Abstract
Seed setting is an important trait that contributes to seed yield and relies greatly on starch accumulation. In this study, a sulfoquinovosyl transferase-like protein, designated as SQD2.2 involved in seed setting and flavonoid accumulation, was identified and characterized in rice. Rice SQD2.2 is localized to the cytoplasm, and the SQD2.2 transcript was highest in leaves. Rice SQD2.2-overexpressing (OE) plants exhibited a decreased seed setting rate and diminished tiller number simultaneously with an increased glycosidic flavonoid level compared with wild-type (WT) plants. SQD2.2 catalyzes the glycosylation of apigenin to produce apigenin 7-O-glucoside using uridine diphosphate-glucose (UDPG) as a sugar donor, but it failed to compensate for sulfoquinovosyldiacylglycerol (SQDG) synthesis in the Arabidopsis sqd2 mutant. Furthermore, apigenin 7-O-glucoside inhibited starch synthase (SS) activity in a concentration-dependent manner, and SQD2.2-OE plants exhibited reduced SS activity accompanied by a significant reduction in starch levels and an elevation in soluble sugar levels relative to WT plants. Both adenosine diphosphate-glucose (ADPG) and UDPG levels in SQD2.2-OE plants were notably lower than those in WT plants. Taken together, rice SQD2.2 exhibits a novel role in flavonoid synthesis and plays an important role in mediating sugar allocation between primary and secondary metabolism in rice.
Introduction
Cereal grain, mostly in the form of starch, is a major product that supplies energy for human activity, feedstock and industrial materials. The seed setting rate is a major factor controlling seed production and is a complicated trait regulated by the combination of genetic, physiological and environmental factors. Seed setting relies on source and sink allocation and is contributed to largely by starch accumulation, which is the result of complex enzymatic processes, with ADP-glucosepyrophosphorylase (AGPase) and starch synthase (SS) being key players1–3. Starch synthesis is catalyzed by SS through the transfer of the glucose moiety from ADPG to an elongated glucan chain in an α-1,4-linked manner. AGPase, a heterotetrameric enzyme consisting of two small subunits and two large subunits, catalyzes a reaction between glucose-1-phosphate and ATP to produce ADPG, which provides a critical substrate fueling starch synthesis1, 4. A deficiency of starch synthesis results in a reduced seed yield in plants5, 6. Increased AGPase activity in the endosperm promotes grain filling and increased seed yield in wheat and rice2, 7. Furthermore, starch also serves as a storage sugar and is important for plant growth. Depleting starch from leaves causes growth inhibition and delayed flowering in Arabidopsis8. These results suggest that starch synthesis in both the sink and the source is important for seed setting and plant growth.
Glycosylation, the process of transferring an active sugar moiety to acceptor molecules, occurs in diverse reactions involved in primary and secondary metabolic processes such as starch synthesis, flavonoid modification and galactolipid synthesis9. Flavonoids belong to a group of secondary metabolites composed largely of 900 molecular species, including flavonols, flavones, anthocyanins and proanthocyanidins. Glycosylation is an important process enabling the solubility and stability of hydrophobic flavonoids, and most flavonoids are present in plant tissues in glycosidic form9, 10. For example, a deficiency in UDP-sugar: 3-O-glycosyltransferase (UF3GT) activity results in a significant reduction in anthocyanin accumulation in Arabidopsis11. Glycosylation is catalyzed by UDP-sugar: glycosyltransferase (UGT), in which the C-terminus contains a conserved plant secondary product glycosyltransferase (PSPG) motif responsible for binding to the UDP moiety of sugar donors12, 13. The genes involved in flavonoid backbone biosynthesis have been extensively characterized in Arabidopsis. In comparison, the UGTs encoded by diverse genes belong to the glycosyltransferase (GT) super-family, which makes it difficult to screen target UGT genes. To date, more than 1500 putative UGT genes have been identified from various plant genomes based on the highly conserved PSPG motif. Many UGTs behave in a more regiospecific manner than a substrate-specific manner9, 14. Glycosylation usually occurs with 3-, 5- or 7-hydroxyl regiospecificity of the flavonoid core structure and is catalyzed by 3-O-glycosyltransferase (F3GT), 5-O-glycosyltransferase (F5GT) and 7-O-glycosyltransferase (F7GT), respectively14, 15.
Flavonoids function as pigments, ultraviolet (UV) protectants, attractants of pollinators, phytoalexins, signaling molecules, regulators of fertility and regulators of auxin transport16–20. The precise relationships between flavonoid species and their physiological functions are still largely unknown due to the huge diversity of structures and distribution in plants. Glycosidic apigenin (4,5,7-trihydroxyflavone), one of the flavonoids widely found in plants, functions as an antioxidant to relieve lipid peroxidation and free radicals and aid in defense21. Flavonoids function as multidrug-resistant proteins20, which are important for organismal growth, development and morphogenic modeling in response to nutrient availability and stimuli16, 19, 22. In addition, flavonoids are also involved in auxin transport and the jasmonic acid (JA) response20, 22, 23. Quercetin inhibits auxin efflux transport and multidrug-resistant proteins20. Pharmacological treatment showed that flavonoids inhibit the activity of various kinases involved in growth and development in eukaryotic cells24. The data suggest that flavonoids have multiple effects, integrating plant growth, development and the stress response. Flavonoid accumulation was found in plant tissues under different stress conditions such as salt, drought, low temperature, nutrient deficiency and UV irradiation, along with growth arrest and seed yield reduction in plants25–28. However, the direct link between flavonoid and sugar metabolism or seed yield is unclear. Moreover, sugars provide the substrates for flavonoid synthesis, and the crosstalk between primary metabolic functions such as starch synthesis and secondary metabolic functions remains to be elucidated in plants.
Sulfoquinovosyldiacylglycerol (SQDG) synthase (SQD) is also a glycosyltransferase that catalyzes the transfer of the sulfoquinovose moiety to diacylglycerol (DAG) to generate SQDG29. SQDG is a sulfur-containing anionic glycerolipid that is exclusively located in thylakoid membranes. The photosystem II complex possesses three molecules of SQDG in addition to monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG) and phosphatidylglycerol (PG)30, 31. SQDG was also found in the cytochrome b6-f complex30. SQDG has a negative charge similar to PG and is capable of replacing PG under phosphate starvation conditions. In plants, SQDG is synthesized via a three-step reaction in chloroplasts. First, UDPG is synthesized from glucose-1-phosphate and uridine-5′-triphosphate (UTP) by UGP3, a plastid UDPG pyrophosphorylase (UDPase)32. Subsequently, SQD1, the SqdB ortholog, catalyzes a reaction between UDPG and sulfite to produce UDP-sulfoquinovose33. Finally, SQDG is produced by SQDG synthase, designated as SQD2, using UDP-sulfoquinovose and DAG as substrates29. Both UGP3 and SQD1 are soluble enzymes and are localized to the chloroplast stroma in Arabidopsis32, 33. SQD2 is localized to the inner envelope membrane of chloroplasts in Arabidopsis29. Genetic mutation in any of these enzymes, UGP3, SQD1 or SQD2, results in a complete loss of SQDG from cells, and Arabidopsis plants with these mutations exhibit growth retardation under phosphate starvation conditions29, 32, 33. Among the three SQDG-deficient mutants of Arabidopsis, the severe growth defect and lack of the novel anionic glycolipid glucuronosyldiacylglycerol (GlcADG) were found only in the sqd2 mutants under phosphate-limited conditions34, suggesting that the function of Arabidopsis SQD2 (AtSQD2) is not only required for SQDG synthesis but also for GlcADG synthesis in Arabidopsis. These results indicate that the SQD2 enzyme may have diverse substrates in plants. In this study, we identified a SQD2-like enzyme, designated as SQD2.2, in rice (Oryza sativa L). Rice SQD2.2 was found to be a UGT enzyme that catalyzes the glycosylation of apigenin to produce apigenin 7-O-glucoside but has no detectable SQDG synthase activity. Overexpression of rice SQD2.2 enhances flavonoid accumulation, resulting in a reduced seed setting rate and diminished tiller number accompanied by a reduced starch content in both source and sink tissues. Apigenin 7-O-glucoside, the product of SQD2.2 activity, inhibited SS activity. Moreover, SQD2.2-overexpressing (OE) plants had a significant reduction in ADPG and UDPG compared with wild-type (WT) plants, suggesting that enhanced production of flavonoid by SQD2.2 results in sugar substrate competition and thus a reduced requirement for ADPG for starch synthesis. These results suggest that rice SQD2 exhibits multifaceted enzymatic properties with different substrate selections and regulates the partitioning of carbon between primary and secondary metabolism in rice.
