Abstract
The escalating problem of antibiotic resistance, specifically cabarpenemase and extended-spectrum β-lacatamase (ESBL) producing K. pneumoniae strains, is directly correlated with increased patient morbidity and mortality and prolonged hospitalization and costs. In this study, a comprehensive genomic analysis encompassing the resistomics, virulence repertoire and mobile genetic elements of an NDM-1 positive ESBL-producing K. pneumoniae EA-MEH ST15 isolated from a urine sample collected from a Syrian refugee was conducted. Illumina paired-end libraries were prepared and sequenced resulting in 892,300 high-quality reads. The initial assembly produced 329 contigs with a combined 5,954,825 bp and a 56.5% G+C content. Resistome analysis revealed the presence of several β-lactamases including NDM-1, SHV-28, CTX-M-15 and OXA-1 in addition to 18 other genes encoding for resistance, among which are aph(3′)-Ia, aac(6′)Ib-cr, armA, strB, strA and aadA2 genes. Additionally, five plasmids IncFIB(Mar), IncHI1B, IncFIB(pKPHS1), IncFIB(K) and IncFII(K) and four integrated phages were detected. In silico MLST analysis revealed that the isolate was of sequence type ST15. To our knowledge this is the first in-depth genomic analysis of a NDM-1 positive K. pneumoniae ST15 in Lebanon associated with the recent population migration. The potential dissemination of such MDR strains is an important public health concern.
Keywords: ESBL, MDR, Klebsiella pneumoniae, draft-genome sequencing, ST15, NDM-1, SHV-28, Middle East
Introduction
Klebsiella pneumoniae are non-motile, encapsulated, lactose-fermenting, Gram-negative, facultative anaerobic rods of the Enterobacteriacae family [1]. They have been linked to both community-acquired and nosocomial infections worldwide, especially in patients with diabetes mellitus, alcoholism [2], immunocompromised patients [3], and in intensive care units [4]. The escalating burden of antibiotic resistance and specifically the dissemination of extended-spectrum β-lactamases (ESBLs) producing K. pneumoniae strains are directly correlated with increased patient morbidity and mortality [5], prolonged hospitalization time and costs [6]. ESBLs confer antimicrobial resistance against a wide range of β-lactams such as penicillins, aztreonam, and second and third generation cephalosporins [7].
Carbapenemases-producing K. pneumoniae is a major worldwide health concern [8,9]. Two particularly alarming carbapenemases are class B New Delhi Metallo-β lactamase-1 (NDM-1) [10] and class A K. pneumoniae carbapenemase (KPC) [11]. NDM-1 is capable of hydrolyzing all β-lactams, including carbapenems, with the exception of monobactams [12]. It is transmitted through mobile genetic elements and has been observed in several Enterobacteriacae species, in at least 40 countries particularly in poor resource settings [13]. KPC-producing K. pneumoniae are associated with mortality rates as high as 50%, due to limited treatment choices [14].
Some K. pneumoniae sequence types (STs) such as ST11, 15, 101, 147, 336 and ST258 have been implicated in worldwide epidemics/endemics. Particularly, ST258 is linked to the global dissemination of K. pneumoniae KPCs [14,15].
With the availability of six complete K. pneumoniae genomes [16–20] and several ongoing sequencing projects [21], the whole-genome sequencing (WGS) of K. pneumoniae has given insights into its pathogenicity, resistomics and genome plasticity [16]. Yet, little evidence exists on the complex relationship between ESBL production and pathogenesis in K. pneumoniae [22].
In this study we report the first comprehensive comparative genome analysis of an NDM-1-producing K. pneumoniae ST15 isolated from a urine sample collected from a Syrian refugee in Lebanon.
Materials and methods
Ethical approval
Ethical approval was not required as the clinical isolate was collected and stored as part of routine clinical care. Clinical isolate and patient record/information were anonymized and de-identified prior to analysis.
Sample collection
The isolate was recovered from the urine sample of an 84-year old Syrian male patient suffering from pneumonia admitted to the Middle East Institute of Health (MEIH) in Lebanon in October 2015. The patient arrived in Lebanon in 2014 when he was hospitalized for having hypertension, atrial fibrillation, moderate chronic pericardial effusion, chronic obstructive pulmonary disease and morbid obesity. He has previously undergone craniotomy and was treated for inguinal hernia.
Antimicrobial testing
Antimicrobial susceptibility test by the disk diffusion method was performed to determine the resistance patterns of the isolate to 27 different antibiotics: Penicillins (Amoxicillin, ticarcillin, amoxicillin + clavulanic acid and piperacillin + tazobactam), cephalosporins (Cefalotin, cefoxitin, cefuroxime, cefixime, cefotaxime, ceftriaxone, ceftazidime and cefepime), monobactams (Aztreonam), carbapenems (Imipenem, meropenem and ertapenem), aminoglycosides (Amikacin, gentamycin and tobramycin), quinolones (Norfloxacin, ciprofloxacin, levofloxacin and ofloxacin), tetracyclines (Tetracycline and tigecycline), sulfamides (sulfamethoxazole + trimethoprim) and nitrofurans (Nitrofurantoïn) (Oxoid, England). All antimicrobial testing was performed on Mueller-Hinton agar by the flooding technique and data interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [23].
DNA isolation and sequencing
DNA extraction was performed using the Nucleospin DNA extraction Kit (Macherey-Nagel, Germany) according to the manufacturer’s instructions.
Genome sequencing
Genomic DNA (gDNA) was used as input for library preparation using the Illumina Nextera XT DNA library preparation kit (Illumina). 1 μg of sample DNA was used as input for library preparation. The gDNA was subjected to end-repair, A-tailing, ligation of adaptors including sample-specific barcodes as recommended in the manufacturer’s protocol. The resulting library was quantified using Qubit. The library was sequenced on an Illumina MiSeq with paired-end 500 cycles protocol to read a length of 250 bp.
