Abstract
Emerging evidence show that long noncoding RNAs (lncRNAs) play critical roles in tumor development. LincRNA-ROR (linc-ROR) is known to promote tumor progress in several human cancers, including hepatocellular carcinoma (HCC). Nevertheless, the roles of linc-ROR in HCC metastasis and its underlying mechanisms remain fully unclear. In the present study, we showed that linc-ROR was upregulated in HCC tissues and high linc-ROR expression level predicted poor prognosis. Functionally, linc-ROR significantly induced epithelial-mesenchymal transition (EMT), and increased in vitro invasion and in vivo metastasis of HCC cells. Mechanistically, linc-ROR acted as a sponge for miR-145 to de-repress the expression of target gene ZEB2, thereby inducing EMT and promoting HCC metastasis. Collectively, our research indicates the potential of linc-ROR as a vital therapeutic target for the treatment of aggressive and metastatic HCC.
Subject terms: Cancer epigenetics, Gastrointestinal cancer
Introduction
Hepatocellular carcinoma (HCC) is one of the most malignant cancers worldwide1. Despite various treatments, the invasive potential and distant metastasis make the prognosis of HCC patients remain poor2. Elucidating the mechanisms underlying HCC metastasis is crucial for establishing new therapeutic targets for successful intervention. Long noncoding RNAs (lncRNAs) are a class of transcripts longer than 200 nucleotides with limited protein-coding ability3. Emerging evidences have revealed that lncRNAs regulate diverse biological processes, such as proliferation, apoptosis and metastasis4. Several lncRNAs with pro- or anti-metastatic functions have been reported to modulate tumor metastasis. For example, lnc-HOTTIP was found promoting metastasis of esophageal squamous cell carcinoma via inducing epithelial-mesenchymal transition (EMT)5. Recently, Liu et al. revealed that lnc-FTX functioned as a tumor suppressor in HCC through binding miR-374a and MCM26.
Long noncoding RNA regulator of reprogramming (lincRNA-ROR), a 2.6 kb lncRNA comprised of four exons, was first discovered in induced pluripotent stem cells (iPSCs), regulated by the key pluripotency factors Oct4, Sox2, and Nanog7, 8. Subsequent studies unveiled that linc-ROR is upregulated in breast cancer, gastric cancer, gallbladder cancer, pancreatic cancer and colon cancer tissues9–13. Most recently, researchers have revealed that linc-ROR could modulate the chemo-sensitivity and hypoxia signaling pathways in HCC14, 15. However, the specific role of linc-ROR in mediating the metastatic process of HCC is not well studied.
In this study, we comprehensively explored the functions of linc-ROR in the metastatic process of HCC. Our data showed that linc-ROR was significantly upregulated in HCC tissues and highly metastatic HCC cells. Functionally, upregulation of linc-ROR significantly induced EMT, and increased in vitro invasion and in vivo metastasis of HCC cells. Also, the interaction among linc-ROR, miR-145 and ZEB2 was studied to reveal the underlying mechanisms in HCC metastasis. It was proved that linc-ROR acted as a sponge for miR-145 to de-repress the expression of the target gene ZEB2, thereby inducing EMT and promoting HCC metastasis. The findings will extend our understanding of the role of lncRNAs in aggressive and metastatic HCC.
Results
Linc-ROR is upregulated in HCC tissues and related to tumor metastasis and poor prognosis of HCC patients
We performed qRT-PCR to detect the expression of linc-ROR in 20 paired of HCC and normal liver tissues. Results showed that linc-ROR was significantly upregulated in HCC tissues (Fig. 1A), which prompts us to investigate the clinical significance of linc-ROR in HCC development. Next, we extended our linc-ROR quantification assay to a cohort of 88 HCC tissues. Higher linc-ROR expression was observed in HCC tissues with lymph node metastasis (LNM) or vascular invasion than those tissues without LNM or vascular invasion (Fig. 1B). Besides, higher linc-ROR expression was related to advanced tumor node metastasis (TNM) stage (Fig. 1C). Most importantly, HCC patients with recurrence showed significant higher linc-ROR expression, comparing to tumors without recurrence (Fig. 1D). Patients were then distributed into high-linc-ROR and low-linc-ROR groups according to the linc-ROR expression in HCC tissues. The correlations between linc-ROR expression and clinic pathological characteristics were presented in Table 1. High linc-ROR expression was observed to be closely correlated with advanced TNM stage, higher incidence of lymph node metastasis and recurrence of HCC patients (p = 0.001, 0.019 and 0.000, respectively). However, there was no significant correlation between linc-ROR expression and other clinical pathological characteristics including gender, age, tumor diameter, alcohol intake, family history and liver function of patients. Furthermore, Kaplan-Meier survival analyses indicated that HCC patients with high linc-ROR expression had poorer prognosis than those with low linc-ROR expression, together with a shorter disease-free survival (DFS) and overall survival (OS) (Fig. 1E). These results indicated that linc-ROR closely correlates with the increase of HCC metastatic potential.
Figure 1.
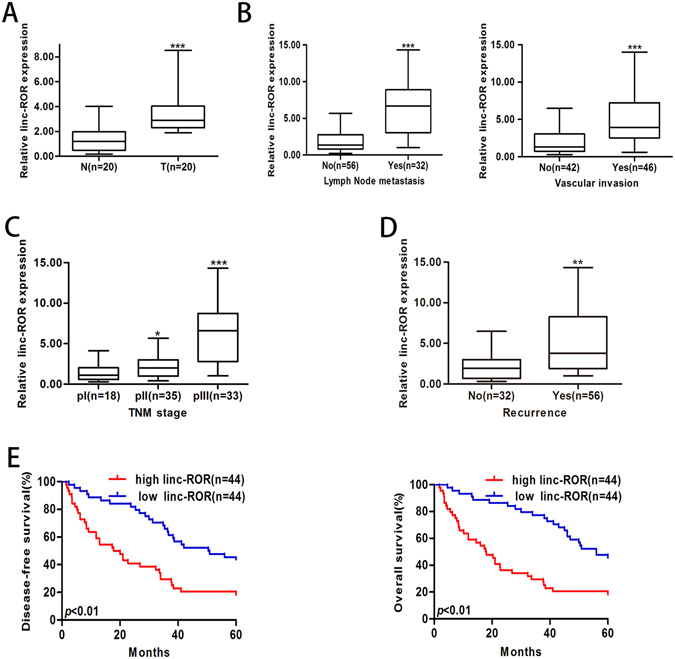
Increased linc-ROR correlates with HCC progression. Expression of linc-ROR was determined by qRT-PCR. GAPDH was used as an endogenous control. (A) Relative linc-ROR expression in HCC and paired adjacent nontumor liver tissues. T: HCC tissues; N: nontumor liver tissues. (B) Relative linc-ROR expression in HCC tissues with or without LNM and HCC tissues with or without vascular invasion. (C) Relative linc-ROR expression in HCC tissues with different TNM stage. (D) Relative linc-ROR expression in HCC tissues with or without recurrence. (E) Kaplan-Meier analysis of DFS or OS of HCC patients. Patients were grouped according to ROR expression in HCC tissues. The median expression level was used as the cutoff. Each experiment was performed at least three times. *p < 0.05, **p < 0.01 and ***p < 0.001.
Table 1.
