Abstract
Genetic drugs such as small interfering RNA (siRNA), mRNA, or plasmid DNA provide potential gene therapies to treat most diseases by silencing pathological genes, expressing therapeutic proteins, or through gene-editing applications. In order for genetic drugs to be used clinically, however, sophisticated delivery systems are required. Lipid nanoparticle (LNP) systems are currently the lead non-viral delivery systems for enabling the clinical potential of genetic drugs. Application will be made to the Food and Drug Administration (FDA) in 2017 for approval of an LNP siRNA drug to treat transthyretin-induced amyloidosis, presently an untreatable disease. Here, we first review research leading to the development of LNP siRNA systems capable of silencing target genes in hepatocytes following systemic administration. Subsequently, progress made to extend LNP technology to mRNA and plasmids for protein replacement, vaccine, and gene-editing applications is summarized. Finally, we address current limitations of LNP technology as applied to genetic drugs and ways in which such limitations may be overcome. It is concluded that LNP technology, by virtue of robust and efficient formulation processes, as well as advantages in potency, payload, and design flexibility, will be a dominant non-viral technology to enable the enormous potential of gene therapy.
Keywords: lipid nanoparticles, gene therapy, genetic drugs, siRNA, mRNA, gene editing
Genetic drugs based on RNA and DNA can potentially treat most diseases by silencing pathological genes, expressing therapeutic proteins, or by editing the human genome. This review summarizes progress made using lipid nanoparticle (LNP) formulations of genetic drugs to enable gene therapy to be practiced.
Main Text
The central problem preventing the widespread implementation of gene therapies based on RNA and DNA polymers is delivery.1 The complexity of the problem is enormous. Naked RNA or DNA molecules are rapidly degraded in biological fluids, do not accumulate in target tissues following systemic administration, and cannot penetrate into target cells even if they get to the target tissue. Further, the immune system is exquisitely designed to recognize and destroy vectors containing genetic information.2
Given these obstacles, it is remarkable that significant progress has been made over the last 30 years to develop delivery systems that enable the therapeutic use of genetic drugs. Here, we focus on non-viral delivery systems that have advantages of ease of manufacture, reduced immune responses, multi-dosing capabilities, larger payloads, and flexibility of design. The lead non-viral delivery systems are lipid nanoparticles (LNPs); more than four LNP small interfering RNA (siRNA) drugs have entered the clinic, one of which is in late stage phase III trials (see Table 1). Here, we describe the origins of LNP technology for delivery of small molecule drugs, summarize the developments leading to encapsulation and delivery of genetic drugs, and indicate the ongoing optimization process3 that is leading to increasingly potent, non-toxic gene therapies with clear clinical potential.
Table 1.
LNP siRNA Drugs in Clinical Trialsa
| Name | LNP-Encapsulated siRNA | Indication | Status | Company |
|---|---|---|---|---|
| Patisiran | siRNA versus transthyretin | transthyretin-induced amyloidosis | phase III | Alnylam |
| ARB 1467 | siRNAs versus 3 HBV proteins | hepatitis b | phase II | Arbutus |
| siRNA versus PLK1 | hepatocellular carcinoma | phase I/II | Arbutus | |
| siRNA versus KSP and VEGF | liver cancer | phase I | Alnylam | |
| siRNA versus PCSK9 | atherosclerosis | phase I | Alnylam |
LNP, lipid nanoparticle; siRNA, small interfering RNA.
As of 2017.
Design of LNP for Delivery of Genetic Drugs
The origins of LNP systems used for genetic drugs lie in the development of liposomal drug delivery systems for small molecule drugs.4 Liposomal systems are a class of LNPs containing lipids organized in a bilayer organization. Many membrane lipids, such as phosphatidylcholine (PC), adopt bilayer structures spontaneously when dispersed in an aqueous medium.5 The particular sub-class of liposomes that have proven useful for drug delivery applications are so-called large unilamellar vesicles (LUVs) exhibiting a size in the range of 100 nm and containing a single bilayer separating the interior aqueous medium from the exterior.6 There are now nine liposome-based drugs for intravenous (i.v.) administration (see Table 2) that have been approved by regulatory authorities worldwide.7 Most of these systems contain small molecule cancer drugs and rely on the ability of small (<100 nm diameter) LNP systems to preferentially extravasate at tumor sites following i.v. administration. This preferential extravasation occurs because tumor neovasculature can contain apertures with diameters up to 200 nm, leading to penetration of small particulate systems into the tumor tissue. The preferential accumulation of small LNP into tumor tissue has been termed the “enhanced penetration and retention” (EPR) effect.8 The EPR effect, in combination with LUVs with long circulation lifetimes, can improve tumor delivery by 10-fold or more compared to equivalent doses of free drug and avoid sensitive tissue, leading to therapeutic benefit.
Table 2.
Approved LNP Drugsa
| Name | Encapsulated Drug | Indication | Year Approved | Company |
|---|---|---|---|---|
| AmBisome | amphotericin B | Fungal infections Leishmanaisis | 1990 (Europe), 1997 (USA) | Gilead |
| Doxil/Caelyx | doxorubicin | Kaposi’s sarcoma | 1995 (USA) | Johnson & Johnson |
| ovarian cancer | 1999 (USA) | |||
| breast cancer | 2003 (Europe, Canada) | |||
| DaunoXome | daunorubicin | Kaposi’s sarcoma | 1996 (Europe), 1996 (USA) | Galen |
| Myocet | doxorubicin | breast cancer | 2000 (Europe) | Cephalon |
| Abelcet | amphotericin B | Aspergillosis | 1995 (USA) | Enzon |
| Amphotec | amphotericin B | Aspergillosis | 1996 (USA) | Intermune |
| Visudyne | verteporphin | macular degeneration | 2000 (USA), 2003 (Japan) | QLT |
| Lipo-Dox | doxorubicin | Kaposi’s sarcoma, breast, and ovarian cancer | 2001 (Taiwan) | Taiwan Liposome |
| Marqibo | vincristine | acute lymphoblastic leukemia | 2012 (USA) | Spectrum Pharma |
LNP, lipid nanoparticle.