Genome assembly and annotation
Genome assembly was performed de novo using Spades Genome Assembler Version 3.6.0 [24]. Quality control checks of raw sequence data was performed using FastQC version 1.0.0 [25]. For comparison purposes, the assembled genome was annotated using two different tools: the RAST server (http://rast.nmpdr.org) [26] using the Glimmer default option for open reading frame (ORF) calling and the Prokka Genome Annotation software v1.0.0 [27]. The number of tRNAs and rRNAs were identified using ARAGORN v1.2.36 [28], and the RNAmmer Prediction Server 1.2 [29], respectively. The classification of predicted genes was performed using the Clusters of Orthologous Groups of proteins database [30]. The obtained ORFs were subjected to BLAST analysis against the Antibiotic Resistance Database [31] and the Comprehensive Antibiotic Resistance Database [32]. Resfinder was also used to identify resistant determinants, using a 98% identity threshold (ID) [33]. Virulence factors (VFs) were identified through BLAST search against VFDB [34]. The multilocus sequence type (MLST) was determined using the MLST 1.8 server [35] available on www.genomicepidemiology.org. The MLST profiles were determined based on PubMLST.org. The clonal group cgMLST was determined using the publically available cgMLST database [36]. The presence of plasmids was determined using PlasmidFinder 1.3 [37]. Plasmid contigs were identified using BLASTn against the nucleotide sequence database. PHAST was used to determine putative phage sequences in the genome [38]. The function linked to annotated genes was inferred by UniProtKB [39] and QuickGO [40]. IslandViewer 3 was used to identify putative genomic islands (GIs) using three independent methods for island prediction: IslandPick, IslandPath-DIMOB and SIGI-HMM [41]. Insertion sequences (ISs) were identified using IS-finder [42].
Comparative genome analysis
For comparative analysis, the following genomes were included: K. pneumoniae EA-MEH, K. pneumoniae Ecl8 (accession # HF536482.1), a streptomycin-resistant reference strain for targeted gene manipulations [43], LAU-KP1 (accession # AYQE00000000.1) producing OXA-1 and SHV-11 isolated previously from a patient in Lebanon [44], K. pneumoniae KGM-IMP216, positive for OXA-1, NDM-1, SHV-28 among other resistance genes and isolated from a urine sample collected from a patient in Lebanon (accession # LJOI01000001.1) [45], and six of the closest matches to K. pneumoniae EA-MEH obtained through BLASTn: K. pneumoniae NTUH-K2044 (accession # AP006725.1), K. pneumoniae strain XH209 (accession # CP009461.1), K. pneumoniae strain U25 (accession # CP012043.1), K. pneumoniae strain NUHL24835 (accession # CP014004.1), K. pneumoniae strain BR (CP015990.1) and K. pneumoniae strain KP36 (accession # CP017385.1). K. pneumoniae PMK-1 (accession # CP008929), a ST15 NDM-1 producing isolate linked to a nosocomial outbreak in Nepal in 2012, was used as a reference for genome comparisons [46].
K. pneumoniae EA-MEH contigs were reordered in ProgressiveMauve [47] using reference strain K. pneumoniae PMK-1 (accession # CP008929) [46]. Phylogenetic analysis based on 37 marker genes was generated using Phylosift [48].
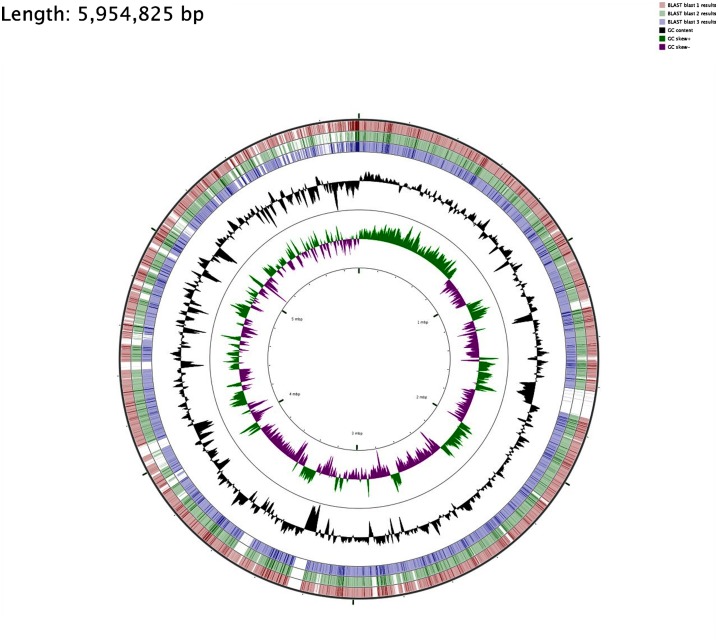
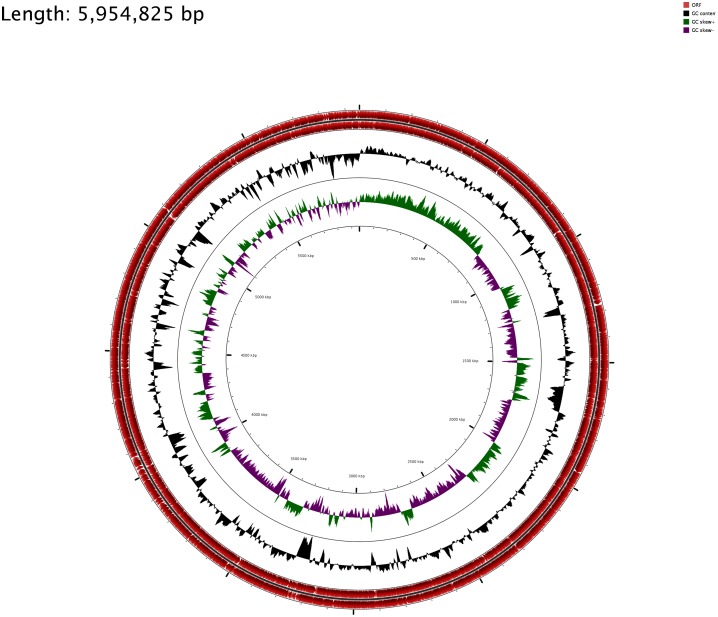
For the visualization of sequence feature information based on the sequence analysis results, circular genome representations of K. pneumoniae EA-MEH was obtained using CGview Server V 1.0 [49] through comparison with K. pneumoniae Ecl8, LAU-KP1 and PMK-1 (Figures 1 and 2).
