Abstract
Multiple therapeutic agents are typically used in concert to effectively control metastatic tumors. Recently, we described microRNAs that are associated with the oligometastatic state, in which a limited number of metastatic tumors progress to more favorable outcomes. Here, we report the effective delivery of an oligometastatic microRNA (miR-655-3p) to colorectal liver metastases using nanoscale coordination polymers (NCPs). The NCPs demonstrated a targeted and prolonged distribution of microRNAs to metastatic liver tumors. Tumor-targeted microRNA miR-655-3p suppressed tumor growth when co-delivered with oxaliplatin, suggesting additive or synergistic interactions between microRNAs and platinum drugs. This is the first known example of systemically administered nanoparticles delivering an oligometastatic microRNA to advanced metastatic liver tumors and demonstrating tumor-suppressive effects. Our results suggest a potential therapeutic strategy for metastatic liver disease by the co-delivery of microRNAs and conventional cytotoxic agents using tumor-specific NCPs.
Keywords: nanoparticles, microRNA, metastases, liver, colorectal cancer
Graphical Abstract
Oshima et al. describe nanoscale coordination polymers (NCPs) with oxaliplatin and metastases-suppressive miR-655-3p detected in oligometastatic patients. Designed NCPs suppressed colorectal liver metastases in preclinical models. Results suggest a therapeutic strategy for metastatic liver disease by the co-delivery of oligometastatic microRNAs and conventional drugs using tumor-specific NCPs.
Introduction
Despite advancements in primary cancer detection and therapy, metastasis remains the primary cause of cancer-related mortality.1 Controlling metastasis is key to improving overall outcomes of patients with cancer. We previously observed a clinical subset of patients exhibiting a limited number of slowly progressing tumors that were curable with localized therapies, such as radiation or surgery, which we defined as oligometastatic.2, 3 We recently found that several microRNAs were differentially expressed in patients with oligometastases as compared with polymetastases and defined microRNAs preferentially expressed in oligometastases as “oligomirs.”4, 5, 6 We further reported that a subgroup of microRNAs associated with oligometastasis is encoded by a polycistronic microRNA gene cluster in the human chromosomal locus 14q32, which regulates adhesion, invasion, and motility pathways (an adhesion, invasion, migration [AIM] phenotype) and suppresses lung metastasis in an experimental model.7 To test the use of microRNAs as a potential therapeutic strategy for metastatic disease, we established a xenogenic hepatic metastasis model of colorectal cancer in which the tumor burden could be quantified by in vivo bioluminescence and ex vivo fluorescence.8, 9
The use of microRNAs to counteract metastasis faces many challenges, including their inherent instability and low penetration into tumor tissues, as well as a lack of effective means of delivering them to the cytosols of tumor cells.10 To overcome these challenges, we constructed a delivery vehicle based on nanoscale coordination polymers (NCPs) that self-assemble from metal ions and organic bridging ligands.11, 12, 13, 14, 15, 16, 17 NCPs are a unique class of hybrid nanomaterials that have been used in biomedical imaging18, 19 and drug delivery.11, 12, 13, 14, 15, 16, 17 NCPs possess many advantages over other existing nanoparticle platforms, including chemical diversity for accommodating different compositions, sizes, shapes, and surface functionalization, high and efficient drug loading, and intrinsic biodegradability because of labile bonds between their metal ions and polydentate bridging ligands. We recently reported the use of NCPs for the co-delivery of chemotherapeutics and small interfering RNAs in ovarian cancer xenograft models.20
In this report, we demonstrate an effective microRNA delivery system in the form of NCPs carrying oligometastases-associated 14q32-encoded miR-655-3p in an experimental model of colorectal hepatic metastases that was quantifiable through in vivo bioluminescence and ex vivo fluorescence.8 Our results suggest that an NCP-based microRNA delivery system can be used as a potential therapeutic strategy for the treatment of metastatic cancer. Such a therapeutic strategy would likely have clinical impact on the treatment of metastatic colorectal cancer.
Results
Human miR-655-3p as a Potential Anti-metastatic Therapeutic Strategy
Recently, we found that several microRNAs encoded by the chromosomal locus 14q32 were upregulated in patients with oligometastases as compared to those with polymetastatic lesions.7 We further found that these microRNAs suppressed lung metastases in an experimental model after ectopic expression in breast cancer cell lines.4, 7 Of these microRNAs, miR-655-3p provided the most robust phenotypic changes in different cell lines. We also found that predicted pathways targeted by miR-655-3p were enriched by genes associated with metastasis development and oncogenesis (Table S1). Based on these findings, we selected human miR-655-3p as a model metastases-suppressive microRNA and tested its delivery to the liver metastases using NCP vehicles.