Results
Identification of the SQDG Synthase in Rice
SQD2 belongs to the GT superfamily that contains a glycosyltransferase catalytic domain (Fig. S1). To functionally characterize the role of rice SQD2, homology searches and protein domain structure prediction were performed using AtSQD2 as a query, and three putative SQD2 genes were identified from the rice genome and were designated as SQD2.1, SQD2.2 and SQD2.3 based on sequence similarity (Fig. S1). Rice SQD2.1, SQD2.2 and SQD2.3 share 72%, 67% and 68% amino acid sequence similarity to Arabidopsis SQD2, respectively. Rice SQD2.1 is related most closely to Arabidopsis SQD2. Phylogenetic analysis showed that SQD2.2 is classified into the group of SQD2 homologs in Arabidopsis and Synechococcus elongatus PCC7924 but is relatively distant from the UGTs involved in flavonoid glycosylation (Fig. S2). Rice SQD2.2 encodes a protein of 515 amino acids with a predicted pI of 7.23 and a molecular weight of 57.9 kDa. The role of SQD2.2 is unknown.
Overexpression of SQD2.2 Results in Reduced Seed Setting
To explore the role of rice SQD2.2 encoding a putative SQDG synthase, a full-length cDNA of rice SQD2.2 was cloned and overexpressed in rice under the control of the maize Ubiquitin promoter. A total of 41 transgenic lines were obtained and were confirmed by PCR. The sequencing result of PCR product in Fig. 1A is identical to the sequence of SQD2.2 cDNA from data base (Fig. S3). Most transgenic lines exhibited a significantly higher transcript level than that of WT plants without transformation. Overexpression of SQD2.2 resulted in a decreased seed setting rate under normal growth conditions in comparison to WT. To further explore the role of SQD2.2, three OE lines, namely, OE14, OE30 and OE40, were randomly selected for detailed characterization (Fig. 1A–C). The seed setting rates of OE14, OE30 and OE40 plants were 27.5%, 27.8% and 23.3%, respectively, whereas that of WT was 82.4% (Fig. 1D and E). The seed yields in OE14, OE30 and OE40 were 47.3%, 52.3% and 57.3% of that in WT plants (Fig. 1F). In addition, the overexpression of SQD2.2 also led to reduced total tiller number and shoot biomass (Fig. 1G and H). The pollen viability in OE plants was not significantly different from that in WT plants (Fig. S4), suggesting that the reduced seed setting rate did not result from pollen fertility.
Figure 1.
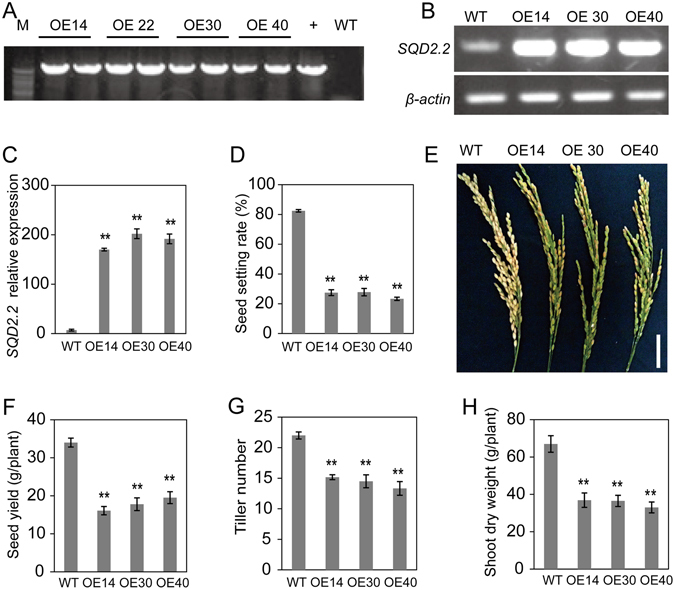
Overexpression of SQD2.2 resulted in a reduced seed setting rate and tiller number. (A) Identification of transgenic plants using PCR. (B) and (C) SQD2.2 overexpression in the transgenic plants was confirmed via semi-quantitative RT-PCR and quantitative real-time PCR. Values are the mean ± standard error (SE) (n = 3). (D) and (E) Seed setting rate. Values are the mean ± SE (n = 10). (F) Seed yield per plant. Values are the mean ± SE (n = 10). (G) The tiller number per plant. Values are the mean ± SE (n = 6). (H) Shoot dry weight per plant. Values are the mean ± SE (n = 10). Student’s t test; **P < 0.01. Scale bar = 5 cm.
SQD2.2 Overexpression Enhances Flavonoid Accumulation
In addition to the reduced seed setting and tiller number, the overexpression of SQD2.2 also conferred plants with enhanced flavonoid levels in various tissues, including hulls, leaves and stems (Fig. 2A–D). Brown pigments were observed in SQD2.2-OE tissues and were much more obvious than those in WT plants after the chlorophyll was removed from the tissues (Fig. 2A–D). To determine the flavonoid molecular species, the brown pigments were extracted from the hulls and leaves of SQD2.2-OE and WT plants using methanol, and the extracts were subjected to liquid chromatography electrospray ionization–tandem mass spectrometry (LC ESI–MS/MS) profiling. The overexpression of SQD2.2 led to a dramatic increase in glycosidic flavonoids, including apigenin 5-O-glucoside, apigenin 7-O-glucoside, naringenin 7-O-glucoside, chrysoeriol 5-O-hexoside and chrysoeriol 7-O-hexoside; in particular, apigenin 7-O-glucoside and apigenin 5-O-glucoside were greatly increased, with 1,000-fold and 500-fold increases in OE plants relative to WT plants, respectively (Fig. 2E and F). By comparison, the flavonoids without glycosylation, such as apigenin and chrysoeriol, exhibited a small increase in SQD2.2-OE plants, and naringenin was also elevated to some extent relative to WT (Fig. 2E and F). These results suggest that the overexpression of SQD2.2 enhances flavonoid accumulation, with a predominant increase in glycosidic flavonoid derivatives in rice. Moreover, the overexpression of rice SQD2.2 in Arabidopsis also led to flavonoid, especially anthocyanin, accumulation in various tissues, including leaves, stems and siliques (Fig. S5), suggesting that SQD2.2 is able to enhance flavonoid accumulation in both monocot and dicot plants.
Figure 2.
