kt. Graubünden, from Achillea millefolium, deposited in CBS Mar. 1951, E. Müller, CBS 304.51. The Netherlands, Emmeloord, from the stem of Rosa multiflora cv. Cathayensis, deposited in CBS Dec. 1975, G.H. Boerema, CBS 615.75 = PD 73/665 = IMI 199779; Emmeloord, from a leaf of Delphinium sp., deposited in CBS Feb. 1996, culture ex-holotype of “Phoma neerlandica” CBS 134.96 = PD 84/676; Naaldwijk, from a stem base of Nerium sp., deposited in CBS Sep. 1991, J. de Gruyter, CBS 502.91 = PD 82/276. UK, from a leg of woman, Apr. 1977, Y.M. Clayton, CBS 377.92 = IMI 213845; near Dumfries, from die-back of Picea excelsa, deposited in CBS Oct. 1937, T.R. Peace, CBS 274.37.
Notes: Phoma neerlandica was regarded distinct from P. herbarum based on its slightly longer and occasionally uniseptate conidia (Chen et al. 2015a). Similar to many other species in Didymellaceae, such an overlapping morphology often caused confusion with regards to species boundaries. Unfortunately, a sequencing error occurred in the tub2 sequence of CBS 134.96, which was not detected in the subsequent control and processing steps. Phoma neerlandica therefore became a name introduced with ambiguous data, and is therefore reduced to synonymy.
Stagonosporopsis Died. emend. Aveskamp et al., Stud. Mycol. 65: 44. 2010.
Stagonosporopsis bomiensis Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818955. Fig. 33.
Fig. 33.
Stagonosporopsis bomiensis (CGMCC 3.18366). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G–H. Pycnidium forming on OA. I. Pycnidia. J. Section of pycnidium. K–N. Conidiogenous cells. O. Conidia. Scale bars: G–H = 40 μm; I–J = 20 μm; K–O = 5 μm.
Etymology: Epithet derived from its location of origin, Bomi in Tibet, China.
Leaf spots amphigenous, circular to irregular, 2–5 mm diam, scattered over the leaf, brown, surrounded by a greenish yellow border, single lesions may coalesce to form larger lesions till the whole leaf and getting dark brown (Fig. 7G). Conidiomata pycnidial, solitary, sometimes aggregated, globose to subglobose, pale brown, glabrous, superficial, ostiolate, 100–200 × 100–180 μm. Ostiole single, with an elongated neck, slightly papillate or non-papillate. Pycnidial wall pseudoparenchymatous, composed of oblong to isodiametric cells, 2–3 layers, 20–30 μm thick, outer wall 1–2-layers pigmented. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 5–8 × 4.5–7 μm. Conidia ovoid to ellipsoidal, smooth- and thin-walled, hyaline, aseptate, 3.5–6.5 × 2–3.5 μm, with 1–2 distinct polar guttules. Conidial matrix buff.
Culture characteristics: Colonies on OA, 35–43 mm diam after 7 d, margin regular, covered by floccose aerial mycelia, dense, white to greyish olivaceous; reverse white to olivaceous. Colonies on MEA 45–47 mm diam after 7 d, margin irregular, olivaceous, with sparse white aerial mycelia near the centre; reverse concolourous. Colonies on PDA, 53–55 mm diam after 7 d, margin regular, aerial mycelia floccose, white to olivaceous, forming concentric rings; reverse olivaceous with pale green margin. NaOH test negative.
Specimens examined: China, Tibet, Bomi, leaves of Boraginaceae, 14 Jun. 2015, Q. Chen (holotype HMAS 247170, dried culture, ex-holotype living culture CGMCC 3.18366 = LC 8167); ibid. LC 8168.
Notes: Stagonosporopsis bomiensis is most closely related to S. papillata, another novel species collected from Tibet. However, S. bomiensis is distinguishable from S. papillata by having slightly shorter and wider conidia (3.5–6.5 × 2–3.5 μm vs. 3.5–9 × 1.5–3 μm), and based on its number of ostioles (1 vs. 2–3).
This is the first record of a Stagonosporopsis species on a member of the Boraginaceae.
Stagonosporopsis papillata Q. Chen, Crous & L. Cai, sp. nov. MycoBank MB818954. Fig. 34.
Fig. 34.