Administered by intravenous injection.
LNP systems for delivery of small molecule drugs represent a relatively mature technology and have led to rigorous design criteria, many of which carry over into the design of LNP systems for delivery of genetic drugs. These criteria include a size range of 100 nm or less, highly efficient encapsulation processes, robust, scalable manufacturing processes, and product stability of at least 1 year at 4°C. A key feature is a relatively neutral surface exterior to avoid extensive adsorption of serum proteins onto the LNP. Such adsorption leads to rapid accumulation by the fixed and free macrophages present in the circulation9 and consequent poor penetration to target tissue.
Application of LNP technology to genetic drugs required the development of methods to achieve efficient encapsulation of RNA and DNA polymers into LNP systems. Techniques used for loading small molecule drugs into LUVs in response to transmembrane pH gradients4 cannot be applied to negatively charged macromolecules because of their size and negative charge. The introduction of cationic lipids10 that have strong electrostatic associations with RNA and DNA polymers provided a way forward. However, as originally practiced, cationic lipids with a permanent positive charge were mixed with nucleic acid polymers to form “complexes.” While these systems have proven useful for in vitro transfection purposes, their utility in vivo is limited due to their large size (>1 μm diameter), instability, positive surface charge, and dose-limiting toxic side effects.11
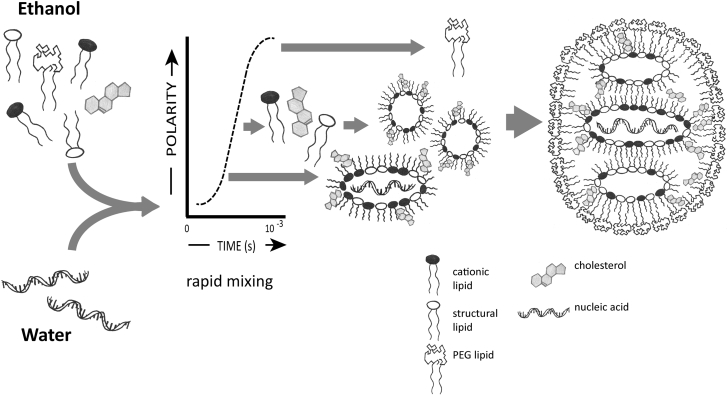
The introduction of ionizable cationic lipids, such as 1,2-dioleoyl-3-dimethylammonium propane (DODAP)12 combined with development of an ethanol-loading procedure, has provided methods to achieve high loading efficiencies for genetic drugs in small (<100 nm diameter) LNP systems with low surface charge, thus overcoming many issues noted for complexes. Specifically, ionizable cationic lipids such as DODAP have an apparent pK (pKa) <7 and can be used to achieve efficient encapsulation of negatively charged polymers into LNP at low pH (e.g., pH 4) where the ionizable lipids are positively charged. Subsequently, the pH can be raised to physiological pH values, leading to LNPs with a relatively neutral surface. The first expression of the ethanol loading technique involved mixing (at pH 4) preformed LNPs containing DODAP and other “structural” lipids with oligonucleotides in the presence of high (∼40% by volume) ethanol concentrations.13 Subsequently, a T-tube mixing process14 was developed where lipids (including ionizable cationic lipids and polyethyleneglycol [PEG] lipids) dissolved in ethanol were rapidly mixed with oligonucleotides in an aqueous buffer (pH 4), again resulting in efficient loading of nucleic acid polymers into small (diameter <100 nm) LNP systems. This was followed by a microfluidic mixing technique to obtain a more rapid and controllable mixing process15 and the production of structurally well-defined systems. A visual depiction of LNP-oligonucleotide formulation using rapid mixing techniques is shown in Figure 1, which also provides an intuitive understanding of how high oligonucleotide encapsulation efficiencies can be achieved using the ethanol loading process.
Figure 1.
Ethanol Loading Formulation Process for LNP Containing Oligonucleotides Such as siRNA
Lipids dissolved in ethanol are rapidly mixed with oligonucleotides in aqueous buffer (pH 4). The head groups of the ionizable cationic lipids are indicated as blue, of the PEG-lipids as white lines and of DSPC as white circles. Cholesterol is indicated as a diamond orange symbol and acyl chains as yellow lines. On mixing, electrostatic interactions drive formation of an inverted micelle containing oligonucleotide (red zig-zag symbol) surrounded by predominantly cationic lipid that falls out of solution as the polarity is raised. If the mixing process occurs rapidly enough, these inverted micelles do not have time to aggregate but rather are coated with PEG-lipid, which precipitates at higher polarity to surround the inverted micelles.
The most common embodiment of the ethanol loading approach to encapsulating oligonucleotides such as siRNA in LNP systems consists of an ethanol solution containing ionizable cationic lipid, cholesterol, distearoyl PC (DSPC), and PEG lipid (molar ratios 40-50/40-30/10/10-1, respectively) that is then mixed rapidly with an aqueous buffer (pH 4) containing the oligonucleotide at an amino lipid-to-oligonucleotide phosphate (N/P) ratio of six.16 A 3:1 ratio of aqueous media to ethanol is usually employed. Following the mixing step, the LNP dispersion is first dialyzed against a pH 4 buffer to remove residual ethanol and then against a pH 7.4 buffer to raise the pH to physiological values.