Synthesis and Functional Validation of NCPs Carrying MicroRNA Mimics
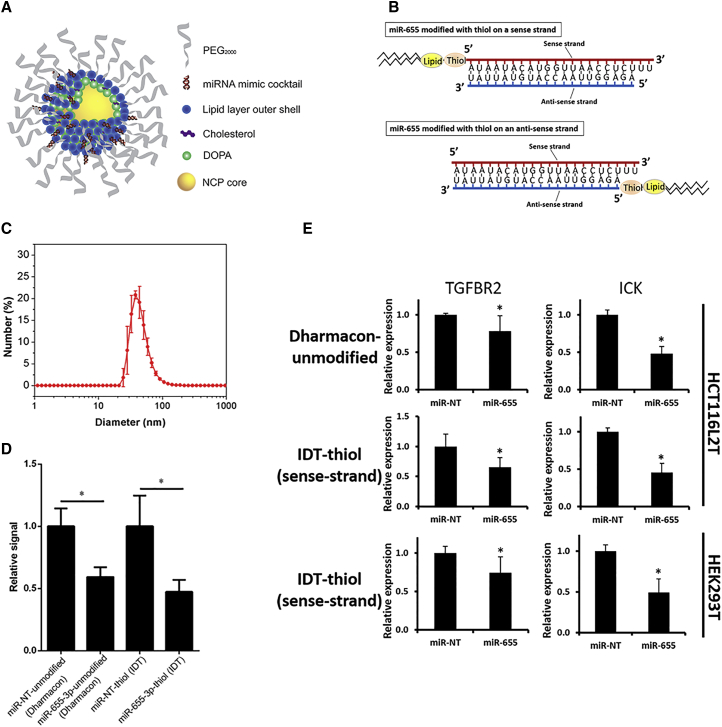
3,4-Dihydroxyphenylalanine (DOPA)-capped NCP particles containing the oxaliplatin prodrug Pt(dach)Cl2(O2CNHPO3H2)2 (dach = R, R-diaminocyclohexane) were synthesized based on previous reports.11 We incorporated microRNAs onto the surface of the particle by conjugating thiol-modified microRNA (IDT) to afford NCP/microRNA (Figure 1A). Thiol-modified custom-made microRNA mimics, provided by IDT, were incorporated into the outer layer of the lipid bilayer on NCPs. We synthesized microRNA mimics with thiol and lipid modifications on either a sense (functional) strand or an anti-sense strand (Figure 1B; Figure S1). Using dynamic light scattering (DLS), we determined that the diameter, polydispersity index (PDI), and zeta potential of NCP/miR-655 were 45.7 ± 2.9 nm, 0.20 ± 0.01, and −2.08 ± 0.56 mV, respectively (Figure 1C). Because the biological behavior of oligonucleotides like microRNAs can be altered by chemical modification, we examined the functions of lipid-/thiol-modified miR-655-3p in its ability to affect post-transcriptional regulation of genes predicted to be targeted by miR-655-3p. In these experiments, we used 3′ UTR constructs of TGFBR2 and ICK, as previously described.7 Unmodified microRNA mimics provided by Dharmacon were used as positive controls. Using 3′ UTR dual-luciferase reporter activity assays, we quantified the inhibitory effect of thiol-modified (IDT) and unmodified microRNA (Dharmacon) mimics in HEK293 cells and found the post-transcriptional level of TGFBR2 was suppressed by thiol-modified miR-655-3p (2.1-fold, p < 0.05) at a level similar to unmodified miR-655-3p microRNA mimics (1.7-fold, p < 0.05) (Figure 1D).
Figure 1.
Nanoscale Coordination Polymers and Thiol-Modified MicroRNA Mimics
(A) Schematic showing self-assembled NCPs. (B) Illustrations of synthesized microRNAs modified with thiol. miR-655-3p mimics modified with thiol and lipid on the 5′ end of the sense strand (top) and on the 5′ end of the anti-sense strand (bottom). (C) Number-weighted diameter of NCP/miR-655 (re-dispersed in PBS) by DLS measurements. Target gene suppression by modified microRNA mimics conjugated with thiol on the 5′ end of a sense strand was validated in (D) and (E), respectively. (D) Dual-luciferase reporter activity assays. HEK293T cells were co-transfected with a luciferase vector containing the 3′ UTR of the TGFBR2 gene and each of the tested microRNA mimics (n = 6, two-tailed unpaired Student t test, *p < 0.05). (E) Expressions of miR-655-3p-targeted genes TGFBR2 and ICK were quantified by qPCR in HCT116L2T and HEK293T cells transfected with each of the tested microRNA mimics (n = 6, two-tailed unpaired Student t test, *p < 0.05). Error bars represent SDs.
We next validated the inhibitory effect of miR-655-3p on target gene levels through expression analysis. We also tested the effects of thiol modifications of both sense and anti-sense strands to ensure that such modifications did not alter the biological activity of miR-655-3p mimics. In these experiments we ectopically expressed double-stranded miR-655-3p mimics with modified sense or anti-sense strands in HEK293 cells and HCT116 colorectal cancer cells double-labeled with luciferase and tdTomato (HCT116L2T8). Seventy-two hours after transfection of miR-655-3p mimics with differentially modified strands, we quantified the expression of targeted genes TGFBR2 and ICK using real-time qPCR. We found that modifications of sense strand preserved biological activity and led to a significant downregulation of targeted genes, compatible with non-modified mimics (Figure 1E). In contrast, modifications of the anti-sense strand of miR-655-3p abolished its biological activity and did not show the inhibitory effect on expression of targeted genes (Figure S1). These results demonstrated that the effects of chemical modifications on biological activity of microRNAs can be strand-specific and should be carefully evaluated to choose an appropriate strand specificity. They also demonstrated that microRNA mimics modified with thiol and lipid in the sense strand suppress the miR-655-3p-targeted gene expression of TGFBR2 and ICK7, 21 similar to unmodified microRNA, which provides an ability to use these modified mimics with NCP as a delivery vehicle.
