Abstract
Streptococcus agalactiae is one of the most common pathogens leading to mastitis in dairy herds worldwide; consequently, the pathogen causes major economic losses for affected farmers. In this study, multilocus sequence typing (MLST), genotypic capsular typing by multiplex polymerase chain reaction (PCR), and virulence gene detection were performed to address the molecular epidemiology of 59 bovine (mastitis) S. agalactiae isolates from 36 dairy farms located in the largest milk-producing mesoregions in Brazil (Minas Gerais, São Paulo, Paraná, and Pernambuco). We screened for the virulence genes bac, bca, bibA, cfb, hylB, fbsA, fbsB, PI-1, PI-2a, and PI-2b, which are associated with adhesion, invasion, tissue damage, and/or immune evasion. Furthermore, five capsular types were identified (Ia, Ib, II, III, and IV), and a few isolates were classified as non-typeable (NT). MLST revealed the following eight sequence types (STs): ST-61, ST-67, ST-103, ST-146, ST-226, ST-314, and ST-570, which were clustered in five clonal complexes (CC64, CC67, CC103, CC17, and CC314), and one singleton, ST-91. Among the virulence genes screened in this study, PI-2b, fbsB, cfb, and hylB appear to be the most important during mastitis development in cattle. Collectively, these results establish the molecular epidemiology of S. agalactiae isolated from cows in Brazilian herds. We believe that the data presented here provide a foundation for future research aimed at developing and implementing new preventative and treatment options for mastitis caused by S. agalactiae.
Keywords: Group B Streptococcus, Multilocus sequence typing, Genotypic capsular typing, Virulence genes, Bovine disease
Introduction
According to the World Food Organization,1 the global production of milk reached 782,000,000 tonnes in 2013. Brazil is among the top producers of milk, ranking 4th globally and producing 34.4 million tonnes. Milk production worldwide is affected by numerous factors, but one of the most important factors is mastitis: the disease can lead to reduced milk production by the affected animals and/or to the production of low-quality milk, which is invariably discarded, resulting in large economic losses.2 Furthermore, mastitis is currently treated with antibiotics, which, when used indiscriminately, can lead to the emergence of antibiotic-resistant bacteria that can be further propagated.3 Thus, the type and the pathogenic nature of various mastitis-related bacterial species must be investigated in order to develop improved treatment and preventative options that will not only limit the health complications for the affected animals but also lessen the impact on farmers, particularly in countries such as Brazil.
The major bovine mastitis-causing pathogenic bacteria are Escherichia coli, Klebsiella pneumoniae, Streptococcus agalactiae, Streptococcus uberis, and Staphylococcus aureus.2, 4, 5 S. agalactiae, also known as Group B Streptococcus (GBS), has been shown to cause both clinical and subclinical mastitis in cattle, and has been detected in 60% of Brazilian dairy herds.2, 6 Moreover, GBS is recognized to be the causative agent of several diseases in various other animal species, including humans, where it is the most common life-threatening disease in newborn humans, causing pneumonia, septic shock syndrome and meningitis with high mortality rates.7, 8, 9 In these various species, GBS also appears to be capable of colonizing different tissues in the body, a characteristic that has been linked to several specific bacterial virulence genes that enable the microorganism to colonize, invade, and spread in the host. In GBS, these genes include the following: bacterial immunogenic adhesin (bibA), fibrinogen-binding protein A (fbsA), and fibrinogen-binding protein B (fbsB), which are related to adhesion; Pilus Island 1 (PI-1), PI-2a, and PI-2b, which are related to adhesion and invasion; cfb and hylFB, which are related to tissue damage; and bac and bca, which are related to immune evasion.9, 10 Furthermore, these virulence genes might also be associated with adaptation and clinical manifestations in various host species.
Because GBSs exert a major effect on both animal and human health, several tools have been developed for epidemiological typing of this pathogen.11, 12, 13 The capsule of this bacterium has been shown to be the first bacterial virulence factor that enables the bacterium to evade the immune system and invade the host. Thus, capsular serotyping is a classic method used in epidemiological studies of GBS. To date, 10 GBS capsular polysaccharide serotypes (CPSs) have been identified (Ia, Ib, and II–IX), and their distribution in humans is directly related to ethnic and geographic regions.8 Furthermore, capsular genotyping is considered to be highly suitable for epidemiological investigations because the serotypes can be identified in the presence or absence of CPS expression.6 Multilocus sequence typing (MLST) is a method based on the amplification and sequencing of bacterial housekeeping genes, and it has been used to investigate, characterize, and distinguish specific clones among GBSs isolated from humans and animals from diverse geographical regions.12, 14, 15 Although MLST and molecular capsular typing have been widely employed in the epidemiological characterization of GBSs, no published study has reported the use of these tools for identifying GBS isolates in mastitis-affected Brazilian dairy herds.
Therefore, in this study, the first aim was to genotypically characterize GBS isolates from bovine mastitis from multiple Brazilian farms located in various regions of the country. The data obtained were then used to address the molecular epidemiology of the strains. Lastly, the presence of 10 GBS virulence genes was analysed in order to identify the genes that have the greatest impact in this population.
Materials and methods
Bacterial strains
We evaluated 65 bacterial strains from the culture collection of the Bacteriology Laboratory/DMVP UFLA, previously identified as S. agalactiae phenotypically by the catalase test, CAMP test, hippurate hydrolysis test and the PYR test, as described by MacFaddin.16 These strains were isolated from the milk of 65 animals from 36 dairy farms located in the largest Brazilian cow milk producing states (Minas Gerais, São Paulo, Paraná, and Pernambuco) in the mesoregions of the country, between 2010 and 2011 (Material supplementary 1). The reference strains NEM 316 and ATCC BAA-611 (also designated 2603V/R) were used as positive controls and a Streptococcus dysgalactiae strain (ATCC 27957) was used as the negative control in the multiplex-polymerase chain reaction (PCR) and PCR assays.