Stagonosporopsis papillata (CGMCC 3.18367). A–B. Colony on OA (front and reverse). C–D. Colony on MEA (front and reverse). E–F. Colony on PDA (front and reverse). G. Pycnidium forming on OA. H. Pycnidium. I. Section of pycnidium. J. Section of pycnidial wall. K–M. Conidiogenous cells. N. Conidia. Scale bars: G = 100 μm; H = 40 μm; I–J = 10 μm; K–N = 5 μm.
Etymology: Name refers to its papillate pycnidia.
Leaf spots amphigenous, angular to irregular, 2–8 mm diam, reddish brown, indefinite border (Fig. 7K). Conidiomata pycnidial, solitary or aggregated, yellowish brown to brown, globose to subglobose or obpyriform, with hyphal outgrowths, semi-immersed in the agar, ostiolate, (130–)200–280 × (100–)150–250 μm. Ostioles 2–3, slightly papillate. Pycnidial wall pseudoparenchymatous, composed of oblong to isodiametric cells, 2–3 layers, 10–15(–20) μm thick, outer wall 1–2-layers pigmented. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 5–8.5 × 4–7.5 μm. Conidia ellipsoidal to oblong, incidentally slightly curved, smooth- and thin-walled, hyaline, aseptate, 3.5–6.5(–9) × 1.5–3 μm, with two large polar guttules. Conidial matrix buff.
Culture characteristics: Colonies on OA, 44–50 mm diam after 7 d, margin regular, covered by white, dense aerial mycelia, grey near the centre, with white margin; reverse olivaceous with white margin. Colonies on MEA 50–52 mm diam after 7 d, margin regular, dull green, aerial mycelia sparsely; reverse concolourous. Colonies on PDA, 55–57 mm diam after 7 d, margin regular, aerial mycelia covering the whole colony, white; reverse olivaceous with white margin. NaOH test negative.
Specimens examined: China, Tibet, Bomi, on leaves of Rumex nepalensis, 14 Jun. 2015, Q. Chen (holotype HMAS 247171, dried culture, ex-holotype living culture CGMCC 3.18367 = LC 8169); ibid. LC 8170; Bomi, on leaves of Boraginaceae, 14 Jun. 2015, Q. Chen, LC 8171.
Notes: Stagonosporopsis papillata is phylogenetically allied to S. bomiensis and S. dorenboschii (Fig. 1). Morphological differences between S. papillata and S. bomiensis are discussed under the latter species. Stagonosporopsis papillata could be differentiated from S. dorenboschii by producing slightly larger conidiogenous cells (5–8.5 × 4–7.5 μm vs. 4–6 × 3–6 μm) and conidia (3.5–9 × 1.5–3 μm vs. 3–5.5 × 1.5–2.5 μm; de Gruyter & Noordeloos 1992).
Discussion
The Didymellaceae has recently undergone extensive revision based on its phylogenetic relationships (Aveskamp et al., 2009a, Aveskamp et al., 2009b, Aveskamp et al., 2010, de Gruyter et al. 2009, Chen et al. 2015a). In this study, 32 new taxa and two new combinations are proposed in nine genera, mostly based on specimens collected from Asia.
The majority of members in Didymellaceae are plant associated fungi. So far, only a few species were reported from other substrates, such as Phoma herbarum, Didymella glomerata, D. pomorum from inorganic materials including asbestos, cement, paint, etc. (Aveskamp et al. 2008), D. eucalyptica from water, D. gardeniae from air, and Leptosphaerulina australis from soil (Aveskamp et al. 2010). In the present study, several new species, namely Allophoma oligotrophica, Didymella aeria, D. aquatica, D. chloroguttulata, D. ellipsoidea, D. suiyangensis were collected from substrates such as air, soil, water and limestone from caves in South-west China, a typical environment with relatively low temperature, low nutrition, high humidity, and absolute darkness (Zhang et al. 2017). All these species are oligotrophic fungi except D. aquatic. It is interesting that many of these new species present pale green to dull green polar guttules which are not often observed in other species, while few other recognizable morphological differences could be observed.