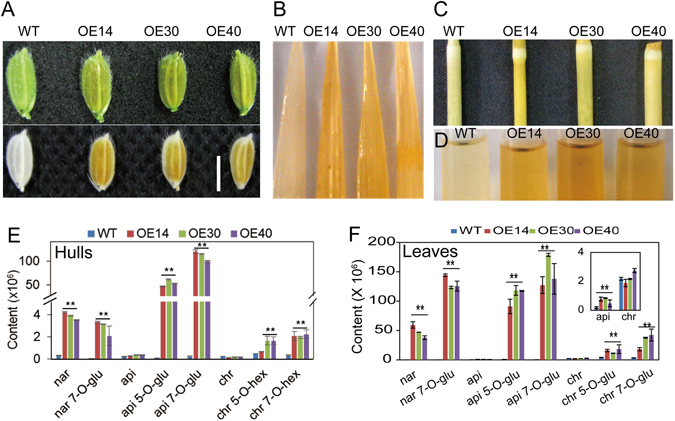
Overexpression of SQD2.2 enhanced flavonoid accumulation in various tissues. (A) to (D) Flavonoid in hulls (A), leaves (B) and stems (C) and methanol extracts from leaves (D). Plants were grown in a paddy field under normal growth conditions, and the samples were photographed after chlorophyll was removed by ethanol. (E) and (F) Overexpression of SQD2.2 increased flavonoid levels, with the predominant accumulation of 7-O-glycosidic and 5-O-glycosidic flavonoids in seed hulls (E) and leaves (F). nar, naringenin; nar 7-O-glu, naringenin 7-O-glucoside; api, apigenin; api 5-O-glu, apigenin 5-O-glucoside; api 7-O-glu, apigenin 7-O-glucoside; chr, chrysoeriol; chr 5-O-hex, chrysoeriol 5-O-hexoside; chr 7-O-hex, chrysoeriol 7-O-hexoside. Values are the mean ± SD (n = 6). Student’s t test; **P < 0.01. Scale bar = 0.5 cm.
SQD2.2 Encodes a Flavonoid Glycosyltransferase
In Arabidopsis, SQDG synthase, designated SQD2, catalyzes the final reaction step in SQDG synthesis by transferring the sulfoquinovose group to DAG29. Rice SQD2.2 also contains a conserved glycosyltransferase catalytic domain and shares 67% sequence similarity with Arabidopsis SQD2 (Fig. S1). To examine whether rice SQD2.2 encodes an SQDG synthase, SQD2.2 was transiently coexpressed with rice SQD1, the gene encoding an enzyme for UDP-sulfoquinovose synthesis, in tobacco leaves. The resultant lipids were extracted and quantitatively analyzed using GC. The SQDG content in the leaves of SQD1/SQD2.2 co-expressing plants was not significantly different from that in the controls, including WT plants, plants transformed with the vector only or plants transformed with either SQD1 or SQD2.2 individually (Fig. 3A). These results suggest that rice SQD2.2 had no detectable SQDG synthase activity under the condition tested. To further confirm this result, rice SQD2.2 was ectopically expressed in an Arabidopsis sqd2 mutant (Fig. 3B). The Arabidopsis sqd2 mutant exhibited defective SQDG, and the ectopic expression of rice SQD2.2 in the mutant failed to restore SQDG synthesis (Fig. 3C). In addition, the overexpression of SQD2.2 did not confer an increased SQDG level in the plants in comparison to WT (Fig. 3D). Taken together, the results indicate that rice SQD2.2 does not exhibit SQDG synthase activity.
Figure 3.
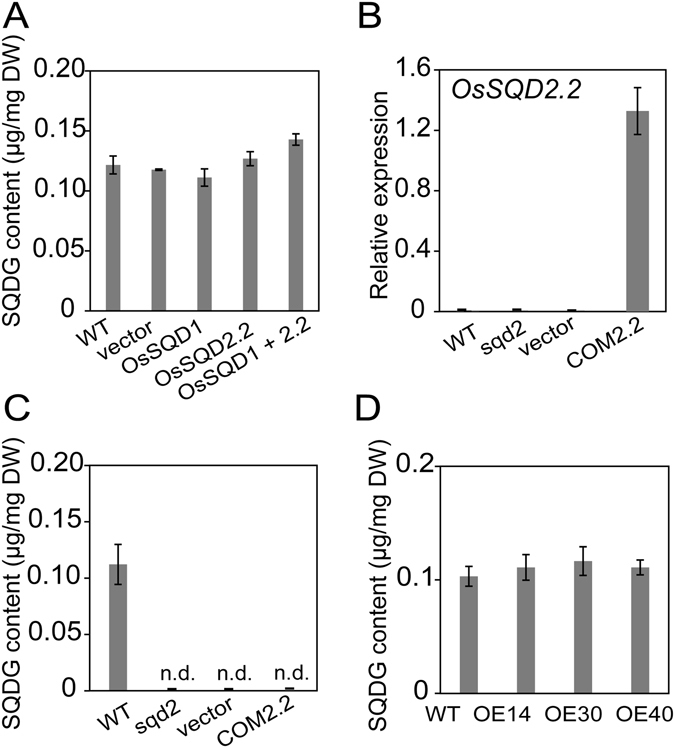
SQD2.2 has no detectable SQDG synthase activity. (A) Co-expression of SQD1 and SQD2.2 in tobacco leaves did not result in elevated SQDG content relative to the controls, WT, vector and leaves transformed with either SQD1 or SQD2.2 individually. Values are the mean ± SE (n = 3). (B) and (C) Genetic complementation with rice SQD2.2 in the Arabidopsis sqd2 mutant failed to restore SQDG synthesis. Lipids were extracted from the leaves of four-week-old seedlings and separated by TLC for GC measurement. Values are the mean ± SE (n = 3). COM2.1, COM2.1, the plants that were genetically complemented with rice SQD2.1 and SQD2.2, respectively, in the Arabidopsis Atsqd2 mutant. n.d., not detectable. (D) Overexpression of SQD2.2 in rice plants did not increase SQDG content relative to WT plants. Lipids were extracted from leaves at the seed filling stage. Values are the mean ± SE (n = 3).
The overexpression of SQD2.2 resulted in a significant increase in flavonoids, with a dramatic elevation in glycosidic flavonoids (Fig. 2), but it had no effect on SQDG content relative to WT plants (Fig. 3D), implicating that rice SQD2.2 may encode a flavonoid glycosyltransferase rather than a SQDG synthase. To test this possibility, full-length rice SQD2.2 cDNA was cloned into the pET28a vector to express the protein. The recombinant SQD2.2-His protein, which has a molecular weight of 57.9 kDa, was expressed in E. coli cells and was purified for an enzymatic activity assay (Fig. 4A). The in vitro assay indicated that SQD2.2 catalyzes the transfer of the glucose moiety from UDPG to 7-hydroxyl of apigenin to produce apigenin 7-O-glucoside (Fig. 4B and E). In comparison, the control cells containing empty vector did not exhibit detectable levels of apigenin 7-O-glucoside (Fig. 4D). To further test the substrate preference, different sugar donors were used for the enzymatic activity assay. The results showed that SQD2.2 catalyzed the transfer of the glucose moiety from UDPG to apigenin but failed to use ADPG and UDP-galactose (UDPGal) as sugar donors for apigenin (Fig. 4B and E). In addition, SQD2.2 was also able to transfer the glucose moiety of UDPG to naringenin to produce naringenin 7-O-glucoside (Fig. 4C and F). These results demonstrate that rice SQD2.2 encodes a flavonoid glycosyltransferase rather than an SQDG synthase. SQD2.2 is directly responsible for flavonoid accumulation, but not for SQDG synthesis, in rice plants.
Figure 4.