Specific PCR, molecular capsular typing and sequencing
Genotypic analyses were then performed to confirm this S. agalactiae classification by using S. agalactiae-specific PCR as previously described.17 All isolates that were positive in this PCR analysis were subjected to CPS typing by using a multiplex-PCR assay, as described by Poyart et al.13 Strains that were not amplified in the multiplex PCR were subjected to serotype IX-specific PCR analysis.18 All reactions were performed twice to ensure data reproducibility. The amplification products were analysed using 1.5% (w/v) agarose gel electrophoresis with 1× Tris–acetate buffer (0.04 M Tris–acetate, pH 8.4, 1 mM EDTA) and were visualized with a UV transilluminator after staining with 0.5× GelRed™ (Biotium, USA). A 100-bp DNA ladder (New England Biolabs, USA) was used as a molecular marker in each electrophoresis assay. PCR products were purified using a Wizard PCR Prep kit (Promega, USA). Sequencing reactions were performed using an Applied Biosystems BigDye Terminator Cycle Sequencing Kit and run on an ABI 3730xl DNA analyser (Applied Biosystems, USA).
Multilocus sequence typing and clonal group assignment
MLST was performed by sequencing the internal fragments of seven housekeeping genes (adhP, alcohol dehydrogenase; pheS, phenylalanyl transfer RNA synthetase; atr, amino acid transporter protein; glnA, glutamine synthetase; sdhA, l-serine dehydratase; glcK, glucose kinase; and tkt, transketolase), according to Jones et al.12 DNA sequences determined on both strands were assembled using the base-calling programme Phred.19 Sequence types (STs) were defined by analysing the seven concatenated sequences in the S. agalactiae MLST database. The eBURST V3 programme (http://eburst.mlst.net)20 was then used to identify clonal complexes among these bovine S. agalactiae strains. In addition, a population snapshot of global S. agalactiae was created in eBURST to show the position of the STs from our study in relation to all other known STs for this bacterial species.
Genetic relationship of S. agalactiae isolated from different hosts in Brazil
Allele sequences found in this study as well as alleles identified previously from different hosts in Brazil21, 22 were used to construct a dendrogram showing the relationships among Brazilian isolates. The sequences were aligned using CLUSTALW,23 and the genetic-distance matrix was obtained using Kimura's two-parameter model.24 Furthermore, an evolutionary tree was created using the neighbour-joining method25 with MEGA4.26 The bootstrap values calculated from 1000 replicates are displayed as percentages.
Detection of virulence genes
Virulence genes were detected by performing PCR screening for the genes bac, bca, bibA, cfb, hylB, fbsA, fbsB, PI-1, PI-2a, and PI-2b, as described previously.21, 27, 28, 29 All the primers used in this study and their respective references can be accessed in supplementary material 2.
Statistical and data analysis
Genetic diversity was calculated using Simpson's index of diversity (SID).30 Fisher's exact tests were performed using SAS1 statistical software (STAT Version 6.12; SAS Institute Inc., USA) in order to determine whether the measured gene frequencies were significantly different. p ≤ 0.05 was considered statistically significant.
Results
A total of 59 S. agalactiae were positive in specific PCRs and used in our subsequent genotypic tests. From these, 12 were isolated from animals diagnosed with clinical bovine mastitis, whereas the remaining 47 were from animals diagnosed with subclinical mastitis.
Table 1 presents all of the data generated during our genotypic analysis, including the CPS and MLST data. Among the 59 Brazilian strains, we identified five CPS types, III (n = 23/38.98%), II (n = 12/20.33%), Ib (n = 9/15.25%), Ia (n = 5/8.47%), and IV (n = 5/8.47%), and a few untypeable strains (NT) (n = 5/8.47%). The SID calculated for this technique was 0.7749, which indicated a high level of discrimination in the case of bovine isolates of S. agalactiae. Notably, in the entire strain set analysed, serotype III was the most frequent, and it appeared to be independent of geographical location and mastitis severity (clinical or subclinical). When analysing only the clinical mastitis strain samples, this distinction was even more apparent: CPS type III was found in 50% of the strains, and this was followed by II (25%), IV (16.66%), and NT (8.33%).
Table 1.
Characteristics and frequency of 59 Group B Streptococcus (GBS) isolates listed according to capsular type, sequence type, and clonal complex.
| STa | Allelic profile | No. of isolates in ST (%) | Capsular type (no. of isolates) | Mastitis type (no. of isolates) | State of originc (no. of isolates) | CCb |
|---|---|---|---|---|---|---|
| 61 | 13,1,1,13,1,1,1 | 11 (18.33%) | III(9), Ia(1), NT(1) | Clinical (3)/Subclinical (8) | MG(7), SP(3), PE(1) | 61 |
| 67 | 13,1,1,13,1,1,5 | 21 (35.59%) | Ia(1), II(12), III(3), IV(3), NT(2) | Clinical (5)/Subclinical (16) | MG(7), PE(6), SP(8) | 67 |
| 91 | 25,1,1,13,15,1 | 9 (15.25%) | III(8), IV(1) | Clinical (4)/Subclinical (5) | MG(8), PE(1) | 91 |
| 103 | 16,1,6,2,9,9,2 | 2 (3.3%) | Ia(2) | Subclinical (2) | MG(1), PE(1) | 103 |
| 146 | 2,1,1,1,1,1,1 | 3 (5%) | III(3) | Subclinical (3) | MG(3) | 17 |
| 226 | 16,1,2,2,9,1,2 | 5 (8.47%) | Ib(4), NT(1) | Subclinical (5) | PR(5) | 314 |
| 314 | 16,1,2,2,9,2,2 | 1 (1.69%) | Ia(1) | Subclinical (1) | PE(1) | 314 |
| 570 | 16,1,1,2,1,1,5 | 7 (11.8%) | Ib(5), IV(1), NT(1) | Subclinical (7) | MG(1), PR(6) | 67 |
ST: sequence type.