The 360 isolates belonging to 194 taxa investigated in this study represent a large collection of Didymellaceae, which occurred on 163 different host genera within 70 families. Our results indicated that Asteraceae, Fabaceae, Poaceae, Ranunculaceae, Rosaceae and Solanaceae were the six most common host families associated with Didymellaceae (Fig. 2). Based on currently available data, several genera exhibited a certain level of host-specificity, i.e. Ascochyta species show relatively high host specificity to Fabaceae, Neoascochyta to Poaceae and Neomicrosphaeropsis to Tamaricaceae. Heterophoma species appear somewhat specific to Scrophulariaceae, as well as Phomatodes to Brassicaceae. Other genera appear to have a rather broad range of host families. Among the five apparently host-specific genera listed above, Neoascochyta is located in the earliest divergent clade in Didymellaceae, followed by Phomatodes, Ascochyta, Neomicrosphaeropsis and Heterophoma. Surprisingly, this evolutionary direction is consistent with that of their respective host families, i.e. Poaceae as earliest, followed by Brassicaceae, Fabaceae, Tamaricaceae and Scrophulariaceae (Bremer et al. 2009). Our data suggest, therefore, a general trend of coevolution in the host-specific groups in Didymellaceae.
Nine new species belonging to Epicoccum and 10 in Didymella are proposed in this paper, which reflect the high diversity of species in these two genera. The most remarkable feature of Epicoccum species is the formation of the darkly pigmented multi-septate conidia (dictyochlamydospores) from sporodochia. Of the nine new taxa, four were only observed as typical Epicoccum conidia, while the pycnidial morphs proved to be absent. These four species could also produce yellowish pigments that diffuse into culture media. In addition, six of the new Epicoccum species showed positive reactions in the NaOH test, that detects the production of metabolite E. Epicoccum camelliae is likely an opportunistic pathogen that could asymptomatically colonise plants as a potential destructive invader, as we obtained two strains, one from a healthy leaf, and another from a diseased leaf. Among the 10 new species described in the sexual genus Didymella, D. sinensis was recorded as sexual morph in all the single ascospore isolates obtained from three different hosts, while the asexual morph was not observed, revealing the homothallic nature of this species.
In spite of the good performance on the resolution of genera and species in Didymellaceae using the combined four loci, LSU, ITS, rpb2 and tub2, there are still several taxa or species complexes that await further assessment, such as the Boeremia exigua varieties (Abeln et al., 2002, Aveskamp et al., 2009b) and the Epicoccum nigrum complex (Fávaro et al. 2011). Additional loci and more isolates are required for a future study to clarify their phylogenetic relationships as well as species boundaries.
Following the 17 genera accepted in Didymellaceae by Chen et al. (2015a), Briansuttonomyces (Crous and Groenewald 2016) and Neomicrosphaeropsis (Thambugala et al. 2017) were subsequently embedded in this family based on the multi-locus phylogenies, which were confirmed in this paper. However, the introduction of several other genera are in need of reassessment. The monotypic genus Heracleicola erected by Ariyawansa et al. (2015) is herewith reduced to synonymy with Ascochyta, according to our combined LSU and ITS sequences analysis (Supplementary Fig. S1). Another genus, Neodidymella, was established in the same paper without any sequence data provided, although a tree was presented which suggested a very close relationship to Boeremia (Ariyawansa et al. 2015). Wijayawardene et al. (2016) proposed another monotypic genus Didymellocamarosporium, typified with Didymelloc. tamaricis, for which only SSU and LSU sequences could be obtained from public sequence repositories. The tree presented in Wijayawardene et al. (2016) provided little information as most of the Didymellaceae members were ignored in their analysis. We conducted a phylogenetic analysis using LSU sequences (Supplementary Fig. S2) and Didymelloc. tamaricis was embedded within the genus Neomicrosphaeropsis that was established later in the same year (Thambugala et al. 2017). Morphologically, Didymellocamarosporium tamaricis produces large brown, muriformly septate conidia (13–21.5 × 7–9.5 μm), which is obviously different from Neomicrosphaeropsis species and other genera in Didymellaceae.