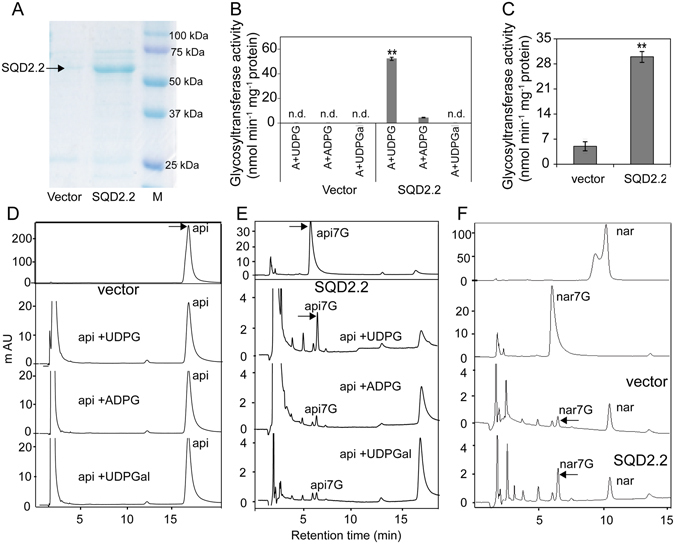
SQD2.2 catalyzed the transfer of glucose from UDPG to apigenin and naringenin to generate apigenin 7-O-glucoside and naringenin 7-O-glucoside. (A) SQD2.2 protein was expressed in E. coli cells and was purified for the enzymatic activity assay. (B), (D) and (E) SQD2.2 activity toward apigenin to produce apigenin 7-O-glucoside using UDPG, but not ADPG or UDPGal, as a sugar donor (B,E). Protein from the cells expressing empty vector only was used as a control (D). Values are the mean ± SE (n = 3). (C) and (F) SQD2.2 activity toward naringenin to produce naringenin 7-O-glucoside using UDPG as a sugar donor. Purified SQD2.2 protein (5 μg) was added to 100 μl of the reaction mixture. Protein from the cells expressing empty vector only was used as a control. n.d., not detectable; M, protein ladder marker; A (api), apigenin; api7G, apigenin 7-O-glucoside; nar, naringenin; nar7G, naringenin 7-O-glucoside; UDPG, UDP-glucose; ADPG, ADP-glucose; UDPGal, UDP-galactose. Values are the mean ± SE (n = 3). Student’s t test; **P < 0.01.
SQD2.2 is Predominantly Expressed in Green Tissues, and SQD2.2 is Predominantly Localized to the Cytoplasm
Quantitative real-time PCR revealed that SQD2.2 is expressed in various organs, including leaves, roots, stems and panicles, with higher levels in leaves and lower levels in roots at the different stages tested; SQD2.2 levels were highest in leaf blades during the mature stage (Fig. 5A). The SQD2.2 transcript level in leaf sheaths was also relatively high during the seedling and mature stages (Fig. 5A). It exhibited a similar pattern by histochemical GUS staining in rice plants containing GUS gene under the control of SQD2.2 promoter (Fig. 5B). To test its subcellular localization, SQD2.2 was fused with GFP and was transiently expressed in the epidermal cells of tobacco leaves. The GFP-SQD2.2 fusion protein was predominantly localized in the cytoplasm (Fig. 5C), which differs from the chloroplast-localized AtSQD229. The cytosolic localization of rice SQD2.2 further demonstrates its role in flavonoid glycosyltransferase rather than SQDG synthesis, which occurs exclusively in chloroplasts in plants.
Figure 5.
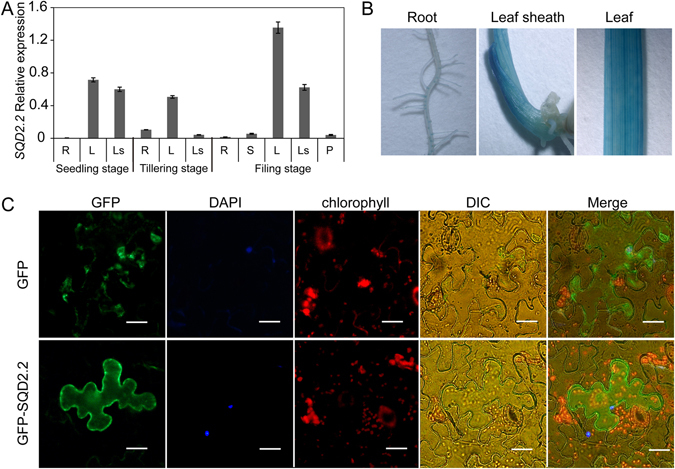
Expression patterns of SQD2.2 in rice and SQD2.2 is localized to the cytoplasm. (A) SQD2.2 is expressed in different tissues. The SQD2.2 transcript level was quantified with real-time PCR using β-actin as an internal standard. R, roots; S, stems; L, leaves; Ls, leaf sheaths; P, panicles. Values are the mean ± SE (n = 3). (B) Histochemical GUS staining patterns of pSQD2.2::GUS transgenic plants. (C) GFP-SQD2.2 was transiently expressed in tobacco epidermal cells, and the subcellular localization was visualized under a fluorescence microscope. Scale bar = 10 μm.
Starch Synthase (SS) Activity is Inhibited by Glycosidic Flavonoids
Overexpression of SQD2.2 led to increased flavonoid levels in various tissues accompanied by a reduced seed setting rate. To test the effect of SQD2.2 on starch synthase activity in vivo, the different classes of enzymes contributing to starch synthesis were measured at the tillering and seed filling stages. The soluble SS activity in the leaves of SQD2.2-OE plants was significantly lower than that in WT and was decreased by 31.1%, 29.6% and 43.6% in the leaves of OE14, OE30 and OE40, respectively (Fig. 6A). Likewise, the soluble SS activity in the filling seeds of SQD2.2-OE plants was significantly decreased at 15 days after anthesis, with 35%, 45.5% and 32.2% reductions in the filling seeds of OE14, OE30 and OE40, respectively, relative to WT (Fig. 6B). In addition, the activities of granule-bound SS (GBSS) and starch branching enzyme (SBE) were reduced in OE plants (Fig. 6C and D). The activity of amylase, the enzyme involved in starch degradation, did not differ between OE and WT plants (Fig. 6E). To test whether glycosidic flavonoids directly affect starch biosynthesis, SS activity was monitored in response to apigenin 7-O-glucoside treatment at various concentrations. The results showed that SS activity was significantly reduced in the presence of apigenin 7-O-glucoside and that the inhibition occurred in a concentration-dependent manner (Fig. 6F). In comparison, the SQD2.2 protein itself did not significantly influence SS activity (Fig. 6G).These data suggest that SQD2.2 catalyzes the synthesis of glycoside flavonoids, which inhibits starch synthesis and thus reduces seed setting.
Figure 6.
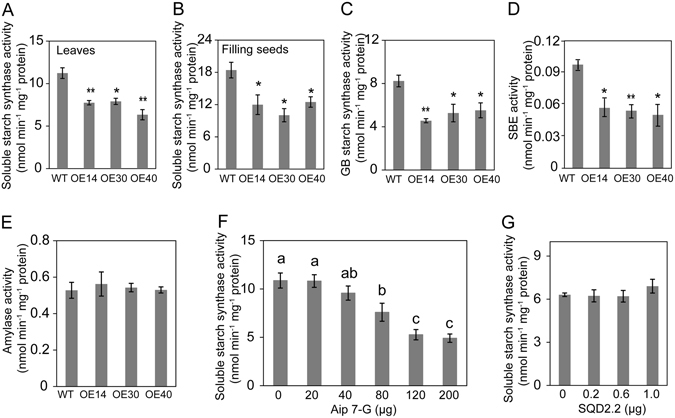
Apigenin 7-O-glucoside inhibited starch synthase activity, and overexpression of SQD2.2 resulted in reduced starch synthase activity. (A) and (B) Soluble starch synthase activity in the leaves and filling seeds of SQD2.2-OE was significantly lower than in that in WT plants. Proteins were extracted from leaves at the tillering stage and extracted from the filling seeds 15 days after anthesis. The same amount of protein (100 μg) from both SQD2.2-OE and WT plants was added to the reaction mixture. Values are the mean ± SE (n = 6). (C) to (E)The activities of granule-bound (GB) starch synthase (C), starch branching enzyme (SBE) (D) and amylase (E) in leaves of SQD2.2-OE and WT plants. Proteins were extracted from leaves at the tillering stage. Values are the mean ± SE (n = 6). (F) Apigenin 7-O-glucoside, the product of SQD2.2, inhibited soluble starch synthase activity in a concentration-dependent manner. Proteins were extracted from the leaves of one-month-old WT plants. Values are the mean ± SE (n = 3). Different letters (a, b and c) indicate significant differences at P < 0.05, according to ANOVA analysis and Duncan’s multiple range test. Api 7-G, apigenin 7-O-glucoside. (G) SQD2.2 itself did not inhibit soluble starch synthase activity. Proteins were extracted from the leaves of one-month-old WT plants and were treated with purified SQD2.2 protein in various concentrations. Values are the mean ± SE (n = 3). Student’s t test; *P < 0.05, **P < 0.01.