Biodistribution of MicroRNA Mimics Carried by NCPs
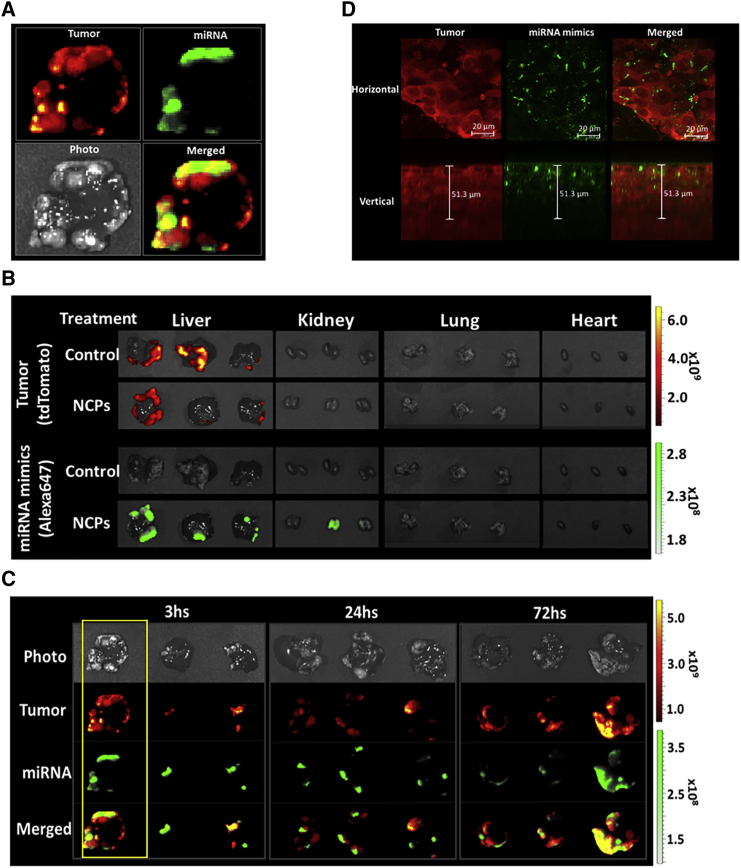
We next investigated the biodistribution of labeled microRNAs mimics carried by NCPs through ex vivo fluorescence and confocal microscopy. Metastatic liver tumors were generated by intrasplenic injection of HCT116L2T cells into athymic nude mice followed by splenectomy, as described in our previous report.8 MicroRNA mimics labeled with Alexa647 were carried by NCPs and injected intraperitoneally 4 weeks after tumor cell injection. Livers, lungs, kidneys, and hearts were harvested 3, 24, and 72 hr after injection of NCPs carrying labeled microRNAs. We detected ex vivo fluorescence signals from labeled microRNA mimics in areas of harvested tissues corresponding to liver tumors, but not in normal liver parenchyma (Figure 2A). Minimal fluorescence intensities were additionally detected in the kidneys at 3 hr after NCPs injection and disappeared at 72 hr after NCPs injection (data not shown). No signals from labeled microRNA mimics were detected in the lungs or the hearts (Figure 2B).
Figure 2.
Accumulation of MicroRNA Mimics in Liver Tumor by NCP Delivery System
Biodistribution of microRNA mimics in a liver metastases model. (A–C) Ex vivo fluorescence images. TdTomato-labeled HCT116L2T colorectal cancer cells are depicted in red (excitation/emission, 535/580 nm). Alexa647-labeled microRNA mimics are depicted in green (excitation/emission, 640/680 nm). (A) Representative images of harvested whole livers at 3 hr after injection of NCPs carrying microRNA mimics demonstrate microRNA accumulation in the liver tumor area. (B) Livers, lungs, kidneys, and hearts harvested 3 hr after injection of NCPs carrying microRNA mimics. Ex vivo fluorescence imaging shows microRNA accumulation localized to liver tumors. (C) Ex vivo fluorescence images of harvested livers 3, 24, and 72 hr after injection of NCPs carrying microRNA mimics demonstrated microRNA accumulation at all time points. (D) Confocal microscopic images of HCT116L2T tumor cells showing microRNA accumulation in cytosol and preferential perinuclear microRNA localization. TdTomato fluorescence from HCT116L2T cells is depicted in red, and Alexa647 fluorescence from microRNA mimics is depicted in green. Scale bars, 20 μm (horizontal images); 51.3 μm (vertical images).
Using confocal microscopy, we detected fluorescence signals from labeled microRNA mimics in the cytosol of tumor cells, where they showed preferential perinuclear distribution (Figure 2D). Taken together, these results indicate that microRNA mimics carried by NCPs accumulate in tumor tissue with a prolonged and stable temporal distribution.
Toxicity of Oxaliplatin-Based NCPs on Liver Metastases Model
To evaluate the toxicity of oxaliplatin-based NCPs in our liver metastases model, we intraperitoneally injected NCPs carrying 0.25, 0.5, 1.0, or 2.0 mg/kg of oxaliplatin twice a week for 2 weeks in athymic nude mice 24 hr after intrasplenic HCT116L2T cell injection. The overall survival rate was 20% in the group of mice injected with 2.0 mg/kg of oxaliplatin 28 days after NCP injection (Figure S3A). No mice died in the other groups. The tumor burden, as quantified by ex vivo fluorescence imaging, revealed decreased levels of tumor signals in groups treated with NCPs carrying 0.5, 1.0, and 2.0 mg/kg of oxaliplatin as compared with 0 or 0.25 mg/kg of oxaliplatin (Figures S3B and S3C). The group treated with NCPs carrying 0.25 mg/kg of oxaliplatin exhibited a similar level of tumor burden as compared with the PBS control group. The results indicate that NCPs carrying 0.25 mg/kg of oxaliplatin cannot suppress liver metastases progression and are nontoxic to mice as well, whereas a concentration of 2.0 mg/kg of oxaliplatin was toxic in our model. Therefore, we selected a dose of 0.25 mg/kg for our following experiments to validate the effect of microRNA.
