Abstract
There is a need for new tools to better quantify intracellular delivery barriers in high-throughput and high-content ways. Here, we synthesized a triple-fluorophore-labeled nucleic acid pH nanosensor for measuring intracellular pH of exogenous DNA at specific time points in a high-throughput manner by flow cytometry following non-viral transfection. By including two pH-sensitive fluorophores and one pH-insensitive fluorophore in the nanosensor, detection of pH was possible over the full physiological range. We further assessed possible correlation between intracellular pH of delivered DNA, cellular uptake of DNA, and DNA reporter gene expression at 24 hr post-transfection for poly-L-lysine and branched polyethylenimine polyplex nanoparticles. While successful transfection was shown to clearly depend on median cellular pH of delivered DNA at the cell population level, surprisingly, on an individual cell basis, there was no significant correlation between intracellular pH and transfection efficacy. To our knowledge, this is the first reported instance of high-throughput single-cell analysis between cellular uptake of DNA, intracellular pH of delivered DNA, and gene expression of the delivered DNA. Using the nanosensor, we demonstrate that the ability of polymeric nanoparticles to avoid an acidic environment is necessary, but not sufficient, for successful transfection.
Keywords: polymer, nanoparticle, non-viral, gene delivery, pH sensor, proton sponge
Graphical Abstract
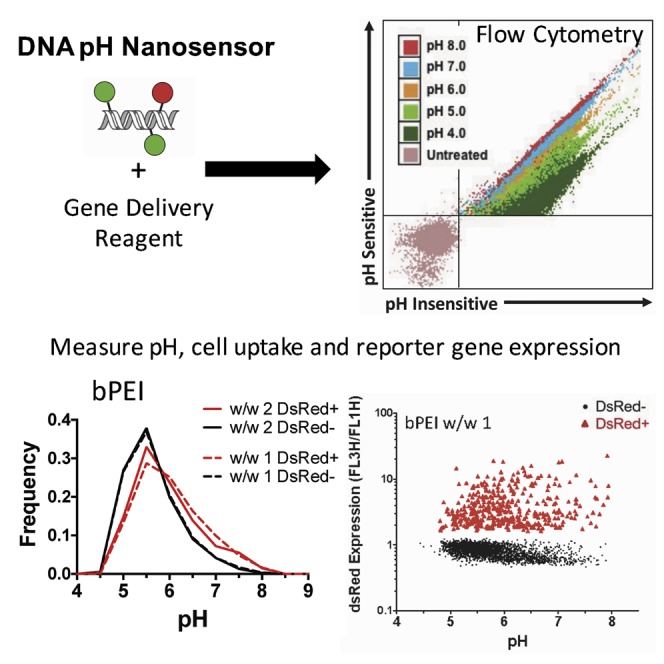
To better investigate endosomal escape of nanoparticles, Wilson et al. synthesized a nanosensor for high-throughput measurements of the intracellular pH of nucleic acids following nanoparticle uptake. Using this sensor, the authors analyzed correlation between nanoparticle pH, uptake, and transfection.
Introduction
Non-viral gene delivery has great promise in clinical applications such as for cancer therapy where transient expression can be sufficient to result in clinical efficacy.1 Compared with DNA-based viral gene therapy, non-viral delivery methods are more amenable to repeat administration due to reduced risk of immunogenicity and insertional mutagenesis, but remain less efficient than viral delivery methods.1, 2 Despite extensive progress in the previous decade, many challenges in engineering successful non-viral gene delivery platforms remain.1, 3 Polymeric gene delivery in particular has made extensive progress toward increased performance through rational engineering of polymer structures as well as screening of broad libraries of polymer structures, but new quantitative bioassays are required to fully understand the mechanisms by which existing nanoparticles achieve transfection.4, 5, 6, 7, 8, 9, 10, 11
Barriers to successful polymeric gene delivery at the level of individual cells include cellular internalization, endosomal escape, nucleic acid unpacking, and nuclear transport.12, 13 Of these barriers, overcoming endosomal escape has been specifically identified as a critical rate-limiting step in polycation nanoparticle-mediated transfection because generally only a minor fraction of endocytosed polyplexes manage to escape to the cytosol.14 Non-endolysosomal trafficking that does not require endosomal escape, but enables nuclear entry through the endoplasmic reticulum, has been shown to occur for certain nanoparticulates, such as specific histone-targeted nanoparticles, but it has not been demonstrated for the majority of nanoparticle formulations.15, 16 The presence of intracellularly delivered nucleic acid at an acidic pH indicates that the nucleic acid is not in the cytosol and is not in the environment required for successful gene expression. Failure to escape the endosome to the cytosol can result in nucleic acid degradation when the early endosome transitions to a late endosome/lysosome, typically within an hour following uptake.17 The shift from early endocytic vesicle to late endosome and eventually lysosome results from fusion of the early endosome with other vesicles. The latter contain hydrogen pump V-ATPases and digestive enzymes that result in acidification and degradation of the nucleic acid contents of polymeric nanoparticles.17, 18
To escape the endosome and avoid lysosomal degradation, polymeric nanoparticles have been designed specifically with either moieties that facilitate membrane pore formation or amine groups designed to enable them to buffer vesicle acidification and consequently escape the endosome via the hypothesized proton sponge mechanism.18 Beginning with branched polyethylenimine (bPEI), many polymeric nanoparticles have been engineered to take advantage of endosome acidification as a means to protect their nucleic acid cargo and enable endosomal escape.19 Despite extensive study in the two decades since the first use of bPEI, the mechanism of endosomal escape for cationic nanoparticles is still not universally agreed upon.19, 20, 21, 22, 23, 24, 25, 26, 27 The ability of the cationic polymers containing tertiary amines to effectively buffer in the physiological pH range between 5 and 7 has been demonstrated in multiple settings,8, 9, 28 yet improved buffering capacity at low, physiologically relevant pH has been shown to not always result in more effective transfection.23 As a means of studying this mechanism universally among cell types and polymer systems, we sought to create an intracellular pH sensor for probing the mean compartmental pH environment of exogenously delivered DNA in individual cells and to design this sensor to be readable in a high-throughput manner by flow cytometry.
Approaches to single-cell and single-compartment pH measurements have previously utilized fluorescence. A ratiometric, fluorophore-based assay utilizing a pH-sensitive fluorophore and a pH-insensitive reference fluorophore was reported by Murphy et al.29 using fluorescein isothiocyanate (FITC) and rhodamine B isothiocyanate (RITC) to measure endosomal pH over time following insulin internalization. Similar assays have been utilized to measure endosomal pH by fluorophore labeling polymeric gene delivery vectors or plasmid DNA.22, 30, 31 More sensitive ratiometric pH probes have recently been developed for non-gene delivery applications using fluorescent polymers and have shown the importance of using multiple pH-sensitive fluorophores to effectively probe the lysosomal pH range.32, 33
Specifically for gene delivery applications, we aimed to investigate the local pH of the exogenously delivered DNA, not the local pH of delivery polymers that may dissociate from the DNA. Here, we report a triple-fluorophore-labeled plasmid DNA-based pH nanosensor with improved sensitivity at lysosomal pH used for probing cellular pH of exogenously delivered DNA to individual cells and cell populations following polymeric gene delivery. This ratiometric pH nanosensor enabled an investigation of endosomal buffering following polymeric gene delivery in a high-throughput manner by flow cytometry. Average pH of the nucleic acid pH nanosensor was monitored at specific time points within individual cells to quantify trends between cellular uptake of DNA, local pH environment of delivered DNA, and successful expression of the DNA in both easy-to-transfect and hard-to-transfect cell lines using bPEI and poly-L-lysine (PLL).
Results
Nucleic Acid pH Nanosensor Synthesis and Validation
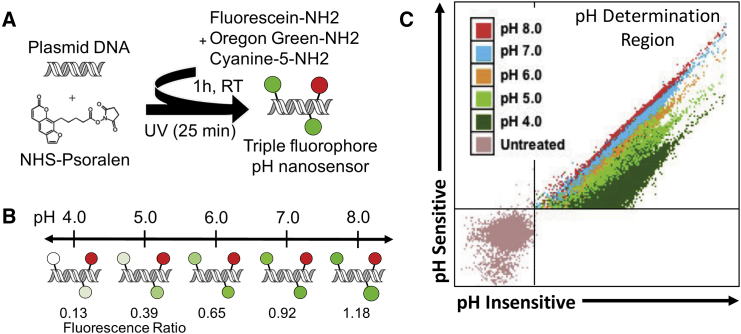
The triple-fluorophore-labeled plasmid pH nanosensor was synthesized in a batch process using the reaction scheme shown in Figure 1A using succinimidyl-[4-(psoralen-8-yloxy)]-butyrate (NHS)-psoralen UV crosslinked into DsRed plasmid DNA prior to reaction with fluorescein cadaverine (FL), Oregon green (OG) cadaverine, and cyanine-5-amine (Cy5).34 It was designed to have a green channel-to-far-red channel fluorescence ratio that would increase with increasing pH as shown in Figures 1B and 1C. The reaction ensured that each plasmid statistically had all three fluorophores with approximately 50 green and 50 far-red fluorophores per 4,700 bp plasmid as determined by spectrophotometry. The labeling efficacy was not particularly sensitive to the amount of NHS-psoralen used, but required sufficient DMSO (∼30%) in the reaction solution to allow the more hydrophobic fluorophores to react effectively.