SQD2.2 Affects Sugar Allocation between Primary and Secondary Metabolism
SQD2.2 used UDPG to participate in the synthesis of glycosidic flavonoids (Fig. 4), while ADPG is a critical substrate for starch biosynthesis35. Both ADPG and UDPG share a common substrate, glucose-1-phosphate. To investigate whether the overexpression of SQD2.2 affects ADPG synthesis due to sugar substrate competition, ADPG and UDPG levels were measured in SQD2.2-OE and WT plants. The ADPG contents in all three SQD2.2-OE lines were significantly lower than that in WT plants, demonstrating decreases of 53.5%, 41.8% and 55.1% in OE14, OE30 and OE40 plants, respectively (Fig. 7A). Likewise, UDPG also exhibited a similar tendency (Fig. 7B). These results suggest that upregulated SQD2.2 in OE plants leads to increased consumption of UDPG, thus significantly reducing the ADPG available for starch synthesis due to substrate competition.
Figure 7.
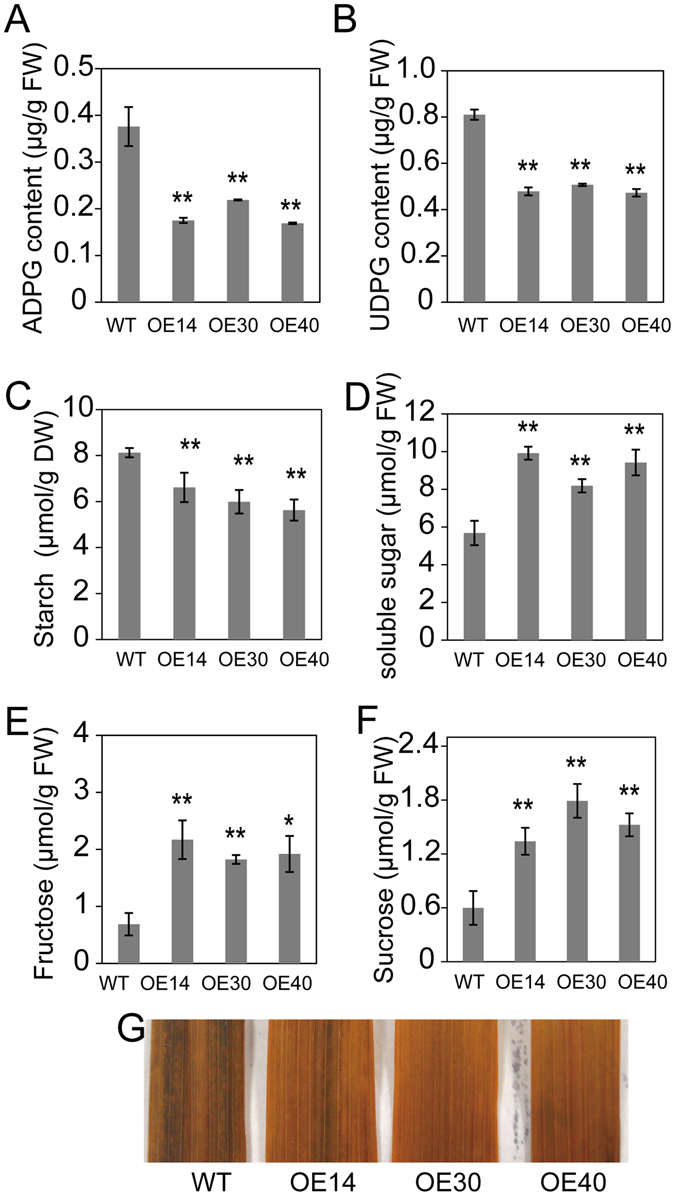
Overexpression of SQD2.2 resulted in reduced UDPG and ADPG content and decreased starch synthesis. (A) and (B) The ADPG and UDPG content in SQD2.2-OE leaves was significantly lower than that in WT plants at the tillering stage. Values are the mean ± SE (n = 6). (C) to (F) Reduced starch and elevated soluble sugar levels in SQD2.2-OE plants compared to WT plants. Samples were taken at the tillering stage. Values are the mean ± SE (n = 6). (G) Starch accumulation in leaves was visualized by KI-I2 staining. Leaf pigments were removed by 96% ethanol, and then leaves were stained with iodine solution (2% KI, 1% I2) for 30 min. Student’s t test; *P < 0.05, **P < 0.01.
To investigate whether overexpression of SQD2.2 influences sugar metabolism, the starch and soluble sugar content were tested in the leaves of SQD2.2-OE and WT plants on a sunny day during the heading stage. The starch content in SQD2.2-OE leaves was significantly lower than that in WT plants; the starch content in OE14, OE30 and OE40 was 81.4%, 73.7% and 69.3% of that in WT, respectively (Fig. 7C and G). In contrast, the level of soluble sugars, including fructose and sucrose, in the leaves of SQD2.2-OE plants exhibited a two-fold increase over the level in WT plants (Fig. 7D–F). These results suggest that overexpression of SQD2.2 led to reduced starch accumulation and an increase in soluble sugars. Taken together, these data suggest that SQD2.2 catalyzes the synthesis of glycosidic flavonoids, which inhibits SS activity and reduces the carbon flux for starch synthesis, thus reducing the seed setting rate and biomass accumulation in rice.
Discussion
Rice SQD2.2 is Involved in Flavonoid Glycosylation Rather than SQDG Synthesis
SQDG belongs to a class of sulfur-containing lipids that are primarily located in the thylakoid membrane and is conditionally important for photosynthetic bacteria, alga and plants due to its ability to replace anionic PG in response to phosphate limitation. Loss of SQDG results in defective growth under phosphate-starvation conditions29, 34, 36, 37. In Arabidopsis and spinach, SQDG is exclusively synthesized in chloroplasts, as demonstrated by the chloroplast localization of all enzymes involved in SQDG synthesis29, 32–34. The Arabidopsis genome contains one gene, SQD2, which encodes an enzyme involved in the final step of the SQDG synthesis pathway29. To date, none of the SQD2 orthologs has been functionally characterized in rice. Through homology searches, we found that the rice genome contains three homologous genes, designated SQD2.1, SQD2.2 and SQD2.3. Their encoding proteins share high sequence similarity and contain a conserved glycosyltransferase domain (Fig. S1), suggesting that rice SQD2 orthologs may be more complicated and functionally diverse. Indeed, the data from this study showed that rice SQD2.2 is predominantly localized to the cytoplasm and failed to restore Arabidopsis sqd2 mutant to a WT phenotype, whereas rice SQD2.1 is localized to chloroplasts and is capable of complementing SQDG synthesis in Arabidopsis sqd2 mutant plants. Rice SQD2.2 shares 67% sequence similarity with AtSQD2. Unexpectedly, rice SQD2.2 was identified to be a glycosyltransferase that acts on flavonoids such as apigenin and naringenin to produce glycosidic flavonoids. Furthermore, overexpression of SQD2.2 had no influence on the SQDG content but caused a notable increase in glycosidic flavonoids (Figs 2 and 4). These data suggest that SQD2.2 is primarily involved in flavonoid accumulation rather than in SQDG synthesis in rice. SQD2 orthologs possess a catalytic domain involved in transferring a sugar moiety to an acceptor, usually DAG, to produce SQDG29. However, a recent study showed that Arabidopsis SQD2 is also involved in GlcADG synthesis, but its precise substrate is unclear34. The current results show that rice SQD2.2, an enzyme homologous to AtSQD2, catalyzes the conversion of apigenin and naringenin to apigenin 7-O-glucoside and naringenin 7-O-glucoside, respectively. These results suggest that plant SQD2 orthologs are capable of using different substrates, including DAG and flavonoids, as sugar moiety acceptors and are involved in diverse metabolic processes, such as sulfolipid biosynthesis and flavonoid glycosylation, in plants.