Clinical characteristics of 88 HCC patients according to linc-ROR expression levels.
| Feature | Relative linc-ROR expression | χ2 | p-value | |
|---|---|---|---|---|
| Low (n = 44) | High (n = 44) | |||
| Gender | 0.063 | 0.812 | ||
| Female | 11 | 10 | ||
| Male | 33 | 34 | ||
| Age (years) | 0.218 | 0.641 | ||
| <55 | 14 | 12 | ||
| ≥55 | 30 | 32 | ||
| Family HCC history | 1.186 | 0.276 | ||
| No | 29 | 24 | ||
| Yes | 15 | 20 | ||
| Alcohol intake | 1.166 | 0.280 | ||
| No | 28 | 23 | ||
| Yes | 16 | 21 | ||
| Tumor diameter (cm) | 1.725 | 0.189 | ||
| ≤5.0 | 30 | 24 | ||
| >5.0 | 14 | 20 | ||
| Liver function | 2.964 | 0.085 | ||
| Child-Pugh A | 23 | 15 | ||
| Child-Pugh B | 21 | 29 | ||
| AFP (ng/L) | 3.777 | 0.520 | ||
| ≤400 | 23 | 14 | ||
| >400 | 21 | 30 | ||
| TNM stage | 7.096 | 0.029 | ||
| I | 20 | 9 | ||
| II | 16 | 19 | ||
| III | 8 | 26 | ||
| Lymph node metastasis | 10.493 | 0.001 | ||
| No | 33 | 18 | ||
| Yes | 11 | 26 | ||
| Vascular invasion | 5.526 | 0.019 | ||
| No | 29 | 18 | ||
| Yes | 15 | 26 | ||
| Recurrence | 17.655 | 0.000 | ||
| No | 35 | 11 | ||
| Yes | 9 | 23 | ||
NOTE: AFP, alpha-fetoprotein; TNM, tumor-node-metastasis. The median expression level was used as the cutoff. Low linc-ROR expression in 44 patients was classified as values below the 50th percentile. High linc-ROR expression in 44 patients was classified as values above the 50th percentile. Results were considered statistically significant at p < 0.05.
Linc-ROR significantly promotes in vitro invasion and in vivo metastasis of HCC cells
To further determine whether linc-ROR is involved in the molecular etiology of metastasis in HCC, we examined the expression of linc-ROR in HCC cell lines with different metastatic potentials. QRT-PCR results showed that linc-ROR increased progressively from normal human hepatocyte cell line (HH) to low metastatic HCC cell lines (HepG2 and SMMC-7721), and finally to highly metastatic HCC cell lines (HCCLM3 and MHCC97-H) (Fig. 2A)16. To explore the biological significance of linc-ROR in HCC, HepG2 or SMMC-7721 cells were stably transfected with linc-ROR overexpression plasmid or control vector respectively and HCCLM3 or MHCC97-H cells were stably transfected with shROR or shCtrl plasmid, respectively. Satisfactory transfection efficiency was obtained at 48 h post-transfection (Fig. 2B). We chose the most efficient shROR plasmid for the forthcoming experiment. Further, we investigated the roles of linc-ROR in migration and invasion of HCC cells. Wound healing assay showed that linc-ROR upregulation significantly increased the capacity of mobility in HepG2 (or SMMC-7721) cells. In contrast, wound healing speed was reduced after linc-ROR knockdown in HCCLM3 (or MHCC97-H) cells (Fig. 2C; Fig. S1A). Similarly, transwell assays confirmed the positive effect of linc-ROR on migration and invasion capacity after linc-ROR overexpression in HepG2 (or SMMC-7721) cells, whereas inhibited migration and invasion occurred after knockdown of linc-ROR in HCCLM3 (or MHCC97-H) cells (Fig. 2D; Fig. S1B). Then, to further assess the effects of linc-ROR in the in vivo metastasis, we made orthotopic implantation tumor models to test HCC cell invasive behavior changes. After 8 weeks, it was observed that the incidence and number of both intrahepatic and lung metastasis in the linc-ROR group was significantly increased, in compared with the control group. Likewise, the effect of linc-ROR downregulation on the in vivo metastasis of HCC cells was also determined. The incidence and number of both intrahepatic and lung metastasis in the shROR group was significantly decreased, as compared to the control group (Fig. 2E). Furthermore, the difference was further confirmed by hematoxylin and eosin (HE) staining of liver and lung sections (Fig. 2F). On the whole, these in vitro and in vivo results verified the role of linc-ROR in HCC metastasis.
Figure 2.
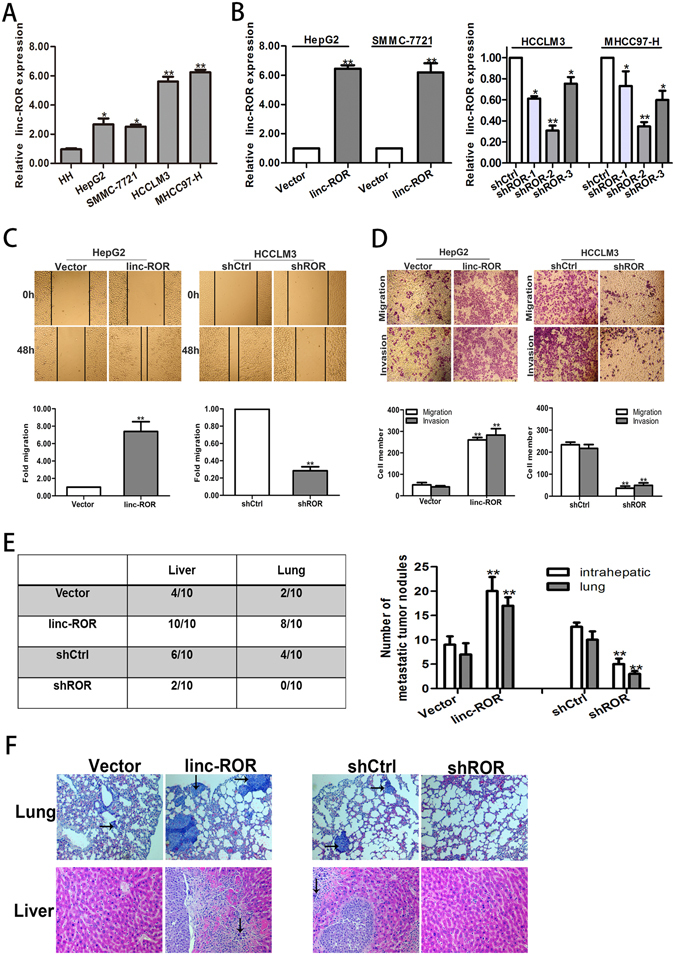
Linc-ROR promotes in vitro migration or invasion and in vivo metastasis in HCC cells. (A) HCC cell lines with different metastatic potentials and a normal human hepatocyte cell line (HH). Expression of linc-ROR was determined by qRT-PCR. (B) qRT-PCR detection of linc-ROR expression in HepG2 cells stably transfected with linc-ROR (or Vector) and HCCLM3 cells stably transfected with shROR (or shCtrl), respectively. (C) Wound healing assay on HepG2 cells stably transfected with linc-ROR (or Vector) and HCCLM3 cells stably transfected with shROR (or shCtrl), respectively. (D) Transwell migration and invasion assays on HepG2 cells stably transfected with linc-ROR (or Vector) and HCCLM3 cells stably transfected with shROR (or shCtrl), respectively. (E) Incidence and metastatic tumor nodules in different groups 8 weeks after orthotopic implantation. (F) Representative histological image of H&E staining. Black arrows: metastatic foci. Each experiment was performed at least three times. *p < 0.05; **p < 0.01.
Linc-ROR induces EMT in HCC cells
EMT, a biological process in which cancer cells lose their epithelial polarity and undergo transition into a mesenchymal phenotype, plays a vital role in tumor invasion and metastasis17. Accumulating evidence indicates that various regulators, including lncRNAs, can motivate EMT to increase cancer migration and invasion18. Although there have been some studies on the contribution of lncRNAs during tumor EMT process, much less is known about the role of linc-ROR during HCC EMT process. Therefore, it is worth investigating whether the EMT phenotype of HCC cells was affected by linc-ROR expression. Western blotting and immunofluorescence indicated that linc-ROR upregulation significantly decreased the expression of epithelial markers (E-cadherin and β-catenin) and increased the expression of mesenchymal markers (N-cadherin and Vimentin) in HepG2 (or SMMC-7721) cells (Fig. 3A,B and Fig. S1C,D). In contrast, linc-ROR downregulation reversed the changes of epithelial and mesenchymal in HCCLM3 (or MHCC97-H) cells (Fig. 3C,D and Fig. S1C,D). Thus, we revealed that linc-ROR could induce EMT phenotype of HCC cells to support migration and invasion.