Figure 1.
pH Nanosensor Synthesis
(A) Synthesis of the triple-labeled pH sensor was achieved via a two-step process of UV photocrosslinking NHS-psoralen to plasmid DNA followed by reaction with three primary amine fluorophores (fluorescein, Oregon green [OG], and Cy5). (B) Far-red channel fluorescence from Cy5 is pH-insensitive over the physiological pH range of interest, whereas OG and fluorescein exhibit pH-sensitive quenching relative to their pKa values of 4.6 and 6.4. (C) The combination of these fluorophores results in a pH-sensitive ratiometric relationship for cells electroporated, fixed, and run through flow cytometry in known pH solutions.
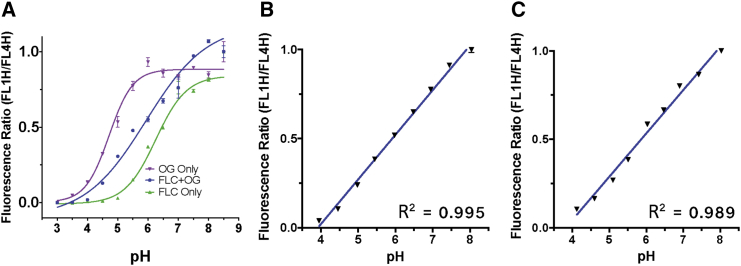
The pH nanosensor sensitivity over the entire pH range of interest was verified to be improved with the inclusion of two pH-sensitive fluorophores and one pH-insensitive fluorophore in comparison with plasmids labeled with only one pH-sensitive fluorophore and one pH-insensitive fluorophore (Figure 2A). This nanosensor was observed to have an approximately linear relationship between pH and fluorescence ratio over the physiological pH range of interest (Figure 2B). For flow cytometry experiments, a calibration curve relating fluorescence ratio to pH for the nanosensor was created by electroporating the pH nanosensor into human embryonic kidney cells (HEK293T) and human primary grade IV glioblastoma cells (GB319),5 fixing the cells to permeabilize them to free flow of ions and running them through the flow cytometer following resuspension in known pH buffer solutions (Figure 2C). The pH nanosensor was further confirmed to be effective in a third cell line and was stable when stored at −20°C over 6 months (Figure S1).
Figure 2.
Fluorescence Ratio pH Calibration Curve
(A) Plate reader normalized fluorescence curves fit with a logarithmic function show that two pH-sensitive fluorophores were necessary for pH sensitivity over the range of interest (all curves also contain Cy5 for pH-insensitive fluorescence on the FL4 channel). (B and C) The triple-labeled pH nanosensor gave an approximately linear relationship over the pH range of interest measured using (B) a fluorescence plate reader and (C) flow cytometry following electroporation into HEK293T cells. Values shown are mean ± SEM of four wells for (A) and (B) and median ratio of >10,000 cells for (C). FLC, fluorescein; OG, Oregon green.
Additionally, fluorophore labeling of plasmid DNA to form the pH nanosensor was confirmed not to affect nanoparticle physicochemical properties in terms of hydrodynamic diameter or zeta potential when combined with cationic polymers to form polyplex nanoparticles (Figures 3A and 3B). Overall, PLL nanoparticles were approximately 50 nm in size with a zeta potential of +20 mV, and PEI nanoparticles were approximately 250 nm in size with a zeta potential of +25 mV. To test the influence of DNA labeling for the pH nanosensor on polymer/DNA binding strength, we performed a heparin competition binding assay using gel electrophoresis35 and a Yo-Pro-1 iodide4 competition binding assay that showed that polymer-DNA interaction was minimally affected by covalent labeling (Figures 3C and 3D).
Figure 3.
Biophysical Properties of Polymeric Gene Delivery Nanoparticles Were Not Affected by pH Nanosensor Labeling
(A and B) Polymeric gene delivery nanoparticles were formed by self-assembly of cationic polymers using the pH nanosensor or unlabeled plasmid DNA and assessed using dynamic light scattering for (A) hydrodynamic diameter and (B) zeta potential, where no statistical differences were observed. Values shown are mean ± SEM of three independently prepared samples. Differences between nanoparticles formulations were assessed using multiple t tests with multiple comparisons corrected for using the Holm-Sidak method. (C and D) Polymer binding strength with labeled pH nanosensor or unlabeled plasmid DNA was compared using a (C) Yo-Pro-1 binding assay (values shown are mean ± SEM of three wells) and (D) heparin binding assay, where only minor differences were observed in polymer-DNA interaction. Values on the heparin binding assay show the w/w ratio of heparin to the mass of polycation per well. A possible increase in binding interaction between pH nanosensor DNA and cationic polymer may be accountable by the presence of carboxylic acid groups in two of the fluorophores used.
Intracellular trafficking of covalently modified plasmid DNA has likewise been a concern in previous studies,36 which we sought to address by utilizing only 20% pH nanosensor DNA in studies involving cellular uptake and expression. Although it is true that the fluorescent labeling method does prevent expression from labeled plasmids, this reduction in reporter gene expression was due to UV exposure that likely results in nicks in the DNA during the labeling process (Figure S2) and not necessarily fluorophore covalent conjugation. Recent studies have shown that bPEI polyplexes have on the order of 10 plasmids per nanoparticle,37 meaning that with 20% pH nanosensor DNA used to form polyplexes in uptake and expression experiments, each polyplex would statistically contain unmodified plasmids.
Cellular Uptake and Transfection
Polymeric nanoparticles have been demonstrated previously in HEK293 cells to traffic via an endolysosomal pathway, and the cell line is particularly amenable as a well-established cell type for non-viral transfection experiments.30, 38 Human GB319 cells, in contrast, are a difficult-to-transfect primary cell line, which show relatively low reporter gene expression for a wide variety of transfection reagents.5 Initial screens of the nanoparticle formulations were done to identify optimal ratios between transfection reagent and DNA that were effective for transfection without being highly cytotoxic (Figures S3C and S3D), where polymer/DNA w/w ratios of 1:1 and 2:1 were selected. These ratios between polymer and DNA were in line with polymer doses used previously20 and can be converted from w/w to nitrogen (polymer) to phosphate (DNA) (N/P) ratio by multiplying by 7.6 or 2.2, respectively, for 25 kDa bPEI and PLL.
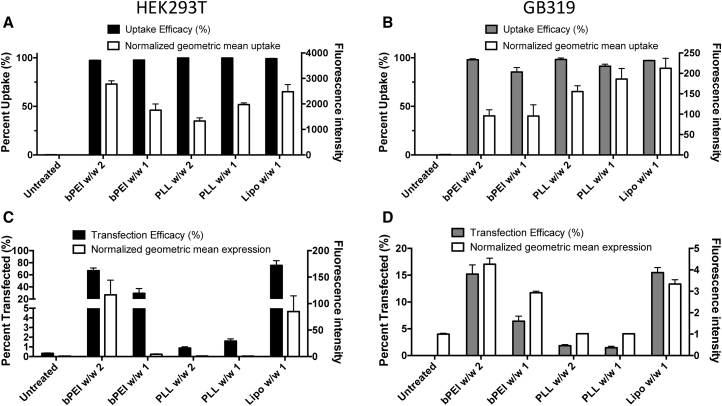
For cellular uptake studies, including for pH measurements, cells were washed with the polyanion heparin prior to flow cytometry to remove any surface-bound but non-internalized nanoparticles (Figure S4).35 Cellular uptake efficacy was determined via flow cytometry by the fluorescence of the pH-insensitive fluorophore (Cy5) at 2 hr after adding nanoparticles to the cells (Figures 4A and 4B) as gated in Figure S5B. All nanoparticle formulations tested demonstrated similar levels of particle uptake within each cell type in terms of percent of uptake-positive cells as well as the normalized geometric mean uptake, which correlates to the average number of nanoparticles taken up by each cell. Due to the high labeling efficacy using NHS-psoralen, and the sensitivity of the nanosensor, the fraction of fluorophore-labeled DNA for pH determination and cellular uptake experiments was able to be reduced to 20% of the total DNA used to form polyplex nanoparticles. This was particularly important, because exposure of the plasmid DNA to UV to promote covalent bond formation in the pH nanosensor synthesis protocol resulted in DNA nicks that eliminated expression from the labeled plasmid DNA (Figure S2).