Figure 1.
Circular genome representation of K. pneumoniae EA-MEH compared with K. pneumoniae Ecl8 (HF536482.1) [43], PMK-1 (CP008929), [46] and LAU-KP1 (AYQE00000000.1) (Tokajian et al., 2015). Starting from the outermost ring the feature rings depict: K. pneumoniae Ecl8 Blast results (red), PMK-1 Blast results (green) and LAU-KP1 Blast results (blue) representing the positions covered by the BLASTN alignment, G+C content (black), G+C positive skew (green) and G+C negative skew (purple). Image created using CGview Server V 1.0 [49].
Figure 2.
K. pneumoniae EA-MEH circular genome. Starting from the outermost rings the tracks represent: Open reading frames (ORF) (red), G+C skew (black), G+C plot (green positive skew and purple negative skew). The image was generated using CGview Server V 1.0 [49].
Capsule typing
The capsular type wzi was determined by a PCR assay followed by sequencing using the primers and PCR conditions described in Table 1. Briefly, the obtained PCR products were purified using ExoSAP-IT (Thermo Fisher Scientific, USA). The amplicons were sequenced using the ABI Prism BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystem, USA). PCR products were sequenced on the Genetic Analyzer 3500 (Life Technologies, USA) using the BigDye XTerminator purification kit (Applied Biosystems, USA). The capsular genotype was determined by uploading the wzi gene sequence to the Institut Pasteur K. pneumoniae database (http://bigsdb.web.pasteur.fr/).
Table 1.
Primers sequences and PCR conditions used for the amplification of blaSHV, blaNDM and wzi genes.
| Gene | Primers | Sequence 5′-3′ | Ta °C | Product size (bp) | References |
|---|---|---|---|---|---|
| wzi | wzi-F | GTG CCG CGA GCG CTT TCT ATC TTG GTA TTC C | 55 | 587 bp | [50] |
| wzi-R | GAG AGC CAC TGG TTC CAG AA[C or T] TT[C or G] ACC GC | ||||
| NDM | NDM-1-F | GAC CGC CCA GAT CCT CAA | 55 | 700 bp | [51] |
| NDM-1-R | CGC GAC CGG CAG GTT | ||||
| SHV | SHV-F | GCC CGG GTT ATT CTT ATT TGT CGC | 62 | 792 bp | [52] |
| SHV-R | TCT TTC CGA TGC CGC CGC CAG TCA |
Resistance genes typing
The presence of blaSHV and blaNDM genes was confirmed by individual PCR assays followed by sequencing, as previously described (Table 1). Sequences were identified using BLASTn.
Nucleotide sequence accession number
This draft genome shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession number: LSBZ00000000.1.
Results
Genomic features
The draft genome sequence of K. pneumoniae EA-MEH was 5,954,825 bp in size with a G+C content of 56.5% in 329 contigs. Annotation of this assembly using RAST identified 5615 coding sequences (CDS), 84 tRNAs and 15 rRNAs. Prokka identified 85 tRNAs, 12 rRNAs, three CRISPRs and 5530 CDS. Genes linked to carbohydrates (873), amino acid derivatives (558), cofactors, vitamins, prosthetic groups and pigments (377) were the most abundant among the SEED subsystem categories. Comparison of K. pneumoniae EA-MEH to the reference strain PMK-1 based on sequence similarity (percent identity ≥ 80) showed that 546 genes are unique for EA-MEH and that most of the functional genes including metal transport are also conserved in the reference strain.
Isolates typing
MLST
In silico MLST analysis based on seven house-keeping genes (gapA, infB, mdh, pgi, phoE, rpoB and tonB) classified the K. pneumoniae EA-MEH as sequence type ST15 belonging to CG15. In silico pMLST based on FII, FIA and FIB loci typed the isolate as [FII+:A−:B−].
Capsular genotyping
The capsular genotype of the clone was determined to be wzi93 associated with serotype K60.
Resistance profile
Antimicrobial susceptibility testing revealed that K. pneumoniae EA-MEH was resistant to all tested antibiotics and had intermediate resistant for two carbapenems: imipenem and meropenem. The minimal inhibitory concentration (MIC) was also determined for the following major antimicrobial agents: 16 mg/l for amoxicillin, 16 mg/l for amoxicillin-clavulanic acid, 32 mg/l for tazobactam, 8 mg/l for imipenem, 6 mg/l for cefotaxime, 16 mg/l for amikacin, 2 mg/l for ciprofloxacin, and 0.125 mg/l for colistin (S). The ResFinder confirmed the observed resistance patterns and identified the presence of six distinct genes conferring resistance to aminoglycosides (aph(3′)-Ia, aac(6′)Ib-cr, armA, strB, strA and aadA2), a number of β-lactamases (blaSHV-28, blaCTX-M-15, blaNDM-1 and blaOXA-1) and genes encoding for fluoroquinolone, macrolide, lincosamide and streptogramin (MLS), phenicol, sulphonamide and trimethoprim resistance. The isolate was susceptible to colistin, fosfomycin, fusidic acid, nitroimidazole, oxazolidinone, rifampicin and glycopeptide (Table 2). Despite the presence of the tetracycline resistance gene tet(D), antimicrobial susceptibility testing showed that K. pneumoniae EA-MEH was sensitive to tetracycline and tigecycline. The Resistance Gene Identifier based on BlastP Hist in the Comprehensive Antibiotic Resistance Database further elucidated the presence of various CTX-M types, OXA types and NDM types and the streptomycin resistance protein B encoded by aph(6′)-Id (Table 2).