Figure 3.
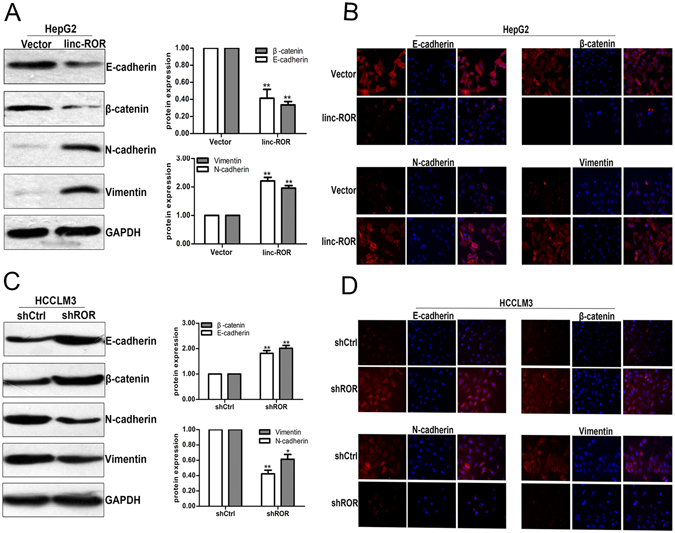
Linc-ROR induces EMT in HCC cells. (A) Western blotting and (B) immunofluorescence detection of epithelial markers (E-cadherin and β-catenin) and mesenchymal markers (N-cadherin and Vimentin) in HepG2 cells stably transfected with linc-ROR or Vector. (C) Western blotting and (D) immunofluorescence detection of epithelial markers (E-cadherin and β-catenin) and mesenchymal markers (N-cadherin and Vimentin) in HCCLM3 cells stably transfected with shROR or shCtrl, respectively. Each experiment was performed at least three times. *p < 0.05; **p < 0.01.
Identification of miR-145 as a target of linc-ROR
LncRNAs are proved to work as sponges to recruit miRNAs, resulting in upregulation of target genes19. To verify whether linc-ROR has a similar mechanism in HCC cells, we used bioinformatics tool Miranda to predict the potential miRNA binding sites in linc-ROR, together considering previously reported miRNAs20. In turn, we measured the expression levels of these 15 predicted miRNAs in linc-ROR downregulated HCCLM3 cells (Supplementary Table 1). We then focused on miR-145, which exhibited the greatest change. Previously studies have revealed that linc-ROR could act as a sponge for miR-145 in various cancers, such as colorectal cancer, pancreatic cancer and breast cancer9, 12, 21. Also, miR-145 has been implicated to function as tumor suppressor and inhibit cell invasion, migration and metastasis in HCC22, 23. We hypothesized that the functions of linc-ROR in HCC cells might be mediated by miR-145. In order to prove the hypothesis, we first examined the expression level of miR-145 in HepG2 and SMMC-7721 cells stably transfected with linc-ROR, finding that miR-145 expression was significantly downregulated (Fig. 4A). Meanwhile, miR-145 expression levels in both cells transfected with shROR were significantly upregulated (Fig. 4B). In a word, the miR-145 expression level correlated negatively with linc-ROR expression level. Additionally, we also found that there was no difference in linc-ROR levels after knockdown of miR-145 or ectopic expression (Fig. 4C,D). To validate the direct binding between linc-ROR and miR-145 at endogenous levels, luciferase reporter assays were constructed, which contain wild-type (WT) or mutated (MUT) miR-145 binding sites. Results showed that miR-145/mimics reduced the luciferase activity of wild-type (WT) linc-ROR reporter vector but not of empty vector and complete mutant reporter vector (Fig. 4E). These data revealed that miR-145 binds to linc-ROR, not inducing the degradation of linc-ROR. It is well known that miRNAs bind their targets and cause translational repression and RNA degradation in an Ago2-dependent manner24. To determine whether linc-ROR was regulated by miR-145 in such a manner, we conducted anti-Ago2 RIP in HepG2 cells transiently overexpressing miR-145. Results showed that endogenous linc-ROR pull-down was specifically enriched in miR-145 transfected cells (Fig. 4F), supporting that miR-145 is a bona fide linc-ROR-targeting miRNA. To serve as a sponge, the abundance of linc-RoR should be comparable to or higher than miR-145. We therefore used quantitative real-time PCR to quantify the exact copy numbers of linc-RoR and miR-145 per cell in normal HH cells and HCCLM3 cells (Fig. 4G). As a result, we found that in HCCLM3 cells, the expression level of miR-145 was only about 30 copies per cell, whereas linc-ROR level was approximately 200 copies per cell. In HH cells, miR-145 was up to about 300 copies per cell, whereas linc-ROR level was no more than 40 copies per cell. This result implied that linc-ROR may be able to function as a sponge for miR-145.
Figure 4.
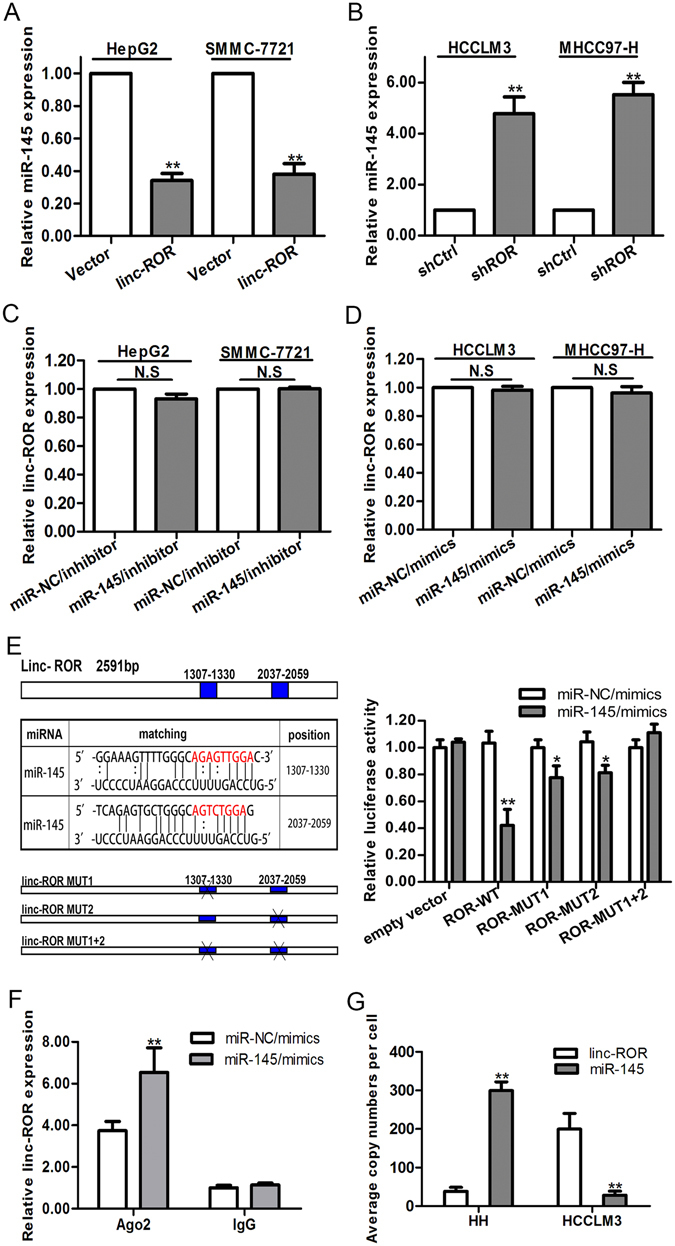
Linc-ROR acts as a molecular sponge for miR-145. (A) and (B) qRT-PCR detection of miR-145 expression in Vector or linc-ROR transfected HepG2 or SMMC-7721 cells and shCtrl or shROR transfected HCCLM3 or MHCC97-H cells. U6 was used as an internal control. (C) and (D) qRT-PCR detection of the expression of linc-ROR in miR-NC/inhibitor or miR-145/inhibitor-transfected HepG2 or SMMC-7721 cells and miR-NC/mimics or miR-145/mimics-transfected HCCLM3 or MHCC97-H cells. GAPDH was used as an internal control. (E) Left: prediction for miRNA-binding elements on linc-ROR by Miranda. The red nucleotides (target sites) were deleted in the mutant constructs. Right: luciferase activity in HepG2 cells co-transfected with miR-145/mimics and luciferase reporters containing empty vector, ROR-WT, ROR-MUT1, ROR-MUT2, ROR-MUT1 + 2. Data are presented as the relative ratio of firefly luciferase activity to Renilla luciferase activity. (F) RNA-IP with anti-antibody was performed in HepG2 cells transfected with miR-NC/mimics and miR-145/mimics. (G) The exact copy numbers of linc-ROR and miR-145 in HH and HCCLM3 cells were quantified with real-time quantitative RT-PCR. For exact quantification of gene copies per cell, linc-ROR and reverse-transcribed miR-145 cDNA were used as standard templates to formulate standard curves with limit dilution approaches, and then the exact copies of linc-ROR and miR-145 per cell were calculated according to their molecular weight and cell counts. Each experiment was performed at least three times. N.S, not significant. *p < 0.05; **p < 0.01.
