Abstract
The effective treatment of glioma is largely hindered by the poor transfer of drug delivery systems across the blood-brain barrier (BBB) and the difficulty in distinguishing healthy and tumorous cells. In this work, for the first time, an interleukin-6 receptor binding I6P7 peptide was exploited as a cascade-targeting ligand in combination with a succinoyl tetraethylene pentamine (Stp)-histidine oligomer-based nonviral gene delivery system (I6P7-Stp-His/DNA). The I6P7 peptide provides multiple functions, including the cascade-targeting potential represented by a combined BBB-crossing and subsequent glioma-targeting ability, as well as a direct tumor-inhibiting effect. I6P7-Stp-His/DNA nanoparticles (NPs) mediated higher gene expression in human glioma U87 cells than in healthy human astrocytes and a deeper penetration into glioma spheroids than scrambled peptide-modified NPs. Transport of I6P7-modified, but not the control, NPs across the BBB was demonstrated in vitro in a transwell bEnd.3 cell model resulting in transfection of underlying U87 cells and also in vivo in glioma-bearing mice. Intravenous administration of I6P7-Stp-His/plasmid DNA (pDNA)-encoding inhibitor of growth 4 (pING4) significantly prolonged the survival time of orthotopic U87 glioma-bearing mice. The results denote that I6P7 peptide is a roborant cascade-targeting ligand, and I6P7-modified NPs might be exploited for efficient glioma therapy.
Keywords: antitumoral cascade-targeting ligand, gene therapy, glioma, interleukin-6 receptor
Graphical Abstract
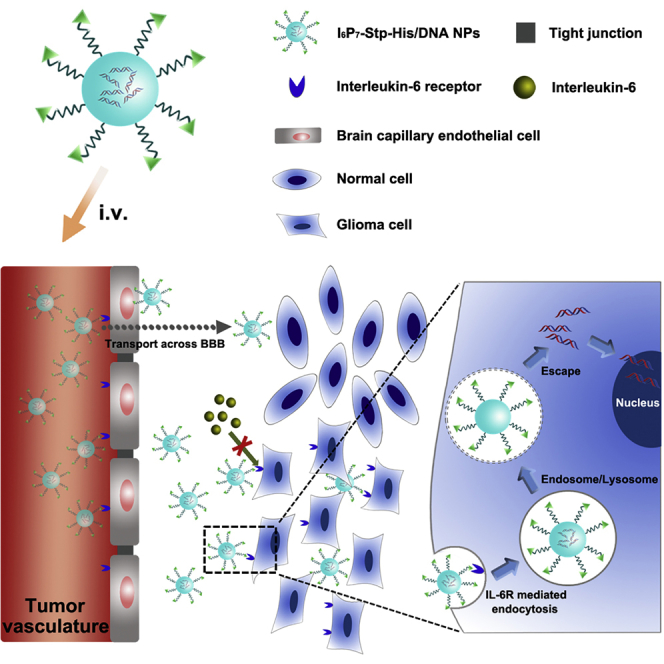
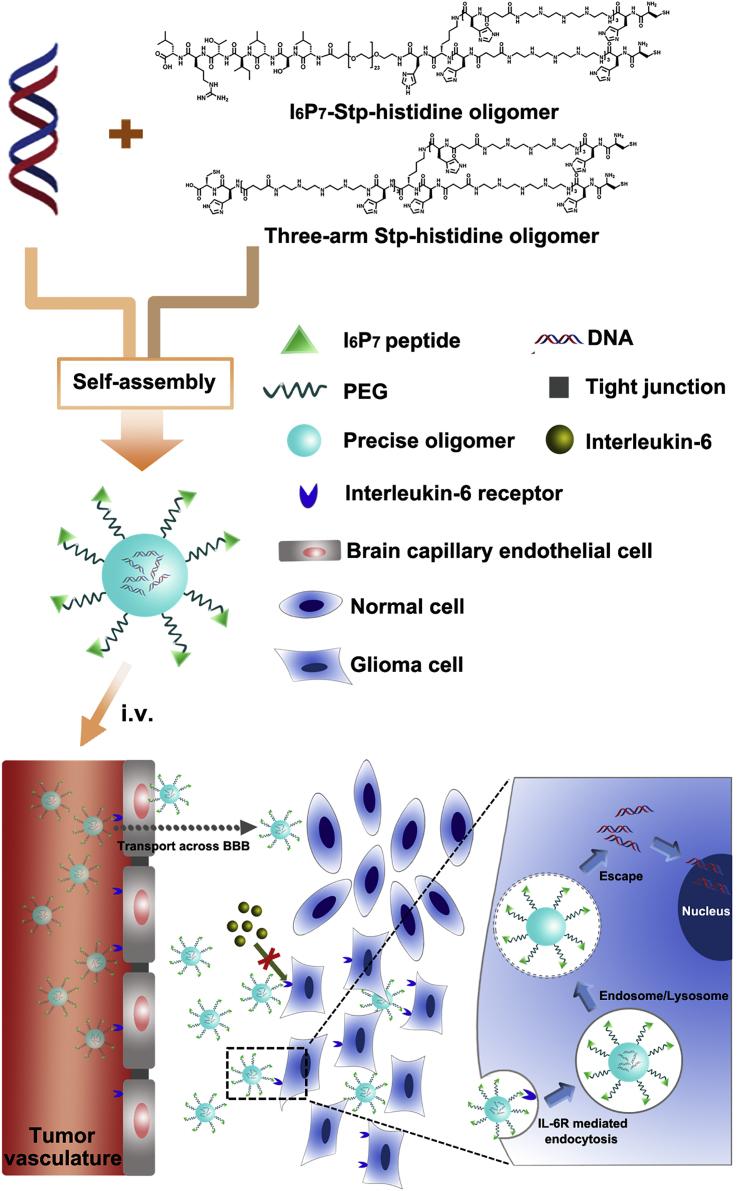
A DNA compacting Stp-histidine oligomer blend modified with I6P7 peptide (I6P7-Stp-His/DNA) was exploited as a cascade-targeting platform for efficient glioma therapy because of its multiple functions including receptor-mediated transportation across the blood-brain barrier and subsequent glioma targeting, gene therapy, and biotherapeutic activity.
Introduction
Glioma has long been considered the most refractory primary brain malignancy because of the difficulty for efficient drug delivery to the tumor without adverse effects on normal tissues.1 The blood-brain barrier (BBB), the natural protective barrier of the brain, plays a pivotal role in restricting the penetration of systemically administrated drugs to glioma, especially at early tumor stage.2 Almost all large molecular drugs (including genes, recombinant proteins, and monoclonal antibodies) and more than 98% of small molecular drugs are excluded from the central nervous system by the BBB.3 Effective glioma therapy is also restricted by potential adverse effects on normal tissues in consequence of required high drug doses.4 Thus, for glioma therapy, ideal drug delivery systems must efficiently transport the therapeutic agents across the BBB and subsequently mediate specific entry into glioma cells; this goal may be achieved by the cascade-targeting strategy presented in the current work.