SQD2.2 Exerts Dual Effects on Starch Synthesis to Mediate Primary and Secondary Metabolic Processes
Cereal grain is mostly composed of starch in the endosperm, and starch synthesis contributes largely to seed setting. Increased AGPase activity resulted in enhanced starch accumulation and increased seed yield in rice and wheat1, 2. Reduced seed setting occurred simultaneously with glycosidic flavonoid accumulation in SQD2.2-OE plants (Figs 1 and 2), suggesting that the glycosidic flavonoids produced by SQD2.2 have a negative effect on seed setting. Glycosylation facilitates the solubility and stability of hydrophobic flavonoids10, 12. Flavonoids are involved in various biological processes such as stress response and hormone signaling16, 23, 28, 38. Whether flavonoids affect starch synthesis remains unknown. An enzymatic assay showed that SQD2.2 directly regulates glycosidic flavonoid accumulation. Moreover, increased flavonoid levels were accompanied by reduced starch content in both the leaves and filling seeds of SQD2.2-OE plants, suggesting that the glycosidic flavonoids produced by SQD2.2 have a negative effect on starch synthesis. Indeed, the SS activity was specifically inhibited by apigenin 7-O-glucoside in a concentration-dependent manner but not by SQD2.2 protein. Furthermore, SQD2.2-OE plants exhibited decreased different classes of SS activity in both leaves and filling seeds relative to WT plants (Fig. 6). The reduced starch and seed setting could be explained, at least in part, by an inhibitory effect of glycosidic flavonoids on SS activity, thus disturbing sugar carbon allocation at the sink and source. Flavonoids also inhibit the activity of kinases that control key processes involved in growth and development in animal cells24, 39.
Nevertheless, the negative effect of flavonoids on starch synthesis may also be due to sugar competition between the glycosidic flavonoid and starch synthesis pathways. SS catalyzes the transfer of the glucose moiety from ADPG to an elongated polymer for starch synthesis40, 41. The enzymatic activity assay showed that SQD2.2 selectively used UDPG as a sugar donor for glycosidic flavonoid synthesis, and SQD2.2-OE plants accumulated a large amount of glycosidic flavonoids, with a 1,000-fold increase in apigenin 7-O-glucoside relative to WT plants (Fig. 2E and F). The results suggest that the increased levels of glycosidic flavonoids in SQD2.2-OE plants may consume a large amount of UDPG, thus reducing the ADPG level in starch synthesis, as both ADPG and UDPG share a common precursor, glucose-1-phosphate, and are generated by AGPase and UDPase, respectively1. ADPG is a critical substrate for starch synthesis. The overexpression of AGPase genes enhanced the starch content in wheat, rice and maize1, 2, 4, 42. The current study found that the levels of both ADPG and UDPG in SQD2.2-OE plants were significantly less than those in WT plants. Increasing the level of glycosidic flavonoids occurs at the expense of starch synthesis. Therefore, the reduced starch synthesis in SQD2.2-OE plants resulted from the dual effects of SQD2.2, the inhibitory effect of glycosidic flavonoids on SS activity and sugar substrate competition due to enhanced SQD2.2 glycosyltransferase activity toward flavonoids.
The Effect of SQD2.2 on Tillering and Biomass
Flavonoids usually accumulate in response to osmotic stress and nutrient deficiency and are important for the adaptation of plants to ever-changing environments due to their roles in regulating ROS homeostasis and mitigating oxidative damage19, 43. In addition to a reduced seed setting rate, SQD2.2-OE plants also exhibited a decreased tiller number and thus decreased biomass compared to WT plants (Fig. 1), suggesting the negative effect of SQD2.2 on vegetative growth. Starch is an essential component required for seed filling as well as functions as an important sugar storage molecule to effectively provide nutrients and energy for plant growth44–46. Reduced starch and increased soluble sugar levels may be responsible for the reduced tiller number and biomass. The starch synthesis deficiency mutants ss4 and ss3 ss4 had a deleterious effect on growth and pale phenotypes in Arabidopsis35, 47. It was shown that flavonoids accumulated in response to various stimuli, such as drought, salt stress, nitrogen limitation and phosphorus deficiency, in plants27, 48. The overexpression of SQD2.2 resulted in alterations in both primary and secondary metabolites, including reduced starch content, enhanced flavonoids and increased soluble sugars relative to WT plants. Thus, the reduced tiller number may be the result of two effects, the diminished starch for energy and nutrient supply and the increased flavonoid and soluble sugar levels that resemble nutrient deficiency and osmotic stress.
Materials and Methods
Plant Materials and Growth Conditions
Rice (Oryza sativa. L) plants were derived from cv Zhonghua 11 (ssp. Japonica). Plants were grown in soil at the experimental field of Huazhong Agriculture University (Wuhan, China) during their natural growing season. Surface-sterilized Arabidopsis seeds (Columbia and Wassilewska) were germinated on 0.8% (w/v) agar-solidified Murashige and Skoog (MS) medium supplemented with 1% (w/v) sucrose. After 10 days, the seedlings were transferred to soil in a growth chamber under conditions of 22 °C (14 h light/10 h dark), a photosynthetic photon flux density of 200–300 μmol·m−2·s−1 and 60% relative humidity.
SQD2.2 Cloning and Plant Transformation
Full-length rice SQD2.2 cDNA was amplified from cDNA synthesized from the total RNA of leaves using the forward primer 5′-GGTACCATGGAATATCCCCCACAATT-3′ paired with the reverse primer 5′-GGATCCGTGTTGTGTGATTCTGTTGA-3′. The purified PCR product was inserted into the pMD18-T vector (TaKaRa). After sequencing confirmation, the fragment was released by BamHI and KpnI digestion and was cloned into the pCAMBIA1301U vector under the control of the maize Ubiquitin promoter. The resultant construct was transformed into Agrobacterium tumefaciens EHA105, which was used to infect the rice callus (cv. Zhonghua 11) according to a protocol described previously49. For genetic complementation tests, the construct was also transformed into the Arabidopsis Atsqd2 mutant via flower dipping with an Agrobacterium cell suspension.
Subcellular Localization of SQD2.2
Full-length SQD2.2 cDNA was fused with a GFP tag at the 5′-end and cloned into the pCAMBIA1301 vector after digestion with BamHI and KpnI. The construct was introduced into Agrobacterium tumefaciens GV3101 and then was infiltrated into tobacco leaves (Nicotiana benthamiana) for transient expression under the control of the 35S promoter. After 2 to 3 days of infection, the GFP-SQD2.2 fusion protein was observed under a fluorescence microscope (Olympus, BX53) using a 488 nm excitation and a 500–530 nm emission. Chlorophyll auto-fluorescence was detected with 570 nm excitation and 640 nm emission.
SQD2.2 Protein Expression, Purification and Activity Assay
Full-length SQD2.2 cDNA was amplified using the forward primer 5′-GGATCCATGGAATATCCCCCACAATT-3′ and the reverse primer 5′-AAGCTTGTGTTGTGTGATTCTGTTGA-3′. The fragment was ligated into the pET28a vector after digestion with BamHI and HindIII. The construct was transformed into E. coli BL21 cells for protein expression. The protein was extracted and was purified using Ni-NTA resin (Sangon, Shanghai, China) according to the manufacturer’s instructions. Purified SQD2.2 was used for activity assays in 100 μl of reaction mixture containing 10 μM flavonoid (apigenin or naringenin [Sigma]), 2 mM nucleotide sugar (UDPG, ADPG, or UDPGal [Sigma]), and 50 mM Tris-HCl, pH 7.5, at 30 °C for 1 h. Reactions were stopped by the addition of 100 μl of methanol. After filtration, the reaction mixture was subjected to high-performance liquid chromatography (HPLC) (Shimadzu, SPD-M20A) equipped with C18-RP columns (5 μm, 4.6 × 150 mm) for analysis. The mixtures were separated by a linear gradient from 40% to 60% methanol containing 0.1% acetic acid with a flow rate of 1 ml min−1 and UV-visible detection was performed at 254 nm.