Figure 4.
Cellular Uptake and Reporter Gene Transfection in Two Cell Lines
(A and B) HEK293T (A) and GB319 (B) cellular uptake as percent of all cells and normalized geometric mean fluorescence of Cy5-labeled DNA were similar between PLL and bPEI. (C and D) Transfection efficacy in (C) HEK293T and (D) GB319 cells as percent of all cells and normalized geometric mean expression showed that bPEI was much more effective than PLL in transfecting cells. Lipofectamine 2000 showed similar levels of cellular uptake and transfection to bPEI with the conditions tested. Bars show mean ± SEM of four wells.
Transfection efficacy was assessed via flow cytometry (Figures 4C and 4D) and fluorescence microscopy (Figures S3A and S3B) in HEK293T and GB319 cells, respectively. Despite the high levels of cellular uptake of PLL/DNA nanoparticles, transfection via PLL polyplexes at either w/w ratio was very ineffective in both HEK293T and GB319, as anticipated. bPEI/DNA nanoparticles were more effective in both cell lines and did so in a polymer-dose-specific fashion with bPEI w/w 2 being superior to bPEI w/w 1 (Figures 4C and 4D).
Calculated pH Measurements
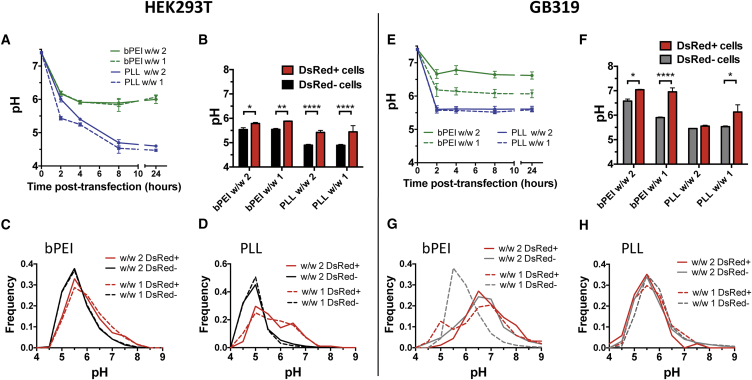
HEK293T and GB319 cells were transfected with the nanoparticle formulations using 20% pH nanosensor plasmid DNA, and average pH of DNA was calculated at specified time points following the initial addition of nanoparticles (Figures 5A and 5E). The fluorescence on the FL1H pH-sensitive and FL4H pH-insensitive channels for individual cells as gated in the third panel of Figure S5A was imported to MATLAB for processing. The ratio between the FL1H and FL4H from individual cells was computed, and input into the linear standard curve relating fluorescence ratio to pH (Figure 2C) for individual cells and the median calculated cell pH per well was determined. For all polyplex nanoparticles studied, acidification rapidly occurred within the first 2 hr of transfection. pH of the nanosensor then remained effectively constant over the subsequent day for all nanoparticles except PLL in HEK293T, where acidification continued to occur at a slower rate until 8 hr post-transfection. PLL/DNA polyplexes at both w/w 1 and w/w 2 acidified to an average pH of approximately 4.5 for HEK293T and 5.5 for GB319. bPEI/DNA polyplexes, in contrast, avoided dramatic acidification in both cell lines, with HEK293T displaying an average pH of 6.0 at both polymer doses. bPEI nanoparticles in GB319s appeared to reduce acidification in a polymer dose-dependent fashion with w/w 1 having an approximate median pH of 6.1 and w/w 2 having an approximate median pH of 6.6.
Figure 5.
Calculated Population pH and Relationship to Transfection at 24 hr
(A and E) Median pH of intracellular DNA calculated at various time points following transfection for the entire cell populations showed PLL failing to avoid a lysosomal fate, whereas bPEI showed a pH closer to neutral in both (A) HEK293T and (E) GB319 cells. (B) Comparison via Holm-Sidak corrected multiple t tests of the median pH values from the DsRed positively transfected populations and negative untransfected populations at 24 hr showed statistically significant differences between the populations for bPEI w/w 1, bPEI w/w 2, PLL w/w 1, and PLL w/w 2 for HEK293T cells. (F) In GB319 cells, statistically significant differences in median pH values between the successfully transfected populations and untransfected populations for bPEI w/w 1, bPEI w/w 2, and PLL w/w 1 were observed. (C) For HEK293T cells transfected with bPEI w/w 2 and w/w 1, less acidic pH of the delivered DNA in individual cells (>5.5) was slightly correlated with an increase in fraction of cells transfected compared with more acidic pH of the delivered DNA in the individual cells (<5.5). (D) Interestingly, HEK293T cells transfected with PLL showed the same behavior to a greater extent. (G) For GB319 cells transfected with bPEI, transfection resulted in a wider distribution of calculated pH than with PLL particles. Following transfection with bPEI, acidic pH (5.0–6.0) correlated with a decreased frequency of successful transfection (DsRed− > DsRed+), whereas neutral pH (∼7.0) correlated with a higher frequency of successful transfection (DsRed+ > DsRed−). (H) For GB319 cells transfected with PLL, there were only minor differences between the DsRed+ and DsRed− populations. All errors bars show the mean ± SEM of four wells. Histograms show the binned flow cytometry data from four wells. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
At the 24 hr time point, DsRed expression was assessed as shown in flow cytometry gating (Figure S5C) to determine characteristics of cells having been successfully transfected compared with those cells that were not successfully transfected. Fluorescence from the pH-sensitive (green), pH-insensitive (far-red), and DsRed (red) channels could also be clearly seen for each nanoparticle formulation using fluorescence microscopy at 24 hr post-transfection (Figure S6). Overall, bPEI and PLL both showed statistically significant differences at the population level for median pH of the DsRed+ and DsRed− populations (Figures 5B and 5F), but the pH distributions (Figures 5C, 5D, 5G, and 5H) were not qualitatively different except in the case for w/w 1 bPEI in GB319 cells. For transfections with bPEI w/w 1 in GB319 cells, acidic pH (5.0–6.0) correlated with a decreased frequency of successful transfection (DsRed− > DsRed+), whereas neutral pH (∼7.0) weakly correlated with a higher frequency of successful transfection (DsRed+ > DsRed−). This trend was not noted in HEK293T cells, however, despite the similar trend in increased transfection efficacy with bPEI w/w 2 particles. For PLL, in both cell types the DsRed− population had a narrow, acidic pH distribution indicative of lysosomal fate (Figures 5D and 5H). For HEK293T cells positively transfected with PLL nanoparticles, the DsRed+ population was shifted to more neutral pH.
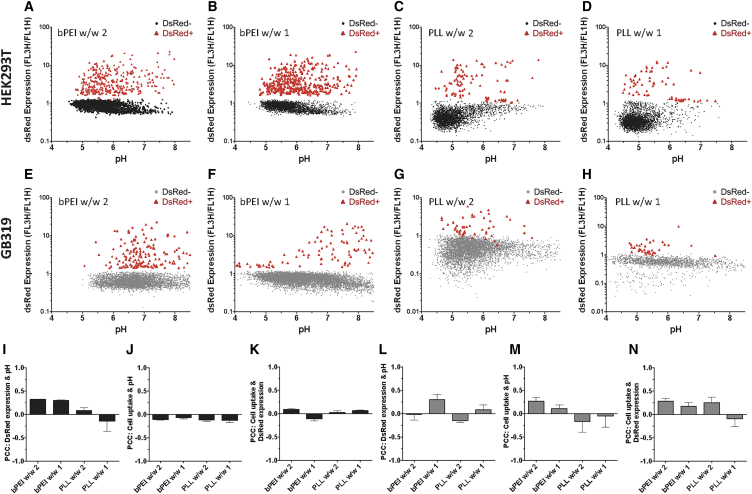
Individual Cell Relationships among Calculated pH, Cellular Uptake, and Exogenous Gene Expression
An advantage of the fluorescence-based nanosensor developed is the capability for high-throughput measurements of cellular uptake of exogenous DNA, measurement of local pH of the exogenous DNA, and expression of the exogenous DNA. Assessment of DsRed reporter gene expression at 24 hr post-transfection as compared with pH nanosensor measurements and cellular uptake measurements on the pH-insensitive channel of the nanosensor enabled the analysis of trends at the single-cell level. To clearly elucidate the DsRed+ population of cells from possible increased autofluorescence, we calculated the ratio of FL3H fluorescence to FL1H fluorescence for each cell, which showed a clear difference for DsRed+ cells (see gating of Figure S6C). Plotting the DsRed expression in terms of FL3H/FL1H versus calculated pH on an individual cell level basis did not show a strong apparent correlation for bPEI or PLL at either w/w ratio or cell line tested (Figures 6A–6H). There did, however, seem to be a weak correlation with bPEI, but only at w/w 1 ratio in GB319 cells (Figure 6F). Quantification of the correlation between variables specifically for the DsRed-expressing cell populations using Pearson’s correlation coefficient (PCC) in MATLAB showed that there was quantitatively minimal correlation between most of the variables (Figures 6I–6N). Weak positive correlation (0.2 < PCC < 0.4) between DsRed expression and pH was observed only for bPEI in either cell line (Figures 6I and 6L), whereas cells transfected with PLL showed no significant correlation between the variables. This result was surprising, given the cases of strong statistical significance between the median pH values of the DsRed+ and DsRed− populations for both bPEI and PLL (Figures 5B and 5F). For other relationships, nanoparticle uptake and cell pH showed weak correlation only for bPEI w/w 2 in GB319 cells (Figure 6M), and nanoparticle uptake and gene expression showed weak correlation only for bPEI w/w 2 and PLL w/w 2 in GB319 cells (Figure 6N).