CC: clonal complex.
States: MG, Minas Gerais; SP, São Paulo; PE, Pernambuco; PR, Parana.
In terms of geographical location, in Minas Gerais, CPS type III was most prevalent (74.07%), followed by IV (18.51%) and Ia (7.4%). By contrast, in São Paulo, CPS type II (54.54%) was the most frequent, followed by III (18.18%) and NT (27.27%). CPS type II was also the most frequent (60%) in the strains isolated from Pernambuco, and it was followed by Ia (30%) and III (10%). Interestingly, a high level of homogeneity was observed in the isolates from Paraná: they were predominantly of type Ib (81.82%), with the remainder being NT (18.18%). Moreover, in 5 of the 36 Brazilian dairy herds sampled (farms: 03 MG, 4 MG, 10 MG, 22 PE, and 25 PE), strains of distinct serotypes were present (supplementary material 1).
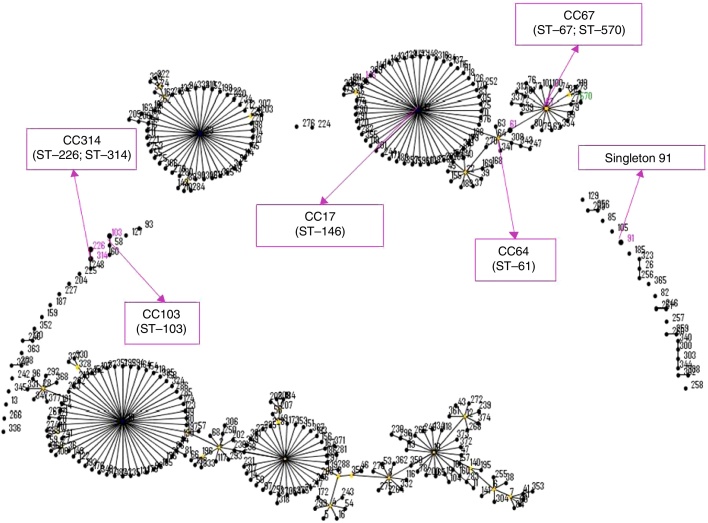
Next, by using MLST, we identified eight unique STs in the 59 strains: ST-61, ST-67, ST-91, ST-103, ST-146, ST-226, ST-314, and ST-570. ST-67 represented the largest fraction of the strains (n = 21/35.59%), followed by ST-61 (n = 11/18.33%), ST-91 (n = 9/15.25%), ST-570 (n = 7/11.86%), and ST-226 (n = 5/8.47%) (Table 1). In ST-103, ST-146, and ST-314, the frequency dropped to ≤5.08%. Intriguingly, the eBURST analysis showed that ST-67 was predicted as the founder of a subgroup originating from CC61, which originated from CC17, whereas ST-91 was a singleton. ST-61 was clustered into clonal complex (CC) 64, and ST-67 and ST-570 were grouped into CC67. ST-146 and ST-103 were clustered into CC17 and CC103, respectively, and ST-226 and ST-314 into CC314. For this MLST analysis, an SID of 0.9088 was calculated, which again demonstrated high-level discrimination among the bovine S. agalactiae isolates analysed in this study.
Using eBURST, a population snapshot of the entire S. agalactiae was created, and showed the position of the STs identified in this study in relation to all known STs for this bacterial species (Fig. 1). The strains classified as ST-226 and ST-570 appear to be related to both their geographic origin and capsular type, whereas the strains belonging to ST-103, ST-146, and ST-314 all appear to be related to their capsular type regardless of their geographic location. Moreover, representative ST-61 and ST-91 strains appear to be mainly associated with CPS III, with one of each group harbouring a distinct capsular type, but the geographic origin of these strains is heterogeneous. Last, strains identified as ST-67 appear to be unrelated to both geographic origin and capsular type. The SID for this relationship analysis performed using MLST and molecular capsular typing was 0.9647, which shows a marginally lower extent of diversity than that observed in the previous analyses, but the level of discrimination is still very high.
Fig. 1.
eBURST diagram of the Streptococcus agalactiae population. Each sequence type (ST) is represented as a dot. The dots positioned centrally in the cluster are predicted founders (blue) or subgroup founders (yellow). Pink and green numbers indicate the STs detected in this study, and pink arrows indicate clonal complexes and predicted founders.