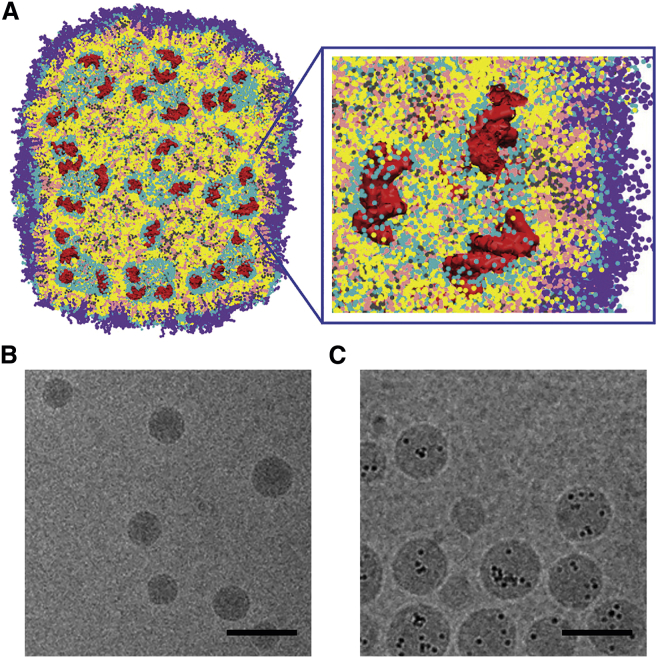
The LNP systems resulting from the rapid mixing ethanol loading process satisfy basic requirements for an in vivo delivery system for genetic drugs in five ways. The formulation process is rapid, reproducible, and scalable, encapsulation efficiencies approaching 100% are readily achievable, the LNP size can be selected by adjusting the PEG-lipid content,15 the resulting systems exhibit adequate stability properties, and the LNP particle exhibits low surface charge as required for in vivo administration. The structure of the LNP systems for siRNA as determined by molecular modeling17 indicates a largely hydrophobic core consisting of inverted micelles of lipid encapsulating oligonucleotides surrounded by a coating of PEG-lipids as shown in Figure 2. This structure is supported by the “solid core” structure detected by cryogenic transmission electron microscopy (cryo-TEM) and the “currant bun” morphology seen for cryo-TEM micrographs of LNP systems containing negatively charged gold nanoparticles, also shown in Figure 2. The structure for LNP encapsulating mRNA is still to be determined; however, it should be noted that the ethanol loading technique is a generally applicable process18 leading to efficient encapsulation of mRNA and plasmid DNA as well as other negatively charged entities such as the gold nanoparticles of Figure 2.
Figure 2.
Structure of LNP-Oligonucleotide Systems
(A) Molecular modeling17 indicates that LNP-nucleic acid systems contain irregular water-filled cavities surrounded by lipid monolayers, with nucleic acids bound to monolayer surfaces: cross-section and zoom views. Yellow, cationic lipid; pink, cholesterol; gray, DSPC; cyan, lipid polar moiety; violet, PEG-lipid; red, nucleic acids (duplex DNA); water not shown for clarity. The lipid composition was cationic lipid/DSPC/cholesterol/PEG-lipid (4:1:4:1; mol/mol) and DNA-to-lipid ratio ∼0.05 wt/wt. Adapted from Leung et al.17 (B) Lipid nanoparticle systems containing siRNA exhibit “solid core” morphology as visualized by cryo-TEM microscopy. Cryo-TEM micrograph obtained from LNP siRNA with lipid composition DLinKC2-DMA/DSPC/Chol/PEG-lipid (40/11.5/47.5/1; mol/mol) and siRNA at a siRNA/lipid ratio of 0.06, wt/wt, corresponding to an siRNA/cationic lipid charge ratio of 0.25. Scale bar, 100 nm. (C) LNP containing gold nanoparticles (5 nm diameter) exhibit a “currant bun” morphology.18 LNP encapsulating negatively charged gold nanoparticles (5 nm diameter) prepared with DLin-KC2-DMA/DOPE/Chol/PEG-lipid (40/11.5/47.5/1; % mol) at an Au-NP-to-lipid ratio of 2.2 × 1013 particles/μmol lipid. Scale bar, 100 nm.
LNP siRNA Systems: Liver Targets
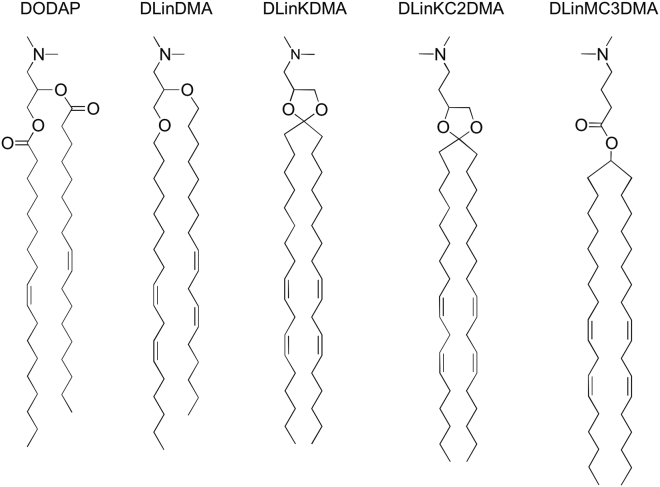
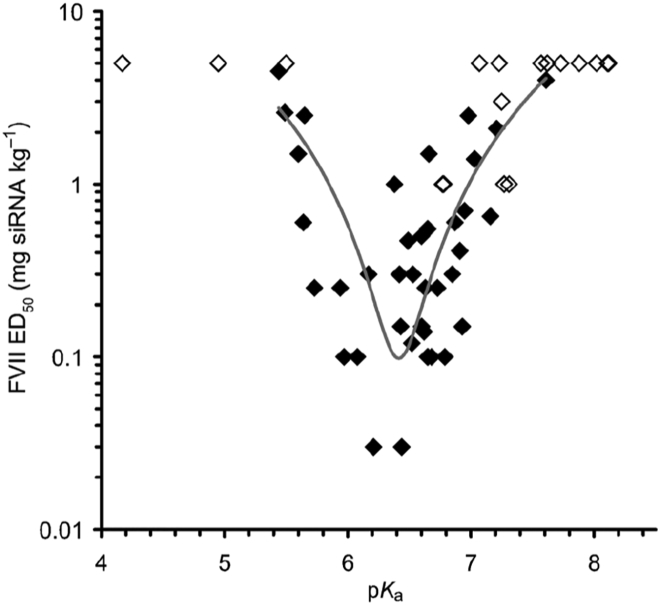
The first demonstration that LNP formulations of genetic drugs containing ionizable cationic lipids could achieve significant in vivo activity was for LNP systems containing siRNA to silence genes in hepatocytes following i.v. administration.19 This study, which employed the ionizable cationic lipid 1,2-dilinoleyloxy-3-dimethylaminopropane (DLinDMA, an ether analog of DODAP containing linoleic acyl chains), was followed by the observation that the activity of these LNP systems was highly sensitive to the particular species of cationic lipid employed.16 This observation led to an intensive lipid synthesis program to identify lipids with optimized in vivo activity when incorporated into LNP siRNA systems, resulting in the identification of dilinoleylmethyl-4-dimethylaminobutyrate (DLin-MC3-DMA, usually abbreviated to MC3), which is currently the “gold standard” cationic lipid for silencing liver targets.3 Remarkably, the gene-silencing activity achieved with MC3 was improved by over three orders of magnitude from that achieved for LNP containing DLinDMA.3 The changes in cationic lipid structure that can so dramatically influence potency appear relatively minor (Figure 3). The dominant factors determining the cationic lipid potency were the unsaturation of the acyl chains, introduction of ether linkages, and most notably, the pKa of the amino function of the cationic lipid. Specifically, LNP siRNA systems containing cationic lipids with amino functions exhibiting a pKa between 6.2 and 6.4 are by far the most effective3 for hepatocyte gene silencing (Figure 4).