Figure 6.
Relationships among Individual Cell pH, Uptake, and Gene Expression at 24 hr
(A–H) Representative scatterplots of DsRed+ (red) and DsRed− (black/gray) populations of cells transfected with (A and E) bPEI w/w 2, (B and F) bPEI w/w 1, (C and G) PLL w/w 2, and (D and H) PLL w/w 1 qualitatively showed minimal correlation between gene expression and calculated pH. (I–N) To quantify correlation between variables, we calculated Pearson’s correlation coefficient (PCC) for each well for variables in the case of HEK293T (I–K) and GB319 (L–N). (I and L) PCC between pH and gene expression showed weak (0.2 < PCC < 0.4) correlation only in the case of HEK293T cells transfected with bPEI at w/w 1 and w/w 2, as well as GB319 cells with bPEI w/w 1. (J and M) pH and nanoparticle uptake showed weakly positive correlation only for GB319 cells transfected with bPEI w/w 2. (K and N) Nanoparticle uptake and gene expression showed weak correlation only for GB319 cells transfected with bPEI w/w 2 and PLL w/w 2. All errors bars show the mean ± SEM of four wells for PCC calculated for the cells in that well.
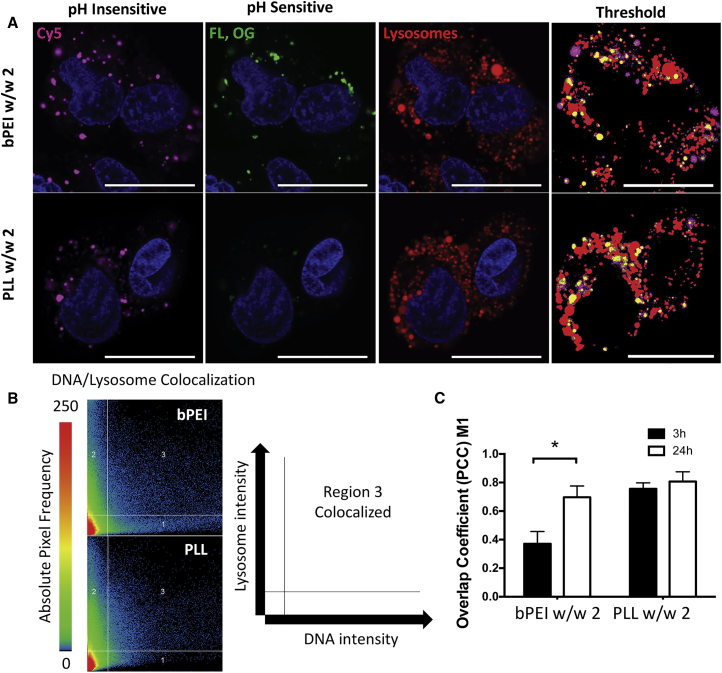
Microscopy Verification and Lysosome Colocalization
The pH nanosensor was verified to function visually using confocal microscopy in the harder to transfect GB319 cell line, where it was noted that bPEI was prone to having high variability in the fluorescence ratio between individual endosomes, spanning both neutral and acidic pH ranges (Figure 7A). Following transfection with bPEI/DNA nanoparticles, within individual cells, some endosomes had a high fluorescence ratio, indicating a near-neutral pH, whereas other endosomes had the pH-sensitive fluorophores effectively quenched, thus indicating their presence in lysosomes. PLL, in contrast, had low pH-sensitive fluorescence and low fluorescence ratios with only the pH-insensitive fluorescence strongly observed. These results were confirmed by staining for lysosomes with a pH-sensitive dye (see Materials and Methods) and analyzing colocalization of the pH-insensitive fluorophore with lysosomes (Figures 7A and 7B). The fraction of DNA colocalized with lysosomes was more than 75% for PLL at both 3 hr and 24 hr post-transfection, whereas for bPEI the fraction of DNA colocalized with lysosomes was less than 40% at 3 hr and increased between the 3 and 24 hr time points (Figure 7C).
Figure 7.
Confocal Microscopy Qualitative Assessment of pH Nanosensor Function and Lysosome Colocalization
(A) Endosomes 3 hr post-transfection containing bPEI w/w 2 or PLL w/w 2 nanoparticles formed with the pH nanosensor show qualitative functioning of the nanosensor for live cell microscopy using GB319 cells, whereby the pH-insensitive fluorophore intensity was not dependent on pH, whereas the pH-sensitive fluorophores were quenched when DNA was present in lysosomes. Images were thresholded for analysis of colocalization of DNA via the pH-insensitive channel with lysosomal tracking dye. Compartments containing DNA non-colocalized with lysosomes are shown in pink, whereas lysosomes not containing DNA are shown in red, and compartments containing DNA colocalized with lysosomes are shown in yellow. Scale bars, 20 μm. (B) Scattergrams show the analysis of the representative microscopy images with distribution of pixels with DNA fluorescence intensity shown on the x axis, lysosome fluorescence intensity shown on the y axis, and region 3 showing thresholded colocalized pixels. (C) Quantification of Pearson’s correlation coefficient M1 for colocalization of DNA and lysosomes showed PLL nanoparticles colocalized with lysosomes at both 3 and 24 hr post-transfection, whereas bPEI nanoparticles partially avoided an initial lysosomal fate at 3 hr, but not at 24 hr. Quantified results were tested using multiple t tests corrected for multiple comparisons using the Holm-Sidak method. Errors bars show mean ± SEM of five images taken with the same microscope settings. *p < 0.05.
Application to Non-polymeric Non-viral Nanoparticles
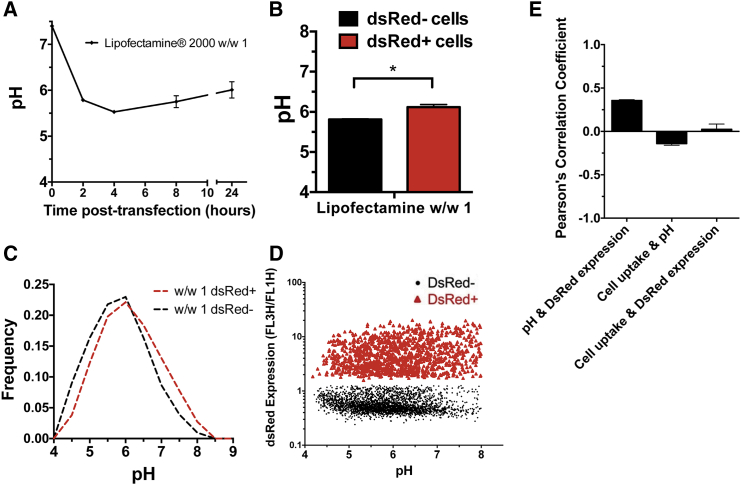
We additionally evaluated the pH nanosensor using the liposomal transfection reagent Lipofectamine 2000 to test its efficacy with a non-polymeric non-viral transfection agent (Figure 8). Lipofectamine 2000 at a w/w 1 ratio showed a median pH of DNA of approximately 6.0, demonstrating that some plasmids fail to fully escape a late endosome or lysosomal fate. The mechanism of endosomal escape of lipid reagents like Lipofectamine 2000 is not fully established, but recent reports have shown that the vast majority of lipid nanoparticles still reach lysosomes and escape is a relatively rare event.39, 40 Overall, we detected only minimal differences in the calculated pH between DsRed+ and DsRed− populations of cells transfected with Lipofectamine 2000 and did not find any correlation between nanoparticle uptake and transfection or nanoparticle uptake and pH. That being said, acidic pH (4.0–6.0) correlated with a slightly decreased frequency of successful transfection (DsRed− > DsRed+), whereas higher pH (7.0–8.0) weakly correlated with a higher frequency of successful transfection (DsRed+ > DsRed−). However, because high transfection was seen with Lipofectamine 2000 across the whole range of pH (4.0–8.0), significant trafficking of the delivered DNA to acidic compartments was not a significant barrier to Lipofectamine 2000 as a non-viral transfection agent, unlike with PLL.