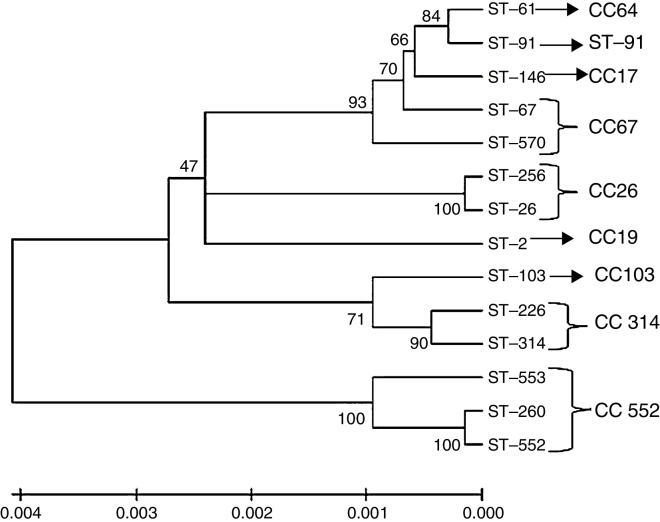
According to the generated dendrogram (Fig. 2), the bovine S. agalactiae strains in this study were clustered into two main branches. Interestingly, the STs described previously were also clustered in distinct branches. In the screening for virulence genes, all isolates were positive for fbsB, cfb, hylB, and at least one PI gene; PI-2b was the most frequent (96.61%), and PI-1 and PI-2a were both considerably less frequent (1.69% and 3.38%, respectively). Furthermore, the frequencies were also low for fbsA (42.37%), bibA (11.86%), and bca (3.38%), and all strains were negative for bac. Based on the combinations of these genes, seven virulence gene profiles were established according to the presence or absence of each gene (Table 2). In this analysis, isolates showing the two highest frequencies were sorted into Profile 1 (52.54%; positive for fbsB, cfb, hylB, and PI-2b) and Profile 7 (28.81%; positive for fbsA, fbsB, cfb, hylB, and PI-2b), which did not differ significantly (p = 0.081). However, these two profiles were significantly different from the other profiles in terms of occurrence (p < 0.05): Profile 4 isolates (positive for bibA, fbsA, fbsB, cfb, hylB, and PI-2b) were observed at a frequency of 10.16%, whereas the other profiles were identified for <3.39% of the strains in this study. Notably, all of the strains isolated from clinical cases of mastitis belong to Profiles 1, 4, or 7. These profiles of virulence genes did not appear to be associated with geographic origin, ST, or capsular type, except in the case of isolates positive for bibA and bca, which appeared to be linked to CPS II and CPS Ia, respectively.
Fig. 2.
Dendrogram constructed by the neighbour joining method based on concatenated sequences from multilocus sequence typing. The dendrogram illustrates the phylogenetic relationship of the ST of strains isolated from bovine mastitis samples (ST-61, ST-67, ST-91, ST-103, ST-146, ST-226, ST-314 and ST-570) that were used in this study and of other S. agalactiae strains isolated in Brazil from fish (ST-103, ST-260, ST-552 and ST-553) and humans (ST-02, ST-26 and ST-256). The clonal complexes are indicated in the figure, and the tree is drawn to scale with the branch lengths representing the evolutionary distances. The scale is shown at the bottom of the tree.
Table 2.
Profiles established according to the relationship between the presence and absence of the genes detected in different farms and states in Brazil.
| Profile | Gene (+/−) |
Farmsa | Stateb | Frequency total (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PI-2a | PI-2b | PI-1 | bibA | fbsA | fbsB | cfB | hylB | bca | bac | ||||
| 1 | − | + | − | − | − | + | + | + | − | − | 1,4,11,12,14,17,21,24,25,27,30,31,32,33,36,38,41,43,45,46,48,49 | MG, PR, PE, SP | 52.54 |
| 2 | + | − | − | − | − | + | + | + | − | − | 50 | PR | 1.7 |
| 3 | + | − | + | − | − | + | + | + | − | − | 52 | PR | 1.7 |
| 4 | − | + | − | + | + | + | + | + | − | − | 34,37,42,44 | SP, PE | 10.16 |
| 5 | − | + | − | + | − | + | + | + | − | − | 38 | PE | 1.7 |
| 6 | − | + | − | − | + | + | + | + | + | − | 42,51 | PR, PE | 3.39 |
| 7 | − | + | − | − | + | + | + | + | − | − | 11,13,16,23,37,40,47,48,52 | MG, PR, PE, SP | 28.81 |
| Strains (+) | 2 | 57 | 1 | 7 | 25 | 59 | 59 | 59 | 2 | 0 | |||
Farms are numbered according to the culture collection of the Bacteriology Laboratory/DMV UFLA.
Brazilian states: MG, Minas Gerais; PR, Paraná; SP, São Paulo; PE, Pernambuco.
Discussion
Of the 65 original strains, only 59 were genotypically confirmed to be S. agalactiae. This result was not completely unexpected because the specific PCR method used here is less prone to error than the phenotypic identification previously used.31 Although genotypically these strains were GBS, they exhibited diversity in their capsular type, ST, and virulence genes.
Notably, the capsular-typing found agrees with a previous report by Duarte et al.,32 who also identified similar capsular types, including a high frequency of CPS III, in bovine mastitis strains isolated in the southeast region of Brazil. However, our results were distinct from another study with a higher frequency of CPS V and NT strains (19.7%) in GBS isolates, although CPS Ia, II, and III were also detected.33 We believe that this discrepancy is due to the origin of the strains from different geographic location. Furthermore, this link between the CPS and the geographic region does not appear to be limited to Brazil. In bovine GBS isolated in Germany, a higher frequency of CPS Ia and NT was detected, while in strains originating from dairy herds in France, NT strains were detected at the highest frequency, followed by CPS III and IV.34, 35 This association between capsular serotype and geographic region is also well defined for GBS strains of human origin. The data show that whereas CPS Ia, Ib, II, III, and IV strains are detected at the highest frequencies in the USA, Brazil, Europe, and Australia, CPS VI and VIII strains are the predominant strains in Japan.8, 33, 36, 37, 38, 39 The bacteria can readily spread through populations, which results in similar capsular types being present in distinct geographical regions. Notably, the presence within the same herd of bacteria of distinct CPS strains might be related to capsular type exchange, a phenomenon common in Streptococcus bacteria.29, 40 Another possible cause for this might be linked to the trafficking/trading of animals among different farms, a common practice in Brazil. In our findings, the genotypic homogeneity observed in the bacteria in Paraná might be associated with limited trade in the region, which would result in diminished transfer of asymptomatic carriers between farms and a reduction in bacterial selection pressure/diversity.