Quantitative Real-time PCR
Total RNA was isolated from different tissues using the TransZol reagent (TransGen Biotech, Beijing). RNA extracts were treated with DNaseI (NEB, UK) to eliminate DNA contamination. First-strand cDNA was produced from the RNA template by reverse transcription using the TIANscript RT Kit according to the manufacturer’s instructions (TransGen Biotech, Beijing). Quantitative real-time PCR was performed as described previously50. The primers for SQD2.2 were 5′-GCCGTGTTCACTGGAATGATGCAA-3′ (forward) and 5′-ATATCAGGAACACCACCAGCACGA-3′ (reverse). β-actin was used as an internal standard with primers 5′-TATGGTCAAGGCTGGGTTCG-3′ (forward) paired with 5′-CCATGCTCGATGGGGTACTT-3′ (reverse).
Flavonoid Measurement by LC–ESI–MS/MS
Fully expanded leaves with a similar position, age and size were sampled and ground into homogenate using liquid nitrogen. After removing chlorophyll, the precipitates were suspended in 2 ml methanol and incubated overnight in the dark at 4 °C to extract flavonoids. The extracted flavonoids were analyzed using LC–ESI–MS/MS (HPLC, Shim-pack UFLC Shimadzu CBM-20A system; MS, Applied Biosystems 4000 Q TRAP) according to the methods described previously51.
SQDG Lipid Analysis
Lipids were extracted and analyzed as described29. Briefly, lipids were extracted from leaves with chloroform: methanol (2:1, v/v) containing 0.01% butylated hydroxytoluene (BHT) at room temperature several times until the samples became completely bleached. The resultant lipids were separated on TLC plates using the developing solvent (acetone: toluene: H2O = 91:30:7.5, v/v) and were visualized with iodine vapor. The spot corresponding to SQDG was scraped out and was methylated with methanol containing 1% H2SO4 and 0.05% BHT at 70 °C for 1 h. The SQDG was quantified according to the methods described previously50.
ADPG and UDPG Measurements
Nucleotide sugars were assayed according to the methods described previously52. Briefly, fresh leaves with similar age, position and size were incubated in a solvent (chloroform: methanol = 3:7, v/v) at −20 °C for 2 h and were vigorously mixed every 30 min. Nucleotide sugars ADPG and UDPG were then extracted by mixing with an equal volume of ddH2O, followed by centrifugation at 13,000 rpm for 5 min. The aqueous phase was transferred to a clean tube and analyzed with HPLC (Shimadzu, SPD-M20A) equipped with C18-RP columns (5 μm, 4.6 × 150 mm). The filtrate was separated by 5% acetonitrile and 95% H2O containing 10 mM tetrabutylammonium bromide (Sigma). The flow rate was 1 ml min−1, and UV-visible detection was performed at 254 nm.
Activity Assay for Starch Synthases and Amylase
The activity of soluble SS was detected according to the procedure described previously53. In brief, pre-weighed fresh leaf and spikelet tissues were ground in liquid nitrogen, and total protein was extracted with a buffer (50 mM N-(2-hydroxyethyl) piperazine-N’-(2-ethanesulfonic acid)-(HEPES-) NaOH, pH 7.5, 5 mM MgCl2, 2 mM DTT, 12% glycerol). For quantitative analysis of soluble SS activity, the extracted proteins (100 μg) were added to a reaction buffer (50 mM HEPES-NaOH, pH 7.5, 1.6 mM ADPG, 15 mM DTT, 1 mg ml−1 amylopectin) with or without apigenin 7-O-Glc. After incubation at 37 °C for 20 min, the reaction was terminated in boiling water and was cooled to room temperature. The produced ADP was further measured by incubating with an equal volume of ADP-ATP reaction buffer (50 mM HEPES-NaOH, pH 7.5, 4 mM phosphoenolpyruvate [Sangon, Shanghai], 200 mM KCl, 100 mM MgCl2, 1.5 U pyruvate kinase [Sigma]) at 37 °C for 30 min. The ATP product was measured with a Luminescent Kinase Assay Kit according to the manufacturer’s instructions (Promega). The measurement of GBSS activity was similar to that of soluble SS, except for using starch granules as a reaction primer. The SBE activity was assayed according to the procedure described previously54. The amylase activity assay was performed as described previously55.
Electronic supplementary material
Acknowledgements
We are grateful to Professor Christoph Benning (Michigan State University) for kindly providing Arabidopsis Atsqd2 mutant. We thank Professor Jie Luo and Dongqin Li (Huazhong Agricultural University) for help in LC–ESI–MS/MS analysis. This work was supported by grants from the Chinese National Key Basic Research Project (2012CB114200), and the National Science Foundation of China (31271514, 31470762).
Author Contributions
X.Z. and Y.H. designed the study, and analyzed the data. X.Z. performed most of the experiments. Q.S. helped some experimental work. Y.H. and X. Z. interpreted the data and wrote the manuscript. X.W. helped interpretation some of the data.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-04002-2
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stark DM, Timmerman KP, Barry GF, Preiss J, Kishore GM. Regulation of the amount of starch in plant tissues by ADP glucose pyrophosphorylase. Science. 1992;258:287–292. doi: 10.1126/science.258.5080.287. [DOI] [PubMed] [Google Scholar]
- 2.Smidansky ED, et al. Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increases seed yield. Proc Natl Acad Sci USA. 2002;99:1724–1729. doi: 10.1073/pnas.022635299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai HL, et al. Starch synthesis in Arabidopsis is achieved by spatial cotranscription of core starch metabolism genes. Plant Physiol. 2009;151:1582–1595. doi: 10.1104/pp.109.144196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li N, Zhang S, Zhao Y, Li B, Zhang J. Over-expression of AGPase genes enhances seed weight and starch content in transgenic maize. Planta. 2011;233:241–250. doi: 10.1007/s00425-010-1296-5. [DOI] [PubMed] [Google Scholar]
- 5.Dumez S, et al. Mutants of Arabidopsis lacking starch branching enzyme II substitute plastidial starch synthesis by cytoplasmic maltose accumulation. Plant Cell. 2006;18:2694–2709. doi: 10.1105/tpc.105.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Streb S, et al. Starch granule biosynthesis in Arabidopsis is abolished by removal of all debranching enzymes but restored by the subsequent removal of an endoamylase. Plant Cell. 2008;20:3448–3466. doi: 10.1105/tpc.108.063487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SK, et al. Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.) Plant Mol Biol. 2007;65:531–546. doi: 10.1007/s11103-007-9153-z. [DOI] [PubMed] [Google Scholar]
- 8.Ventriglia T, et al. Two Arabidopsis ADP-glucose pyrophosphorylase large subunits (APL1 and APL2) are catalytic. Plant Physiol. 2008;148:65–76. doi: 10.1104/pp.108.122846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogt T, Jones P. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 2000;5:380–386. doi: 10.1016/S1360-1385(00)01720-9. [DOI] [PubMed] [Google Scholar]
- 10.Bowles D, Isayenkova J, Lim EK, Poppenberger B. Glycosyltransferases: managers of small molecules. Curr Opin Plant Biol. 2005;8:254–263. doi: 10.1016/j.pbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Tohge T, et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005;42:218–235. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- 12.Yonekura-Sakakibara K, Hanada K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011;66:182–193. doi: 10.1111/j.1365-313X.2011.04493.x. [DOI] [PubMed] [Google Scholar]
- 13.Caputi L, Malnoy M, Goremykin V, Nikiforova S, Martens S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012;69:1030–1042. doi: 10.1111/j.1365-313X.2011.04853.x. [DOI] [PubMed] [Google Scholar]
- 14.He XZ, Wang X, Dixon RA. Mutational analysis of the Medicago glycosyltransferase UGT71G1 reveals residues that control regioselectivity for (iso) flavonoid glycosylation. J Biol Chem. 2006;281:34441–34447. doi: 10.1074/jbc.M605767200. [DOI] [PubMed] [Google Scholar]
- 15.Rojas Rodas F, et al. Linkage mapping, molecular cloning and functional analysis of soybean gene Fg2 encoding flavonol 3-O-glucoside (1- > 6) rhamnosyltransferase. Plant Mol Biol. 2014;84:287–300. doi: 10.1007/s11103-013-0133-1. [DOI] [PubMed] [Google Scholar]