Figure 8.
Lipid Nanoparticle pH Nanosensor Measurements
(A) pH of DNA in Lipofectamine 2000 nanoparticles introduced to HEK293T cells was determined to average approximately 6.0 at all time points following transfection. (B and C) There was only a minor statistical difference between the (B) the median pH of cells that were DsRed+ and DsRed− at 24 hr post-transfection, which was only slightly evident when looking at a (C) representative distribution of the populations. (D) Likewise to bPEI and PLL there was no highly significant correlation between DsRed expression and measured pH for the cell at 24 hr post-transfection. (E) Cell pH and DsRed gene expression showed weak (0.2 < PCC < 0.4) correlation using PCC, and no significant correlation was observed between the other relationships. Error bars show mean ± SEM of four wells.
Discussion
In this work, we have created a nucleic-acid-based pH nanosensor for investigating the local pH of plasmid DNA following transfection with non-viral carriers. Notably, the pH nanosensor synthesized here utilized two pH-sensitive fluorophores, fluorescein and OG, with respective pKa values of 4.6 and 6.4 for improved sensitivity at lower pH ranges of interest (Figure 2A), which was required to probe lysosomal pH levels.32 Using the ratio of the fluorescence of the pH-sensitive fluorophores to the pH-insensitive fluorophore, we calibrated a standard curve for intracellular pH readings (Figure 2C) that could be quantified in a high-throughput manner through flow cytometry.
Previous studies have focused on labeling polymers directly as a pH probe, whereas less research has been done constructing pH sensors from nucleic acids to specifically investigate vesicles containing DNA.20, 24, 30, 31 Labeling of polymers directly has been shown to influence the size and properties of the particles studied.30 In this study, nanoparticles formed from cationic polymers complexed with 100% pH nanosensor DNA were not statistically different in size, zeta potential, or polymer binding strength from nanoparticles formed with unlabeled DNA (Figure 3). Additionally, given that free polymer has been shown to play a large role in transfection and that some polymers may form particles in the absence of DNA, studying the trafficking of a fluorescently labeled polymer is a less direct method to assess the behavior of those particles and endosomes specifically containing plasmids.20, 41 Labeling plasmids directly with fluorophores also enabled a single ratiometric curve to be generated for the specific batch of labeled plasmid as nanosensors, eliminating the need to label and characterize multiple polymers individually when using the nanosensor to probe biomaterial structure-function relationships.
PLL and bPEI were optimal polymers to use as a proof of concept, because they are established gene delivery materials and were shown to have a similar degree of nanoparticle uptake (Figures 4A and 4B). As expected from previously published results in other cell types, bPEI was much more effective than PLL for overall transfection in both cell lines tested (Figures 4C and 4D). Using the pH nanosensor, we observed that plasmid DNA delivered with bPEI maintained a near-neutral pH, whereas DNA delivered with PLL was trafficked to acidic compartments (Figures 5A and 5E).24, 31 Interestingly, the dose-dependent difference in median pH of the GB319 cell population for bPEI was not observed in the HEK293T cells and disappeared when examining only the DsRed+ population (Figure 5F). Examining differences between DsRed+ and DsRed− populations at 24 hr post-transfection showed significant differences in the median pH of the populations for both bPEI and PLL, although the significance was greatest for bPEI at a w/w 1 dose (Figures 5B and 5F). Analysis of the DsRed+ and DsRed− populations (Figure 5) showed that for both polymers in both cell lines, when the average pH was close to neutrality, the relative frequency of successful transfection (DsRed+/DsRed−) increased. Intriguingly, when moving from a population perspective to a single-cell perspective, the strong positive correlation between increased intracellular pH and increased transfection is no longer observed (Figure 6). These results are in agreement with previous claims that only a small fraction of polyplexes ever manage to escape endolysosomes to the cytosol.20 Thus, the many polyplexes that do not escape the endolysosomes and the many cells that do not become successfully transfected obscure the potential correlation of those that do. Nonetheless, with the right polyplex material, these non-escaping polyplexes and these non-transfected cells can still all exhibit an increase in pH sensed by the nanosensor that correlates to a transfection advantage for the cell population overall.
To further investigate functioning of the pH nanosensor following transfection and to evaluate polyplex activity in GB319 human glioblastoma cells in the context of previous reports that have observed the vast majority of polyplexes taken up by a cell fail to escape the endosomes, we performed confocal microscopy at 24 hr post-transfection (Figure 7). The pH sensor was confirmed to function as expected, further validating results from flow cytometry, with the pH-sensitive channel showing much greater fluorescence in proportion to the pH-insensitive channel for bPEI-mediated transfection than for PLL. Interestingly, endosomes containing the pH nanosensor and bPEI appeared to exhibit a wide variance in pH, which was apparent from confocal images and was further confirmed by assessing the specific fraction of DNA in lysosomes via colocalization analysis (Figures 7B and 7C). For nucleic-acid-containing nanoparticles that manage to escape the endosome, they will no longer appear as punctate points in microscopy images, and imaging would be hindered by the much brighter concentrated pixels of nanoparticles in endolysosomes; these same nanoparticles that have successfully escaped the endosomes would, however, be accounted for in total fluorescence with flow cytometry. This could explain in part the possible discrepancies between flow cytometry and confocal microscopy analysis, whereby nucleic acids delivered with bPEI have been reported in other cell types to maintain a median pH greater than 5 when assessed with flow cytometry, but less than 5 with confocal microscopy.20, 24, 31
Our studies set out to investigate to what extent certain non-viral gene delivery nanoparticles reach a state of acidification in endolysosomes or else manage to avoid that fate through the use of a DNA-based nanobiosensor. An important aspect to note is that there is a variety of means for a nanoparticle to avoid acidification. Although preventing acidification through endosomal buffering and titratable amine groups has been demonstrated in part by some biomaterials such as PEI, this is not the only method by which particles escape a lysosomal fate.22 An alternative possibility is avoidance of these acidic pathways altogether through uptake into non-acidifying vesicles13, 16 or direct entry to the cytosol through induced nanopores.42 A related aspect is that the type of endocytosis, such as clathrin-mediated versus caveolae-mediated, can also make a significant difference to the efficiency of transfection, and that this can occur in a polymer structure-specific manner.43 Nonetheless, it is clear that sequestration of delivered DNA into acidic late endosomes and lysosomes means that those DNA molecules are not available to be transcribed in the pH-neutral nucleus, the target of the gene delivery. Thus, this nanobiosensor can play an important role in probing this barrier in a high-throughput, single-cell, and dynamic fashion.
Even given successful and canonical endocytosis of a nanoparticle, there are additional intracellular barriers beyond endosomal escape to successful transfection including DNA unpacking, nuclear import, and nuclear retention of DNA that must be considered for a delivery approach, but the endosomal barrier remains in many cases the rate-limiting step in expression following nanoparticle uptake.14 In recent studies, the rarity of endosomal escape of lipid nanoparticles as well as polymeric nanoparticles has been likewise well documented.39, 40 These studies in particular have noted the difficulty in using fluorescence-based imaging for tracking labeled molecules or particles that have successfully escaped the endosome, because the fraction of total fluorescence detectable in the cytosol is very low. With this in mind, we have chosen to focus on the specific barrier to transfection of endosomal escape in our creation of a new tool.
The proton sponge hypothesis posits that because of endosomal buffering by a protonatable base, such as PEI (the “proton sponge”), there will be a concomitant flux of anions into the endosome to maintain electroneutrality and consequent buildup of osmotic pressure that can lead to endosomal rupture. The results of this study do not confirm or deny the existence of the proton sponge mechanism as it was first proposed by Boussif et al.,19 but do argue against a simple buffering-limited endosomal escape path for polymeric vectors. If transfection of cells were to be limited by the buffering capacity of the polymeric vectors used, including bPEI, buffering capacity of nanoparticles in the pH 5–7 range would be expected to show strong direct correlation with transfection efficacy, which is not what we observe when we look at the cells on a single-cell level in our study (Figure 6).