Inhibition of miR-145 mimics the biological functions of linc-ROR upregulation in HCC cells
To further investigate the biological function of miR-145 in HCC cells, HCCLM3 and MHCC97-H cells were transfected with miR-145/mimics, miR-NC/mimics as a negative control. And miR-145/inhibitor was transfected into HepG2 and SMMC-7721 cells, miR-NC/inhibitor as a negative control. The upregulation or downregulation of miR-145 in those transfected cells was confirmed by qRT-PCR (Fig. 5A). Wound healing assay revealed enhanced mobility of HepG2 (or SMMC-7721) cells transfected with miR-145/inhibitor and the opposite effects for HCCLM3 (or MHCC97-H) cells transfected with miR-145/mimics (Fig. 5B and Fig. S1A). As expected, the in vitro migration and invasion of HepG2 (or SMMC-7721) cells in the presence of miR-145/inhibitor could be significantly increased. In contrast, the opposite results were obtained for HCCLM3 (or MHCC97-H) cells treated with miR-145/mimics (Fig. 5C and Fig. S1B). Moreover, western blotting analysis was performed to observe the function of miR-145 on EMT phenotypes of HCC cells, and results showed that the expression of epithelial markers were decreased while the mesenchymal markers were increased following transfection of the miR-145/inhibitor into HepG2 (or SMMC-7721) cells. The opposite results were obtained for HCCLM3 (or MHCC97-H) cells transfected with miR-145/mimics (Fig. 5D and Fig. S1C). Meanwhile, immunofluorescence studies further suggested that miR-145 reversed EMT in HCC cells (Fig. 5E and Fig. S1D). The above results suggest that inhibition of miR-145 mimics the functions of linc-ROR overexpression in HCC cells.
Figure 5.
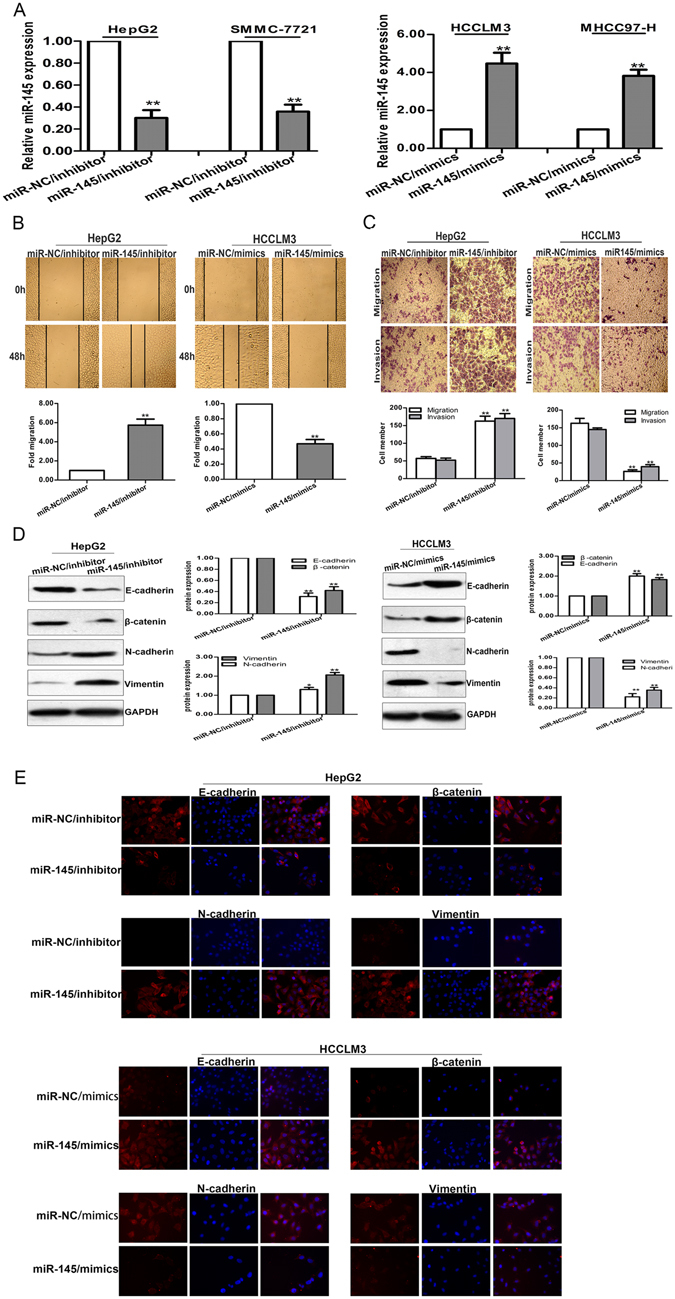
miR-145 negatively regulates invasion, migration and EMT phenotype of HCC cells. (A) qRT-PCR detection of miR-145 expression in miR-145/inhibitor transfected HepG2 or SMMC-7721 cells and miR-145/mimics transfected HCCLM3 or MHCC97-H cells. (B) Wound healing assay, and (C) transwell assays on HepG2 cells transfected with miR-145/inhibitor (or miR-NC/inhibitor) and HCCLM3 cells transfected with miR-145/mimics (or miR-NC/mimics), respectively. (D) Western blotting and (E) immunofluorescence analysis of changes in epithelial and mesenchymal markers of miR-145/inhibitor transfected HepG2 cells and miR-145/mimics transfected HCCLM3 cells. Each experiment was performed at least three times. *p < 0.05; **p < 0.01.
The function of linc-ROR in HCC cells in vitro was partially reversed by miR-145
Then, we performed rescue experiments to determine whether linc-ROR influenced HCC cell invasion, migration, and the induction of EMT in a miR-145-dependent manner. MiR-145/inhibitor or miR-NC/inhibitor was transfected into HCCLM3 or MHCC97-H cells stably transfected with shROR, while miR-145/mimics or miR-NC/mimics was transfected into HepG2 or SMMC-7721 cells stably transfected with linc-ROR. Wound healing assays showed that the enhanced mobility induced by linc-ROR in HepG2 (or SMMC-7721) cells was in part abolished by the introduction of miR-145/mimics, and vice versa in the transfected HCCLM3 (or MHCC97-H) cells (Fig. 6A and Fig. S1A). The migratory and invasive effect of linc-ROR overexpression in HepG2 (or SMMC-7721) cells was also partially rescued by co-transfection with miR-14/mimics, whereas the anti-metastasis effect induced by linc-ROR downregulation in HCCLM3 (or MHCC97-H) cells could be partially reversed by miR-145/inhibitors (Fig. 6B and Fig. S1B). Furthermore, western blotting assays indicated that the effect of linc-ROR overexpression on EMT phenotypes of HCC cells could be partially rescued by miR-145/mimics, and re-expression of miR-145/inhibitor could induce EMT in HCCLM3/shROR (or MHCC97-H/shROR) cells by downregulating epithelial markers and upregulating mesenchymal markers (Fig. 6C and Fig. S1C). Meanwhile, immunofluorescence analysis showed that downregulated epithelial markers and upregulated mesenchymal markers induced by linc-ROR overexpression in HepG2 (or SMMC-7721) cells was partially reversed by miR-145/mimics, and vice versa in the transfected HCCLM3 (or MHCC97-H) cells (Fig. 6D and Fig. S1D). These results showed that linc-ROR influences EMT, invasion and metastasis in HCC cells in vitro, at least partially in a miR-145-dependent manner.