Figure 3.
Evolution of Ionizable Cationic Lipids
DODAP, originally used for encapsulating antisense oligonucleotides,10 was replaced by DLinDMA in LNP siRNA systems to enhance potency.16 Subsequently DLinKC2DMA was found to further enhance potency,13 leading to a large synthesis effort to arrive at the gold standard ionizable cationic lipid DLinMC3DMA.3
Figure 4.
Optimized Ionizable Cationic Lipids Exhibit pKa Values in the Range pKa = 6.2–6.5
The figure presents a plot of the dose required to achieve 50% gene silencing (ED50) in hepatocytes in mice versus the pKa of the cationic lipids employed in the LNP.3 Fifty-six amino lipids were synthesized and formulated in LNPs and the ED50 measured and plotted against their pKa. For 15 lipids the ED50 dose was not achieved, these are indicated by the open diamonds representing the highest dose tested for that lipid. For the remaining lipids (closed diamonds), a polynomial best-fit curve highlights the most active compounds, which exhibit an optimal pKa between 6.2 and 6.6. Each data point is derived from a four-dose response curve with groups of n = 4 mice per dose. Reproduced with permission from Jayaraman et al.3
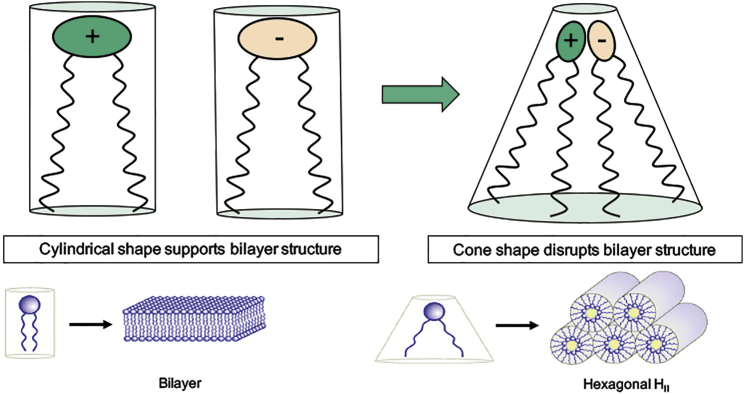
The optimization process for ionizable cationic lipids has been guided by considerations concerning the potential role of non-bilayer lipid structures in facilitating intracellular delivery of macromolecules such as siRNA. Nearly 40 years ago, we showed that membrane lipid species such as unsaturated phosphatidlyethanolamines (PEs) adopted non-bilayer structures such as the hexagonal HII phase in aqueous solution20 and that so-called fusogens that induce inter-cellular fusion also induce HII structure in biological membranes21, leading to the proposal that membrane fusion proceeds via non-lamellar intermediates. Some 20 years later, we made the related observation that permanently positively charged cationic lipids used as transfection reagents induce the HII phase in dispersions with naturally occurring anionic lipids.22 This behavior is rationalized by the “shape” hypothesis depicted in Figure 5 and led to the proposal that transfection mediated by cationic lipids results from the association of exogenous cationic lipids with endogenous anionic lipids to form membrane disruptive non-lamellar structures, allowing associated DNA to access the cell interior.16
Figure 5.