Theoretical approaches to assessing the feasibility of the proton sponge mechanism are inconclusive. Purely theoretical assessment of the feasibility of the proton sponge mechanism based on the combination of osmotic pressure and pressure from the charged polymer has been used to argue that buffering of endosomal pH with enough free (uncomplexed) cationic polymer was sufficient for rupturing endosomes.26 Assessment of the feasibility of the proton sponge mechanism via quantification of the amount of fluorescently labeled bPEI that accumulated in endosomes in combination with the theoretical buffering capacity and estimated critical lysosomal membrane tension of 10 mJ/m2 has been used to argue that osmotic pressure could not lead to endosomal escape on its own.20 Likewise, the maximum endosomal osmotic pressure possible based on the buffering capacity of bPEI has been estimated to be below the level necessary for full endosomal membrane disruption.25
Perhaps buffering capacity of polymers for gene delivery may make endosomal escape more likely without directly causing endosomal escape via the hypothesized proton sponge mechanism on its own. Polymer buffering capacity may cause an increase in osmotic pressure across the membrane in such a way as to make it more vulnerable to disruption by other attributes of the polymer, such as its increasingly high charge, similarly to how an overly inflated balloon is easier to pop with a pin than a partially inflated balloon. This hypothesis is supported by experimental results from multiple groups demonstrating interaction of polymer directly with cellular membranes, because charged cationic polyplexes have been demonstrated to lead to membrane disruption directly.14, 18 Further, the spacing of cationic charges in polymer structure has been demonstrated to affect both transfection efficacy and cellular viability, as observed in the odd-even effect with polyaspartamides.44 Coupled with results from groups demonstrating efficacy of amphiphilic polymers for gene delivery that have relatively low charge densities, this balance between ability to possibly generate osmotic pressure because of buffering capability and ability to disrupt membranes by physical means may be critical for polymeric vector cytosolic delivery.45
In conclusion, we have demonstrated that a triple-fluorophore-labeled nucleic acid can function as a sensitive pH nanosensor to probe the environment of exogenous nucleic acid delivered by polymeric and liposomal non-viral carriers through the whole range of physiological pH. bPEI, in comparison to PLL, exhibited a higher average intracellular pH, as well as a higher variance in intracellular pH. Population average measurements of pH were found to correlate well to successful transfection of human cells, whereas single-cell measurements were found to be only weakly correlative. These results suggest limitations to the proton sponge hypothesis and also that the ability of polymeric nanoparticles to avoid an acidic environment is necessary, but not sufficient, for successful transfection. To our knowledge, this is the first reported instance of high-throughput single-cell analysis between cellular uptake of DNA, intracellular pH of delivered DNA, and gene expression of the delivered DNA. This nanosensor technology can be of benefit to increase fundamental quantitative understanding of how biomaterial properties affect intracellular delivery and to glean high-content and high-throughput information on the local environment of DNA as it transports through the cell.
Materials and Methods
Materials
bPEI 25 kDa (408727), PLL sodium chloride 15–30 kDa (P2658), buffer salts, and organic solvents were purchased from Sigma-Aldrich. NHS-psoralen (23013), OG cadaverine (O10465), and FL (A10466) were purchased from Thermo Fisher. Cy5-amine (230C0) was purchased from Lumiprobe. All fluorophores were dissolved in DMSO at a concentration of 10 μg/μL and stored at −20°C in small aliquots. Plasmid pDsRed-Max-N1 (Addgene 21718) was used for construction of the pH nanosensor, whereas peGFP-N1 (Addgene 2491) was used for initial transfection efficacy screens.
Nucleic Acid pH Nanosensor Synthesis
DsRed plasmid DNA at a concentration of 1 μg/μL was mixed in a mass ratio of 16:1 with NHS-psoralen in DMSO at a concentration of 1 μg/μL.46 The solution was distributed to a 96-well round-bottom plate with 50 μg DNA/well and placed on ice. Psoralen was then crosslinked into the DNA by 25 min of UV exposure using a 0.16 A 365 nm lamp placed directly over the plate. For each well, 8 μL of 10× PBS, 17 μL of DMSO, 25 μg of FL, 25 μg of OG cadaverine, and 2.5 μg of Cy5-amine were added, well mixed, and then incubated at room temperature for 1 hr while protected from light to prevent photobleaching. It was necessary to fine-tune DMSO volume percent of the solution to 30% to facilitate reaction of the more hydrophobic fluorophores. Labeled DNA was then ethanol precipitated two times to remove excess reactants and purify the DNA. The purified pH nanosensor was then resuspended in ultrapure water at 75% of the original volume. The concentration and labeling efficacy were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific), and the volume was increased to make the pH nanosensor concentration 1 μg/μL. DNA was then divided into aliquots and stored at −20°C protected from light. Additional DNA was labeled using only fluorescein or OG and Cy5 for comparison of the triple-labeled pH nanosensor to more commonly utilized dual-labeling techniques with only one pH-sensitive fluorophore. Fluorescence over the pH range from 3.0 to 9 for the different versions of the pH nanosensor was assessed using a Synergy 2 multiplate reader (Biotek) with four replicates.
Cell Culture
Human glioblastoma astrocytes (GB319) were grown as adherent cells as previously described on tissue culture flasks in DMEM/F12 (11330057; Thermo Fisher) with 10% heat-inactivated fetal bovine serum (16140071; Thermo Fisher) and 1% antimycotic/antibiotic (15240062; Thermo Fisher).5 HEK293T were purchased from ATCC and cultured in DMEM (11965092; Thermo Fisher) with 10% fetal bovine serum and 1% penicillin/streptomycin.
pH Standard Curves
The pH nanosensor was introduced to cells for creation of the standard curve relating fluorescence ratio to pH by electroporation using the Neon Transfection System (Thermo Fisher). In detail, cells were passaged to individual tubes with 200,000 cells/tube, then fully aspirated and kept on ice for 20 min. The cells were then resuspended in 10 μL of the Neon System-supplied R-buffer containing 1 μg of 100% labeled DNA and electroporated with one pulse at 1,300 V for 20 ms. For the flow-cytometry-based pH curve, the electroporated cells were transferred to a solution of PBS with 2% FBS, centrifuged, and aspirated to remove R-buffer. Cells were then resuspended in PBS containing 4% formalin for 15 min for simultaneous fixation and permeabilization, centrifuged, and aspirated to remove formalin, then resuspended in known pH point buffers prior to flow cytometry. Known pH point 150 mM buffer solutions were made for pH values between 4.0 and 9.0 with a final concentration of 120 mM NaCl and 30 mM citrate phosphate buffer with 2% v/v FBS. Precise pH was measured using a SevenEasy pH Meter (Mettler Toledo) following addition of FBS.31 A BD Accuri C6 (BD Biosciences) flow cytometer with two lasers (488 and 633 nm) with four channels corresponding to green, yellow, red, and far-red fluorescence (FL1 at 530 ± 15 nm, FL2 at 565 ± 10 nm, FL3 at 610 ± 10 nm, and FL4 at 675 ± 12.5 nm, respectively) was used for all flow cytometry experiments in combination with a HyperCyt autosampler (IntelliCyt). For plate reader experiments, pH nanosensor was diluted to 0.01 μg/μL in known pH buffer solution in 96-well plates with four replicates. Fluorescence measurements were taken with a Synergy 2 plate reader (Biotek).
Transfection
For transfection efficacy experiments, cells were plated in CytoOne 96-well tissue culture plates (USA Scientific) 24 hr prior to transfection with 15,000 cells/well in 100 μL of media. Nanoparticles were formed by mixing eGFP-N1 DNA and polymer solutions at 1:1 v/v ratio followed by 10 min of incubation to allow for particle formation. PLL and bPEI from stock aqueous concentrations of 10 μg/μL were diluted to their necessary concentrations in 10 mM HEPES or 150 mM NaCl, respectively, before being mixed 1:1 with DNA diluted to 0.06 μg/μL in the equivalent buffer. For notation, w/w 1 or w/w 2 for bPEI and PLL denotes a 1:1 or 2:1 w/w ratio with plasmid DNA, respectively. Lipofectamine 2000 was used according to manufacturer’s instructions, prepared in Opti-MEM at w/w 2 and w/w 1 ratios. Twenty microliters of the nanoparticle solution was added to each well of cells containing 100 μL of complete media and allowed to incubate for 2 hr, at which point the media were replaced. Transfection efficacy was assessed for percent transfected cells and geometric mean expression approximately 48 hr following transfection. Cell viability was assessed using MTS CellTiter 96 Aqueous One (Promega) cell proliferation assay approximately 24 hr following transfection.
Cellular Uptake and pH Measurements
Cells were again plated at a density of 15,000 cells/well in 96-well plates in 100 μL of media and allowed to adhere for 24 hr. Nanoparticles were formed identically to the procedure for determination of transfection efficacy other than the use of 20% plasmid pH nanosensor pre-mixed with 80% unlabeled DsRed DNA. Following 2 hr of incubation, media were aspirated and the cells were washed two times with 50 μg/μL heparin in 150 mM PBS with five triturations followed by a single PBS wash to remove surface-bound polyplexes. PBS was then aspirated and replaced with fresh complete media. At specified time points, cells were lifted and resuspended in 150 mM PBS containing 2% v/v FBS before being run through a BD Accuri C6 flow cytometer with HyperCyt CFlow autosampler as previously detailed. Flow cytometry data were gated using FlowJo as shown in Figure S4 to determine cellular uptake and exclude cells with fluorescence in either channel of the pH nanosensor below background autofluorescence levels. Cellular uptake was quantified using Cy5 from the pH nanosensor for the percent of cells having uptaken polyplexes and geometric mean fluorescence. Data from the cells above background level for both FL1H and FL4H were exported and analyzed in MATLAB. The ratio of FL1H/FL4H for pH-sensitive/-insensitive fluorescence (FL1H/FL4H) was calculated for each cell. Then the median FL1H/FL4H ratio for each well was determined and used to calculate the average pH for that well using the linear standard pH curve (Figure 2C).