Receptor-targeted drug delivery has been demonstrated as a potential approach to achieve drug accumulation in glioma. A series of receptors that are expressed on the endothelium of the BBB, as well as glioma cells, including transferrin receptor, lactoferrin receptor, low-density lipoprotein receptor, and folate receptor, have been exploited for this purpose.5, 6, 7, 8 Research has continued to identify new receptors and their specific ligands. Interleukin-6 receptor (IL-6R), a membrane-bound glycoprotein, has been detected to be expressed on the BBB and various brain tumors including glioblastoma, astrocytoma, and pituitary tumors, but not in healthy astrocytes.9, 10, 11 The natural ligand of IL-6R is interleukin-6 (IL-6), a member of the inflammatory cytokine family, which was initially described as a central mediator of the immune system, hematopoiesis, and inflammation.12, 13 Furthermore, it is well established that the interaction of IL-6 and IL-6R triggers a biological activity, which promotes glioma growth and progression.14, 15, 16 Thus, IL-6 itself is not suitable as a ligand in glioma pharmacotherapy. Recently, a heptapeptide (N to C terminus: LSLITRL), designated as I6P7, which can specifically bind to IL-6R, was shown to inhibit the binding between IL-6 and IL-6R, and thereby retard tumor growth.11, 16 In this regard, the interaction of IL-6R with I6P7 as targeting ligand might not only increase the BBB transport of therapeutic agents toward glioma, but also inhibit glioma growth and progression. Also, compared with protein or antibody ligands, the I6P7 peptide has several advantages, such as small size, simple chemical synthesis, and sufficient stability during additional synthetic manipulations. In addition, compared with previously reported two ligand-mediated targeting strategies, I6P7 could act as both a BBB-crossing and a glioma-targeting ligand to realize a cascade-targeting effect of drug delivery.17, 18
Current treatment regimens have only a modest therapeutic effect on the progressive course of glioma, despite recent advances in surgical intervention, radiotherapy, and chemotherapy.19 Effective gene therapy presents an encouraging new modality, but it has been greatly hindered by the development of suitable gene vectors. Non-viral gene transfer has been widely studied because of its inherent safety compared with virus-based strategies. Efficient vectors for gene therapy should possess three important features: nucleic acid binding, cellular uptake, and subsequent endolysosomal escape.20 Different non-viral vectors, especially cationic polymers, have been most intensely studied.21, 22, 23 However, classical polymers such as polyethylenimine, because of the nature of polymer chemistry, generally exhibit a certain degree of heterogeneity.23 From the clinical point of view, more defined molecules would be highly appreciated. To meet this demand, precise sequence-defined and well-tolerated oligo(ethanamino)amide-based oligomers were generated utilizing solid-phase chemistry to act as nanocarriers.24, 25 Members of this oligomer compound class containing repeats of the building block succinoyl tetraethylene pentamine (Stp) and histidine have been shown to efficiently deliver nucleic acid cargos into tumor cells.20 Therefore, we fabricated optimized histidine and Stp-based oligo(ethanamino)amide oligomers containing a shielding polyethylene glycol (PEG) and the multifunctional I6P7 peptide to mediate effective and receptor-specific gene transfer. In addition, a PEG-free three-arm Stp-histidine oligomer has been assembled to compensate complex disturbance by a high PEG content, further enhancing endolysosomal escape and condensing DNA into rod- or toroid-like structures.25 Hence, the PEGylated two-arm targeting oligomer and the PEG-free three-arm DNA-compacted oligomer were mixed together at suitable ratio to form a bi-component oligomer blend in order to get a promising gene vector.
The nanocarrier based on the Stp-histidine oligomers containing I6P7 and complexed DNA was exploited to form a cascade-targeting gene delivery system (I6P7-Stp-His/DNA) (Figure 1). The therapeutic gene construct used here was a plasmid DNA-encoding inhibitor of growth 4 (pING4). The tumor suppressor ING4 had been proven to inhibit the tumorigenesis and progression of glioma.26 Therefore, the ING4 gene was successfully applied in several vector-mediated gene therapy systems.26, 27, 28 In addition, another analogous nanoparticle (NP) modified with a scrambled I6P7 sequence was prepared as control group (I6P7scr-Stp-His/DNA). Overall properties of the different NPs including formulation optimization, glioma cell targeting efficiency, cellular uptake mechanisms, and BBB-crossing potential were systematically investigated in vitro, and the anti-glioma effects were evaluated in vivo in a murine orthotopic glioma model.
Figure 1.
Schematic Diagram of the Formation of I6P7-Stp-His/DNA NPs and Their In Vivo Delivery Process
Results and Discussion
Functionalization of the Precise Oligomers
A peptide ligand I6P7, which can specifically bind to IL-6R, was selected to functionalize a gene delivery system for cascade targeting to glioma. To achieve this objective, we assembled a PEGylated two-arm Stp-histidine oligomer with I6P7 via a fully controlled and precise solid-phase assisted methodology.29, 30 The chemical structures of the precise oligomers are shown in Figure 1. Besides the multifunctional peptide, a monodisperse PEG moiety with 24 oxyethylene units was introduced to the oligomer for surface shielding. Repeats of the Stp and histidine in the oligomer sequence were used for improving the nucleic acid packaging and endosomal buffer capacity. The non-targeting control oligomer, I6P7scr-Stp-His, has the analogous PEG-containing structure to I6P7-Stp-His except the scrambled ligand sequence. Importantly, a cationic three-arm oligomer was synthesized and utilized in combination with the PEG-containing oligomers for improving the DNA compaction efficiency and endolysosomal escape of NPs. The physicochemical characterizations of the oligomers are described in the Supplemental Information.
NP Formation, Optimization, and Physicochemical Characterizations
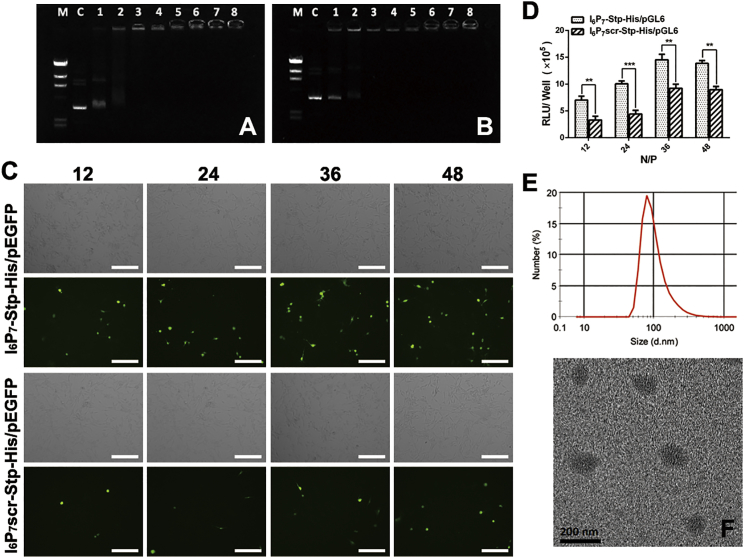
First, the precise oligomer/DNA complexes with different N/P ratios were tested via agarose gel electrophoresis to monitor the NP formation. As shown in Figures 2A and 2B, two kinds of NPs with an oligomer nitrogen-to-DNA phosphate (N/P) ratio greater than 1.5 could encapsulate DNA completely without migration in the gel as compared with naked DNA. NPs with the N/P ratios of 12, 24, 36, and 48 were used in the following experiments.