The genera Endocoryneum and Pseudohendersonia were established by Petrak (1922) and Crous & Palm (1999) respectively, with their familial placements undetermined. Recently two new species, Endocoryneum festucae and Pseudohendersonia galiorum were introduced and they were placed in Didymellaceae by Wijayawardene et al. (2016), in which however, only LSU sequences were provided. Our analysis on the basis of LSU sequences revealed that E. festucae belonged to Stagonosporopsis, and formed an unexpected very long terminal branch (Supplementary Fig. S2). The sequence of E. festucae (access number: KU848203) needs to be verified. The phylogenetic relationships of Pseudohendersonia galiorum can also not be clarified solely based on the LSU sequence provided in Wijayawardene et al. (2016). From the morphological aspect, E. festucae and P. galiorum both produce large brown conidia (30–37 × 9–12 μm, and 9–15 × 3.5–5.5 μm), with 3–4 transverse septa (Wijayawardene et al. 2016), obviously atypical for Didymellaceae. Due to the lack of sequences and the morphological divergence, their taxonomic placement remains unresolved, and these genera can thus not be accepted in Didymellaceae.
Our study, together with that of Aveskamp et al. (2010) and Chen et al. (2015a), have set the foundation for the systematics and taxonomy of Didymellaceae. Most of the genera included in this family have been well circumscribed, and useful molecular loci have been identified for species delimitation. The present study adds further indication that a large number of unknown Didymellaceae species exist in nature, especially from previously ignored ecosystems.
Acknowledgements
This study was financially supported by the Project for Fundamental Research on Science and Technology, MOST (2014FY120100) and NSFC 31600023. Jiang-Rui Jiang, Zhi-Feng Zhang, Nan Zhou and Fang Liu are thanked for help with sample collection, as well as providing cultures. Roger G. Shivas and Bevan S. Weir are acknowledged for providing fungal cultures. Da-Chuan Bao and Bing Liu are thanked for helping in identifying the various host plants. Xiao-Ling Zhang is thanked for helping with the statistical analysis. Ling-Wei Hou acknowledges CAS GJHZ1310 for supporting her visit to the Westerdijk Fungal Biodiversity Institute.
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.simyco.2017.06.002.
Contributor Information
P.W. Crous, Email: p.crous@westerdijkinstitute.nl.
L. Cai, Email: cail@im.ac.cn.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Supplementary Fig. S1. Phylogenetic tree inferred from a Maximum likelihood analysis based on a concatenated alignment of LSU and ITS sequences of 363 strains. New taxa and new combinations introduced in this study are formatted in bold, of which the ex-type strains are marked by an asterisk (*). The tree was rooted to Leptosphaeria conoidea (CBS 616.75) and L. doliolum (CBS 505.75).
Supplementary Fig. S2. Phylogenetic tree inferred from a Maximum likelihood analysis based on LSU sequences of 365 strains. New taxa and new combinations introduced in this study are formatted in bold, of which the ex-type strains are marked by an asterisk (*). The tree was rooted to Leptosphaeria conoidea (CBS 616.75) and L. doliolum (CBS 505.75).
References
- Abeln E.C.A., Stax A.M., de Gruyter J. Genetic differentiation of Phoma exigua varieties by means of AFLP fingerprints. Mycological Research. 2002;106:419–427. [Google Scholar]
- Ariyawansa H.A., Hyde K.D., Jayasiri S.C. Fungal diversity notes 111–252–taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity. 2015;75:27–274. [Google Scholar]
- Aveskamp M.M., de Gruyter J., Crous P.W. Biology and recent developments in the systematics of Phoma, a complex genus of major quarantine significance. Fungal Diversity. 2008;31:1–18. [Google Scholar]
- Aveskamp M.M., de Gruyter J., Woudenberg J.H.C. Highlights of the Didymellaceae: A polyphasic approach to characterise Phoma and related pleosporalean genera. Studies in Mycology. 2010;65:1–60. doi: 10.3114/sim.2010.65.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aveskamp M.M., Verkley G.J.M., de Gruyter J. DNA phylogeny reveals polyphyly of Phoma section Peyronellaea and multiple taxonomic novelties. Mycologia. 2009;101:363–382. doi: 10.3852/08-199. [DOI] [PubMed] [Google Scholar]