Proposed Mechanism of Action for Membrane Disruptive Effects of Cationic Lipids
In isolation, cationic lipids and anionic lipids present in the endosome (such as lyso-bis phosphatidic acid) adopt a cylindrical molecular shape, which is compatible with packing in a bilayer configuration. However, when cationic and anionic lipids are mixed together, they combine to form ion pairs where the cross-sectional area of the combined head group is less than that of the sum of individual head group areas in isolation. The ion pair therefore adopts a molecular “cone” shape, which promotes the formation of inverted, non-bilayer phases such as the hexagonal HII phase illustrated. Inverted phases do not support bilayer structure and are associated with membrane fusion and membrane disruption. Adapted from Semple et al.16
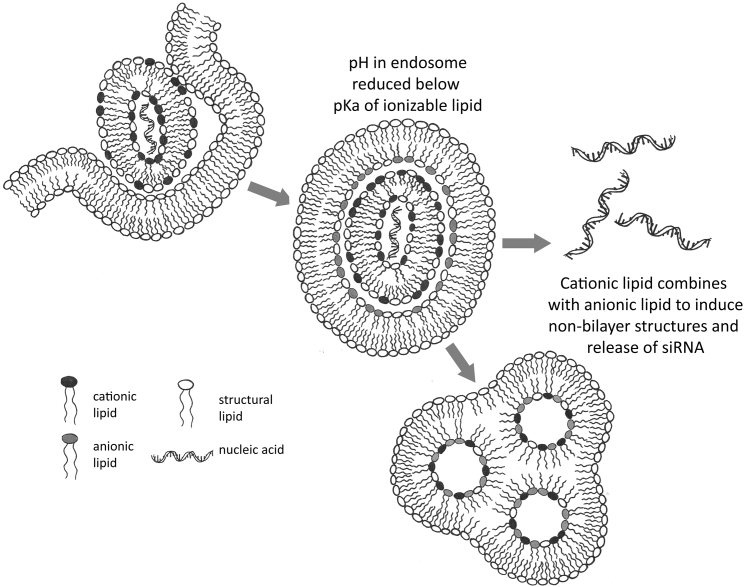
There is considerable evidence that LNP siRNA systems containing ionizable cationic lipids are accumulated into target hepatocyte cells in vivo via endocytosis.23 Thus, the rationale for optimization of ionizable cationic lipids in LNP siRNA systems was that the pKa had to be sufficiently high so that, at endosomal pH values, a large proportion was protonated (positively charged) in order to combine with endogenous anionic lipid to induce endosome-disrupting non-bilayer structures, but also sufficiently low so that the LNP surface charge did not result in clearance by the immune system prior to accumulation by target cells. The need for polyunsaturated acyl chains could also be rationalized by this non-bilayer hypothesis as more unsaturated acyl chains assume non-bilayer structure more readily.5 Escape of genetic material resulting from non-bilayer disruption of endosomes is depicted in Figure 6.
Figure 6.
Proposed Mechanism Whereby LNP Containing Ionizable Cationic Lipids and Genetic Drugs Deliver Encapsulated RNA or DNA to the Cell Cytoplasm
The LNP is accumulated into target cells by endocytosis following adsorption of ApoE.23 As the pH of the endosome is reduced by inwardly directed proton pumps the ionizable cationic lipid becomes progressively protonated (positively charged). Endogenous anionic lipids displace the cationic lipids from the genetic drug22 and combine with the cationic lipids to produce non-bilayer structures resulting in disruption of the endosomal membrane and release of the genetic drug into the cell cytoplasm.
The non-bilayer hypothesis proved a productive rationale for improving the potency of the cationic lipids, however, the relative specificity of these LNP siRNA systems for hepatocytes suggested a targeting mechanism is also operating. It is here that a benefit of conducting LNP optimization using an in vivo gene-silencing model became apparent, it was found that the LNP systems containing ionizable cationic lipids take advantage of a “natural” targeting process that would have been difficult to predict a priori. Specifically, the ability of these systems to adsorb apolipoprotein E (ApoE) is vital to hepatocyte gene-silencing potency. LNP siRNA systems giving rise to essentially complete hepatocyte gene silencing in wild-type mice result in little or no gene silencing in ApoE knockout mice23, however, potency can be restored by pre-incubation with ApoE. These results suggest that LNP siRNA systems containing ionizable cationic lipids are processed much like chylomicrons, which adsorb endogenous ApoE to trigger uptake into hepatocytes via several receptors that contain ApoE binding ligands.24
The presence and relative amounts of LNP components other than cationic lipids also determine the potency of LNP siRNA systems. For example, while relatively complete encapsulation of siRNA can be achieved at N/P values as low as 1, the resulting systems are not effective gene-silencing agents.15 Optimized LNP systems exhibit N/P values in the range of 63, 15, 16, indicating a need for non-siRNA-associated cationic lipids to achieve endosomal escape. In addition, the type and amount of PEG-lipid employed determines the size15 and strongly influences the gene-silencing potency of LNP siRNA systems.25, 26 Higher levels of PEG-lipid lead to smaller LNP systems,15 but more importantly, the presence of a long-lived PEG coating dramatically reduces gene-silencing potency.25, 26 This presumably arises from a reduced potential to interacting with cells and/or a reduced ability to adsorb ApoE. The use of a PEG lipid containing short (C14) acyl chains that dissociates from LNP in vivo with a halftime <30 min results in optimum hepatocyte gene-silencing potency.25
The combination of the optimized cationic lipid MC3, cholesterol, and DSPC, together with the rapidly dissociating PEGC14-lipid, has led to LNP systems that can silence any gene in hepatocytes at dose levels as low as 0.005 mg siRNA/kg body weight in mice.3 The high potency and excellent tolerability properties observed for MC3 in preclinical models resulted in the clinical development of an LNP siRNA drug to treat transthyretin (TTR) induced amyloidosis, a condition that affects ∼50,000 people in the western world. There is currently no treatment for this disease that leads to cardiotoxicity and neurotoxicity and is eventually lethal. The LNP TTR-siRNA drug (Patisiran) is now in phase III clinical trials27 and is likely to become the first non-viral systemically administered gene therapy drug to be approved by the Food and Drug Administration (FDA).
LNP siRNA Systems: Non-hepatic Tissues
There are many potential clinical applications of LNP siRNA drugs for silencing genes in hepatocytes, however, ApoE-mediated uptake of lipoprotein particles can also occur in other tissues such as the brain. Cholesterol in low density lipoproteins (LDL) does not reach the brain due to the blood brain barrier. Consequently, the brain synthesizes its own cholesterol and uses ApoE-containing lipoproteins to transport the sterol to neurons where they are taken up via ApoE-mediated endocytosis similar to hepatocytes.28 It has been demonstrated that LNP siRNA systems effectively identical to those employed for hepatocyte gene silencing are extremely effective gene-silencing agents for neuronal targets29 when administered intra-cortically. The utility of these systems for target validation applications has been demonstrated by the identification of a protein associated with neuronal swelling following brain injury.30 The potential for global gene silencing in the brain by LNP siRNA systems following intrathecal or interventricular administration clearly exists for the treatment of serious neurological diseases.