Table 2.
Resistance genes, plasmids and phages identified in K. pneumoniae EA-MEH.
| Predicted phenotype | Resistance gene | Position in contig |
|---|---|---|
| Aminoglycoside resistance | aph(3′)-Ia | 143..956 |
| aac(6′)Ib-cr | 1878..2477 | |
| armA | 3442..4215 | |
| strB | 3710..4546 | |
| strA | 4546..5349 | |
| aadA2 | 634..1425 | |
| Beta-lactam resistance | blaSHV-28 | 12372..13232 |
| blaCTX-M-15 | 176..1051 | |
| blaNDM-1 | 4496..5308 | |
| blaOXA-1 | 917..1747 | |
| Quinolone resistance | QnrB66 | 13226..13870 |
| aac(6′)Ib-cr | 1878..2477 | |
| oqxB | 77466..79915 | |
| oqxA | 79939..81114 | |
| MLS - Macrolide, Lincosamide and Streptogramin B resistance | msr(E) | 6514..7989 |
| mph(E) | 8045..8929 | |
| Phenicol resistance | catB3 | 338..779 |
| catA1 | 750..1409 | |
| Sulphonamide resistance | sul1 | 25..861 |
| Tetracycline resistance | tet(D) | 1979..3163 |
| Trimethoprim resistance | dfrA12 | 1833..2330 |
| dfrA14 | 5978..6460 | |
| Others | Elongation factor G | 2687…4789 |
| Fluoroquinolone resistant (gyrA) (Ser83 → Phe & Asp87 → Ala) | 46525…44111 | |
| Fosfomycin resistance protein (FosA) | 15585…16004 | |
| Plasmids | IncFIB(Mar) | 1579..2017 |
| IncHI1B | 2932..3501 | |
| IncFIB(pKPHS1) | 32186..32745 | |
| IncFIB(K) | 4153..4712 | |
| IncFII(K) | 8938..9085 | |
| Phages | Vibrio phage henriette 12B8 (NC_021073) | 111576-120734 |
| Sphingomonas phage PAU (NC_019521) | 57324-63593 | |
| Bacillus phage G (NC_023719) | 94476-106610 | |
| Salisaeta icosahedral phage 1 (NC_017983) | 73831-84962 |
Mobile genetic elements and phage-associated regions
Five plasmids of the IncF family were identified: IncFIB(Mar), IncHI1B, IncFIB(pKPHS1), IncFIB(K) and IncFII(K) (Table 2).
PHAST identified four phage-associated regions in K. pneumoniae EA-MEH genome: Vibrio phage henriette 12B8 (NC_021073), Sphingomonas phage PAU (NC_019521), Bacillus phage G (NC_023719) and Salisaeta icosahedral phage 1 (NC_017983) (Table 2), with many phage-related proteins unique to EA-MEH compared to the reference; Only two phage-associated regions were identified in PMK1 (Entero P1 (NC_005856); Entero P88 (NC_026014)).
A total of 38 GIs were also detected ranging in size from 4,095 bp to 186,289 bp (Average: 19,274 ± 31,298 bp) with at least three, seven and nine islands conferring resistance to mercury, antimicrobial agents and heavy metals, respectively. Two GIs were related to defective phages, one to cell division and one to sugar transport and metabolism. GIs encoded ABC-type multidrug transport system, bleomycin resistance protein (ble), streptomycin resistance, aminoglycosides resistance and β-lactamases (Table S1).
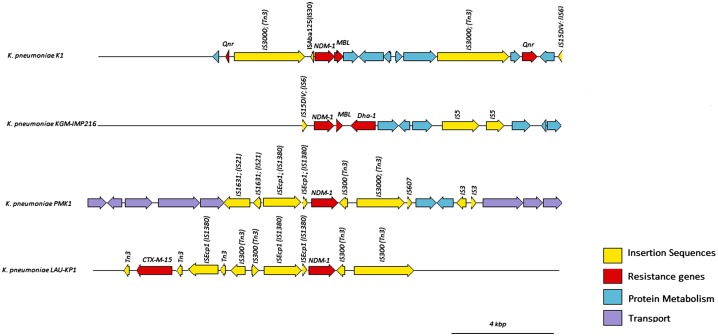
A total of 105 ISs were identified in K. pneumoniae EA-MEH using IS-finder with 120 open-reading frames (ORFs) related to ISs. Some of the detected ISs included: IS1 (IS1B, IS1D, IS1G, IS1R, IS1SD, IS1X2, IS1X3, IS1X4), IS3 (ISEc36, ISKpn8, ISSen4, ISYen3), ISKpn21, ISKpn25, IS6 and others. Tn3 was detected downstream of ISAba125, which in turn was downstream of blaNDM-1. Another Tn3 resided further upstream of blaNDM-1. The downstream Tn3 was not detected in PMK-1. This order was interrupted in other isolates as well (Figure 3).
Figure 3.
Structure of the blaNDM-1 locus in K. pneumoniae EA-MEH compared to PMK1, KGM-IMP216 and LAU-KP1 reference strains as inferred from RAST. Transposases were identified and labeled using the IS Finder database.
Virulence factors
A BLAST search revealed 173 VFs in K. pneumoniae EA-MEH, classified into 18 different categories. Among the identified VFs, two were related to capsular polysaccharide (wcbF; siaD/synD) (Table S2).