- 16.Taylor LP, Grotewold E. Flavonoids as developmental regulators. Curr Opin Plant Biol. 2005;8:317–323. doi: 10.1016/j.pbi.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Hsieh K, Huang AH. Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell. 2007;19:582–596. doi: 10.1105/tpc.106.049049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agati G, Tattini M. Multiple functional roles of flavonoids in photoprotection. New Phytol. 2010;186:786–793. doi: 10.1111/j.1469-8137.2010.03269.x. [DOI] [PubMed] [Google Scholar]
- 20.Peer WA, Murphy AS. Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci. 2007;12:556–563. doi: 10.1016/j.tplants.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Singh JP, Selvendiran K, Banu SM, Padmavathi R, Sakthisekaran D. Protective role of Apigenin on the status of lipid peroxidation and antioxidant defense against hepatocarcinogenesis in Wistar albino rats. Phytomedicine. 2004;11:309–314. doi: 10.1078/0944711041495254. [DOI] [PubMed] [Google Scholar]
- 22.Pourcel L, et al. A chemical complementation approach reveals genes and interactions of flavonoids with other pathways. Plant J. 2013;74:383–397. doi: 10.1111/tpj.12129. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs M, Rubery PH. Naturally occurring auxin transport regulators. Science. 1988;241:346–349. doi: 10.1126/science.241.4863.346. [DOI] [PubMed] [Google Scholar]
- 24.Lamoral-Theys D, et al. Natural polyphenols that display anticancer properties through inhibition of kinase activity. Curr Med Chem. 2010;17:812–825. doi: 10.2174/092986710790712183. [DOI] [PubMed] [Google Scholar]
- 25.Rowan DD, et al. Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol. 2009;182:102–115. doi: 10.1111/j.1469-8137.2008.02737.x. [DOI] [PubMed] [Google Scholar]
- 26.Catala R, Medina J, Salinas J. Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:16475–16480. doi: 10.1073/pnas.1107161108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakabayashi R, Saito K. Integrated metabolomics for abiotic stress responses in plants. Curr Opin Plant Biol. 2015;24:10–16. doi: 10.1016/j.pbi.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Nakabayashi R, et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014;77:367–379. doi: 10.1111/tpj.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu B, Xu C, Benning C. Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc Natl Acad Sci USA. 2002;99:5732–5737. doi: 10.1073/pnas.082696499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones MR. Lipids in photosynthetic reaction centres: structural roles and functional holes. Prog Lipid Res. 2007;46:56–87. doi: 10.1016/j.plipres.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 31.Shimojima M. Biosynthesis and functions of the plant sulfolipid. Prog Lipid Res. 2011;50:234–239. doi: 10.1016/j.plipres.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Okazaki Y, et al. A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell. 2009;21:892–909. doi: 10.1105/tpc.108.063925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Essigmann B, Guler S, Narang RA, Linke D, Benning C. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1998;95:1950–1955. doi: 10.1073/pnas.95.4.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okazaki Y, et al. A new class of plant lipid is essential for protection against phosphorus depletion. Nat Commun. 2013;4:1510. doi: 10.1038/ncomms2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ragel P, et al. Loss of starch granule initiation has a deleterious effect on the growth of arabidopsis plants due to an accumulation of ADP-glucose. Plant Physiol. 2013;163:75–85. doi: 10.1104/pp.113.223420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benning C, Somerville CR. Isolation and genetic complementation of a sulfolipid-deficient mutant of Rhodobacter sphaeroides. J Bacteriol. 1992;174:2352–2360. doi: 10.1128/jb.174.7.2352-2360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato N, Sonoike K, Tsuzuki M, Kawaguchi A. Impaired photosystem II in a mutant of Chlamydomonas reinhardtii defective in sulfoquinovosyl diacylglycerol. Eur J Biochem. 1995;234:16–23. doi: 10.1111/j.1432-1033.1995.016_c.x. [DOI] [PubMed] [Google Scholar]
- 38.Peer WA, et al. Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell. 2004;16:1898–1911. doi: 10.1105/tpc.021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li C, et al. Quercetin attenuates cardiomyocyte apoptosis via inhibition of JNK and p38 mitogen-activated protein kinase signaling pathways. Gene. 2016;577:275–280. doi: 10.1016/j.gene.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Delvalle D, et al. Soluble starch synthase I: a major determinant for the synthesis of amylopectin in Arabidopsis thaliana leaves. Plant J. 2005;43:398–412. doi: 10.1111/j.1365-313X.2005.02462.x. [DOI] [PubMed] [Google Scholar]
- 41.Seung D, et al. PROTEIN TARGETING TO STARCH is required for localising GRANULE-BOUND STARCH SYNTHASE to starch granules and for normal amylose synthesis in Arabidopsis. PLoS Biol. 2015;13:e1002080. doi: 10.1371/journal.pbio.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hannah LC, et al. A shrunken-2 transgene increases maize yield by acting in maternal tissues to increase the frequency of seed development. Plant Cell. 2012;24:2352–2363. doi: 10.1105/tpc.112.100602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai X, et al. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007;143:1739–1751. doi: 10.1104/pp.106.094532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith AM, Stitt M. Coordination of carbon supply and plant growth. Plant Cell Environ. 2007;30:1126–1149. doi: 10.1111/j.1365-3040.2007.01708.x. [DOI] [PubMed] [Google Scholar]
- 45.Graf A, Smith AM. Starch and the clock: the dark side of plant productivity. Trends Plant Sci. 2011;16:169–175. doi: 10.1016/j.tplants.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 46.Stitt M, Zeeman SC. Starch turnover: pathways, regulation and role in growth. Curr Opin Plant Biol. 2012;15:282–292. doi: 10.1016/j.pbi.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 47.Usadel B, et al. Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol. 2008;146:1834–1861. doi: 10.1104/pp.107.115592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winkel-Shirley B. Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol. 2002;5:218–223. doi: 10.1016/S1369-5266(02)00256-X. [DOI] [PubMed] [Google Scholar]
- 49.Wu C, et al. Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 2003;35:418–427. doi: 10.1046/j.1365-313X.2003.01808.x. [DOI] [PubMed] [Google Scholar]
- 50.Sun L, et al. Ribosomal protein S6 kinase1 coordinates with TOR-Raptor2 to regulate thylakoid membrane biosynthesis in rice. Biochim Biophys Acta. 2016;1861:639–649. doi: 10.1016/j.bbalip.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 51.Gong L, et al. Genetic analysis of the metabolome exemplified using a rice population. Proc Natl Acad Sci USA. 2013;110:20320–20325. doi: 10.1073/pnas.1319681110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Behmuller R, Forstenlehner IC, Tenhaken R, Huber CG. Quantitative HPLC-MS analysis of nucleotide sugars in plant cells following off-line SPE sample preparation. Anal Bioanal Chem. 2014;406:3229–3237. doi: 10.1007/s00216-014-7746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakamura T, Vrinten P, Hayakawa K, Ikeda J. Characterization of a granule-bound starch synthase isoform found in the pericarp of wheat. Plant Physiol. 1998;118:451–459. doi: 10.1104/pp.118.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guan HP, Preiss J. Differentiation of the properties of the branching isozymes from maize (Zea mays) Plant Physiol. 1993;102:1269–1273. doi: 10.1104/pp.102.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie Z, Zhang ZL, Hanzlik S, Cook E, Shen QJ. Salicylic acid inhibits gibberellin-induced alpha-amylase expression and seed germination via a pathway involving an abscisic-acid-inducible WRKY gene. Plant Mol Biol. 2007;64:293–303. doi: 10.1007/s11103-007-9152-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