Other tissues for which gene silencing following i.v. administration of LNP siRNA systems have been demonstrated include macrophages,31 osteoclast and osteoblasts in “hard” bone,32 and distal tumor cells.33 The rational for targeting immune cells is that they avidly accumulate LNP systems in the circulation. Bone marrow and tumor tissues may be expected to preferentially accumulate long-circulating LNP due to EPR effects noted for both sites. However, the dose levels required to see appreciable gene silencing in macrophages, osteoclasts, and osteoblasts as well as distal tumor cells are at least two orders of magnitude higher than required for hepatocyte gene silencing, leading to low therapeutic indices that do not warrant clinical development. It is here that significant improvements in LNP siRNA potency will be required before therapeutically relevant doses can be achieved.
LNP mRNA and Plasmid DNA Systems
As noted,18 the ethanol-loading LNP formulation techniques used to generate LNP siRNA systems can be applied with equal facility to load mRNA and plasmid DNA to achieve LNP mRNA and LNP plasmid DNA (pDNA) systems. Interestingly, ionizable cationic lipids (such as MC3) optimized to achieve maximum gene silencing in hepatocytes for siRNA cargoes are not necessarily optimized for maximum gene expression from mRNA or plasmid cargoes. However, similar tissue affinities apply, likely due to ApoE-dependent uptake of LNP systems into target cells.23 Thus, while LNP mRNA systems containing MC3 exhibit appreciable gene expression in hepatocytes following i.v. administration, cationic lipids optimized for maximum mRNA delivery can result in 20-fold higher expression levels (see https://acuitastx.com). These expression levels are sufficiently high that therapeutic levels of proteins such as erythropoietin can be achieved at dose levels as low as 0.03 mg mRNA/kg body weight in non-human primates.34 These results indicate the potential for utilizing the liver as a factory for protein replacement protocols or as a way to introduce therapeutic proteins, such as monoclonal antibodies, to target malignancies or other pathologies. This has recently been demonstrated by the systemic administration of LNP encapsulating mRNA encoding the light and heavy chains of the anti-HIV-1 antibody VRC01. A dose of 1 mg/kg of mRNA in mice resulted in ∼170 μg/mL VRC01 antibody concentrations in the plasma at 24 hr post-injection. Remarkably, the translated antibody from a single injection of VRC01 mRNA fully protected humanized mice from intravenous HIV-1 challenge.35
The ability to use the liver as a bioreactor in this manner opens up new therapeutic approaches to multiple diseases. For example, enabling the transient, rapid secretion of neutralizing antibodies from the liver could provide a novel method to induce rapid “passive” immunization following exposure to potentially lethal viral infections such as Ebola or rabies. In addition to the HIV example referenced above, the authors are aware of several manuscripts in submission that clearly show this is possible in preclinical models. Equally impressive is the potential application of LNP mRNA systems in vaccines for “active” immunization. The subcutaneous, intramuscular, or intradermal administration of LNP systems containing mRNA coding for protein antigens associated with multiple infectious diseases are yielding impressive immune responses in preclinical models, often resulting in complete protection from disease in rodents and non-human primates as demonstrated recently for Zika virus infection.36
Given the demonstrated ability of the LNP delivery platform to enable mRNA protein expression in vivo, particularly in liver tissue, it will be fascinating to see applications in the emerging field of targeted gene editing. Currently, the various gene-editing technologies rely upon viral vectors to deliver site-specific targeted nucleases into cells. A major limitation of viral vectors is the induction of neutralizing antibodies that prevent repeat administrations, a problem that has not been observed to-date for LNP systems delivering siRNA.27 As with the application of this platform in vaccines, we predict examples of its utility in the gene-editing field will soon be appearing in the literature.
LNPs containing plasmid DNA have also demonstrated in vivo activity. The ability of the ethanol loading approach, in combination with ionizable cationic lipids, to achieve efficient encapsulation of plasmids as large as 7 kb has been demonstrated.37 Further, these systems can be highly effective transfection reagents for a variety of (dividing) cells in tissue culture, including primary cells. Injection of these systems into developing chick embryos has resulted in robust transfection of a marker protein (GFP) in local tissues with no apparent toxicity, suggesting therapeutic applications are possible.
LNP Formulations of Genetic Drugs: Issues and Solutions
LNP systems clearly have the potential to enable the promise of gene therapy. However, significant issues remain. Two major issues concern the immune response to LNP formulations of RNA and DNA polymers and the need for LNP systems with improved potencies so that siRNA, mRNA, and plasmid-based therapies can be extended to non-hepatic tissues. It is here that the innate flexibility of LNP technology comes to the fore. For example, the dominant cause of immune reactions to LNP formulations containing RNA- and DNA-based genetic drugs are the presence of multiple pattern recognition receptors, tuned to sense non-self-nucleic acids, that make up the complex innate safety mechanisms protecting cells from infection2. Such immune reactions can be reduced by altering nucleic acid chemistry,38 however, it is difficult to completely eliminate the potential for significant immune responses especially in susceptible individuals.39 As a result, the clinical administration of LNP siRNA, for example, is often accompanied by co-dosing with potent immunosuppressors such as dexamethasone.39 This complicates the clinical development of LNP formulations of genetic drugs. However, the inherent ability of LNP systems to act as delivery systems can be used to advantage. Recent work has shown, for example, that inclusion of as little as 4 mol% (with respect to total lipid) of a hydrophobic pro-drug of dexamethasone into LNP systems can essentially abrogate the response to immunostimulatory CpG motifs present in encapsulated oligonucleotides (S. Chen and P.R.C., unpublished data).
Similar approaches can potentially be used to enhance the potency of LNP systems containing genetic drugs in tissues other than the liver. The potential for improvement is clear in that <5% of endocytosed LNP siRNA is actually released into the cell cytosol.40 Up to 60% is recycled to the extracellular medium,41 and much of the remaining endocytosed LNP siRNA proceeds to other non-productive endpoints such as degradation in lysosomes. Inhibition of the proteins orchestrating endosome recycling or progression to endosomes can therefore potentially enhance the opportunity of siRNA and other genetic drugs to escape into the cytoplasm. For example, it has been shown that inhibition of the Niemann-Pick protein by the small molecule NP3.47 results in enhanced LNP siRNA retention in cells in vitro, leading to enhanced gene-silencing potency.42 Creation of hydrophobic prodrug versions of NP3.47 and other inhibitors of proteins leading to non-productive endpoints may be therefore be expected to improve potency significantly.