Suppression of Liver Metastases by NCPs Carrying miR-655-3P
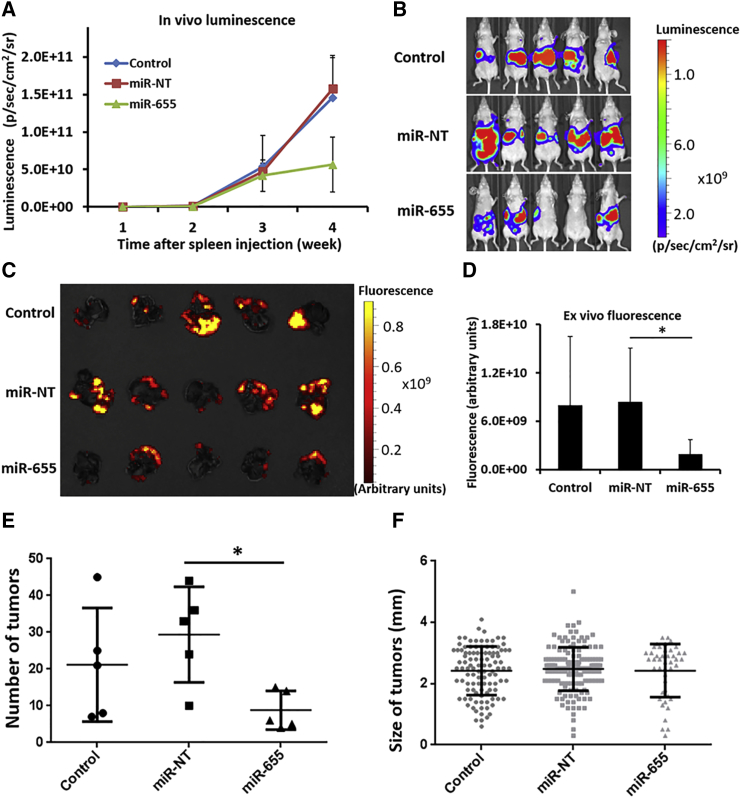
After evaluating the toxicity of NCPs carrying oxaliplatin in our model (Figure S3), 0.25 mg/kg of oxaliplatin and 62.5 μg/kg of microRNA were loaded onto NCPs as the standard dose for efficacy experiments. One group was injected with PBS as a negative control, the second group was injected with NCPs carrying a nontargeting microRNA (miR-NT), and the final group was injected with NCPs carrying only miR-655-3p. MicroRNAs carried by NCPs were injected intraperitoneally in each group twice a week for 2 weeks 24 hr after injection of HCT116L2T cells. Tumor burden, as quantified by in vivo luminescence imaging, demonstrated suppression of liver tumor development in mice injected with NCPs carrying miR-655-3p at 4 weeks following tumor cell injection. On the other hand, NCPs carrying miR-NT did not suppress tumor growth, and tumor burden in mice treated with miR-NT was similar to that in mice injected with PBS (Figures 3A and 3B). We further quantified tumor burden in harvested whole livers through ex vivo fluorescence imaging (Figure 3C). The fluorescence intensities from liver tumors were 4.3-fold lower in the miR-655-3p-treated group as compared with the miR-NT-treated group (p < 0.05) (Figure 3D). The number of tumor colonies in the miR-655-3p-treated group significantly decreased as compared with the miR-NT-treated group (p < 0.05) (Figure 3E), whereas tumor sizes were similar in these two groups (Figure 3F).
Figure 3.
NCPs Carrying miR-655-3p Suppress Liver Metastasis
NCPs carrying 0.25 mg/kg of oxaliplatin and either 62.5 μg/kg of miR-655-3p or nontargeting microRNAs (miR-NT) were injected intraperitoneally into mice twice a week for 2 weeks starting at 24 hr after HCT116L2T cell injection. (A) Tumor burden quantified weekly for PBS-containing controls, NCPs carrying miR-NT, and NCPs carrying miR-655-3p by in vivo bioluminescent imaging. Data are expressed as means ± SEM (n = 5). (B) In vivo bioluminescent images of mice injected with NCPs carrying miR-NT or miR-655-3p 4 weeks after cell injection. (C) Ex vivo fluorescence imaging of harvested whole livers with metastatic tumors 4 weeks after cell injection. (D) Quantification of ex vivo fluorescence imaging of harvested whole livers 4 weeks after cell injection (n = 5, two-tailed unpaired Student t test, *p < 0.05). Error bars represent SDs. (E) Macroscopic findings of the number of tumors colonies (n = 5, two-tailed unpaired Student t test, *p < 0.05). (F) Macroscopic findings of the sizes of tumor colonies in liver (n ≥ 44).
Suppression of Target Gene Expression by NCPs Carrying miR-655-3p
We further validated the inhibitory effect of miR-655-3p on target genes in vivo. Athymic nude mice were given splenic injections of HCT116L2T cells, and their spleens were then removed. Six weeks after tumor cell injection, miR-655-3p or miR-NT carried by NCPs was injected intraperitoneally into mice. The liver tumor colonies and normal liver tissues were harvested 48 hr after the microRNA injection; then the expression levels of targeted genes TGFBR2 and ICK in tumor colonies and liver tissues were detected through qPCR analysis. We found that the expression of TGFBR2 and ICK in liver tumor colonies significantly decreased by 63% (p < 0.0001) and 73% (p < 0.0001), respectively, in the miR-655-3p-treated group as compared with the miR-NT-treated group (Figure 4). This result suggests that miR-655-3p administration effectively inhibits the expression levels of target genes in liver tumor colonies. However, we did not detect the suppressed targeted gene expression at 16 days after the last NCP injection (Figure S3). Together, these results indicate that delivery of miR-655-3p by NCP leads to the immediate suppression of corresponding target genes and inhibits metastases growth.
Figure 4.