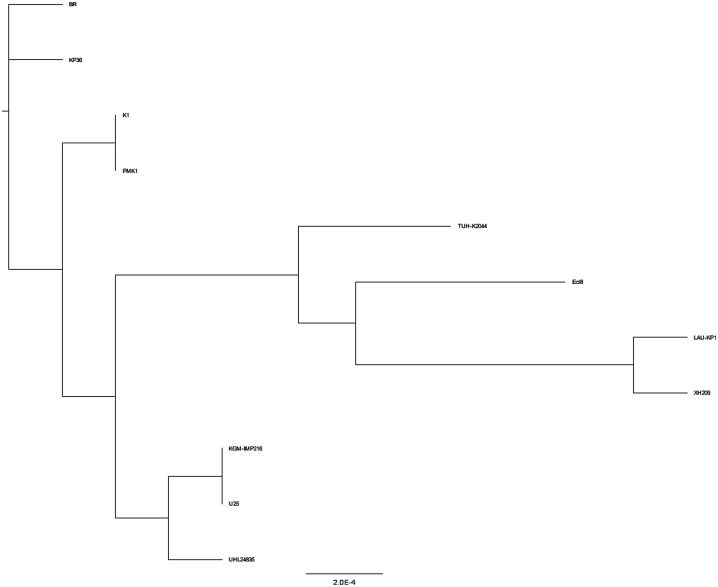
Phylogenetic tree
Phylogenetic analysis indicated a close association between K. pneumoniae EA-MEH and PMK-1 (Figure 4). LAU-KP1 clustered with XH209, Ecl8 and TUH-K2044 forming a single clade. KGM-IMP216 clustered more closely to U25 and UHL24835. K. pneumoniae strain BR and KP36 formed two branches distinct from the other isolates.
Figure 4.
Phylogenetic tree of K. pneumoniae EA-MEH compared to the reference strains.
Discussion
To the best of our knowledge, this is the first in-depth comparative genomic analysis of ST15 NDM-1 and ESBL-producing K. pneumoniae in Lebanon. ST15 is part of the epidemic CG15 within the MLST clonal complex 14 (CC14) [53]. ST15 has been previously implicated in numerous hospital outbreaks worldwide [13, 46]. Its success is partly attributed to the accumulation of various resistance genes with no fitness cost, including several β-lactamases [54]. Additionally, K. pneumoniae harbors all three drug resistance mechanisms found in Gram-negative bacteria [55], which are the acquisition of antibiotic catalytic genes, mutations in antibiotic targets and membrane proteins and the differential expression of efflux systems and pumps [56], and all were detected in K. pneumoniae EA-MEH.
K. pneumoniae EA-MEH was found to harbor several plasmid-borne resistance genes including four conferring β-lactam resistance (blaSHV-28, blaCTX-M-15, blaNDM-1, blaOXA-1) that were previously identified in Enterobacteriacae disseminating in Europe [57,58]. Additionally, 18 other enzymes conferring resistance to aminoglycosides, quinolone, MLS, phenicol, sulfonamide, tetracycline and trimethoprim were detected. K. pneumoniae is intrinsically resistant to ampicillin with the blaSHV being part of its core genome [59]. Despite the presence of the tetracycline resistance gene tet(D), antimicrobial susceptibility testing showed that the isolate was sensitive to tetracycline and tigecycline. Previous studies showed no correlation between the presence of tetracycline resistance determinants tet(A) or tet(E) and the MIC of tigecycline in Enterobacteriaceae [60]. The transcription repressor of the multidrug efflux pump acrAB operon, acrR, and which was found in EA-MEH, could be accounted for this genotype/phenotype discrepancy [61]. Although nitroimidazole, oxazolidinone and glycopeptides have poor activity against Gram-negative bacteria [62], these antimicrobials are not commonly used in Lebanon. Susceptibility to glycopeptides was also previously described Escherichia coli isolated from Lebanon [63].
blaNDM-1 is frequently coupled with several antibiotic resistance genes located on the same conjugative plasmids [13], narrowing and sometimes excluding all therapeutic options [14]. Concomitant detection of blaCTX-M-15 and blaSHV in MDR K. pneumoniae was linked to nosocomial outbreaks involving ST15, ST147 and ST336 [15]. K. pneumoniae EA-MEH also carried oqxA and oqxB genes as part of its core genome, encoding for quinolone/fluoroquinone resistance and armA encoding for aminoglycoside resistance [59]. All are among the major resistance determinants in MDR K. pneumoniae causing severe infections in hospitals and communities worldwide [13].
Plasmids of the IncF family are known to acquire novel virulence traits through replicon diversification and horizontal gene transfer [64], which contributes to the adaptability of K. pneumoniae to different health care setting [65]. IncFIB(K) plasmid was detected with high homology to plasmid pKPN3, conferring copper and silver resistance [66], additionally IncFIB(K) and IncFII(K) were previously linked with the global dissemination of CTX-M-15 [64].
The numerous Tn3 ISs detected upstream and/or downstream of blaNDM-1 in K. pneumoniae EA-MEH, LAU-KP1 and PMK1 mediate genomic plasticity and the acquisition of resistance genes through homologous recombination [67]. blaNDM-1 detected in this study has a chimeric structure forming one blaNDM-bleMBL operon with the bleomycin resistance gene bleMBL [68]. The blaNDM-bleMBL operon along with few neighboring genes, were preassembled in Acinetobacter spp. before transmission to Enterobacteriaceae. The blaNDM-bleMBL operon was previously detected on transposon Tn125 having two ISs (ISAba125) [65]. The absence of the upstream ISAba125 in K. pneumoniae EA-MEH could suggest deletion linked to Tn3 insertion, which is common in Enterobacteriaceae and is in contrast to A. baumannii [69].
Prophages contribute to genome plasticity and favor the transfer of antimicrobial resistance genes and VFs [70]. They were additionally used as potential therapeutic agents and as markers for diagnosis and epidemiological typing in K. pneumoniae [71]. Although K. pneumoniae PMK1 was the closest to EA-MEH on BLASTn showing the highest sequence similarity (Figures 1 and 4), their prophage DNA profile did not match.