Confocal Microscopy
GB319 cells were plated on Nunc Lab-Tek chambered borosilicate coverglass well plates (155411; Thermo Fisher) at 37,500 cells/well for the same seeding density as transfections in 96-well plates. Cells were incubated for 2 hr with nanoparticles formed from 1,500 ng of the plasmid pH sensor and the respective polymer. Cells were then washed three times with 50 μg/mL heparin (379059; Sigma-Aldrich) in 150 mM PBS. At 24 hr post-transfection, cells were stained for 10 min with Hoechst 33342 (H3570; Thermo Fisher) for nuclei visualization, as well as Cell Navigator Lysosome Staining dye (AAT Bioquest), then washed three times with PBS and imaged in live cell imaging solution (A14291DJ; Thermo Fisher) at 37°C in 5% CO2. Images were acquired using a Zeiss LSM 780 microscope with Zen software and 63× oil immersion lens. Specific laser channels used were 405 nm diode, 488 nm argon, 561 nm solid-state, and 639 nm diode lasers. Laser intensity and detector gain settings were optimized for cells transfected with PEI w/w 2 and maintained for all imaging experiments.
Nanoparticle Characterization
Three samples were independently prepared for each nanoparticle formulation at the same concentrations as outlined earlier in the Transfection section. Nanoparticles for pH nanosensor measurements were formed from 100% labeled DNA. Nanoparticle hydrodynamic diameters were determined by dynamic light scattering (DLS) in disposable micro-cuvettes using a Malvern Zetasizer NanoZS (Malvern Instruments) with a detection angle of 173°. Each sample was then diluted in 10 mM NaCl by a dilution factor of 5, and zeta potential was measured by electrophoretic light scattering in disposable zeta cuvettes at 25°C using the same Malvern Zetasizer NanoZS (Malvern Instruments).
Heparin binding competition assay between polyplex nanoparticles and DNA was run similarly to previously published.35 In brief, polyplex nanoparticles were formed between unlabeled or 100% labeled plasmid DNA and either bPEI or PLL at a w/w ratio of 2 and concentration of 0.03 μg/μL in 10 mM NaCl. The nanoparticles were then added to separate solutions of heparin sodium salt (Sigma H3393) diluted in 10 mM NaCl to give the specified amounts of heparin per well in nanograms. The solutions of polyplex nanoparticles with heparin were then diluted with 30% glycerol to give a 5% glycerol solution, after which 15 μL was loaded into a 1% agarose gel.
Yo-Pro-1 iodide binding assays were run similarly to previously published results,4 where DNA and Yo-Pro-1 iodide (Thermo Fisher) were both diluted to a concentration of 1 μM in 10 mM NaCl. For pH nanosensor trials, 100% Cy5-only-labeled DNA was used to avoid fluorescence from the green channel fluorophores. The solution of Yo-Pro-1 and DNA was then mixed in a 1:1 ratio with a polymer solution to give the specified polymer concentration per well. Green channel fluorescence was then measured using a plate reader (Biotek Synergy 2).
Data Analysis, Statistics, and Figures
Prism 6 (GraphPad) was used for all statistical analyses and curve plotting. Unless otherwise specified, statistical tests were performed with a global alpha value of 0.05. ChemDraw (Perkin Elmer) was used for chemical structures and schematics. Unless otherwise stated, absence of statistical significance markings where a test was stated to have been performed signified no statistical significance. Figures were structured such that red lines, bars, or dots specifically denote DsRed+ populations, whereas DsRed− populations were denoted with black (HEK293T) or gray (GB319).
Author Contributions
Conceptualization, D.R.W. and J.J.G.; Methodology, D.R.W. and J.J.G.; Investigation, D.R.W., D.R., Y.R., A.M., and K.J.W.; Resources and Funding Acquisition, J.J.G., A.Q.-H, and D.J.Z; Writing and Editing, D.R.W., D.R., Y.R., K.J.W., J.J.G., A.Q.-H., and D.J.Z; Supervision and Administration, J.J.G.
Acknowledgments
The authors thank Scot Kuo, Barbara Smith, and Tyler Stephens at the JHU SOM Microscopy Facility for training and guidance for microscopy experiments. This work was supported by NSF Graduate Research Fellowship DGE-0707427 to D.R.W.; NSF Graduate Research Fellowship DGE-1232825 to Y.R.; NIH grants R01EB016721, R01EB022148, R01CA195503, and S10 OD016374; and Wilmer Core Grant P30EY001765. This work was supported by generous gifts from the Guerrieri Family Foundation and from Mr. and Mrs. Robert and Clarice Smith.
Footnotes
Supplemental Information includes Supplemental Materials and Methods and six figures and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.04.008.
Supplemental Information
References
- 1.Yin H., Kanasty R.L., Eltoukhy A.A., Vegas A.J., Dorkin J.R., Anderson D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y., Gao D.-Y., Huang L. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv. Drug Deliv. Rev. 2015;81:128–141. doi: 10.1016/j.addr.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones C.H., Chen C.-K., Ravikrishnan A., Rane S., Pfeifer B.A. Overcoming nonviral gene delivery barriers: perspective and future. Mol. Pharm. 2013;10:4082–4098. doi: 10.1021/mp400467x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tzeng S.Y., Green J.J. Subtle changes to polymer structure and degradation mechanism enable highly effective nanoparticles for siRNA and DNA delivery to human brain cancer. Adv. Healthc. Mater. 2013;2:468–480. doi: 10.1002/adhm.201200257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tzeng S.Y., Guerrero-Cázares H., Martinez E.E., Sunshine J.C., Quiñones-Hinojosa A., Green J.J. Non-viral gene delivery nanoparticles based on poly(β-amino esters) for treatment of glioblastoma. Biomaterials. 2011;32:5402–5410. doi: 10.1016/j.biomaterials.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sunshine J.C., Akanda M.I., Li D., Kozielski K.L., Green J.J. Effects of base polymer hydrophobicity and end-group modification on polymeric gene delivery. Biomacromolecules. 2011;12:3592–3600. doi: 10.1021/bm200807s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sunshine J., Green J.J., Mahon K.P., Yang F., Eltoukhy A.A., Nguyen D.N., Langer R., Anderson D.G. Small molecule end group of linear polymer determines cell-type gene delivery efficacy. Adv. Mater. 2009;21:4947–4951. doi: 10.1002/adma.200901718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lächelt U., Kos P., Wagner E. Sequence-defined nucleic acid carriers combining distinct modules for complexation, shielding, receptor-targeting and endosomal escape. J. Control. Release. 2015;213:e106–e107. doi: 10.1016/j.jconrel.2015.05.178. [DOI] [PubMed] [Google Scholar]
- 9.Lächelt U., Kos P., Mickler F.M., Herrmann A., Salcher E.E., Rödl W., Badgujar N., Bräuchle C., Wagner E. Fine-tuning of proton sponges by precise diaminoethanes and histidines in pDNA polyplexes. Nanomedicine (Lond.) 2014;10:35–44. doi: 10.1016/j.nano.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Eltoukhy A.A., Sahay G., Cunningham J.M., Anderson D.G. Niemann-Pick C1 affects the gene delivery efficacy of degradable polymeric nanoparticles. ACS Nano. 2014;8:7905–7913. doi: 10.1021/nn501630h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eltoukhy A.A., Siegwart D.J., Alabi C.A., Rajan J.S., Langer R., Anderson D.G. Effect of molecular weight of amine end-modified poly(β-amino ester)s on gene delivery efficiency and toxicity. Biomaterials. 2012;33:3594–3603. doi: 10.1016/j.biomaterials.2012.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J., Wilson D.R., Zamboni C.G., Green J.J. Targeted polymeric nanoparticles for cancer gene therapy. J. Drug Target. 2015;23:627–641. doi: 10.3109/1061186X.2015.1048519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vercauteren D., Rejman J., Martens T.F., Demeester J., De Smedt S.C., Braeckmans K. On the cellular processing of non-viral nanomedicines for nucleic acid delivery: mechanisms and methods. J. Control. Release. 2012;161:566–581. doi: 10.1016/j.jconrel.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 14.Bieber T., Meissner W., Kostin S., Niemann A., Elsasser H.-P. Intracellular route and transcriptional competence of polyethylenimine-DNA complexes. J. Control. Release. 2002;82:441–454. doi: 10.1016/s0168-3659(02)00129-3. [DOI] [PubMed] [Google Scholar]