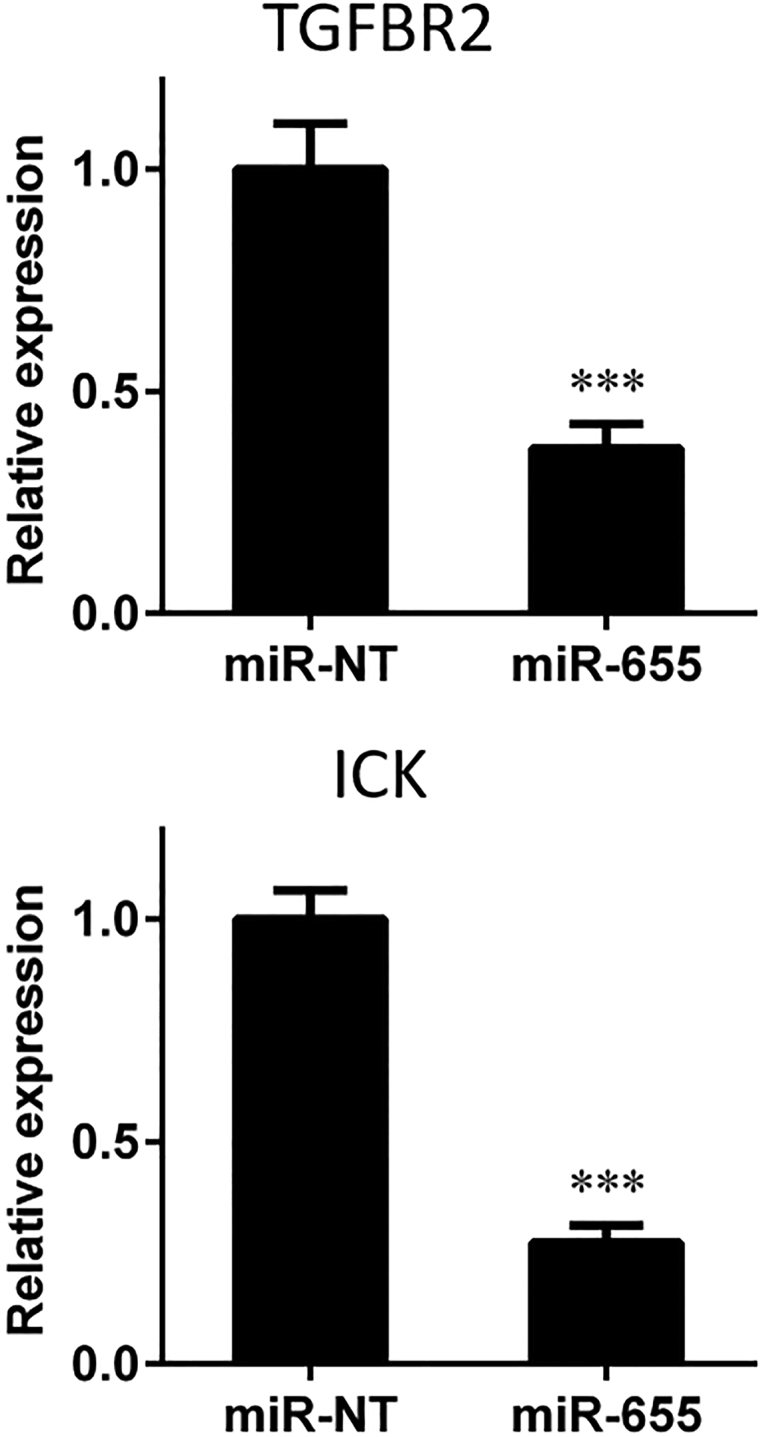
NCPs Carrying miR-655-3p Suppress the Expression Level of Target Genes in Liver Tumor Colonies
Athymic mice were given splenic injections of HCT116L2T cells. After 6 weeks, NCPs carrying 0.25 mg/kg of oxaliplatin and miR-655-3p or nontargeting microRNAs (miR-NT) were injected intraperitoneally. Liver tumor colonies were harvested 48 hr post-microRNA injection. Expressions of miR-655-3p-targeted genes TGFBR2 and ICK were quantified by qPCR in liver tumor colonies (n ≥ 5, two-tailed unpaired Student t test, ***p < 0.001). Error bars represent SDs.
Discussion
Previously, we reported that a subset of microRNAs was differentially expressed between oligo- and poly-metastatic patients who were either treated with high-dose radiotherapy5 or underwent lung metastasectomy,4 indicating a potential role of these microRNAs in the regulation of metastasis development and response to treatments. We also found that a subgroup of these microRNAs encoded by a polycistronic microRNA gene cluster in the human chromosome loci 14q32 was involved in the regulation of adhesion, migration, and invasion (AIM phenotype) and was able to suppress metastasis development in vivo after ectopic expression in highly metastatic tumor clones.6, 7 In the current study, we used one of these microRNAs (miR-655-3p) to explore the ability of targeted delivery of such “anti-metastatic” microRNAs in the liver using NCP-based nanoparticles. Our results demonstrated that miR-655-3p delivery significantly decreased the number of metastatic colonies without significant reduction of tumor sizes (Figure 4). These results suggest that reduction in tumor size is connected with inhibition of colonization ability, rather than growth propensity, and is consistent with our previous observations.6, 7, 8 Upregulation of miR-655-3p has previously been shown to suppress cell invasion in esophageal squamous cell carcinoma22 and epithelial-to-mesenchymal transition in breast cancer,23 thus supporting our findings. Further studies could take advantage of our NCP system to determine the effects of a combination of multiple oligo-microRNAs on tumor metastasis progression and prevention. Modifying the doses of oxaliplatin and microRNA delivered, as well as the timing of administration, could further elucidate optimal doses and timing for maximal tumor control. We hypothesize that further experiments regarding pretreatment with microRNAs encapsulated in NCPs could open possibilities for metastasis prevention in patients with primary colorectal tumors.