Figure 2.
Optimization and Physical Characterizations of I6P7-Stp-His/DNA and I6P7scr-Stp-His/DNA NPs
DNA binding capacity of I6P7-Stp-His (A) and I6P7scr-Stp-His (B) oligomers was determined by an agarose gel shift assay at a range of N/P ratios. Lane M: DNA marker, Hind III digested; lane C: naked DNA; lane 1: 0.75 (N/P); lane 2: 1.5 (N/P); lane 3: 3 (N/P); lane 4: 6 (N/P); lane 5: 12 (N/P); lane 6: 24 (N/P); lane 7: 36 (N/P); lane 8: 48 (N/P). (C) EGFP gene expression of I6P7-Stp-His/pEGFP and I6P7scr-Stp-His/pEGFP NPs in U87 cells at an N/P ratio of 12, 24, 36, and 48, respectively. Green represents EGFP. (D) Luciferase expression levels of I6P7-Stp-His/pGL6 and I6P7scr-Stp-His/pGL6 NPs in U87 cells at different N/P ratios (12, 24, 36, 48). (E and F) The particle size distribution of I6P7-Stp-His/pGL6 at an N/P ratio of 36 was analyzed by dynamic light scattering (E) and transmission electron microscope (F). Scale bars, 200 μm. **p < 0.01; ***p < 0.001.
Second, the in vitro gene expression of different NPs was assessed in U87 cells, using EGFP expression and fluorescence microscopy qualitatively, as well as luciferase expression and bioluminescence assay quantitatively, to further optimize the formulation of NPs. As shown in Figure 2C, fluorescent images showed that U87 cells treated with different NPs exhibited increasing gene expression levels with increasing N/P ratios up to 36, where expression saturated. Furthermore, EGFP expression of I6P7-modified NPs was significantly higher than that of scrambled peptide-modified NPs. Consistent with the results of EGFP expression, the luciferase expression efficiency of the I6P7-Stp-His/pGL6 NPs was 2.2-, 2.3-, 1.5-, and 1.6-fold higher than that of I6P7scr-Stp-His/pGL6 at the N/P ratios 12, 24, 36, and 48, respectively (Figure 2D). In vitro transfection, however, represents a static cell culture model, where NPs are in close proximity to cells over a long period of time and, especially regarding receptor interactions, the relevance for dynamic in vivo conditions is limited. Considering the obtained gene expression levels, the N/P ratio was fixed at 36 to prepare the precise oligomer/DNA NPs in the subsequent experiments.
Particle sizes and zeta potentials of different NPs were further determined (Table 1). As the N/P ratio increased, the sizes of all NPs significantly reduced, whereas zeta potentials increased. The positive zeta potentials demonstrated that all NPs were capable of condensing DNA effectively. A transmission electron microscopy (TEM) image showed that the I6P7-Stp-His/DNA NPs were generally spherical and well dispersed at N/P = 36 (Figure 2F). The particle size observed by TEM was consistent with the results obtained by dynamic light scattering (DLS) (Figure 2E).
Table 1.
Sizes and Zeta Potentials of NPs with Different N/P Ratios
| N/P | I6P7-Stp-His/DNA |
I6P7scr-Stp-His/DNA |
||
|---|---|---|---|---|
| Mean Size (nm) | Zeta Potential (mV) | Mean Size (nm) | Zeta Potential (mV) | |
| 12 | 283.5 ± 11.2 | 13.2 ± 0.53 | 298.7 ± 9.9 | 14.9 ± 0.31 |
| 24 | 132.8 ± 25.6 | 15.3 ± 0.39 | 124.6 ± 5.2 | 15.9 ± 0.55 |
| 36 | 99.0 ± 15.0 | 15.9 ± 0.56 | 111.6 ± 10.6 | 17.5 ± 0.38 |
| 48 | 96.9 ± 17.2 | 19.0 ± 0.29 | 96.0 ± 8.7 | 19.6 ± 0.42 |
Data represent mean ± SD. n = 3.
Targeting and Spheroid-Penetrating Ability
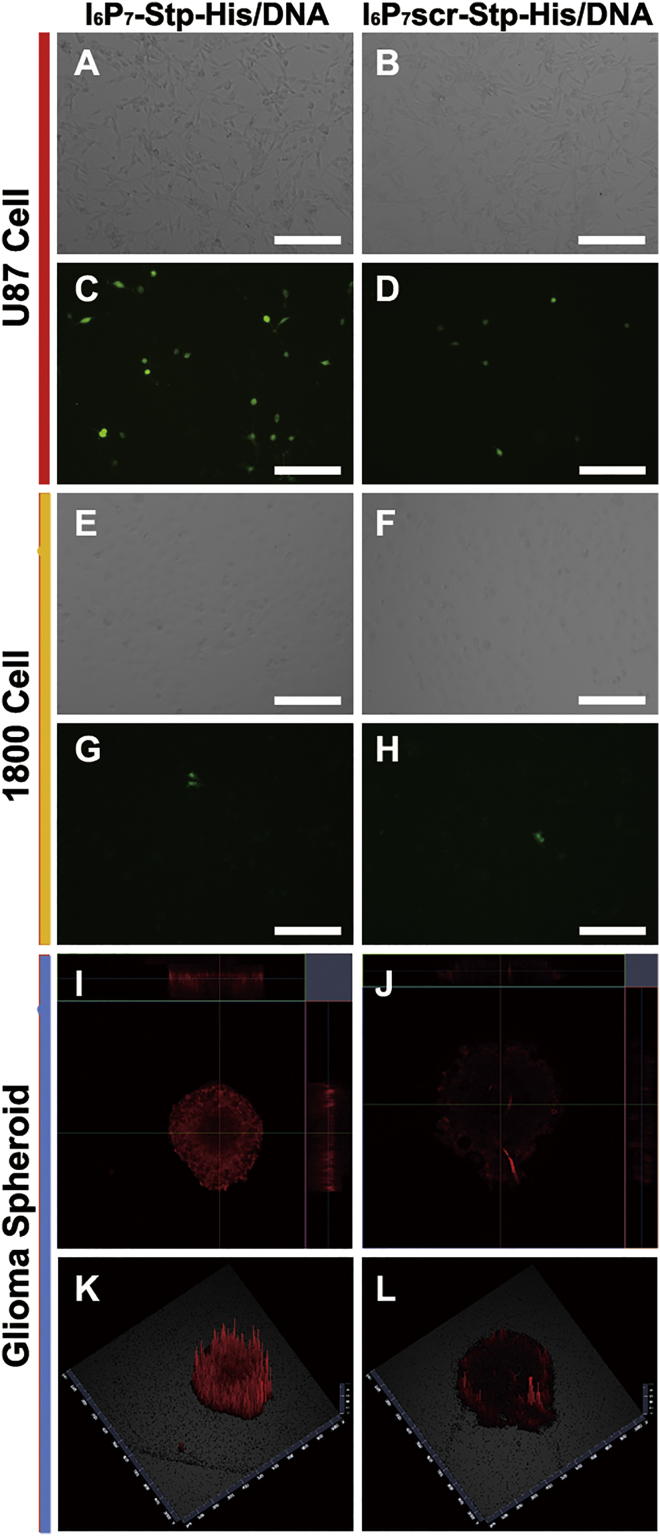
In order to assess glioma-targeting efficiency in vitro, we applied U87 cells and astrocyte line 1800 cells as the cell culture models for EGFP expression. As shown in Figures 3A–3H, higher transfection efficiency was found in U87 cells for I6P7-Stp-His/plasmid EGFP (pEGFP) as compared with I6P7scr-Stp-His/pEGFP NPs. Also, the I6P7-modified NPs mediated higher gene expression levels in U87 glioma than in 1800 astrocyte cells, demonstrating the tumor-targeting ability of I6P7. This was probably attributed to the apparently higher expression of IL-6R on U87 glioma cells than in healthy astrocytes.11, 14 A better transfection of dividing tumor cells during the stage of mitosis as compared with nondividing healthy cells may also contribute.31
Figure 3.