Among the 173 VFs identified in K. pneumoniae EA-MEH, 40 genes were related to metal uptake. Siderophores and iron-metabolism are critical for virulence in K. pneumoniae [72], playing an important role in modulating the immune response and protecting against reactive oxygen species [73]. Moreover, the capsule is an essential virulence determinant in K. pneumoniae. The polysaccharide capsule helps the pathogen to evade phagocytosis [74] and complement-mediated killing [75], and to suppress β-defensin expression [76]. Different capsular serotypes were linked to biofilm formation in carbapenem-resistant K. pneumoniae [72]. The capsular genotype of K. pneumoniae EA-MEH was wzi93 related to serotype K60, and was recently connected to the transmission of a CTX-M-15-producing ST15K. pneumoniae between patients treated in a single center in the Netherlands followed by inter-institutional spread [58]. The role of the capsular serotypes in the pathogenesis of K. pneumoniae requires further attention and could be considered as a novel therapy target [58].
Phylogenetic analysis revealed the clustering of related STs into three major clades. K. pneumoniae EA-MEH ST15 formed a separate clade with PMK1 ST15. K. pneumoniae LAU-KP1 ST336 clustered with XH209 ST17, an MDR strain isolated from the blood of a patient in China [57]. The latter two STs are closely associated, since ST336 is a single-locus variant (SLV) of ST17 with both belonging to the same clonal group (CG) 17 [77]. Similarly, K. pneumoniae KGM-IMP216 ST14 clustered with U25 ST14, an MDR strain isolated from a tertiary care hospital in India [77], and with NUHL24835, NDM-5-producing K. pneumoniae ST14 strain from China [78]. The observed phylogenetic heterogeneity among the three isolates recovered from Lebanon (K. pneumoniae EA-MEH ST15, LAU-KP1 ST336 and KGM-IMP216 ST14) underlined the diversity in their molecular features.
The identification of NDM-1 producing ST15 K. pneumoniae in a hospital setting in Lebanon is alarming and poses a public health concern. It is compulsory to expand our current understanding of the genome plasticity in K. pneumoniae spp. through in-depth functional genomic analysis. This requires the effective cooperation between clinicians and public health practitioners in the region for adequate monitoring coupled with the implementation of effective control measure. Understanding the dynamics of transmission and shifting disease control away from policies focused on geopolitical borders would help in better understanding and controlling communicable diseases especially the ones associated with population displacement.
Supplemental data
Supplemental data for this article can be accessed at http://dx.doi.org/10.1080/20477724.2017.1314069.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
Acknowledgements
We thank the staff of the Middle East Institute of Health (MIEH) for their cooperation in sample collection.
References
- [1].Fang CT, Lai SY, Yi WC, et al. . Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis. 2007;45:284–293. 10.1086/519262 [DOI] [PubMed] [Google Scholar]
- [2].Tsay RW, Siu LK, Fung CP, et al. . Characteristics of bacteremia between community-acquired and nosocomial Klebsiella pneumoniae infection: risk factor for mortality and the impact of capsular serotypes as a herald for community-acquired infection. Arch Intern Med. 2002;162:1021–1027. 10.1001/archinte.162.9.1021 [DOI] [PubMed] [Google Scholar]
- [3].Snitkin ES, Zelazny AM, Thomas PJ, et al. . Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Trans Med. 2012;4:148ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lin MY, Lyles-Banks RD, Lolans K, et al. . The importance of long-term acute care hospitals in the regional epidemiology of Klebsiella pneumoniae carbapenemase–producing Enterobacteriaceae. Clin Infect Dis. 2013;14:cit500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gupta N, Limbago BM, Patel JB, et al. . Carbapenem-resistant enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011;53:60–67. 10.1093/cid/cir202 [DOI] [PubMed] [Google Scholar]
- [6].Giske CG, Cornaglia G. Supranational surveillance of antimicrobial resistance: the legacy of the last decade and proposals for the future. Drug Resis Update. 2010;13:93–98. 10.1016/j.drup.2010.08.002 [DOI] [PubMed] [Google Scholar]
- [7].Bradford PA. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14:933–951. 10.1128/CMR.14.4.933-951.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9:228–236. 10.1016/S1473-3099(09)70054-4 [DOI] [PubMed] [Google Scholar]
- [9].Richter SN, Frasson I, Franchin E, et al. . KPC-mediated resistance in Klebsiella pneumoniae in two hospitals in Padua, Italy, June 2009-December 2011: massive spreading of a KPC-3-encoding plasmid and involvement of non-intensive care units. Gut Pathog. 2012;4:1 DOI: 10.1186/1757-4749-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yong D, Toleman MA, Giske CG, et al. . Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yigit H, Queenan AM, Anderson GJ, et al. . Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2009;45:1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Li H, Zhang J, Liu Y, et al. . Molecular characteristics of carbapenemase-producing Enterobacteriaceae in China from 2008 to 2011: predominance of KPC-2 enzyme. Diagn Microbiol Infect Dis. 2014;78:63–65. 10.1016/j.diagmicrobio.2013.10.002 [DOI] [PubMed] [Google Scholar]
- [13].Chung PY. The emerging problems of Klebsiella pneumoniae infections: carbapenem resistance and biofilm formation. FEMS Microbiol Lett. 2016;363:fnw219 DOI: 10.1093/femsle/fnw219 [DOI] [PubMed] [Google Scholar]
- [14].Munoz-Price LS, Poirel L, Bonomo RA, et al. . Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13:785–796. 10.1016/S1473-3099(13)70190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rodrigues C, Machado E, Ramos H, et al. . Expansion of ESBL-producing Klebsiella pneumoniae in hospitalized patients: a successful story of international clones (ST15, ST147, ST336) and epidemic plasmids (IncR, IncFII K). Int J Med Microbiol. 2014;304:1100–1108. 10.1016/j.ijmm.2014.08.003 [DOI] [PubMed] [Google Scholar]