The lower frequency of strains classified as NT in this study compared to previous Brazilian studies33, 39 could be due to the more specific and sensitive molecular-typing technique used.41 NT S. agalactiae strains have been detected in samples originating from humans, bovines, and fish,8, 21, 39 and this might be related to mutation events in the alignment region of the primer in the gene, an unspecified capsular type, or the absence/switching of the capsule.29 The last, also explain the occurrence of the single ST-570 CPS IV strain in Minas Gerais, which is distinct from the ST-570 strains isolated in Paraná.
The MLST screen showed a distinct heterogeneity among the isolates, which were divided into five CCs. In this work, no new ST was identified; the STs found in this study (ST-67, ST-61, ST-91, ST-570, and ST-226) were described in other herds.6, 35, 42 The diversity of STs in S. agalactiae Brazilian strains might be related to the wide geographic spread and national and international trade of animals, as discussed above for CPS typing. The three most frequent STs, ST-61, ST-91, and ST-67, have been shown to be genetically related, and these strains have adapted themselves for colonizing the bovine mammary gland, probably by their sharing of a common recombinant event in virulence genes that play a crucial role in host–parasite interactions.42 This hypothesis is supported by our results, which led to the classification of all of the clinical mastitis strains as one of these STs (Table 1). The presence of ST-103 CPS Ia among our Brazilian isolates, even at a low frequency, was not unexpected because it has been shown to be prevalent in bovine mastitis in Denmark and Eastern China.4, 6 This strain also appears to be capable of infecting diverse host species, being described as a disease-causing bacterium in Brazilian fisheries.21, 35 The ST-103 CPS Ia strain might play a major role in this ability to colonize distinct hosts and to potentially be anthropozoonotic and zooanthroponotic when animals and handlers are in close contact. However, additional studies are necessary to confirm this hypothesis.
Our analysis is the first to identify the ST-146 strains in association with bovine mastitis, although they were detected at a low frequency, similar to ST-103 strains and ST-314, which was found in low frequency in bulk tank milk in Denmark by Zadoks et al.4 ST-146 is clustered in CC17, which is mainly composed of strains isolated in humans, suggesting high potential for zoonotic transfer and spread through populations, as well as ST-103. ST-314 and ST-146 strains, similar to ST-103 strains, might have emerged in bovine mastitis due to the accumulation of short indels that have been selected during evolution and might be advantageous for survival in the bovine udder. However, this phenomenon is currently under investigation.4, 43
The populations obtained in the eBURST snapshot (last analysed on November 04, 2015) showed a difference in the CC related to ST-91 and ST-67, which were previously clustered in CC61.21, 42 This discrepancy could be explained by the dynamic nature of S. agalactiae in the MLST database and by the high degree of genetic diversity observed for this pathogen.44 Previously, Morozumi et al.45 observed a similar relationship between CC17 and CC61; however, according to Sorensen et al.,46 the structure of the S. agalactiae genome invalidates phylogenetic inferences based on MLST. Nevertheless, the results of MLST classification are supported by a study showing that the evolution of GBS is marked by an accumulation of short indels, and when the whole-genome sequences of S. agalactiae were evaluated, CC17 and CC61 were clustered into two distinct subgroups, similar to the CCs obtained in the MLST.43 Moreover, Richards et al.47 found that although the pangenome might be affected by lateral gene transfer, several Streptococcus species have retained distinct characteristics since their formation.
In the dendrogram constructed using MEGA software, ST-61, ST-67, ST-146, ST-570, and ST-91 were within clusters that appeared to originate from the same root. A discrepancy between eBURST results and the dendrogram was observed for strain ST-91, which currently is disconnected from the group that is formed by the subgroups that all originated from ST-17. The other three bovine isolates (ST-103, ST-226, and ST-314) were clustered in the same branch and were located among the fish and human isolates, which shows the phylogenetic proximity among these strains. This suggests that these strains have the capacity to cause disease in different hosts, as discussed in the preceding paragraphs, but additional studies addressing bacterium-host interactions are necessary because the MLST analysis cannot provide an answer to this question. The SID calculated in this study showed that capsular typing and MLST are effective tools for evaluating genetic diversity in S. agalactiae strains isolated from bovines. Although capsular typing was considered to be an inefficient method in a previous study,44 we found that its discriminatory power differed only slightly from that of MLST and both were lower than that calculated for the two techniques used in combination. This could be because the S. agalactiae bovine isolates appear to be highly adapted to the host, probably because they face weak selective pressure. Therefore, addressing the diversity of these bacteria might not require studies that examine the lateral gene transfer of virulence genes.