- Aveskamp M.M., Woudenberg J.H.C., de Gruyter J. Development of taxon-specific sequence characterized amplified region (SCAR) markers based on actin sequences and DNA amplification fingerprinting (DAF): a case study in the Phoma exigua species complex. Molecular Plant Pathology. 2009;10:403–414. doi: 10.1111/j.1364-3703.2009.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berner D., Cavin C., Woudenberg J.H.C. Assessment of Boeremia exigua var. rhapontica, as a biological control agent of Russian knapweed (Rhaponticum repens) Biological Control. 2015;81:65–75. [Google Scholar]
- Boerema G.H. Contributions towards a monograph of Phoma (Coelomycetes) – II. Section Peyronellaea. Persoonia. 1993;15:197–221. [Google Scholar]
- Boerema G.H., de Gruyer J., Noordeloos M.E. CABI Publishing; Wallingford, UK.: 2004. Phoma identification manual. Differentiation of specific and infra-specific taxa in culture. [Google Scholar]
- Bremer B., Bremer K., Chase M. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society. 2009;161:105–121. [Google Scholar]
- Cai L., Hyde K.D., Taylor P.W.J. A polyphasic approach for studying Colletotrichum. Fungal Diversity. 2009;39:183–204. [Google Scholar]
- Chen Q., Jiang J.R., Zhang G.Z. Resolving the Phoma enigma. Studies in Mycology. 2015;82:137–217. doi: 10.1016/j.simyco.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Zhang K., Zhang G.Z. A polyphasic approach to characterise two novel species of Phoma (Didymellaceae) from China. Phytotaxa. 2015;197:267–281. [Google Scholar]
- Choi Y.W., Hyde K.D., Ho W.H. Single spore isolation of fungi. Fungal Diversity. 1999;3:29–38. [Google Scholar]
- Crous P.W., Gams W., Stalpers J.A. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology. 2004;50:19–22. [Google Scholar]
- Crous P.W., Groenewald J.Z. They seldom occur alone. Fungal Biology. 2016;120:1392–1415. doi: 10.1016/j.funbio.2016.05.009. [DOI] [PubMed] [Google Scholar]
- Crous P.W., Palm M.E. Systematics of selected foliicolous fungi associated with leaf spots of Proteaceae. Mycological Research. 1999;103:1299–1304. [Google Scholar]
- Crous P.W., Verkley G.J.M., Groenewald J.Z. Vol. 1. Westerdijk Fungal Biodiversity Institute; Utrecht, Netherlands: 2009. Fungal biodiversity. (CBS Laboratory Manual Series). [Google Scholar]
- Cubero O.F., Crespo A., Fatehi J. DNA extraction and PCR amplification method suitable for fresh, herbarium stored, lichenized, and other fungi. Plant Systematics and Evolution. 1999;216:243–249. [Google Scholar]
- De Gruyter J., Aveskamp M.M., Woudenberg J.H.C. Molecular phylogeny of Phoma and allied anamorph genera: Towards a reclassification of the Phoma complex. Mycological Research. 2009;113:508–519. doi: 10.1016/j.mycres.2009.01.002. [DOI] [PubMed] [Google Scholar]
- De Gruyter J., Boerema G.H., van der Aa H.A. Contributions towards a monograph of Phoma (Coelomycetes) VI – 2. Section Phyllostictoides: Outline of its taxa. Persoonia. 2002;18:1–53. [Google Scholar]
- De Gruyter J., Noordeloos M.E. Contributions towards a monograph of Phoma (Coelomycetes) – I. 1. Section Phoma: taxa with very small conidia in vitro. Persoonia. 1992;15:71–92. [Google Scholar]
- De Gruyter J., Noordeloos M.E., Boerema G.H. Contributions towards a monograph of Phoma (Coelomycetes) – I. 2. Section Phoma: additional taxa with very small conidia and taxa with conidia up to 7 μm long. Persoonia. 1993;15:369–400. [Google Scholar]
- De Gruyter J., Noordeloos M.E., Boerema G.H. Contributions towards a monograph of Phoma (Coelomycetes) – I. 3. Section Phoma: taxa with conidia longer than 7 μm. Persoonia. 1998;16:471–490. [Google Scholar]
- De Hoog G.S., Gerrits van den Ende A.H.G. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses. 1998;41:183–189. doi: 10.1111/j.1439-0507.1998.tb00321.x. [DOI] [PubMed] [Google Scholar]
- Dearness J. New or noteworthy species of fungi. Mycologia. 1916;8:98–107. [Google Scholar]