I6P7-Stp-His/DNA NPs Improve Gene Expression in U87 Cells and Glioma Spheroid Penetration In Vitro
(A–H) Gene expression of I6P7-Stp-His/pEGFP and I6P7scr-Stp-His/pEGFP NPs in U87 glioma cells (A–D) and 1800 astrocyte cells (E–H). (A, C, E, and G) I6P7-Stp-His/pEGFP NPs. (B, D, F, and H) I6P7scr-Stp-His/pEGFP NPs. (A), (B), (E), and (F) are corresponding bright-field images of fluorescent images (C), (D), (G), and (H). Green represents EGFP. (I–L) Glioma spheroid penetration of I6P7-Stp-His/DNA and I6P7scr-Stp-His/DNA NPs after 12 hr incubation. (I and K) I6P7-Stp-His/DNA NPs. (J and L) I6P7scr-Stp-His/DNA NPs. (I) and (J) are the corresponding 2D images of glioma spheroid. (K) and (L) are the corresponding 2.5D images of glioma spheroid obtained from the middle of z planes. DNA was pre-labeled by YOYO-3. Red represents YOYO-3. Scale bars, 200 μm.
Owing to the difference between the two-dimensional (2D) cultured monolayers and three-dimensional (3D) solid tumors, in vitro glioma spheroid penetration might reflect more accurately the glioma-accumulating effect of different NPs.32 Thus, the distribution of the different NPs in glioma spheroids was observed (Figures 3I–3L). The results demonstrated that I6P7-Stp-His/DNA NPs possess better spheroid-penetrating efficiency than I6P7scr-Stp-His/DNA NPs.
Cellular Uptake Mechanisms
The cellular uptake mechanisms of the oligomer/DNA NPs were studied in U87 cells. First, the fluorescence intensity of cells treated with YOYO-3-labeled I6P7-Stp-His/DNA NPs was far higher than for I6P7scr-Stp-His/DNA NP-treated cells, both after 37°C and 4°C incubation (Figures S1A–S1D). This proved that I6P7 possesses significantly enhanced ability to deliver NPs into U87 cells. Furthermore, both NPs showed higher cellular uptake at 37°C compared with 4°C. The temperature dependency indicates the involvement of energy-dependent processes during the cellular internalization. Second, the fluorescent intensity within cells decreased significantly after being treated with excessive free I6P7 peptide (100× excess) (Figure S1E) compared with that of the untreated group (Figure S1A). This competitive inhibition indicates a receptor-mediated process and that I6P7 has sufficient affinity for IL-6R as mediator for cellular uptake into U87 cells.26 To further elucidate the endocytosis pathways involved in the internalization of I6P7-Stp-His/DNA NPs, we investigated the cellular uptake of NPs after pretreatment with various endocytosis inhibitors. Phenylarsine oxide (PhAsO), filipin, and colchicine were utilized as inhibitors of clathrin-dependent endocytosis, caveolae-mediated endocytosis, and macropinocytosis, respectively.26 As shown in Figures S1F–S1H, the fluorescent intensity of the I6P7-Stp-His/DNA NPs declined to some extent in all three inhibitors. This suggests that all investigated pathways, the clathrin-mediated endocytosis, the caveolae-mediated endocytosis, and macropinocytosis may contribute to the cellular internalization process of I6P7-modified NPs. These results are also consistent with other reports about receptor-mediated transport processes.26, 33
BBB-Penetrating Efficiency and Cascade-Targeting Expression
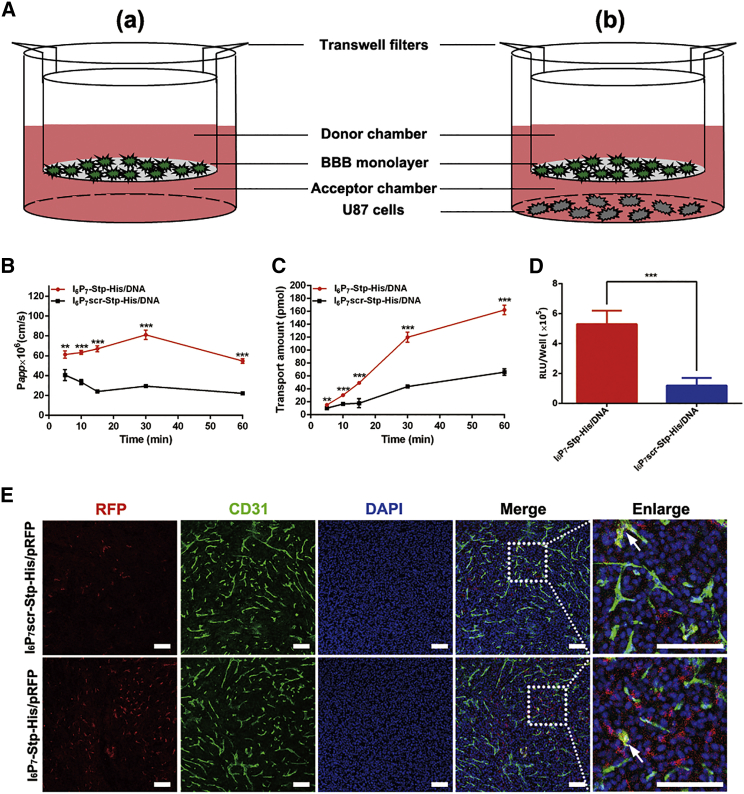
In order to verify the BBB-penetrating efficiency of the gene delivery system, we established an in vitro BBB model (Figure 4A), and NPs were labeled with YOYO-3 for their detection. The results of transport studies were shown in Figure 4B. The apparent permeability (Papp) of I6P7-Stp-His/DNA NPs, which reached (81.10 ± 4.66) × 10−6 cm/s at 30 min, was approximately 2.8-fold higher than that of I6P7scr-Stp-His/DNA. This indicated that the modification of I6P7 could facilitate the BBB penetration of NPs because of the specific binding of I6P7 to IL-6R, which possesses transport function across the BBB. The transendothelial electrical resistance (TEER) showed no obvious reduction compared with that of a control (data not shown), indicating the integrity of the BBB and that the transport of I6P7 did not disrupt the cell layer. Also, the cumulative transport amount of I6P7-Stp-His/DNA NPs was much higher than that of the I6P7scr counterpart (Figure 4C). These results revealed that I6P7 modification exerted its effect on the transport of NPs across the BBB.