Targeting tumors with oligo-microRNAs is a novel therapy for patients with metastatic diseases. However, targeting tumors with systemic microRNAs is challenging because of their inherent instability due to degradation by RNases during blood circulation and the endosome/lysosome system inside cells, poor penetration into tumor tissues, and unpredictable immunologic toxicities.10 Moreover, enhancement of endosomal escape is an important step in allowing microRNAs to function once they have been introduced into tumor cells. To overcome these difficulties and effectively utilize oligo-miRNAs as a potential therapeutic strategy, we established a unique NCP-based nanoparticle system delivering both microRNAs and a platinum drug. As other researchers have demonstrated, nanoparticles represent one method for the successful delivery of microRNAs as an anticancer therapy in combination with conventional cytotoxic agents.24, 25, 26 The NCPs are self-assembled from metal ions and organic bridging ligands, and can overcome many drawbacks of existing drug delivery systems by virtue of tunable compositions, sizes, and shapes, high drug loading, ease of surface modification, and decreased biodegradability, all of these leading to enhancement of delivery efficacy and reduction of toxicity. We should also note that in the current research we used intraperitoneal delivery of NCPs, but further improvements may be achieved using different delivery routes, such as intrahepatic artery administration.27, 28
In summary, we demonstrated the successful development of an NCP-based system for delivery of microRNAs to tumor tissues, leading to suppression of metastatic development in our experimental colorectal liver metastasis model. Our microRNA delivery system provides the tool for systemic administration of microRNAs that can inhibit liver metastasis. Our findings suggest a potential therapeutic strategy that combines tumor-suppressive microRNA with conventional cytotoxic chemotherapies for the treatment of liver metastatic disease. We are conducting additional preclinical studies in order to justify early-phase clinical trials to assess potential clinical impact of such NCP-assisted microRNA therapy.
Materials and Methods
Cell Lines, Chemicals, and Reagents
The Luc2-tdTomato plasmid and HCT116 cell line were graciously provided by Dr. Geoffrey Greene at the University of Chicago. HCT116 human colorectal cancer cells were stably transfected with luciferase and tdTomato fluorescent protein genes using lentivirus-based gene delivery.29 The cell line was maintained in DMEM (GIBCO) with 10% fetal bovine serum and 1% penicillin/streptomycin (GIBCO).
Colorectal Cancer Animal Model of Liver Metastases
The establishment of the colorectal cancer mouse model with liver metastases was described previously.8 In summary, 6- to 8-week-old female athymic nude mice (Envigo) were anesthetized with 2% isoflurane in oxygen. The spleen was exposed through a flank incision. A total of 1.6–2 × 106 HCT116 colorectal cancer cells with stably transfected luciferin and tdTomato (HCT116L2T) was slowly injected into the spleen. After 5 min, the spleen was removed to avoid carcinomatous peritonitis and residual tumor growth in spleen. According to the experimental design, NCPs were delivered by intraperitoneal injection either 24 hr after tumor cell injection or 4–6 weeks after tumor cells injection. The tumor burden on liver was quantified by in vivo bioluminescence imaging and ex vivo fluorescence imaging of whole livers using an IVIS Spectrum 200 (Xenogen) in the Integrated Small Animal Imaging Research Resource at the University of Chicago. The measurement of in vivo bioluminescence intensity of liver metastases was performed weekly, and the ex vivo fluorescence imaging of whole livers was performed at the endpoint of experiments according to the experimental design. Data were analyzed using Living Image 4.0 Software (Caliper Life Sciences). All animal procedures were carried out in accordance with the Protocol 72213 approved by the Institutional Animal Care and Use Committee of the University of Chicago.
Procedures for Nanoparticle Synthesis
DOPA-capped NCPs containing the oxaliplatin prodrug were synthesized based on the previous literature.11 We prepared NCP/miR-655-3p by combining a tetrahydrofuran (THF) solution of DOPC, cholesterol (DOPC/cholesterol = 2:1 molar ratio), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (DSPE-PEG2k; 20 mol%), and DOPA-capped NCP with DSPE-miR-655 in 30% (v/v) ethanol/water at 50°C. The mixture was stirred vigorously for 1 min. THF and ethanol were completely evaporated, and the solution was allowed to cool to room temperature.
Sequences and Modifications of Custom-Made Oligonucleotides
miR-655-3p mimics contain sense/anti-sense sequences of 5′-AUAAUACAUGGUUAACCUCUUU-3′/5′-AGAGGUUAACCAUGUAUUAU-3′. As negative control, we used miRIDIAN microRNA Mimic Negative Control 1: 5′-UCACAACCUCCUAGAAAGAGUAGA-3′/5′-UACUCUUUCUAGGAGGUUGUGA-3′. In biodistribution experiments, we used sense and anti-sense sequences of survivin small interefering SRNA (siRNA) mimic as we described in previous literature.20 Each oligonucleotide was modified with thiol at the 5′ end of functional (miR-655-3p and miR-NT) or anti-sense strand. All custom-made oligonucleotides were provided by IDT.
MicroRNA Transfection
HCT116 cells were forward transfected with RNAiMAX (Invitrogen) and microRNA mimics (Dharmacon) at a concentration of 50 nM per the manufacturer’s instructions. miR-NT were used as negative controls. Transfection medium was replaced after 24 hr. Cells were harvested at 48 and 72 hr post-transfection.
3′ UTR Dual-Glo Luciferase Report Assay
Twenty-four hours prior to transfection, HEK293T cells were plated in a 96-well plate at a density of 15,000 cells per well. Cells were subject to two sequential transfections: (1) microRNA mimics or a nontargeting control (50 nM); and (2) after 24 hr, Renilla luciferase vector (pLightSwitch_3UTR, 100 ng per well; Switchgear Genomics) and Luc2 plasmid (100 ng per well; Promega). Ten hours after the second transfection, cell lysis reagent was added and reacted using a Dual-Glo luciferase assay kit (Promega) according to the manufacturer’s guidelines. Luciferase activity was quantified using a plate reader. Values represent percentage of activity relative to control-treated cells. All experiments were performed in triplicate.