Figure 6.
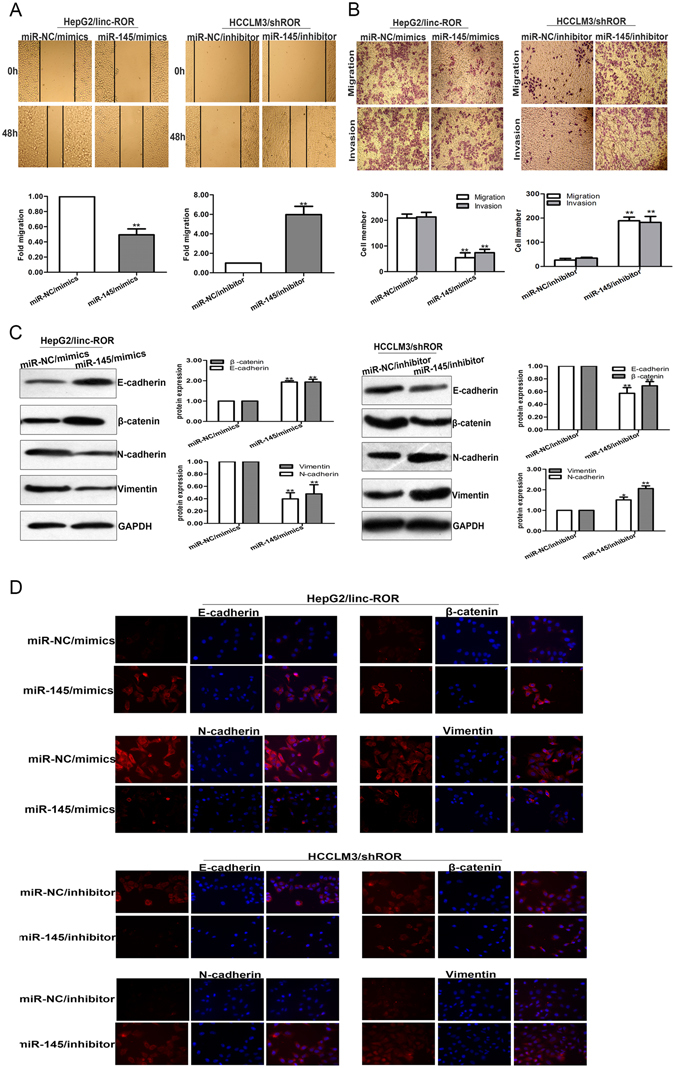
The function of linc-ROR in invasion and migration of HCC cells in vitro was dependent on miR-145. (A) Wound healing assay was performed to determine the mobility capacity of HepG2 cells stably transfected with linc-ROR and HCCLM3 cells stably transfected with shROR in the presence of miR-145/mimics or miR-145/inhibitor, respectively. (B) The metastasis capacity of HepG2 cells stably transfected with linc-ROR and HCCLM3 cells stably transfected with shROR was partially rescued by miR-145/mimics and miR-145/inhibitor, respectively. (C) Western blotting and (D) immunofluorescence analysis of changes in epithelial markers and mesenchymal markers in HepG2 cells stably transfected with linc-ROR and HCCLM3 cells stably transfected with shROR after treatment with miR-145/mimics or miR-145/inhibitor, respectively. Each experiment was performed at least three times. *p < 0.05; **p < 0.01.
Linc-ROR positively regulates the miR-145 target gene ZEB2 in HCC cells
It has been reported that miR-145 represses EMT, tumor migration and invasion by directly targeting the 3′-UTRs of ZEB2 in tumors25. Besides, ZEB2 is reported having the ability to stimulate the initiation of EMT by binding to E-boxes on the E-cadherin promoter, thus repressing its promoter activity26. We proposed that linc-ROR and ZEB2 interact with miR-145 by functioning as competing endogenous RNAs (ceRNAs). Here, to confirm such model, we measured the level of ZEB2 in response to different levels of linc-ROR. We observed that in HepG2 and SMMC-7721 cells, the ectopic expression of linc-ROR upregulated ZEB2 at the transcript and protein levels. Co-transfection of linc-ROR overexpression plasmid and miR-145/mimics could significantly reduce ZEB2 expression level (Fig. 7A). In HCCLM3 and MHCC97-H cells, shROR decreased ZEB2 at the transcript and protein levels. Similarly, miR-145/inhibitors restored the decrease of ZEB2 expression in the HCC cells transfected with shROR (Fig. 7B). Furthermore, we demonstrated that miR-145 expression level in HCC tissues was significantly lower than those in the adjacent normal tissues. Additionally, by linear regression analysis, miR-145 level and linc-ROR level were negatively correlated (Fig. 7C). Meanwhile, the expression of ZEB2 in HCC and corresponding nontumor tissues was also measured by qRT-PCR. It was showed that ZEB2 had high expression in HCC tissues compared with those in the corresponding normal tissues. Besides, ZEB2 and linc-ROR expression levels were positively correlated (Fig. 7D). Collectively, these findings indicated that there is a regulatory signaling pathway in which linc-ROR regulates ZEB2 by competitively sponging up miR-145, inducing increased invasion, metastasis and an EMT phenotype in HCC cells (Fig. 7E).
Figure 7.
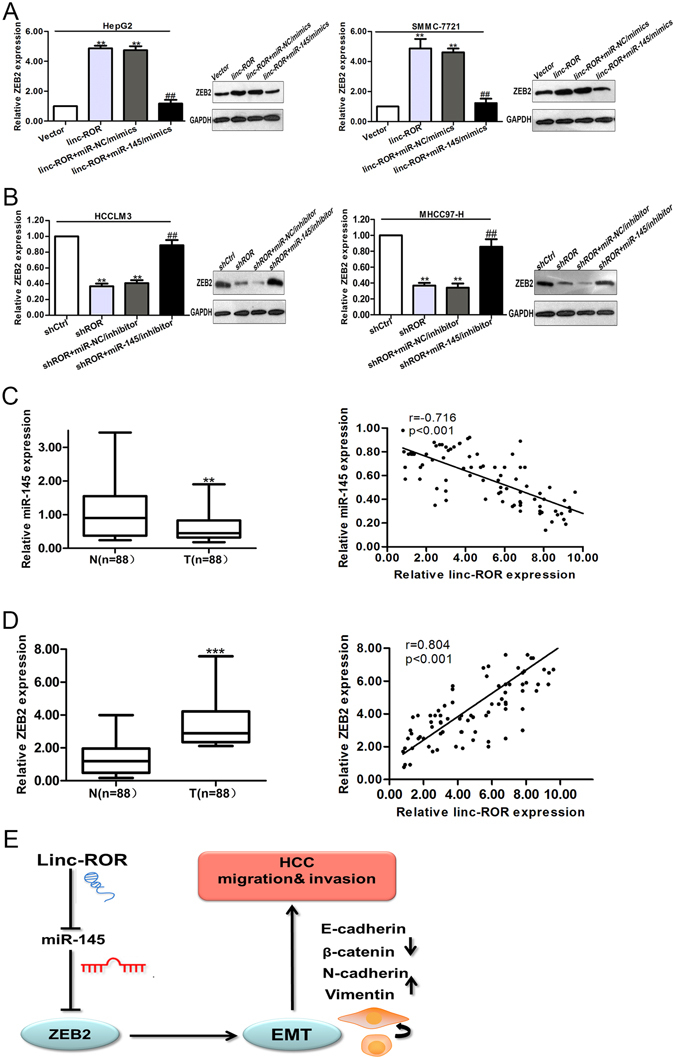
Linc-ROR acts as a ceRNA for ZEB2 by releasing miR-145 in HCC cells. (A) qRT-PCR and Western blotting assays were performed to detect the mRNA and protein levels of ZEB2 after cells transfected with linc-ROR and miR-145/mimics. **p < 0.01 vs. Vector group; ##p < 0.01 vs. linc-ROR + miR-NC/mimics group. (B) qRT-PCR and Western blotting assays were performed to detect the mRNA and protein levels of ZEB2 after cells transfected with shROR and miR-145/inhibitor. **p < 0.01 vs. shCtrl group; ##p < 0.01 vs. shROR + miR-NC/inhibitor group. (C) qRT-PCR detection of miR-145 expression in HCC and adjacent nontumor liver tissues (p < 0.01), showing negative correlation between linc-ROR and miR-145 expression levels in HCC tissues (Spearman’s correlation, r = −0.716, p < 0.01). (D) The expression of ZEB2 in paired samples of primary HCC and adjacent nontumor liver tissues, showing positive correlation between ROR expression and ZEB2 expression in HCC tissues (Spearman’s correlation, r = 0.804, p < 0.01). (E) Schematic overview of linc-ROR regulatory signaling. T: HCC tissues; N: nontumor liver tissues. Data are presented as mean ± SD from three independent experiments. ***p < 0.001.