Our study showed that fbsB, hylB, cfb, and PI-2b are crucial during GBS infection: these genes were detected at high frequencies. The protein encoded by fbsB binds to fibrinogen in different species independently of strain origin, and this binding ability appears be enhanced by Ca2+.48 Thus, because milk secretions are rich in Ca2+, colonizing the mammary gland might serve as a favourable factor for the presence of fbsB in strains originating from bovines. The other screened gene that encodes a fibrinogen-binding protein, fbsA, was less abundant in our isolates compared to frequencies previously reported for this gene in S. agalactiae of 92.2% and 70.7%.35, 49 This discrepancy can be explained by the replacement of this gene by other adhesion factors, such as proteins encoded by fbsB and other genes that were not screened in this study. The detection of hylB here was similar to that by other researchers in both human and bovine strains, but distinct from fish strains, suggesting that this gene might be related to mammalian hosts.8, 21, 27 Furthermore, the frequency of the bac gene was similar to that previously described in bovine strains.27, 46, 50 Perhaps during the infectious process in bovines, the S. agalactiae strains use other surface-associated proteins that were not screened here. In the case of bca, Duarte et al.32 showed that 64.7% of the S. agalactiae strains isolated from bovines in Brazil were positive for this gene. This is in contrast to the considerably lower frequency detected in this study; this disparity could be associated with differences in farm origin or period of bacterial isolation. Notably, the few strains that were positive for bca also tended to be associated with CPS Ia and Ib, as reported previously; thus, the GBS strains that are bca positive and bac negative appear to be frequently associated with these capsular types.51, 52 The findings for bibA showed that the adhesion protein encoded by this gene is not essential for S. agalactiae infection of bovines, and the low frequency of this gene indicates that other genes likely perform a similar function. Interestingly, most of the isolates that were positive for bibA were CPS II strains, which suggests that this is an allelic variant of the type II or III bibA previously described to be related to this capsular type.28
Finally, this is the first study to screen for PI genes in isolates from bovine herds in Brazil; however, our results were similar to those described in other bovine populations, with the high frequency of PI-2b.6, 46, 53 Our findings suggest the requirement for at least one PI gene, as well as studies of isolates from humans,29, 38 but the frequencies of the PI genes are reversed: PI-1 and PI-2a were found in a greater percentage of the strains than PI-2b. Intriguingly, this appears to be associated with disease in distinct hosts; human infections caused by GBS strains are typically systemic and invasive, and the PI-1 and PI-2a genes have been related to invasion and biofilm formation.54, 55 By contrast, PI-2b appears to be related to intracellular survival in macrophages,56 which might result in the host harbouring the bacteria without presenting clinical signs. This would result in subclinical mastitis, the major infection caused by GBS in bovines.
In conclusion, the S. agalactiae bovine strains studied here were genetically diverse and the strains included five different capsular types and six distinct populations (five CCs and one singleton) according to STs. However, the results suggest that infections can evolve into clinical mastitis in cattle only in the case of CC61, CC67, and ST-91. Furthermore, our results enabled the establishment of the molecular epidemiology of S. agalactiae isolated from Brazilian bovines, which is essential for planning and implementing bacterial species-specific prevention and treatment of mastitis. Last, our data highlighting the presence of the genes fbsB, hylB, cfb, and PI-2b can facilitate future studies designed to develop a global vaccine against this pathogen.
Funding
This work was supported by grants FAPEMIG APQ 02025/13. We thank FAPEMIG (BPD-00261-14) and CAPES for the student fellowship.
Conflict of interest
The authors declare no competing financial interests.
Associate Editor: Miliane Moreira Soares de Souza
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bjm.2017.02.004.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.FAO . 2013. Food and Agriculture Organization of the United Nations. http://faostat3.fao.org Accessed 02.09.2016. [PubMed] [Google Scholar]
- 2.Keefe G. Update on control of Staphylococcus aureus and Streptococcus agalactiae for management of mastitis. Vet Clin North Am Food Anim Pract. 2012;28:203–216. doi: 10.1016/j.cvfa.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 3.de Vliegher S., Fox L.K., Piepers S., McDougall S., Barkema H.W. Invited review: mastitis in dairy heifers: nature of the disease, potential impact, prevention, and control. J Dairy Sci. 2012;95:1025–1040. doi: 10.3168/jds.2010-4074. [DOI] [PubMed] [Google Scholar]
- 4.Zadoks R.N., Middleton J.R., McDougall S., Katholm J., Schukken Y.H. Molecular epidemiology of mastitis pathogens of dairy cattle and comparative relevance to humans. J Mammary Gland Biol Neoplasia. 2011;16:357–372. doi: 10.1007/s10911-011-9236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rato M.G., Bexiga R., Florindo C., Cavaco L.M., Vilela C.L., Santos-Sanches I. Antimicrobial resistance and molecular epidemiology of Streptococci from bovine mastitis. Vet Microbiol. 2013;161:286–294. doi: 10.1016/j.vetmic.2012.07.043. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y., Liu Y., Ding Y. Molecular characterization of Streptococcus agalactiae isolated from bovine mastitis in eastern China. PLOS ONE. 2013;8:e67755. doi: 10.1371/journal.pone.0067755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans J.J., Bohnsack J.F., Klesius P.H. Phylogenetic relationships among Streptococcus agalactiae isolated from piscine, dolphin, bovine and human sources: a dolphin and piscine lineage associated with a fish epidemic in Kuwait is also associated with human neonatal infections in Japan. J Med Microbiol. 2008;57:1369–1376. doi: 10.1099/jmm.0.47815-0. [DOI] [PubMed] [Google Scholar]