DNA and RNA Isolation
Genomic DNA was extracted from cells using the Gentra Puregene cell and tissue kit (QIAGEN) according to the manufacturer’s instructions. Total RNA, including microRNAs, was isolated from cells using TRIzol (Invitrogen) or the mirVana microRNA Isolation Kit (Ambion) per manufacturers’ protocols. The concentration and quality of RNA was measured by UV absorbance at 260 and 280 nm (260/280 nm) using Nanodrop 2000 spectrophotometry (Thermo Scientific) and Qubit fluorometer (Invitrogen).
RNA Extraction and Quantitative Real-Time PCR
Total RNA from cultured cells and tumors was extracted with TRIzol (Invitrogen) and reverse transcribed with the High Capacity cDNA Synthesis Kit (Applied Biosystems). Tumor samples were mechanically homogenized using Omni TH Tissue Homogenizer (Omni International) for extraction with TRIzol. Real-time PCR was performed with primers (see Table S2) using iCycler thermal cycler with an iQ5 real-time qPCR detection system attached (Bio-Rad Laboratories) according to the manufacturers’ instructions. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the endogenous normalization control. The 2-ΔΔCt method was used to calculate relative expression changes.
Modeling Liver Metastases and Procedures for Imaging
We generated liver metastases and quantified tumor burden as previously described.8 In brief, HCT116 cells double-labeled with luciferase and tdTomato fluorescent protein were splenically injected followed by splenectomy. The tumor burden was quantified by in vivo bioluminescent imaging and ex vivo fluorescence imaging of whole livers. These imaging procedures were performed in the Integrated Small Animal Imaging Research Resource at the University of Chicago on an IVIS Spectrum (PerkinElmer).
Confocal Imaging
Images were collected with a Leica SP5 II AOBS tandem scanner spectral confocal system on a DMI6000 microscope and controlled by LASAF software (version 2.7.3.9723) in the Integrated Light Microscopy Facility at the University of Chicago.
Statistical Analysis
Data analyses were performed using JMP 10 software (SAS Institute) and GraphPad Prism 6 software (GraphPad Software). Data were represented as the mean ± SD for all figure panels in which error bars were shown, unless otherwise indicated. The p values were assessed using two-tailed Student t tests, and p < 0.05 was considered statistically significant. Overall survival curves were estimated by the Kaplan-Meier method and compared with use of the two-sided log rank test.
Author Contributions
G.O., C.H., M.C.P., W.L., N.N.K., and R.R.W. conceived the project and designed the experiments; G.O., N.G., C.H., M.E.S., C.P., K.B.S., S.C.W., and A.U. performed the experiments and analyzed the data; G.O., A.P., M.C.P., W.L., N.N.K., and R.R.W. interpreted data and wrote the manuscript.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
We thank Dr. Geoffrey L. Greene (University of Chicago) for the Luc2-tdTomato plasmid and HCT116 cell line, Mr. Ani Solanki (Animal Resource Center) for mice management, and Dr. Lara Leoni for assistance with IVIS imaging. This work was supported in part by the National Cancer Institute (grant U01-CA198989), Virginia and D.K. Ludwig Fund for Cancer Research, Lung Cancer Research Foundation (LCRF), Prostate Cancer Foundation (PCF), and Cancer Center Support grant P30CA014599.
Footnotes
Supplemental Information includes Supplemental Materials and Methods, three figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.04.005.
Contributor Information
Wenbin Lin, Email: wenbinlin@uchicago.edu.
Nikolai N. Khodarev, Email: n-khodarev@uchicago.edu.
Ralph R. Weichselbaum, Email: rrw@radonc.uchicago.edu.
Supplemental Information
References
- 1.Chaffer C.L., Weinberg R.A. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.Hellman S., Weichselbaum R.R. Oligometastases. J. Clin. Oncol. 1995;13:8–10. doi: 10.1200/JCO.1995.13.1.8. [DOI] [PubMed] [Google Scholar]
- 3.Weichselbaum R.R., Hellman S. Oligometastases revisited. Nat. Rev. Clin. Oncol. 2011;8:378–382. doi: 10.1038/nrclinonc.2011.44. [DOI] [PubMed] [Google Scholar]
- 4.Lussier Y.A., Khodarev N.N., Regan K., Corbin K., Li H., Ganai S., Khan S.A., Gnerlich J.L., Darga T.E., Fan H. Oligo- and polymetastatic progression in lung metastasis(es) patients is associated with specific microRNAs. PLoS ONE. 2012;7:e50141. doi: 10.1371/journal.pone.0050141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lussier Y.A., Xing H.R., Salama J.K., Khodarev N.N., Huang Y., Zhang Q., Khan S.A., Yang X., Hasselle M.D., Darga T.E. MicroRNA expression characterizes oligometastasis(es) PLoS ONE. 2011;6:e28650. doi: 10.1371/journal.pone.0028650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uppal A., Ferguson M.K., Posner M.C., Hellman S., Khodarev N.N., Weichselbaum R.R. Towards a molecular basis of oligometastatic disease: potential role of micro-RNAs. Clin. Exp. Metastasis. 2014;31:735–748. doi: 10.1007/s10585-014-9664-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uppal A., Wightman S.C., Mallon S., Oshima G., Pitroda S.P., Zhang Q., Huang X., Darga T.E., Huang L., Andrade J. 14q32-encoded microRNAs mediate an oligometastatic phenotype. Oncotarget. 2015;6:3540–3552. doi: 10.18632/oncotarget.