- Fávaro L.C.dL, de Melo F.L., Aguilar-Vildoso C.I. Polyphasic analysis of intraspecific diversity in Epicoccum nigrum warrants reclassification into separate species. PLoS One. 2011;6:e14828. doi: 10.1371/journal.pone.0014828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes R.R., Glienke C., Videira S.I.R. Diaporthe: a genus of endophytic, saprobic and plant pathogenic fungi. Persoonia. 2013;31:1–41. doi: 10.3767/003158513X666844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J.R., Chen Q., Cai L. Polyphasic characterisation of three novel species of Paraboeremia. Mycological Progress. 2017;16:285–295. [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes evidence from an RNA polymerase II subunit. Molecular Biology and Evolution. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Lombard L., Houbraken J., Decock C. Generic hyper-diversity in Stachybotriaceae. Persoonia. 2016;36:156–246. doi: 10.3767/003158516X691582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller E. Kulturversuche mit Ascomyceten I. Sydowia. 1953;7:325–334. [Google Scholar]
- Nylander J.A.A. Evolutionary Biology Centre, Uppsala University; 2004. MrModeltest v2. Program distributed by the author. [Google Scholar]
- Petrak F. Mykologische Notizen. IV. Annales Mycologici. 1922;20:300–345. [Google Scholar]
- Punithalingam E. Graminicolous Ascochyta species. Mycological Papers. 1979;142:1–214. [Google Scholar]
- Punithalingam E., Tulloch M., Leach C.M. Phoma epicoccina sp. nov. on Dactylis glomerata. Transactions of the British Mycological Society. 1972;59:341–345. [Google Scholar]
- Rayner R.W. Commonwealth Mycological Institute and British Mycological Society; Kew, Surrey, UK: 1970. A mycological colour chart. [Google Scholar]
- Rehner S.A., Samuels G.J. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycological Research. 1994;98:625–634. [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccardo P.A. Vol. 16. 1902. pp. 1–1291. (Sylloge Fungorum omnium hucusque cognitorum: Supplementum Universale, Pars V). Padova, Italy. [Google Scholar]
- Saccardo P.A., Trotter A. Vol. 22. 1913. pp. 1–1612. (Sylloge Fungorum omnium hucusque cognitorum: Supplementum Universale, Pars IX). Padova, Italy. [Google Scholar]
- Smith H., Wingfield M.J., Coutinho T.A. Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. South African Journal of Botany. 1996;62:86–88. [Google Scholar]
- Stamatakis A., Alachiotis N. Time and memory efficient likelihood-based tree searched on phylogenomic alignments with missing data. Bioinformatics. 2010;26:i132–i139. doi: 10.1093/bioinformatics/btq205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung G.-H., Sung J.-M., Hywel-Jones N.L. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Molecular Phylogenetics and Evolution. 2007;44:1204–1223. doi: 10.1016/j.ympev.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolutionary. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thambugala K.M., Daranagama D.A., Phillips A.J.L. Microfungi on Tamarix. Fungal Diversity. 2017;82:239–306. [Google Scholar]
- Van der Aa H.A., Boerema G.H., de Gruyer J. Contributions towards a monograph of Phoma (Coelomycetes) – VI-1. Section Phyllostictoides: characteristics and nomenclature of its type species Phoma exigua. Persoonia. 2000;17:435–456. [Google Scholar]
- Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright M., Al-Talhi A. Selective isolation and oligotrophic growth of Candida on nutrient-free silica gel medium. Journal of Medical Microbiology. 1999;48 doi: 10.1099/00222615-48-12-1130. 1130–1130. [DOI] [PubMed] [Google Scholar]
- White T.J., Bruns T., Lee S. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: a guide to methods and applications. Academic Press; San Diego, California, USA: 1990. pp. 315–322. [Google Scholar]
- Wijayawardene N.N., Hyde K.D., Wanasinghe D.N. Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Diversity. 2016;77:1–316. [Google Scholar]
- Woudenberg J.H.C., Aveskamp M.M., de Gruyter J. Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia. 2009;22:56–62. doi: 10.3767/003158509X427808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Su Y.Y., Cai L. An optimized protocol of single spore isolation for fungi. Cryptogamie, Mycologie. 2013;34:349–356. [Google Scholar]
- Zhang Z.F., Liu F., Zhou X. Culturable mycobiota from Karst caves in China, with descriptions of 20 new species. Persoonia. 2017;39:1–31. doi: 10.3767/persoonia.2017.39.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.