- [16].Wu KM, Li LH, Yan JJ, et al. . Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol. 2009;191:4492–4501. 10.1128/JB.00315-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ogawa W, Li DW, Yu P, et al. . Multidrug resistance in Klebsiella pneumoniae MGH78578 and cloning of genes responsible for the resistance. Biol Pharm Bull. 2005;28:1505–1508. 10.1248/bpb.28.1505 [DOI] [PubMed] [Google Scholar]
- [18].Liu P, Li P, Jiang X, et al. . Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J Bacteriol. 2012;194:1841–1842. 10.1128/JB.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shin SH, Kim S, Kim JY, et al. . Complete genome sequence of the 2, 3-butanediol-producing Klebsiella pneumoniae strain KCTC 2242. J Bacteriol. 2012;194:2736–2737. 10.1128/JB.00027-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lin AC, Liao TL, Lin YC, et al. . Complete genome sequence of Klebsiella pneumoniae 1084, a hypermucoviscosity-negative K1 clinical strain. J Bacteriol. 2012;194:6316. 10.1128/JB.01548-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ramos PI, Picão RC, de Almeida LG, et al. . Comparative analysis of the complete genome of KPC-2-producing Klebsiella pneumoniae Kp13 reveals remarkable genome plasticity and a wide repertoire of virulence and resistance mechanisms. BMC Genomics. 2014;15:1 DOI: 10.1186/1471-2164-15-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Carasso E, Salmon-Divon M, Carmeli Y, et al. . Draft Genome sequences of two multidrug-resistant extended-Spectrum-β-Lactamase-producing Klebsiella pneumoniae strains causing bloodstream infections. Genome A. 2016;4:e01533–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing. 24th informational supplement. CLSI document M100-S24 Wayne (PA): CLSI; 2014. [Google Scholar]
- [24].Bankevich A, Nurk S, Antipov D, et al. . Spades: a new genome assembly algorithm and its applications to single-cell sequencing. J Comp Biol. 2012;19:455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk
- [26].Aziz RK, Devoid S, Disz T, et al. . SEED servers: high-performance access to the SEED genomes, annotations, and metabolic models. PLOS One. 2012;7:e48053. 10.1371/journal.pone.0048053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. [DOI] [PubMed] [Google Scholar]
- [28].Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. 10.1093/nar/gkh152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lagesen K, Hallin P, Rødland EA, et al. . RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tatusov RL, Fedorova ND, Jackson JD, et al. . The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:1 DOI: 10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Liu B, Pop M. ARDB – antibiotic resistance genes database. Nucleic Acids Res. 2009;37:D443–D447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jia B, Raphenya AR, Alcock B, et al.. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zankari E, Hasman H, Cosentino S, et al. . Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chen L, Zheng D, Liu B, et al. . VFDB 2016: hierarchical and refined dataset for big data analysis – 10 years on. Nucleic Acids Res. 2016;44:D694–7. 10.1093/nar/gkv1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Larsen MV, Cosentino S, Rasmussen S, et al. . Multilocus sequence typing of total genome sequenced bacteria. J Clin Microbiol. 2012;50:1355–1361. 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jolley KA, Maiden MC. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Carattoli A, Zankari E, García-Fernández A, et al. . In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhou Y, Liang Y, Lynch KH, et al. . PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–W352. 10.1093/nar/gkr485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].UniProt Consortium Activities at the universal protein resource (UniProt). Nucleic Acids Res. 2013;42:D191–D198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Binns D, Dimmer E, Huntley R, et al. . QuickGO: a web-based tool for gene ontology searching. Bioinformatics. 2009;25:3045–3046. 10.1093/bioinformatics/btp536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Dhillon BK, Laird MR, Shay JA, et al.. IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res. 2015;43:W104–W108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Siguier P, Pérochon J, Lestrade L, et al. . ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fookes M, Yu J, De Majumdar S, et al. . Genome sequence of Klebsiella pneumoniae Ecl8, a reference strain for targeted genetic manipulation. Genome A. 2013;1:e00027–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tokajian S, Eisen JA, Jospin G, et al.. Whole genome sequencing of extended-spectrum β-lactamase producing Klebsiella pneumoniae isolated from a patient in Lebanon. Front Cell Infect Microbiol. 2015;5:32. DOI: 10.3389/fcimb.2015.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tokajian S, Eisen JA, Jospin G, et al. . Draft Genome sequence of Klebsiella pneumoniae KGM-IMP216 Harboring blaCTX-M-15, blaDHA-1, blaTEM-1B, blaNDM-1, blaSHV-28, and blaOXA-1 isolated from a patient in Lebanon. Genome A. 2016;4:e01632–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Stoesser N, Giess A, Batty EM, et al.. Genome sequencing of an extended series of NDM-Klebsiella pneumoniae neonatal infections in a Nepali hospital characterizes the extent of community versus hospital-associated transmission in an endemic setting. Antimicrob Agents Chemother. 2014;58:7347–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Darling AE, Mau B, Perna NT. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLOS One. 2010;5:e11147. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Darling AE, Jospin G, Lowe E, et al. . PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ. 2014;2:e243. 10.7717/peerj.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Grant JR, Stothard P.. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–W184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Brisse S, Passet V, Haugaard A, et al. . wzi gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol. 2013;4073–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Centre for Disease Control and Prevention Multiplex Real-Time PCR Detection of Klebsiella pneumoniae Carbapenemase (KPC) and New Delhi metallo-β-lactamase (NDM-1). https://www.cdc.gov/hai/settings/lab/kpc-ndm1-lab-protocol.html [Google Scholar]