Discussion
In spite of the encouraging advances in therapies for HCC patients, the invasive potential and distant metastasis make the prognosis remain dismal27. Hence, it is of great importance to explain the underlying mechanisms on the development and progression of HCC metastasis. Emerging evidence has demonstrated a key regulatory role of lncRNAs in tumorigenesis and metastasis of multiple cancers, including HCC4. For example, HBx-induced lncRNA Unigene561591 has an oncogenic effect on cell migration/invasion and EMT by acting as a ceRNA of miR-140-5p in HCC cells1. Another lncRNA, DANCR, could increase stemness features of HCC by derepression of CTNNB128. Linc-ROR was first described in iPSCs and can be regulated by the key pluripotency factors Oct4, Sox2, and Nanog7. Most recently, scientists pointed out that linc-ROR could be regarded as a novel biomarker, indicating a poor prognosis in gallbladder cancer and pancreatic cancer patients13, 21. However, the specific role of linc-ROR in mediating the metastatic process of HCC is not well studied.
In the present study, we first demonstrated that linc-ROR was upregulated in HCC tissues compared with pair-matched noncancerous tissues. Patients with high linc-ROR expression levels tended to have more advanced TNM stage and higher incidence of lymph node metastasis or vascular invasion and tumor recurrence. Moreover, HCC patients with high linc-ROR expression levels had a poorer prognosis than those with low linc-ROR expression. Then, we comprehensively investigated the function of linc-ROR in HCC metastasis by employing gain-of and loss-of-function approaches. Indeed, our data showed that linc-ROR upregulation significantly promoted migration and invasion of HCC cells. Besides, orthotopic implantation tumor models provided additional support for the involvement of linc-ROR in promoting hepatic and lung metastasis in vivo.
The acquisition of invasive capabilities, including degradation of the cell matrix and turnover of cell-cell junctions, is one of the most important steps in the metastatic cascade of tumors29. EMT is a biological process where epithelial cells lose their polarity and undergo transition into a mesenchymal phenotype30. Accumulating evidence indicates that lncRNAs can trigger EMT to increase cancer migration and invasion. Therefore, we proposed that whether the EMT phenotype of HCC cells was affected by linc-ROR expression. Western blotting and immunofluorescence assays revealed that linc-ROR upregulation could exert obvious effects on EMT phenotypes of HCC cells by decreasing epithelial markers and increasing mesenchymal markers. In contrast, linc-ROR downregulation could cause opposite changes of those markers. These data demonstrated that linc-ROR functions as a metastasis inducer in HCC cells through increasing invasion and promoting EMT.
Growing number of research confirmed that lncRNAs can antagonize miRNA function by competing with miRNAs for binding to shared target mRNAs, and then to silence target mRNAs31. For example, Jia el al found that lncRNA H19 acts as a sponge for miR-29a to modulate VASH2 expression in glioma-associated endothelial cells32. LncRNA MEG3 enhanced cell proliferation by competitively binding miR-29 in HCC33. To clarify whether linc-ROR functions as a sponge for miRNA, bioinformatics was used to analyze the potential miRNA binding sites contained in linc-ROR. Besides, based on previously studies and function analysis of linc-ROR, we chose miR-145 for further study. qRT-PCR results showed that miR-145 expression level was downregulated by linc-ROR in HCC cells. Also, we found that there was no difference in linc-ROR levels after ectopic expression or knockdown of miR-145. Fluorescent reporter assay and RIP analysis showed that linc-ROR was a direct target of miR-145 and could act as a sponge for miR-145.
Previous reports have proved that miR-145 possesses a tumor suppressive role in many cancers, including lung cancer, pancreatic cancer, breast cancer, HCC and colorectal cancer34–38. Moreover, miR-145 widely participates in cell proliferation, apoptosis, migration and invasion39–42. In the present study, our findings showed that miR-145 suppression yielded very similar effects to ectopic linc-ROR expression in HCC cells. Besides, rescue experiments showed that linc-ROR influences cell invasion, metastasis and EMT in HCC cells in vitro, partially in a miR-145-dependent manner. Taken together, these data strongly suggested that miR-145 is a bona fide linc-ROR-targeting miRNA. MiR-145 has been reported to inhibit EMT and tumor metastasis by directly targeting 3′-UTRs of ZEB2, which is a key transcription factor of EMT25. We considered whether linc-ROR and ZEB2 interact with miR-145 by functioning as ceRNAs. Our study demonstrated that miR-145 could reverse the reduction of ZEB2 caused by linc-ROR knockdown. Besides, linc-ROR expression was negatively correlated with miR-145, but positively associated with ZEB2 in HCC tissues. These results revealed that linc-ROR could function as a ceRNA to regulate the expression of ZEB2 in a miR-145-dependent manner.
In conclusion, we discovered that linc-ROR was highly expressed in HCC tissues and played a key role in regulating HCC metastasis. Linc-ROR competitively binds to miR-145, and subsequently up-regulates the expression of its target gene ZEB2 to promote the migration and invasion of HCC cells. Collectively, our research implicates the relevance of linc-ROR/miR-145/ZEB2 regulatory network as a potential therapeutic target for the highly aggressive and malignant HCC cancers.
Materials
Patients and tissue samples
HCC tissues and pair-matched noncancerous tissues were obtained from the Department of Hepatobiliary Surgery of First Hospital Affiliated to the Chinese PLA General Hospital and the Liver Disease Center of the 81th Hospital of PLA during 2005 to 2007. Each sample was rapidly frozen in liquid nitrogen and stored at −80 °C. Details of clinical characteristics of the patients were listed in Table 1. The data does not contain any information that can identify patients. All patients were accepted a close follow-up observation for disease recurrence at 1 month intervals during the first 2 postoperative years, and every 3 months after that. Informed consent was obtained from patients, and ethical approval was granted by the Review Board of Hospital Ethics Committee (Jingling Hospital, Nanjing University). All experiments were performed in accordance with the approved guidelines and regulations, including any relevant details.
Cell lines and mice
The cell lines HH, HCCLM3, MHCC97-H, HepG2 and SMMC-7721 were cultured in Dulbecco’s modified Eagle’s medium (GIBCO-BRL) supplemented with 10% fetal bovine serum (FBS), streptomycin (100 μg/ml), penicillin (100 U/ml). All cells were fostered at 37 °C in an atmosphere containing 5% CO2. We purchased all female BALB/C nude mice (4–6 weeks old) from the Animal Core Facility of Nanjing Medical University and housed these mice in laminar flow cabinets under specific pathogen-free conditions. All animal experiments were performed with the approval of the Institutional Committee for Animal Research. The study protocol was also approved by the Committee on the Use of Live Animals in research, Nanjing University (Nanjing, China).