Future Perspectives
It is our contention that the third generation of therapeutics, following first generation small molecule drugs and second generation biologics, will be “smart” nanomedicines tailored to take maximum advantage of biological processes and to minimize toxic side effects by employing highly targeted mechanisms. LNP systems containing genetic drugs are clearly poised to play a major role in this evolution. A remarkable emerging feature is that the development of LNP systems capable of delivering negatively charged macromolecules the size of siRNA (MW ∼13 kD) in vivo is also leading to LNP systems that can deliver much larger mRNA and plasmid DNA constructs, potentially allowing many variants of precise gene therapies to be practiced. It is notable that these advances are being facilitated by taking advantage of “natural” targeting processes such as ApoE-dependent endocytic pathways in hepatocytes or the inherent targeting of nanoparticles to immune cells, rather than the presence of targeting ligands. Further, we note that LNP systems contain a variety of modifiable components in addition to the genetic drug payload that clearly offer a considerable toolbox to maximize potency and minimize toxicity.
Author Contributions
P.R.C. and M.J.H. contributed equally to planning and writing this paper and to the many joint publications referenced herein.
Conflicts of Interest
P.R.C. and M.J.H. have financial interests in Acuitas and P.R.C. also has financial interests in Precision.
Acknowledgments
The authors wish to acknowledge collaborations extending over many years with colleagues based at Alnylam Pharmaceuticals, Arbutus Biopharma, Precision Nanosystems, Acuitas Therapeutics, and the University of BC. Continuous funding for this work has been provided through the Canadian Institutes for Health Research and its precursor, the Canadian Medical Research Council, for the past 39 years. Additional funding has been provided by the National Science and Engineering Research Council (NSERC) and Genome BC.
References
- 1.Whitehead K.A., Langer R., Anderson D.G. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowie A.G., Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. Engl. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cullis P.R., Mayer L.D., Bally M.B., Madden T.D., Hope M.J. Generating and loading of liposomal systems for drug-delivery applications. Adv. Drug Deliv. Rev. 1989;3:267–282. [Google Scholar]
- 5.Cullis P.R., de Kruijff B. Lipid polymorphism and the functional roles of lipids in biological membranes. Biochim. Biophys. Acta. 1979;559:399–420. doi: 10.1016/0304-4157(79)90012-1. [DOI] [PubMed] [Google Scholar]
- 6.Hope M.J., Bally M.B., Webb G., Cullis P.R. Production of large unilamellar vesicles by a rapid extrusion procedure: characterization of size distribution, trapped volume and ability to maintain a membrane potential. Biochim. Biophys. Acta. 1985;812:55–65. doi: 10.1016/0005-2736(85)90521-8. [DOI] [PubMed] [Google Scholar]
- 7.Allen T.M., Cullis P.R. Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug Deliv. Rev. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 8.Maeda H. Macromolecular therapeutics in cancer treatment: the EPR effect and beyond. J. Control. Release. 2012;164:138–144. doi: 10.1016/j.jconrel.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 9.Chonn A., Semple S.C., Cullis P.R. Association of blood proteins with large unilamellar liposomes in vivo. Relation to circulation lifetimes. J. Biol. Chem. 1992;267:18759–18765. [PubMed] [Google Scholar]
- 10.Felgner P.L., Gadek T.R., Holm M., Roman R., Chan H.W., Wenz M., Northrop J.P., Ringold G.M., Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lv H., Zhang S., Wang B., Cui S., Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Bailey A.L., Cullis P.R. Modulation of membrane fusion by asymmetric transbilayer distributions of amino lipids. Biochemistry. 1994;33:12573–12580. doi: 10.1021/bi00208a007. [DOI] [PubMed] [Google Scholar]
- 13.Semple S.C., Klimuk S.K., Harasym T.O., Dos Santos N., Ansell S.M., Wong K.F., Maurer N., Stark H., Cullis P.R., Hope M.J., Scherrer P. Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: formation of novel small multilamellar vesicle structures. Biochim. Biophys. Acta. 2001;1510:152–166. doi: 10.1016/s0005-2736(00)00343-6. [DOI] [PubMed] [Google Scholar]
- 14.Jeffs L.B., Palmer L.R., Ambegia E.G., Giesbrecht C., Ewanick S., MacLachlan I. A scalable, extrusion-free method for efficient liposomal encapsulation of plasmid DNA. Pharm. Res. 2005;22:362–372. doi: 10.1007/s11095-004-1873-z. [DOI] [PubMed] [Google Scholar]
- 15.Belliveau N.M., Huft J., Lin P.J., Chen S., Leung A.K., Leaver T.J., Wild A.W., Lee J.B., Taylor R.J., Tam Y.K. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol. Ther. Nucleic Acids. 2012;1:e37. doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., Sah D.W., Stebbing D., Crosley E.J., Yaworski E. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 17.Leung A.K., Hafez I.M., Baoukina S., Belliveau N.M., Zhigaltsev I.V., Afshinmanesh E., Tieleman D.P., Hansen C.L., Hope M.J., Cullis P.R. Lipid nanoparticles containing siRNA synthesized by microfluidic mixing exhibit an electron-dense nanostructured core. J. Phys. Chem. C Nanomater Interfaces. 2012;116:18440–18450. doi: 10.1021/jp303267y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung A.K., Tam Y.Y., Chen S., Hafez I.M., Cullis P.R. Microfluidic mixing: a general method for encapsulating macromolecules in lipid nanoparticle systems. J. Phys. Chem. B. 2015;119:8698–8706. doi: 10.1021/acs.jpcb.5b02891. [DOI] [PubMed] [Google Scholar]