Figure 4.
Effect of I6P7-Stp-His/DNA NPs on BBB-Penetrating Efficiency and Cascade-Targeting Expression in U87 Cells
(A) Schematic illustration of the in vitro BBB model (a) and the b.End3/U87 cell co-culture model (b). (B and C) The apparent permeability (B) and cumulative transport amount (C) of different NPs across BBB monolayer at various incubation times. DNA was pre-labeled by YOYO-3. (D) Luciferase expression levels of different NPs in U87 cells after transporting across the BBB. Results are presented as mean ± SD (n = 3). **p < 0.01; ***p < 0.001. (E) Immunofluorescence images of glioma sections from glioma-bearing mice injected with I6P7scr-Stp-His/pRFP and I6P7-Stp-His/pRFP NPs. Red represents red fluorescent protein (RFP); green represents anti-CD31-labeled brain capillary; blue represents DAPI-labeled cell nuclei. White arrows: co-localization of RFP and brain capillary. Scale bars, 100 μm.
In order to verify cascade-targeting effects, using a luciferase assay, we further evaluated the gene expression of NPs in glioma cells after crossing the BBB monolayer. A schematic illustration of the bEnd.3/U87 cell co-culture model is shown in Figure 4A. As depicted in Figure 4D, I6P7-Stp-His/DNA NPs, which had crossed the bEnd.3 layer, exhibited a higher luciferase expression in U87 glioma cells than control NPs, consistent with the data from the transport studies. These results demonstrate that NPs modified with I6P7 exhibit favorable cascade-targeting potential: crossing the artificial BBB and then targeting and transfecting glioma cells.
In order to evaluate BBB-penetrating efficiency and transgene expression of proteins in the glioma in vivo, we obtained frozen sections 2 days after intravenous administration of NPs encoding RFP (red fluorescent protein), and brain capillaries were stained with anti-CD31 antibody. As demonstrated in Figure 4E, compared with injection of I6P7scr-Stp-His/pRFP, the injection of I6P7-Stp-His/pRFP NPs remarkably improved the gene expression product RFP in the glioma site, indicating a favorable glioma-targeting ability of I6P7 peptide that was consistent with the previous results in vitro. The red fluorescence of RFP partially co-localized with the green fluorescence of brain capillaries, suggesting that a small part of NPs remained within the brain capillary and mediated gene expression there. However, the stronger red fluorescent signal demonstrated that most of the I6P7-modified NPs mediated the gene expression in glioma tissue.
In Vivo Biodistribution
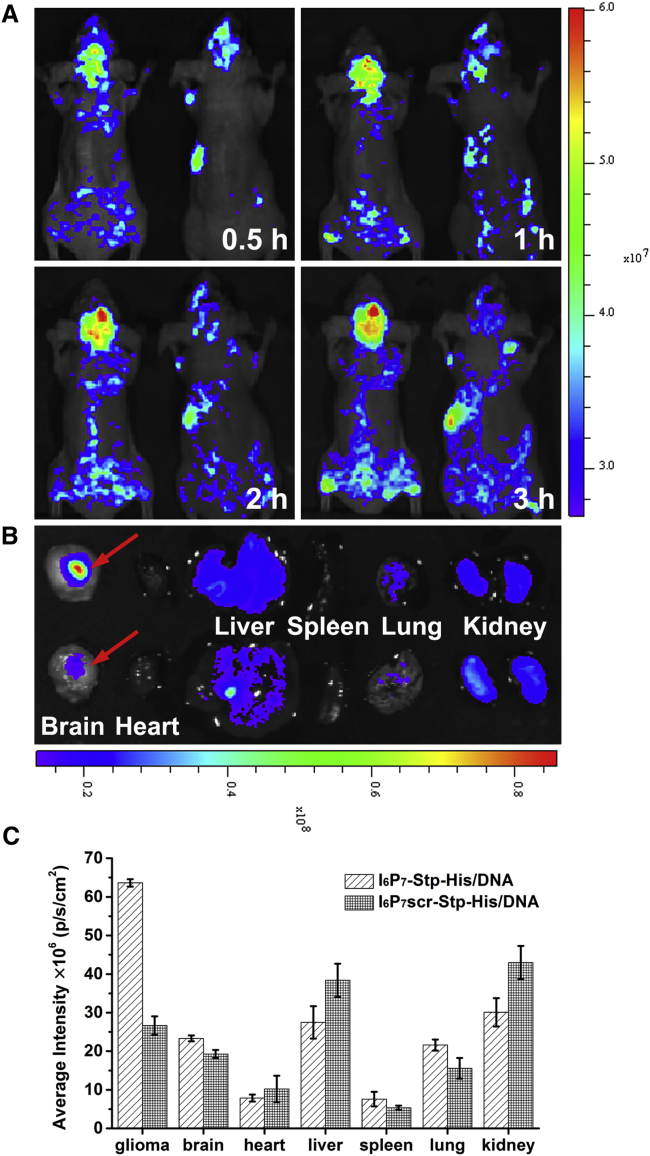
To investigate the in vivo targeting behavior of I6P7-Stp-His/DNA NPs, we carried out in vivo imaging in glioma-bearing mice administrated with YOYO-3-labeled precise oligomer/DNA NPs. As shown in Figure 5A, the fluorescence intensity in the brain of I6P7-Stp-His/DNA-treated mice was significantly higher than in I6P7scr-Stp-His/DNA-treated mice at 0.5 hr after injection; this pattern was maintained for the entire study (from 0.5 to 3 hr postinjection). The ex vivo imaging of brains at 3 hr and the fluorescence quantitative analysis results showed that the I6P7-Stp-His/DNA NPs were obviously accumulated in glioma tissue rather than normal brain tissue (Figures 5B and 5C). In contrast, glioma tissue accumulation of the control NPs was much lower compared with I6P7-Stp-His/DNA, but displayed higher accumulation in liver and kidney (Figure 5B). These results indicate that I6P7 is an effective targeting ligand that can facilitate transport of NPs across the BBB and improve their accumulation in glioma via the IL-6R-mediated pathway.