Quantitative real-time PCR (qRT-PCR)
Total RNA from tissues and cells was isolated using Trizol reagent (Invitrogen, CA, USA). Real-time PCR was performed with SYBR Prime Script RT-PCR Kits (Takara, Japan) based on the manufacturer’s instructions. The linc-ROR, ZEB2, or miR-145 level were calculated with the 2ΔΔCt method. GAPDH mRNA was employed as an endogenous control for mRNA and lncRNA, while U6 RNA as a miRNA internal control. For exact quantification of gene copies per cell, linc-ROR and reverse-transcribed miR-145 cDNA were used as standard templates to formulate standard curves with limit dilution approaches, and then the exact copies of linc-ROR and miR-145 per cell were calculated according to molecular weights and cell counts. All assays were performed in triplicate. PCR primers were designed as follows: linc-ROR forward: 5′-CAGAAGCTGGAGTGGACAGGATTA-3′, and reverse: 5′-GTTCTATTTGTGGCCTGAAGATGTG-3′; ZEB2 forward: 5′-CGCTTGACATCACTGAAGGA-3′ and reverse: 5′-CTTGCCACACTCTGTGCATT-3′; miR-145 forward: 5′-GTCCAGTTTTCCCAGGAATC-3′, and reverse: 5′- AGAACAG-TATTTCCAGGAAT-3′; GAPDH forward: 5′-CTGGGCTACACTGAGCACC-3′, and reverse: 5′- AAGTGGTCGTTGAGGGCAATG-3′; U6 forward: 5′-CGCTTCGGCAGCACATATACTA-3′, and reverse: 5′-CGCTTCACGAATTTGCGTGTCA-3.
Cell transfection
The linc-ROR overexpression plasmid was a gift from Dr Wang Yue (College of Basic Medicine, Second Military Medical University, China). The shROR expression plasmid was a gift from Dr Hou P (The Institute of Genetics and Cytology, Northeast Normal University, Changchun, China). Hsa-miR-145 mimic/negative control mimics and hsa-miR-145 inhibitor/negative control inhibitor were purchased from Applied Biological Materials (ABM, Canada). The cell transfection was performed using Lipofectamine 2000 (Invitrogen, CA, USA) following the manufacturer’s instructions. HepG2 or SMMC-7721 cells were transfected with the plasmid linc-ROR to constructed cell lines stably expressing linc-ROR and were screened with Puromycin (2.0 μg/ml) for four weeks. HCCLM3 or MHCC97-H cell lines stably suppressing linc-ROR were constructed by transfection with a lentivirus construct containing the desired vector and screened with Puromycin (2.0 μg/ml) for four weeks.
Wound healing assay
Cell migration capacity was measured using a wound healing assay. Briefly, cells were planted in 6-well plates and cultured to confluence. Once cells were attached completely, they were scraped by a plastic scriber to form a wound in the middle of the plates. The cells were washed, and the medium was replaced with serum-free medium. After incubation for 48 h, we observed the cultures under a microscope. A minimum of five randomly chosen areas was measured and the distance of cell migration to the wound area was determined.
Cell migration and invasion assays
Cell migration and invasion were measured by transwell chamber (8.0 μm pore size, Corning) and Matrigel invasion (Becton Dickinson) according to the manufacturer’s instructions. In short, the upper chamber was filled with cells in serum-free media, coating with or without 10 μg/ml Matrigel. Media containing 10% FBS was added to the lower chamber. After 48 h of incubation, cells on the lower chamber membrane were fixed with 4% polyoxymethylene and stained with 0.1% crystal violet. Five predetermined fields were counted under a microscope.
Luciferase reporter assays
Luciferase activities were detected with the Dual-Luciferase Assay Kit (Promega, Madison, WI), according to the manufacturer’s instructions. Linc-ROR fragment containing the predicted miR-145 binding site was cloned into a pmirGlO Dual-luciferase miRNA Target Expression Vector (Promega, Madison, WI, USA) to form the reporter vector pmiRGLO-ROR-wild-type (ROR-WT). We mutagenized the miR-145 binding site on linc-ROR at nucleotide positions to form the pmiRGLO-ROR-mutated-type (ROR-MUT1, ROR-MUT2, ROR-MUT1 + 2). Vector ROR-MUT1 was mutagenized at 1307–1330 nucleotide positions and ROR-MUT2 at 2037–2059 nucleotide positions. The two miR-145 binding sites on linc-ROR were both mutagenized to form the vector ROR-MUT1 + 2. PmirGLO vector, ROR-WT, ROR-MUT1, ROR-MUT2, ROR-MUT1 + 2 were co-transfected with miR-145 mimics or miRNA-NC into HEK293 cells by Lipofectamine 2000 (Invitrogen, USA) mediated gene transfer. The relative luciferase activity was normalized to Renilla luciferase activity 48 h after transfection. The data was relative to the fold change of the corresponding control groups defied as 1.0.
RNA immunoprecipitation (RIP)
RNA immunoprecipitation was performed using thermo fisher RIP kit (Thermo, USA) according to the manufacturer’s protocol. The Ago2 antibodies were purchased from Abcam (USA). Normal mouse IgG (Abcam, USA) was applied as the negative control. Purified RNA was subjected to qRT-PCR analysis to demonstrate the presence of the binding targets using respective primers.
Western blotting assay
Total protein lysates were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and were transferred to polyvinylidene difluoride membranes (Roche). Antibody dilutions of 1:2000 were used for rabbit anti-human E-cadherin, β-catenin, N-cadherin, Vimentin, ZEB2, and 1:3000 for GAPDH. The proteins were detected using an enhanced chemiluminescence system and exposed to x-ray firm. All antibodies were purchased from Abcam (USA).
Immunofluorescence
Immunofluorescence staining assay was performed following the standard protocol. In brief, the transfected cells were fixed with 4% paraformaldehyde and permeabilized in 0.1% Triton X-100. 5% BSA was used to blocked cells for 1 h at room temperature. Finally, cells were incubated with primary antibody at 4 °C overnight, followed by incubation with florescent-dye conjugated secondary antibody (Invitrogen) for 1 h, and then stained with DAPI. Images were taken with a Zeiss photomicroscope (Carl Zeiss, Oberkochen, Germany).
In vivo tumor metastasis assay
For in vivo metastasis study, HCCLM3 cells transfected with shROR (or shCtrl) or HepG2 cells transfected with linc-ROR (Vector) were suspended in 100 μl PBS and injected subcutaneously into the right side of the posterior flank of female BALB/C athymic nude mice at 4 to 6 weeks of age. After 4 weeks, the subcutaneous tumors were resected and diced into 1.0 mm3 cubes, which were then implanted into the left lobes of the livers of the nude mice (10/group). After 8 weeks, the animals were sacrificed for intrahepatic and lung metastases assessment. All animal studies were conducted according to protocols that were approved by the Animal Care and Use Committee of Jinling Hospital (Nanjing University, Nanjing, China).
Statistical analysis
Experimental data were presented as means ± standard deviation (SD). The SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) was used for all statistical analyses. Quantitative variables for nonparametric analyses were performed using Wilcoxon text for paired and Mann-Whitney U test for unpaired analyses. The disease-free survival (DFS) and over survival (OS) curves were plotted using the Kaplan-Meier method. The correlation was evaluated by Spearman’s rank correlation coefficients. Differences were considered significant if p < 0.05. Corresponding significance levels are indicated in the figures.
Electronic supplementary material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81472266 and 81641100) and the Excellent Youth Foundation of Jiangsu Province, China (BK20140032).