- 19.Zimmermann T.S., Lee A.C., Akinc A., Bramlage B., Bumcrot D., Fedoruk M.N., Harborth J., Heyes J.A., Jeffs L.B., John M. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 20.Cullis P.R., de Kruijff B. The polymorphic phase behaviour of phosphatidylethanolamines of natural and synthetic origin. A 31P NMR study. Biochim. Biophys. Acta. 1978;513:31–42. doi: 10.1016/0005-2736(78)90109-8. [DOI] [PubMed] [Google Scholar]
- 21.Cullis P.R., Hope M.J. Effects of fusogenic agent on membrane structure of erythrocyte ghosts and the mechanism of membrane fusion. Nature. 1978;271:672–674. doi: 10.1038/271672a0. [DOI] [PubMed] [Google Scholar]
- 22.Hafez I.M., Maurer N., Cullis P.R. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8:1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- 23.Akinc A., Querbes W., De S., Qin J., Frank-Kamenetsky M., Jayaprakash K.N., Jayaraman M., Rajeev K.G., Cantley W.L., Dorkin J.R. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams K.J., Chen K. Recent insights into factors affecting remnant lipoprotein uptake. Curr. Opin. Lipidol. 2010;21:218–228. doi: 10.1097/MOL.0b013e328338cabc. [DOI] [PubMed] [Google Scholar]
- 25.Mui B.L., Tam Y.K., Jayaraman M., Ansell S.M., Du X., Tam Y.Y., Lin P.J., Chen S., Narayanannair J.K., Rajeev K.G. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol. Ther. Nucleic Acids. 2013;2:e139. doi: 10.1038/mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S., Tam Y.Y., Lin P.J., Sung M.M., Tam Y.K., Cullis P.R. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J. Control. Release. 2016;235:236–244. doi: 10.1016/j.jconrel.2016.05.059. [DOI] [PubMed] [Google Scholar]
- 27.Coelho T., Adams D., Silva A., Lozeron P., Hawkins P.N., Mant T., Perez J., Chiesa J., Warrington S., Tranter E. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 2013;369:819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 28.Vance J.E., Hayashi H. Formation and function of apolipoprotein E-containing lipoproteins in the nervous system. Biochim. Biophys. Acta. 2010;1801:806–818. doi: 10.1016/j.bbalip.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Rungta R.L., Choi H.B., Lin P.J., Ko R.W., Ashby D., Nair J., Manoharan M., Cullis P.R., Macvicar B.A. Lipid nanoparticle delivery of siRNA to silence neuronal gene expression in the brain. Mol. Ther. Nucleic Acids. 2013;2:e136. doi: 10.1038/mtna.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rungta R.L., Choi H.B., Tyson J.R., Malik A., Dissing-Olesen L., Lin P.J., Cain S.M., Cullis P.R., Snutch T.P., MacVicar B.A. The cellular mechanisms of neuronal swelling underlying cytotoxic edema. Cell. 2015;161:610–621. doi: 10.1016/j.cell.2015.03.029. [DOI] [PubMed] [Google Scholar]
- 31.Basha G., Novobrantseva T.I., Rosin N., Tam Y.Y., Hafez I.M., Wong M.K., Sugo T., Ruda V.M., Qin J., Klebanov B. Influence of cationic lipid composition on gene silencing properties of lipid nanoparticle formulations of siRNA in antigen-presenting cells. Mol.Ther. 2011;19:2186–2200. doi: 10.1038/mt.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basha G., Ordobadi M., Scott W.R., Cottle A., Liu Y., Wang H., Cullis P.R. Lipid nanoparticle delivery of siRNA to osteocytes leads to effective silencing of SOST and inhibition of sclerostin in vivo. Mol. Ther. Nucleic Acids. 2016;5:e363. doi: 10.1038/mtna.2016.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J.B., Zhang K., Tam Y.Y., Tam Y.K., Belliveau N.M., Sung V.Y., Lin P.J., LeBlanc E., Ciufolini M.A., Rennie P.S., Cullis P.R. Lipid nanoparticle siRNA systems for silencing the androgen receptor in human prostate cancer in vivo. Int. J. Cancer. 2012;131:E781–E790. doi: 10.1002/ijc.27361. [DOI] [PubMed] [Google Scholar]
- 34.Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlake T. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pardi N., Secreto A.J., Shan X., Debonera F., Glover J., Yi Y., Muramatsu H., Ni H., Mui B.K., Tam Y.K. Administration of broadly neutralizing antibody-encoding nucleoside-modified mRNA protects humanized mice from HIV-1 infection. Nat. Comm. 2017;8 doi: 10.1038/ncomms14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulkarni J.A., Myhre J.L., Chen S., Tam Y.Y., Danescu A., Richman J.M., Cullis P.R. Design of lipid nanoparticles for in vitro and in vivo delivery of plasmid DNA. Nanomedicine. 2016 doi: 10.1016/j.nano.2016.12.014. Published online December 28, 2016. [DOI] [PubMed] [Google Scholar]
- 38.Karikó K., Weissman D. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Curr. Opin. Drug Discov. Devel. 2007;10:523–532. [PubMed] [Google Scholar]
- 39.Szebeni J. Complement activation-related pseudoallergy: a stress reaction in blood triggered by nanomedicines and biologicals. Mol. Immunol. 2014;61:163–173. doi: 10.1016/j.molimm.2014.06.038. [DOI] [PubMed] [Google Scholar]
- 40.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C., Stöter M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 41.Sahay G., Querbes W., Alabi C., Eltoukhy A., Sarkar S., Zurenko C., Karagiannis E., Love K., Chen D., Zoncu R. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013;31:653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H., Tam Y.Y., Chen S., Zaifman J., van der Meel R., Ciufolini M.A., Cullis P.R. The Niemann-Pick C1 inhibitor NP3.47 enhances gene silencing potency of lipid nanoparticles containing siRNA. Mol. Ther. 2016;24:2100–2108. doi: 10.1038/mt.2016.179. [DOI] [PMC free article] [PubMed] [Google Scholar]