- 8.Kong F., Lambertsen L.M., Slotved H.C., Ko D., Wang H., Gilbert G.L. Use of phenotypic and molecular serotype identification methods to characterize previously nonserotypeable group B Streptococci. J Clin Microbiol. 2008;46:2745–2750. doi: 10.1128/JCM.00189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajagopal L. Understanding the regulation of Group B streptococcal virulence factors. Future Microbiol. 2009;4:201–221. doi: 10.2217/17460913.4.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma P., Lata H., Arya D.K. Role of pilus proteins in adherence and invasion of Streptococcus agalactiae to the lung and cervical epithelial cells. J Biol Chem. 2013;288:4023–4034. doi: 10.1074/jbc.M112.425728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakib M.A., Longo M., Tazi A. Comparison of the diversilab® system with multi-locus sequence typing and pulsed-field gel electrophoresis for the characterization of Streptococcus agalactiae invasive strains. J Microbiol Methods. 2011;85:137–142. doi: 10.1016/j.mimet.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Jones N., Bohnsack J.F., Takahashi S. Multilocus sequence typing system for group B Streptococcus. J Clin Microbiol. 2003;41:2530–2536. doi: 10.1128/JCM.41.6.2530-2536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poyart C., Tazi A., Reglier-Poupet H. Multiplex PCR assay for rapid and accurate capsular typing of group B Streptococci. J Clin Microbiol. 2007;45:1985–1988. doi: 10.1128/JCM.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maiden M.C.J. Multilocus sequence typing of bacteria. Annu Rev Microbiol. 2006;60:561–588. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- 15.Pérez-Losada M., Cabezas P., Castro-Nallar E., Crandall K.A. Pathogen typing in the genomics era: MLST and the future of molecular epidemiology. Infect Genet Evol. 2013;16:38–53. doi: 10.1016/j.meegid.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 16.MacFaddin J. Lippincott Williams & Wilkins; Philadelphia, PA: 2000. Biochemical Tests for Identification of Medical Bacteria. [Google Scholar]
- 17.Mata A.I., Gibello A., Casamayor A., Blanco M.M., Dominguez L., Fernandez-Garayzabal J.F. Multiplex PCR assay for detection of bacterial pathogens associated with warm-water streptococcosis in fish. Appl Environ Microbiol. 2004;70:3183–3187. doi: 10.1128/AEM.70.5.3183-3187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slotved H.C., Kong F., Lambertsen L., Sauer S., Gilbert G.L., Serotype I.X. a proposed new Streptococcus agalactiae serotype. J Clin Microbiol. 2007;45:2929–2936. doi: 10.1128/JCM.00117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ewing B., Hillier L., Wendl M.C., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 20.Feil E.J., Li B.C., Aanensen D.M., Hanage W.P., Spratt B.G. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godoy D.T., Carvalho-Castro G.A., Leal C.A.G., Pereira U.P., Leite R.C., Figueiredo H.C.P. Genetic diversity and new genotyping scheme for fish pathogenic Streptococcus agalactiae. Lett Appl Microbiol. 2013;57:476–483. doi: 10.1111/lam.12138. [DOI] [PubMed] [Google Scholar]
- 22.Oliveira I.C.M., de Mattos M.C., Pinto T.A. Genetic relatedness between group B Streptococci originating from bovine mastitis and a human group B streptococcus type V cluster displaying an identical pulsed-field gel electrophoresis pattern. Clin Microbiol Infect. 2006;12:887–893. doi: 10.1111/j.1469-0691.2006.01508.x. [DOI] [PubMed] [Google Scholar]
- 23.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 25.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 27.Corrêa A.B.D.A., Oliveira I.C.M.D., Pinto T.D.C.A., Mattos M.C.D., Benchetrit L.C. Pulsed-field gel electrophoresis, virulence determinants and antimicrobial susceptibility profiles of type Ia group B Streptococci isolated from humans in Brazil. Mem Inst Oswaldo Cruz. 2009;104:599–603. doi: 10.1590/s0074-02762009000400011. [DOI] [PubMed] [Google Scholar]
- 28.Santi I., Maione D., Cesira L.G., Grandi G., John L.T., Soriani M. BibA induces opsonizing antibodies conferring in vivo protection against group B Streptococcus. J Infect Dis. 2009;200:564–570. doi: 10.1086/603540. [DOI] [PubMed] [Google Scholar]
- 29.Martins E.R., Melo-Cristino J., Ramirez M. Evidence for rare capsular switching in Streptococcus agalactiae. J Bacteriol. 2010;192:1361–1369. doi: 10.1128/JB.01130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter P.R., Gaston M.A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braga P.A.C., Tata A., dos Santos V.G. Bacterial identification: from the agar plate to the mass spectrometer. RSC Adv. 2013;3:994–1008. [Google Scholar]
- 32.Duarte R.S., Miranda O.P., Bellei B.C., Brito M.A.V.P., Teixeira L.M. Phenotypic and molecular characteristics of Streptococcus agalactiae isolates recovered from milk of dairy cows in Brazil. J Clin Microbiol. 2004;42:4214–4222. doi: 10.1128/JCM.42.9.4214-4222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinto T.C.A., Costa N.S., Souza A.R.V. Distribution of serotypes and evaluation of antimicrobial susceptibility among human and bovine Streptococcus agalactiae strains isolated in Brazil between 1980 and 2006. Braz J Infect Dis. 2013;17:131–136. doi: 10.1016/j.bjid.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merl K., Abdulmawjood A., Lämmler C., Zschöck M. Determination of epidemiological relationships of Streptococcus agalactiae isolated from bovine mastitis. FEMS Microbiol Lett. 2003;226:87–92. doi: 10.1016/S0378-1097(03)00564-0. [DOI] [PubMed] [Google Scholar]