- [52].Kaur M, Aggarwal A. Occurrence of the CTX-M, SHV and TEM genes among the extended spectrum β-lactamase producing isolates of Enterobacteriaceae in a tertiary care hospital of north India. J Clin Diag Res. 2013;4:642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Bialek-Davenet S, Criscuolo A, Ailloud F, et al. . Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Diseases. 2014;20:1812–1820. 10.3201/eid2011.140206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Toth A, Kocsis B, Damjanova I, et al. . Fitness cost associated with resistance to fluoroquinolones is diverse across clones of Klebsiella pneumoniae and may select for CTX-M-15 type extended-spectrum β-lactamase. Eur J Clin Microbiol Infect Dis. 2014;33:837–843. 10.1007/s10096-013-2022-6 [DOI] [PubMed] [Google Scholar]
- [55].Kumar V, Sun P, Vamathevan J, et al. . Comparative genomics of Klebsiella pneumoniae strains with different antibiotic resistance profiles. Antimicrob Agents Chemother. 2011;55:4267–4276. 10.1128/AAC.00052-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010;362:1804–1813. 10.1056/NEJMra0904124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hua X, Chen Q, Li X, et al. . Complete genome sequence of Klebsiella pneumoniae sequence type 17, a multidrug-resistant strain isolated during tigecycline treatment. Genome A. 2014;2:e01337–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhou K, Lokate M, Deurenberg RH, et al.. Use of whole-genome sequencing to trace, control and characterize the regional expansion of extended-spectrum β-lactamase producing ST15 Klebsiella pneumoniae. Sci Rep. 2016;6:20840. DOI: 10.1038/srep20840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wyres KL, Holt KE. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol. 2016;24:944–956. 10.1016/j.tim.2016.09.007 [DOI] [PubMed] [Google Scholar]
- [60].Fluit AC, Florijn A, Verhoef J, et al. . Presence of tetracycline resistance determinants and susceptibility to tigecycline and minocycline. Antimicrob Agents Chemother. 2005;49:1636–1638. 10.1128/AAC.49.4.1636-1638.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Li R, Han Y, Zhou Y, et al. . Tigecycline susceptibility and molecular resistance mechanisms among clinical Klebsiella pneumoniae strains isolated during Non-Tigecycline treatment. Microb Drug Res. 2017;23:139–146 DOI: 10.1089/mdr.2015.0258. [DOI] [PubMed] [Google Scholar]
- [62].Zhanel GG, Schroeder C, Vercaigne L, et al. . A critical review of oxazolidinones: an alternative or replacement for glycopeptides and streptogramins? Can J Infect Dis Med Microbio. 2001;12:379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Tokajian S, Salloum T, Eisen JA, et al. . Genomic attributes of extended-spectrum β-lactamase-producing Escherichia coli isolated from patients in Lebanon. Future Microbiol. 2017;3:213–226. 10.2217/fmb-2017-0171 [DOI] [PubMed] [Google Scholar]
- [64].Coelho A, González-López JJ, Miró E, et al. . Characterisation of the CTX-M-15-encoding gene in Klebsiella pneumoniae strains from the Barcelona metropolitan area: plasmid diversity and chromosomal integration. Int J Antimicrob Agents. 2010;36:73–78. 10.1016/j.ijantimicag.2010.03.005 [DOI] [PubMed] [Google Scholar]
- [65].Poirel L, Labarca J, Bello H, et al. . Emergence of the 16S rRNA methylase RmtG in an extended-spectrum-β-lactamase-producing and colistin-resistant Klebsiella pneumoniae isolate in Chile. Antimicrob Agents Chemother. 2014;58:618–619. 10.1128/AAC.02059-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Villa L, García-Fernández A, Fortini D, et al. . Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. Antimicrob Agents Chemother. 2010;65:2518–2529. 10.1093/jac/dkq347 [DOI] [PubMed] [Google Scholar]
- [67].Poirel L, Bonnin RA, Boulanger A, et al. . Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob Agents Chemother. 2012;56:1087–1089. 10.1128/AAC.05620-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Fiett J, Baraniak A, Izdebski R, et al. . The first NDM metallo-β-lactamase-producing Enterobacteriaceae isolate in Poland: evolution of IncFII-type plasmids carrying the blaNDM-1 gene. Antimicrob Agents Chemother. 2014;58:1203–1207. 10.1128/AAC.01197-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012;18:263–272. 10.1016/j.molmed.2012.03.003 [DOI] [PubMed] [Google Scholar]
- [70].Kutateladze M, Adamia R. Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol. 2010;28:591–595. 10.1016/j.tibtech.2010.08.001 [DOI] [PubMed] [Google Scholar]
- [71].Hsu CR, Lin TL, Pan YJ, et al. . Isolation of a bacteriophage specific for a new capsular type of Klebsiella pneumoniae and characterization of its polysaccharide depolymerase. PLOS ONE. 2013;8:e70092. 10.1371/journal.pone.0070092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Holt KE, Wertheim H, Zadoks RN, et al. . Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Nat Acad Sci. 2015;112:E3574–E3581. 10.1073/pnas.1501049112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Holden VI, Bachman MA. Diverging roles of bacterial siderophores during infection. Metallomics. 2015;7:986–995. 10.1039/C4MT00333K [DOI] [PubMed] [Google Scholar]
- [74].Lee CH, Chang CC, Liu JW, et al. . Sialic acid involved in hypermucoviscosity phenotype of Klebsiella pneumoniae and associated with resistance to neutrophil phagocytosis. Virulence. 2014;5:673–679. 10.4161/viru.32076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Clements A, Gaboriaud F, Duval JF, et al. . The major surface-associated saccharides of Klebsiella pneumoniae contribute to host cell association. PLoS One. 2008;3:e3817. 10.1371/journal.pone.0003817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Moranta D, Regueiro V, March C, et al. . Klebsiella pneumoniae capsule polysaccharide impedes the expression of β-defensins by airway epithelial cells. Infect Immun. 2010;78:1135–1146. 10.1128/IAI.00940-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Rafiq Z, Sam N, Vaidyanathan R. Whole genome sequence of Klebsiella pneumoniae U25, a hypermucoviscous, multidrug resistant, biofilm producing isolate from India. Mem Inst Oswaldo Cruz. 2016;111:144–146. 10.1590/0074-02760150423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Liu PP, Liu Y, Wang LH, et al. . Draft genome sequence of an NDM-5-producing Klebsiella pneumoniae sequence type 14 strain of serotype K2. Genome A. 2016;4:e01610–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.