- 15.Ross N.L., Munsell E.V., Sabanayagam C., Sullivan M.O. Histone-targeted polyplexes avoid endosomal escape and enter the nucleus during postmitotic redistribution of ER membranes. Mol. Ther. Nucleic Acids. 2015;4:e226. doi: 10.1038/mtna.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reilly M.J., Larsen J.D., Sullivan M.O. Polyplexes traffic through caveolae to the Golgi and endoplasmic reticulum en route to the nucleus. Mol. Pharm. 2012;9:1280–1290. doi: 10.1021/mp200583d. [DOI] [PubMed] [Google Scholar]
- 17.Duncan R., Richardson S.C.W. Endocytosis and intracellular trafficking as gateways for nanomedicine delivery: opportunities and challenges. Mol. Pharm. 2012;9:2380–2402. doi: 10.1021/mp300293n. [DOI] [PubMed] [Google Scholar]
- 18.Varkouhi A.K., Scholte M., Storm G., Haisma H.J. Endosomal escape pathways for delivery of biologicals. J. Control. Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Boussif O., Lezoualc’h F., Zanta M.A., Mergny M.D., Scherman D., Demeneix B., Behr J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjaminsen R.V., Mattebjerg M.A., Henriksen J.R., Moghimi S.M., Andresen T.L. The possible “proton sponge ” effect of polyethylenimine (PEI) does not include change in lysosomal pH. Mol. Ther. 2013;21:149–157. doi: 10.1038/mt.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behr J.-P. The proton sponge: a trick to enter cells the viruses did not exploit. CHIMIA Int. J. Chem. 1997;51:34–36. [Google Scholar]
- 22.Sonawane N.D., Szoka F.C., Jr., Verkman A.S. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J. Biol. Chem. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 23.Funhoff A.M., van Nostrum C.F., Koning G.A., Schuurmans-Nieuwenbroek N.M.E., Crommelin D.J.A., Hennink W.E. Endosomal escape of polymeric gene delivery complexes is not always enhanced by polymers buffering at low pH. Biomacromolecules. 2004;5:32–39. doi: 10.1021/bm034041+. [DOI] [PubMed] [Google Scholar]
- 24.Akinc A., Thomas M., Klibanov A.M., Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J. Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 25.Won Y.-Y., Sharma R., Konieczny S.F. Missing pieces in understanding the intracellular trafficking of polycation/DNA complexes. J. Control. Release. 2009;139:88–93. doi: 10.1016/j.jconrel.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang S., May S. Release of cationic polymer-DNA complexes from the endosome: a theoretical investigation of the proton sponge hypothesis. J. Chem. Phys. 2008;129:185105. doi: 10.1063/1.3009263. [DOI] [PubMed] [Google Scholar]
- 27.Parhamifar L., Larsen A.K., Hunter A.C., Andresen T.L., Moghimi S.M. Polycation cytotoxicity: a delicate matter for nucleic acid therapy—focus on polyethylenimine. Soft Matter. 2010;6:4001–4009. [Google Scholar]
- 28.Sunshine J.C., Peng D.Y., Green J.J. Uptake and transfection with polymeric nanoparticles are dependent on polymer end-group structure, but largely independent of nanoparticle physical and chemical properties. Mol. Pharm. 2012;9:3375–3383. doi: 10.1021/mp3004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy R.F., Powers S., Cantor C.R. Endosome pH measured in single cells by dual fluorescence flow cytometry: rapid acidification of insulin to pH 6. J. Cell Biol. 1984;98:1757–1762. doi: 10.1083/jcb.98.5.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forrest M.L., Pack D.W. On the kinetics of polyplex endocytic trafficking: implications for gene delivery vector design. Mol. Ther. 2002;6:57–66. doi: 10.1006/mthe.2002.0631. [DOI] [PubMed] [Google Scholar]
- 31.Akinc A., Langer R. Measuring the pH environment of DNA delivered using nonviral vectors: implications for lysosomal trafficking. Biotechnol. Bioeng. 2002;78:503–508. doi: 10.1002/bit.20215. [DOI] [PubMed] [Google Scholar]
- 32.Benjaminsen R.V., Sun H., Henriksen J.R., Christensen N.M., Almdal K., Andresen T.L. Evaluating nanoparticle sensor design for intracellular pH measurements. ACS Nano. 2011;5:5864–5873. doi: 10.1021/nn201643f. [DOI] [PubMed] [Google Scholar]
- 33.Ma X., Wang Y., Zhao T., Li Y., Su L.-C., Wang Z., Huang G., Sumer B.D., Gao J. Ultra-pH-sensitive nanoprobe library with broad pH tunability and fluorescence emissions. J. Am. Chem. Soc. 2014;136:11085–11092. doi: 10.1021/ja5053158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson D.R., Routkevitch D., Wahlin K.J., Zack D.J., Quinones-Hinojosa A., Green J.J. Development of a pH sensor to probe endosomal buffering of polymeric nanoparticles effective for gene delivery. Mol. Ther. 2016;24(Suppl 1):S196. [Google Scholar]
- 35.Bishop C.J., Majewski R.L., Guiriba T.-R.M., Wilson D.R., Bhise N.S., Quiñones-Hinojosa A., Green J.J. Quantification of cellular and nuclear uptake rates of polymeric gene delivery nanoparticles and DNA plasmids via flow cytometry. Acta Biomater. 2016;37:120–130. doi: 10.1016/j.actbio.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gasiorowski J.Z., Dean D.A. Postmitotic nuclear retention of episomal plasmids is altered by DNA labeling and detection methods. Mol. Ther. 2005;12:460–467. doi: 10.1016/j.ymthe.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beh C.W., Pan D., Lee J., Jiang X., Liu K.J., Mao H.-Q., Wang T.H. Direct interrogation of DNA content distribution in nanoparticles by a novel microfluidics-based single-particle analysis. Nano Lett. 2014;14:4729–4735. doi: 10.1021/nl5018404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tripathi S.K., Goyal R., Kumar P., Gupta K.C. Linear polyethylenimine-graft-chitosan copolymers as efficient DNA/siRNA delivery vectors in vitro and in vivo. Nanomedicine (Lond.) 2012;8:337–345. doi: 10.1016/j.nano.2011.06.022. [DOI] [PubMed] [Google Scholar]
- 39.Wittrup A., Ai A., Liu X., Hamar P., Trifonova R., Charisse K., Manoharan M., Kirchhausen T., Lieberman J. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 2015;33:870–876. doi: 10.1038/nbt.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C., Stöter M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 41.Kozielski K.L., Tzeng S.Y., De Mendoza B.A.H., Green J.J. Bioreducible cationic polymer-based nanoparticles for efficient and environmentally triggered cytoplasmic siRNA delivery to primary human brain cancer cells. ACS Nano. 2014;8:3232–3241. doi: 10.1021/nn500704t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong S., Leroueil P.R., Janus E.K., Peters J.L., Kober M.-M., Islam M.T., Orr B.G., Baker J.R., Jr., Banaszak Holl M.M. Interaction of polycationic polymers with supported lipid bilayers and cells: nanoscale hole formation and enhanced membrane permeability. Bioconjug. Chem. 2006;17:728–734. doi: 10.1021/bc060077y. [DOI] [PubMed] [Google Scholar]
- 43.Kim J., Sunshine J.C., Green J.J. Differential polymer structure tunes mechanism of cellular uptake and transfection routes of poly(β-amino ester) polyplexes in human breast cancer cells. Bioconjug. Chem. 2014;25:43–51. doi: 10.1021/bc4002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uchida H., Miyata K., Oba M., Ishii T., Suma T., Itaka K., Nishiyama N., Kataoka K. Odd-even effect of repeating aminoethylene units in the side chain of N-substituted polyaspartamides on gene transfection profiles. J. Am. Chem. Soc. 2011;133:15524–15532. doi: 10.1021/ja204466y. [DOI] [PubMed] [Google Scholar]
- 45.Eltoukhy A.A., Chen D., Alabi C.A., Langer R., Anderson D.G. Degradable terpolymers with alkyl side chains demonstrate enhanced gene delivery potency and nanoparticle stability. Adv. Mater. 2013;25:1487–1493. doi: 10.1002/adma.201204346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson D.R., Mosenia A., Suprenant M.P., Upadhya R., Routkevitch D., Meyer R.A., Quinones-Hinojosa A., Green J.J. Continuous microfluidic assembly of biodegradable poly(beta-amino ester)/DNA nanoparticles for enhanced gene delivery. J. Biomed. Mater. Res. A. 2017 doi: 10.1002/jbm.a.36033. Published online April 12, 2017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.