Figure 5.
In Vivo Biodistribution of I6P7-Stp-His/DNA NPs
(A) In vivo fluorescent images of glioma-bearing mice at different time points after intravenous (i.v.) administration of I6P7-Stp-His/DNA (left) and I6P7scr-Stp-His/DNA (right). (B) Ex vivo fluorescent images of the organs harvested at 3 hr after i.v. administration of I6P7-Stp-His/DNA (top) and I6P7scr-Stp-His/DNA NPs (bottom). (C) Fluorescence quantitative analysis of ex vivo organs of U87 tumor-bearing mice at different time intervals after intravenous injection. DNA was pre-labeled by YOYO-3.
Anti-glioma Effect
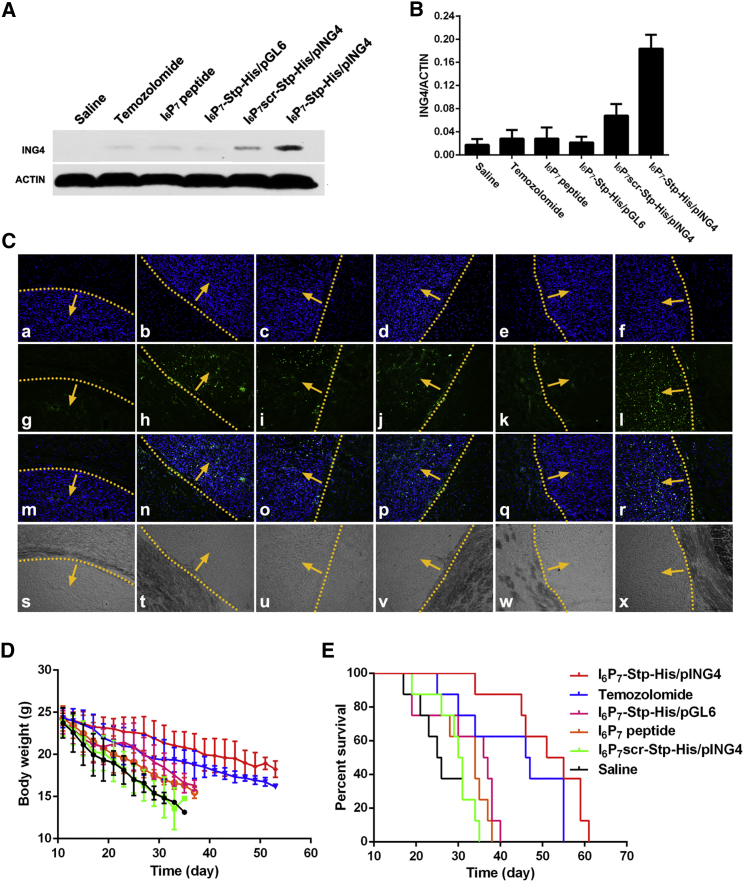
In order to evaluate the effect of I6P7 peptide on cell growth, we performed a Cell Counting Kit-8 (CCK-8) assay. As shown in Figure S2, incubation with I6P7-Stp-His/DNA NPs for 48 or 72 hr resulted in lower U87 cell viability than in the case of I6P7scr-Stp-His/DNA NPs, although none of the formulations affected cell viability upon 24 hr of incubation. Free I6P7 peptide also led to lower U87 cell viability with extended incubation time. These results show that free I6P7 peptide and I6P7-modified NPs both can inhibit U87 cell growth, and that the inhibitory effect is related to the incubation time and the amount of I6P7 peptide. To some modest extent, I6P7scr-Stp-His/DNA NPs can also cause cytotoxicity, presumably because of unspecific adverse effects of the cationic oligomer backbone. Further assessment of the in vivo anti-glioma effect is presented in Figure 6. Western blot assay was performed to analyze the ING4 expression level of different tissues after intravenous administration (Figures 6A, 6B, and S4). Compared with healthy brain tissues, glioma exhibits decreased expression levels of ING4.34 After intravenous administration, ING4 expression in glioma tissues significantly increased in the I6P7-Stp-His/pING4 group compared with the other treatment groups. This suggests that the increased inhibition of glioma by I6P7-Stp-His/pING4 was partly caused by a higher expression level of ING4 protein; the data further verify the efficiency of I6P7-Stp-His/pING4. As demonstrated in Figure S4, ING4 protein could be detected by western blot analysis in all tissues, which is consistent with previous studies.34 Besides, the I6P7-Stp-His/pING4 and I6P7scr-Stp-His/pING4 group enhanced the ING4 gene expression level in liver and kidney, which might be because of a significant accumulation of NPs. As shown in Figure 6C, TdT-mediated dUTP nick end labeling (TUNEL) assay was used to examine the induction of apoptosis by different treatments. Groups of glioma-bearing mice treated with either temozolomide or saline were used as the positive or negative control, respectively. Remarkably, the largest number of apoptotic cells in tumor tissue was induced by the treatment with I6P7-Stp-His/pING4. Furthermore, this treatment showed the presence of apoptotic cells mainly in the tumor tissue, possibly by an IL-6R-mediated effect. Even more interesting, it was found that intravenous injection of I6P7 peptide or I6P7-Stp-His/pGL6 NPs (luciferase transgene as control) could also induce apoptosis in glioma. This might be attributed to a blockade of the IL-6-mediated JAK/STAT tumor growth pathway (glioma cell growth factor IL-6 is overexpressed in vivo) by I6P7 peptide. Other treatment groups, except the saline negative control, also mediated apoptosis to some extent, but not as pronounced. The antitumor effect was also evaluated via monitoring the body weight and the median survival time of nude mice bearing glioma. As shown in Figure 6D, glioma-bearing mice treated with I6P7-Stp-His/pING4 maintained the body weight compared with other groups’ best. The median survival times for the saline, temozolomide, I6P7 peptide, I6P7-Stp-His/pGL6, I6P7scr-Stp-His/pING4, and I6P7-Stp-His/pING4 groups were 25.5, 46.5, 34, 36.5, 30.5, and 53 days, respectively. Compared with the control groups, I6P7-Stp-His/pING4 treatment significantly prolonged the survival time (Figure 6E). This might be because of the multiple functions of I6P7 peptide, increasing the BBB crossing and glioma cellular uptake of NPs and retarding the glioma growth by interfering with the interaction between IL-6 and IL-6R. All results indicate that I6P7-Stp-His/pING4 could be a promising nanomedicine for targeted glioma therapy.
Figure 6.