- 35.Brochet M., Couvé E., Zouine M. Genomic diversity and evolution within the species Streptococcus agalactiae. Microbes Infect. 2006;8:1227–1243. doi: 10.1016/j.micinf.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Corrêa A.B.D.A., Silva L.G.D., Pinto T.D.C.A. The genetic diversity and phenotypic characterisation of Streptococcus agalactiae isolates from Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz. 2011;106:1002–1006. doi: 10.1590/s0074-02762011000800017. [DOI] [PubMed] [Google Scholar]
- 37.Dutra V.G., Alves V.M.N., Olendzki A.N. Streptococcus agalactiae in Brazil: serotype distribution, virulence determinants and antimicrobial susceptibility. BMC Infect Dis. 2014;14:323–329. doi: 10.1186/1471-2334-14-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otaguiri E., Morguette A.E., Tavares E. Commensal Streptococcus agalactiae isolated from patients seen at University Hospital of Londrina, Paraná, Brazil: capsular types, genotyping, antimicrobial susceptibility and virulence determinants. BMC Microbiol. 2013;13:297. doi: 10.1186/1471-2180-13-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmeiro J.K., Dalla-Costa L.M., Fracalanzza S.E.L. Phenotypic and genotypic characterization of group B streptococcal isolates in southern Brazil. J Clin Microbiol. 2010;48:4397–4403. doi: 10.1128/JCM.00419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cieslewicz M.J., Chaffin D., Glusman G. Structural and genetic diversity of group B Streptococcus capsular polysaccharides. Infect Immun. 2005;73:3096–3103. doi: 10.1128/IAI.73.5.3096-3103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao K., Poulsen K., Maione D. Capsular gene typing of Streptococcus agalactiae compared to serotyping by latex agglutination. J Clin Microbiol. 2013;51:503–507. doi: 10.1128/JCM.02417-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Springman A.C., Lacher D.W., Wu G. Selection, recombination, and virulence gene diversity among group B streptococcal genotypes. J Bacteriol. 2009;191:5419–5427. doi: 10.1128/JB.00369-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosinski-Chupin I., Sauvage E., Mairey B. Reductive evolution in Streptococcus agalactiae and the emergence of a host adapted lineage. BMC Genomics. 2013;14:252. doi: 10.1186/1471-2164-14-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tettelin H., Masignani V., Cieslewicz M.J. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial pan-genome. Proc Natl Acad Sci U S A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morozumi M., Wajima T., Kuwata Y. Associations between capsular serotype, multilocus sequence type, and macrolide resistance in Streptococcus agalactiae isolates from Japanese infants with invasive infections. Epidemiol Infect. 2014;142:812–819. doi: 10.1017/S0950268813001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorensen U.B.S., Poulsen K., Ghezzo C., Margarit I., Kilian M. Emergence and global dissemination of host-specific Streptococcus agalactiae clones. mBio. 2010;1 doi: 10.1128/mBio.00178-10. e00178–00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richards V.P., Palmer S.R., Bitar P.D.P. Phylogenomics and the dynamic genome evolution of the genus Streptococcus. Genome Biol Evol. 2014;6:741–753. doi: 10.1093/gbe/evu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Devi A.S., Ponnuraj K. Cloning, expression, purification and ligand binding studies of novel fibrinogen-binding protein FbsB of Streptococcus agalactiae. Protein Expr Purif. 2010;74:148–155. doi: 10.1016/j.pep.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 49.Rosenau A., Martins K., Amor S. Evaluation of the ability of Streptococcus agalactiae strains isolated from genital and neonatal specimens to bind to human fibrinogen and correlation with characteristics of the fbsA and fbsB genes. Infect Immun. 2007;75:1310–1317. doi: 10.1128/IAI.00996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duarte R.S., Bellei B.C., Miranda O.P., Brito M.A.V.P., Teixeira L.M. Distribution of antimicrobial resistance and virulence-related genes among Brazilian group B Streptococci recovered from bovine and human sources. Antimicrob Agents Chemother. 2005;49:97–103. doi: 10.1128/AAC.49.1.97-103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dore N., Bennett D., Kaliszer M., Cafferkey M., Smyth C.J. Molecular epidemiology of group B Streptococci in Ireland: associations between serotype, invasive status and presence of genes encoding putative virulence factors. Epidemiol Infect. 2003;131:823–833. doi: 10.1017/s0950268803008847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kong F., Gowan S., Martin D., James G., Gilbert G.L. Serotype identification of group B Streptococci by PCR and sequencing. J Clin Microbiol. 2002;40:216–226. doi: 10.1128/JCM.40.1.216-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Springman A., Lacher D.W., Waymire E.A. Pilus distribution among lineages of group B Streptococcus: an evolutionary and clinical perspective. BMC Microbiol. 2014;14:159. doi: 10.1186/1471-2180-14-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Konto-Ghiorghi Y., Mairey E., Mallet A. Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 2009;5:e1000422. doi: 10.1371/journal.ppat.1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rinaudo C.D., Rosini R., Galeotti C.L. Specific involvement of pilus type 2 a in biofilm formation in group B Streptococcus. PLoS ONE. 2010;5:e9216. doi: 10.1371/journal.pone.0009216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chattopadhyay D., Carey A.J., Caliot E. Phylogenetic lineage and pilus protein Spb1/SAN1518 affect opsonin-independent phagocytosis and intracellular survival of Group B Streptococcus. Microbes Infect. 2011;13:369–382. doi: 10.1016/j.micinf.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.