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oshima G., Wightman S.C., Uppal A., Stack M.E., Pitroda S.P., Oskvarek J.J., Huang X., Posner M.C., Hellman S., Weichselbaum R.R., Khodarev N.N. Imaging of tumor clones with differential liver colonization. Sci. Rep. 2015;5:10946. doi: 10.1038/srep10946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oshima G., Stack M.E., Wightman S.C., Bryan D., Poli E., Xue L., Skowron K.B., Uppal A., Pitroda S.P., Huang X. Advanced animal model of colorectal metastasis in liver: imaging techniques and properties of metastatic clones. J. Vis. Exp. 2016;117:e54657. doi: 10.3791/54657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y., Gao D.Y., Huang L. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv. Drug Deliv. Rev. 2015;81:128–141. doi: 10.1016/j.addr.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu D., Poon C., Lu K., He C., Lin W. Self-assembled nanoscale coordination polymers with trigger release properties for effective anticancer therapy. Nat. Commun. 2014;5:4182. doi: 10.1038/ncomms5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He C., Liu D., Lin W. Self-assembled nanoscale coordination polymers carrying siRNAs and cisplatin for effective treatment of resistant ovarian cancer. Biomaterials. 2015;36:124–133. doi: 10.1016/j.biomaterials.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huxford-Phillips R.C., Russell S.R., Liu D., Lin W. Lipid-coated nanoscale coordination polymers for targeted cisplatin delivery. RSC Advances. 2013;3:14438–14443. doi: 10.1039/C3RA42033G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rieter W.J., Pott K.M., Taylor K.M., Lin W. Nanoscale coordination polymers for platinum-based anticancer drug delivery. J. Am. Chem. Soc. 2008;130:11584–11585. doi: 10.1021/ja803383k. [DOI] [PubMed] [Google Scholar]
- 15.Huxford R.C., Dekrafft K.E., Boyle W.S., Liu D., Lin W. Lipid-coated nanoscale coordination polymers for targeted delivery of antifolates to cancer cells. Chem. Sci. (Camb.) 2012;3:198–204. doi: 10.1039/C1SC00499A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu D., Kramer S.A., Huxford-Phillips R.C., Wang S., Della Rocca J., Lin W. Coercing bisphosphonates to kill cancer cells with nanoscale coordination polymers. Chem. Commun. (Camb.) 2012;48:2668–2670. doi: 10.1039/c2cc17635a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poon C., He C., Liu D., Lu K., Lin W. Self-assembled nanoscale coordination polymers carrying oxaliplatin and gemcitabine for synergistic combination therapy of pancreatic cancer. J. Control. Release. 2015;201:90–99. doi: 10.1016/j.jconrel.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.deKrafft K.E., Xie Z., Cao G., Tran S., Ma L., Zhou O.Z., Lin W. Iodinated nanoscale coordination polymers as potential contrast agents for computed tomography. Angew. Chem. Int. Ed. Engl. 2009;48:9901–9904. doi: 10.1002/anie.200904958. [DOI] [PubMed] [Google Scholar]
- 19.Liu D., He C., Poon C. Theranostic nanoscale coordination polymers for magnetic resonance imaging and bisphosphonate delivery. J. Mater. Chem. B Mater. Biol. Med. 2014;2:8249–8255. doi: 10.1039/c4tb00751d. [DOI] [PubMed] [Google Scholar]
- 20.He C., Poon C., Chan C., Yamada S.D., Lin W. Nanoscale coordination polymers codeliver chemotherapeutics and siRNAs to eradicate tumors of cisplatin-resistant ovarian cancer. J. Am. Chem. Soc. 2016;138:6010–6019. doi: 10.1021/jacs.6b02486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harazono Y., Muramatsu T., Endo H., Uzawa N., Kawano T., Harada K., Inazawa J., Kozaki K. miR-655 Is an EMT-suppressive microRNA targeting ZEB1 and TGFBR2. PLoS ONE. 2013;8:e62757. doi: 10.1371/journal.pone.0062757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Zang W., Du Y., Ma Y., Li M., Li P., Chen X., Wang T., Dong Z., Zhao G. Mir-655 up-regulation suppresses cell invasion by targeting pituitary tumor-transforming gene-1 in esophageal squamous cell carcinoma. J. Transl. Med. 2013;11:301. doi: 10.1186/1479-5876-11-301. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Lv Z.D., Kong B., Liu X.P., Jin L.Y., Dong Q., Li F.N., Wang H.B. miR-655 suppresses epithelial-to-mesenchymal transition by targeting Prrx1 in triple-negative breast cancer. J. Cell. Mol. Med. 2016;20:864–873. doi: 10.1111/jcmm.12770. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Mi Y., Mu C., Wolfram J., Deng Z., Hu T.Y., Liu X., Blanco E., Shen H., Ferrari M. A micro/nano composite for combination treatment of melanoma lung metastasis. Adv. Healthc. Mater. 2016;5:936–946. doi: 10.1002/adhm.201500910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng C.J., Bahal R., Babar I.A., Pincus Z., Barrera F., Liu C., Svoronos A., Braddock D.T., Glazer P.M., Engelman D.M. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang P., Wang C., Zhao J., Xiao A., Shen Q., Li L., Li J., Zhang J., Min Q., Chen J. Near infrared-guided smart nanocarriers for microRNA-controlled release of doxorubicin/siRNA with intracellular ATP as fuel. ACS Nano. 2016;10:3637–3647. doi: 10.1021/acsnano.5b08145. [DOI] [PubMed] [Google Scholar]
- 27.Hallaj-Nezhadi S., Dass C.R., Lotfipour F. Intraperitoneal delivery of nanoparticles for cancer gene therapy. Future Oncol. 2013;9:59–68. doi: 10.2217/fon.12.171. [DOI] [PubMed] [Google Scholar]
- 28.Cheng C.J., Tietjen G.T., Saucier-Sawyer J.K., Saltzman W.M. A holistic approach to targeting disease with polymeric nanoparticles. Nat. Rev. Drug Discov. 2015;14:239–247. doi: 10.1038/nrd4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H., Patel M.R., Prescher J.A., Patsialou A., Qian D., Lin J., Wen S., Chang Y.F., Bachmann M.H., Shimono Y. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc. Natl. Acad. Sci. USA. 2010;107:18115–18120. doi: 10.1073/pnas.1006732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.