Antitumor Effect of I6P7-Stp-His/DNA NPs
(A and B) Western blot analysis for ING4 expression in glioma tissues for different experimental animal groups (A) and quantitative analysis of the protein level (B). Data are expressed as mean ± SD (n = 3). (C) Apoptosis of glioma-bearing mice treated with saline (a, g, m, and s), temozolomide (b, h, n, and t), I6P7 peptide (c, i, o, and u), I6P7-Stp-His/pGL6 (d, j, p, and v), I6P7scr-Stp-His/pING4 (e, k, q, and w), and I6P7-Stp-His/pING4 (f, l, r, and x). Slides were subjected to TUNEL apoptosis detection kit (FITC-labeled). Blue represents DAPI-labeled cell nuclei; green represents FITC-labeled apoptotic cells. Yellow dotted lines indicate borders of glioma, and arrows point to the inner sides of glioma. (s–x) Corresponding bright-field images. (D) The average change in body weight of glioma-bearing mice after different treatments. (E) Kaplan-Meier survival curves of glioma-bearing mice after different treatments.
Safety Assessment
The histological examination of the tissue slices after different treatments was performed to evaluate the in vivo toxicity of NPs (Figure S5). The H&E staining images of normal brain tissue, heart, liver, spleen, lung, and kidney revealed that none of the treatments caused apparent inflammatory response or damage in main organs, which suggests that the NPs of this study exhibited low toxicity.
In order to further evaluate the safety of all preparations in vivo, we monitored the hematopoietic cell counts and important cytokines. As shown in Figure S6, although individual differences were noted, the hematopoietic cell counts of mice treated with NPs were on a similar level as in control mice (saline group) and within normal ranges. ELISA results clearly indicated IL-6 and IL-10 serum levels had no obvious change in the NPs groups compared with the saline control group. The above results demonstrated that these formulations including I6P7-modified NPs had no obvious side effects on mice upon systemic administration.
Conclusions
In this study, the IL-6R was used as receptor for targeted drug delivery in glioma therapy, with the multifunctional peptide I6P7 being exploited as corresponding targeting ligand moiety. The cascade-targeting gene transfer system could not only demonstrate high transfection efficiency and low toxicity, but also improved brain-penetrating and glioma-targeting efficiency, as well as elevated transgene expression. In vivo experiments indicate that the I6P7 modification of NPs could enhance the BBB-crossing and glioma-targeting efficiency. Intravenous administration of I6P7 NPs encoding ING4 reduced the growth of orthotopic glioma without any obvious sign of toxicity to normal tissues. In sum, the I6P7-modified NPs display an encouraging potential for future therapeutic development against glioma.
Materials and Methods
Materials
The plasmid pEGFP-N2 (Clontech), the plasmid-encoding luciferase pGL6-control vector (Genomeditech), and pcDNA3.1-ING4 (ING4; Boshang Biotech) were purified using QIAGEN Plasmid Mega Kit (QIAGEN) according to the manufacturer’s specifications. HEPES and glucose were purchased from GIBCO. Fluorescent dye YOYO-3 iodide (YOYO-3) was purchased from Molecular Probes. Bright-Glo luciferase assay buffer and substrate were obtained from Promega. CCK-8 was purchased from Dojindo Molecular Technologies. TUNEL apoptosis detection kit (fluorescein isothiocyanate [FITC]-labeled) was purchased from KeyGEN Biotech. Temozolomide was purchased from Meilun Biotech. Dichloromethane (DCM) and n-hexane were purchased from Brenntag. Methanol (MeOH), 1-hydroxy-benzotriazole (HOBt), triisopropylsilane (TIS), 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU), fluorescein sodium (NaF), PhAsO, filipin complex (from Streptomyces filipinesis), colchicine, and other reagents, if not specified, were purchased from Sigma-Aldrich. All of the chemicals were used without further purification.
Synthesis and Characterizations of Precise Oligomers
The polymer, three-arm Stp-His was synthesized as described previously.25 The synthesis of I6P7-Stp-His and I6P7scr-Stp-His was carried out with a SyroWave synthesizer (Biotage) as described in the Supplemental Information.
After synthesis, the oligomers were solubilized in D2O and analyzed by 1H nuclear magnetic resonance (NMR) spectrometry. In addition, the compounds were further analyzed by mass spectrometry.
Preparation of Oligomer/DNA NPs
The precise oligomer/DNA NPs in this study are formed with a bi-component oligomer blend composed of the PEGylated oligomer and the three-arm PEG-free oligomer. The calculated amounts of oligomer blend were mixed together in a total volume of 10 μL of HBG (20 mM HEPES-buffered 5% glucose [pH 7.4]), and 200 ng of DNA was diluted to the same volume of 10 μL of HBG in a separate tube. The oligomer blend was added to the DNA solution and rapidly vortexed for 30 s. NP formation was accomplished after incubation at room temperature for 40 min.
Plasmid DNA was labeled with the fluorescent dye YOYO-3 according to the manufacturer’s protocol. Fluorescent NPs were prepared using YOYO-3-labeled plasmid DNA for specific experiments where indicated.
Agarose Gel Retardation Assay
An agarose gel retardation assay was performed to investigate the binding of DNA by the oligomer blend. The precise oligomer/DNA NPs at various oligomer blend-to-DNA ratios (nitrogen/phosphate [N/P] ratios) were mixed with appropriate amounts of 1× Tris-acetate-EDTA (TAE) loading buffer and then subjected to electrophoresis on 0.7% (w/v) agarose gel containing ethidium bromide (0.25 mg/mL) at a voltage of 80 V for 30 min. The DNA in the gel was visualized by ethidium bromide staining and imaged using the Fluor Chem Imaging System (Alpha Innotech).
Cell Culture
Brain capillary endothelial cells (bEnd.3) and healthy human astrocytes (1800 cells) were both obtained from American Type Culture Collection (ATCC) and routinely cultured in special DMEM supplemented with 20% heat-inactivated fetal bovine serum (FBS), 2 mM L-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, and cultured in incubators maintained at 37°C under a humidified atmosphere containing 5% CO2. Human glioma cells (U87) were obtained from Shanghai Cell Bank, Chinese Academy of Sciences and cultured in DMEM with the addition of 10% FBS, 1% L-glutamine, 1% penicillin, and 1% streptomycin, at 37°C and in 5% CO2 atmosphere.
Quantitative and Qualitative Gene Expression In Vitro
For qualitative analysis of gene expression, U87 cells were seeded in 24-well plates at a density of 4 × 104 cells/well and allowed to grow for 48 hr. After rinsing once with PBS, cells were treated with different NPs containing the plasmid DNA-encoding EGFP (pEGFP), together with oligomer blend at an N/P ratio of 36, at 37°C for 8 hr in the presence of serum-free medium. Subsequently, the transfection agents were replaced by fresh complete culture medium, and the cells were incubated for another 48 hr. Cells were photographed under a DMI4000B inverted fluorescence microscope to acquire the fluorescent images of GFP. Under the same condition, 1800 cells were treated with different NPs at an N/P ratio of 36 as controls to examine the targeting efficiency in vitro.