Author Contributions
R.W. formulated the idea of the paper and supervised the research, reviewed and revised the manuscript. C.L. performed the research and wrote the manuscript. L.C. and X.C. provided comments and technical advice. B.F., K.Z. and S.H. participated in preparing figures, Tables and data analyzing. D.H. revised the manuscript and provided comments. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41598-023-32251-x"
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
3/29/2023
This article has been retracted. Please see the Retraction Notice for more detail: 10.1038/s41598-023-32251-x
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-04113-w
References
- 1.Lv J, et al. Long non-coding RNA Unigene56159 promotes epithelial-mesenchymal transition by acting as a ceRNA of miR-140-5p in hepatocellular carcinoma cells. Cancer letters. 2016;382:166–175. doi: 10.1016/j.canlet.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 2.Hou, G. et al. Aldehyde Dehydrogenase-2 (ALDH2) Opposes HCC Progression By Regulating AMPK Signaling. Hepatology (Baltimore, Md.), doi:10.1002/hep.29006 (2016). [DOI] [PubMed]
- 3.Zhang M, Du X. Noncoding RNAs in gastric cancer: Research progress and prospects. World journal of gastroenterology. 2016;22:6610–6618. doi: 10.3748/wjg.v22.i29.6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, X. et al. Upregulation of long noncoding RNA HOTTIP promotes metastasis of esophageal squamous cell carcinoma via induction of EMT. Oncotarget, doi:10.18632/oncotarget.12995 (2016). [DOI] [PMC free article] [PubMed]
- 6.Liu F, et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene. 2016;35:5422–5434. doi: 10.1038/onc.2016.80. [DOI] [PubMed] [Google Scholar]
- 7.Loewer S, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nature genetics. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Developmental cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Eades G, et al. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Molecular cancer research: MCR. 2015;13:330–338. doi: 10.1158/1541-7786.mcr-14-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhan HX, et al. LincRNA-ROR promotes invasion, metastasis and tumor growth in pancreatic cancer through activating ZEB1 pathway. Cancer letters. 2016;374:261–271. doi: 10.1016/j.canlet.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 11.Wang S, et al. Long Noncoding RNA ROR Regulates Proliferation, Invasion, and Stemness of Gastric Cancer Stem Cell. Cellular reprogramming. 2016;18:319–326. doi: 10.1089/cell.2016.0001. [DOI] [PubMed] [Google Scholar]
- 12.Yang, P. et al. The long non-coding RNA-ROR promotes the resistance of radiotherapy for human colorectal cancer cells by targeting the P53/miR-145 pathway. Journal of gastroenterology and hepatology, doi:10.1111/jgh.13606 (2016). [DOI] [PubMed]
- 13.Wang SH, et al. Overexpression of LncRNA-ROR predicts a poor outcome in gallbladder cancer patients and promotes the tumor cells proliferation, migration, and invasion. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:12867–12875. doi: 10.1007/s13277-016-5210-z. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi K, Yan IK, Kogure T, Haga H, Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS open bio. 2014;4:458–467. doi: 10.1016/j.fob.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi K, Yan IK, Haga H, Patel T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. Journal of cell science. 2014;127:1585–1594. doi: 10.1242/jcs.141069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tu K, et al. Fibulin-5 inhibits hepatocellular carcinoma cell migration and invasion by down-regulating matrix metalloproteinase-7 expression. BMC cancer. 2014;14:938. doi: 10.1186/1471-2407-14-938. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. Journal of hepatology. 2016;65:798–808. doi: 10.1016/j.jhep.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. International journal of cancer. 2016;139:269–280. doi: 10.1002/ijc.30039. [DOI] [PubMed] [Google Scholar]
- 19.Duru N, Wolfson B, Zhou Q. Mechanisms of the alternative activation of macrophages and non-coding RNAs in the development of radiation-induced lung fibrosis. World journal of biological chemistry. 2016;7:231–239. doi: 10.4331/wjbc.v7.i4.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enright AJ, et al. MicroRNA targets in Drosophila. Genome biology. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao S, et al. ROR functions as a ceRNA to regulate Nanog expression by sponging miR-145 and predicts poor prognosis in pancreatic cancer. Oncotarget. 2016;7:1608–1618. doi: 10.18632/oncotarget.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noh JH, et al. MiR-145 functions as a tumor suppressor by directly targeting histone deacetylase 2 in liver cancer. Cancer letters. 2013;335:455–462. doi: 10.1016/j.canlet.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 23.Yang XW, et al. miR-145 suppresses cell invasion in hepatocellular carcinoma cells: miR-145 targets ADAM17. Hepatology research: the official journal of the Japan Society of Hepatology. 2014;44:551–559. doi: 10.1111/hepr.12152. [DOI] [PubMed] [Google Scholar]
- 24.Hammond SM. An overview of microRNAs. Advanced drug delivery reviews. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan J, Qiu K, Li M, Liang Y. Double-negative feedback loop between long non-coding RNA TUG1 and miR-145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS letters. 2015;589:3175–3181. doi: 10.1016/j.febslet.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 26.Hegarty SV, Sullivan AM, O’Keeffe GW. Zeb2: A multifunctional regulator of nervous system development. Progress in neurobiology. 2015;132:81–95. doi: 10.1016/j.pneurobio.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Schlachterman A, Craft WW, Jr., Hilgenfeldt E, Mitra A, Cabrera R. Current and future treatments for hepatocellular carcinoma. World journal of gastroenterology. 2015;21:8478–8491. doi: 10.3748/wjg.v21.i28.8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan SX, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology (Baltimore, Md.) 2016;63:499–511. doi: 10.1002/hep.27893. [DOI] [PubMed] [Google Scholar]
- 29.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nature reviews. Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 30.Jayachandran A, Dhungel B, Steel JC. Epithelial-to-mesenchymal plasticity of cancer stem cells: therapeutic targets in hepatocellular carcinoma. Journal of hematology & oncology. 2016;9:74. doi: 10.1186/s13045-016-0307-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bayoumi AS, et al. Crosstalk between Long Noncoding RNAs and MicroRNAs in Health and Disease. International journal of molecular sciences. 2016;17:356. doi: 10.3390/ijms17030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia P, et al. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer letters. 2016;381:359–369. doi: 10.1016/j.canlet.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 33.Braconi C, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mataki H, et al. Dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p) coordinately targeted MTDH in lung squamous cell carcinoma. Oncotarget. 2016;7:72084–72098. doi: 10.18632/oncotarget.12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang L, Wei D, Yan F. MicroRNA-145 functions as a tumor suppressor by targeting matrix metalloproteinase 11 and Rab GTPase family 27a in triple-negative breast cancer. Cancer gene therapy. 2016;23:258–265. doi: 10.1038/cgt.2016.27. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, et al. MiR-145 functions as a tumor suppressor via regulating angiopoietin-2 in pancreatic cancer cells. Cancer cell international. 2016;16:65. doi: 10.1186/s12935-016-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu RL, Dong Y, Deng YZ, Wang WJ, Li WD. Tumor suppressor miR-145 reverses drug resistance by directly targeting DNA damage-related gene RAD18 in colorectal cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:5011–5019. doi: 10.1007/s13277-015-3152-5. [DOI] [PubMed] [Google Scholar]
- 38.Duan X, et al. MicroRNA-145: a promising biomarker for hepatocellular carcinoma (HCC) Gene. 2014;541:67–68. doi: 10.1016/j.gene.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Yuan Wang T, et al. Acquisition cancer stemness, mesenchymal transdifferentiation, and chemoresistance properties by chronic exposure of oral epithelial cells to arecoline. Oncotarget. 2016 doi: 10.18632/oncotarget.11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W, et al. Epigenetically regulated miR-145 suppresses colon cancer invasion and metastasis by targeting LASP1. Oncotarget. 2016;7:68674–68687. doi: 10.18632/oncotarget.11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao H, et al. miR-145 suppresses breast cancer cell migration by targeting FSCN-1 and inhibiting epithelial-mesenchymal transition. American journal of translational research. 2016;8:3106–3114. [PMC free article] [PubMed] [Google Scholar]
- 42.Li S, et al. miR-145 suppresses colorectal cancer cell migration and invasion by targeting an ETS-related gene. Oncology reports. 2016;36:1917–1926. doi: 10.3892/or.2016.5042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