To quantify the gene expression level, we performed luciferase gene transfections with different NPs on U87 cells. In brief, U87 cells were seeded in 96-well plates at a density of 1 × 104 cells/well and allowed to grow for 48 hr. The transfection procedure was the same as described above, except that the reporter gene construct was changed to pGL6. Two days later, 100 μL of Bright-Glo luciferase assay reagent was added to cells grown in 100 μL of medium. After at least 2 min to allow complete cell lysis, luciferase expression was measured in a Tecan Infinite M1000 Pro microplate reader.
Physicochemical Characterizations of NPs
The mean diameter and zeta potentials of the precise oligomer/DNA NPs at various N/P ratios (12, 24, 36, and 48) were measured by DLS using a Nano-ZS instrument. The shape and morphology of precise oligomer/DNA NPs were observed using TEM (JEOL).
Penetration of Different NPs into U87 Glioma Spheroid
For the construction of in vitro 3D glioma spheroids, U87 cells were seeded onto 2% (w/v) sterilized agarose-coated 48-well plate at a density of 4 × 103 cells/well and incubated for 7 days. Afterward, different YOYO-3-labeled fluorescent NPs at the DNA concentration of 50 μg/mL were added to the wells with U87 spheroids. After 12 hr, the glioma spheroids were rinsed with PBS twice and fixed with 4% paraformaldehyde. Fluorescent images were taken with a laser scanning confocal microscope (Carl Zeiss LSM710; Zeiss).
Exploration of Cellular Uptake Mechanism
U87 cells were incubated with YOYO-3-labeled fluorescent NPs at different temperatures to explore cellular uptake mechanism. In brief, U87 cells were seeded in 24-well plates at a density of 4 × 104 cells/well and allowed to grow for 48 hr. After rinsing once with PBS, cells were treated with different NPs at an N/P ratio of 36 for 1 hr at 4°C and 37°C, respectively. Then cells were rinsed with PBS twice and were photographed with a DMI4000B inverted fluorescent microscope to acquire the red fluorescence images.
To further identify the endocytosis pathways involved in the cellular internalization of I6P7-Stp-His/DNA NPs, we performed cellular association and uptake studies of NPs at an N/P ratio of 36 in the presence of excessive I6P7 peptide or various endocytosis inhibitors. U87 cells were seeded in 24-well plates at a density of 4 × 104 cells/well and incubated for 48 hr. The medium was removed and the cells were pretreated with 100× excess I6P7 peptide for 20 min before YOYO-3-labeled I6P7-Stp-His/DNA NPs. After 1 hr incubation, cells were rinsed twice with PBS and then photographed under a DMI4000B inverted fluorescent microscope to acquire the red fluorescent images. In the case of inhibition groups, different inhibitors including PhAsO, filipin, and colchicine were pre-incubated with cells for 10 min, following the addition of the YOYO-3-labeled I6P7-Stp-His/DNA NPs treated at the condition mentioned above.
Evaluation of Cascade Targeting
In order to establish an in vitro BBB model, we seeded bEnd.3 cells at a density of 4 × 104 cells/cm2 onto polycarbonate 24-well transwell filters (Falcon Cell Culture Insert).26 The culture medium was changed every other day. Seven days later, complete confluency of the BBB model was observed under the microscope. The integrity was monitored using an epithelial volt-ohmmeter (Millicell ERS) to measure the TEER. Only the in vitro BBB model with TEER above 200 Ω cm2 could be used for the following experiment.
Different NPs, in which DNA was labeled by YOYO-3, were added into the donor chamber at the DNA concentration of 50 μg/mL. Simultaneously, NaF (100 μg/mL) was added to monitor the integrity of the BBB model during the whole experiment. Then cells were incubated on a 37°C shaking platform. The volume of a 300 μL sample was taken from each acceptor chamber at 5, 10, 15, 30, and 60 min, and 300 μL of fresh medium was added immediately after each sampling. The BBB transport ratios of NPs were determined by a Tecan Infinite M1000 Pro microplate reader. The Papp was calculated according to Irvine et al.29 as follows:
where is the permeability rate (nmol/s), C0 is the initial concentration (nmol/mL) in the donor chamber, and A is the surface area (cm2) of the membrane filter.35 The data of cumulative transport amount of NPs were also presented.
In order to verify cascade-targeting effects, we established a BBB/U87 cells co-culture model. U87 cells were seeded on the basolateral compartment of the transwell filters at a density of 4 × 104 cells/well, co-cultured with the BBB model for 24 hr when the TEER was sustained over 200 Ω cm2. Different NPs containing pGL6 were added into the donor chamber in the presence of serum-free medium. Eight hours later, all transwell filters were removed and the U87 cells were cultured for another 48 hr. A total of 400 μL of Bright-Glo luciferase assay reagent was added to cells grown in 400 μL of medium. After at least 2 min to allow complete cell lysis, luciferase expression was measured in a Tecan Infinite M1000 Pro microplate reader.
Orthotopic Glioma-Bearing Mouse Model
Nude mice of 20–24 g body weight were purchased from Shanghai Laboratory Animal Center and maintained under standard conditions. All animal experiments were performed in accordance with protocols evaluated and approved by the ethics committee of Fudan University.
The orthotopic glioma-bearing mouse model was established using human U87 glioma cells. In brief, a 5 μL suspension containing 5 × 105 U87 cells was slowly implanted into the right corpus striatum of the nude mice using a stereotactic fixation device with a mouse adaptor. After surgery, the skin incision closed, and mice were further maintained under standard housing conditions.
Immunofluorescence Assay
I6P7scr-Stp-His/pRFP and I6P7-Stp-His/pRFP NPs were injected into the tail veins of orthotopic glioma-bearing mice at a dose of 50 μg of DNA/mouse. Two days after injection, the mice were humanely sacrificed. The brains were harvested, carefully washed with saline, fixed at 4% paraformaldehyde overnight, and dehydrated with 15% sucrose PBS solution until subsidence and then 30% sucrose PBS solution until subsidence. Afterward, the brains were coated with OCT embedding medium, frozen at −80°C, and cut into 20 μm sections. Then the sections were blocked with 10% goat serum at room temperature. One hour later, the sections were incubated with the primary antibody (1:30 dilution of rabbit anti-CD31 polyclonal antibody) overnight at 4°C. After being washed with immunology staining wash buffer for three times, the sections were immediately incubated with the secondary antibody (1:200 dilution of Alexa Fluor 488-conjugated goat anti-rabbit IgG secondary antibody) for 2 hr at room temperature. Followed by three washes with immunology staining wash buffer, the sections were stained with 1 μg/mL DAPI for 15 min. Finally, fluorescent images of the sections were captured through a laser scanning confocal microscope.
In Vivo Biodistribution
The YOYO-3-labeled precise oligomer/DNA NPs were injected into the tail veins of orthotopic glioma-bearing mice at dose of 50 μg of DNA/mouse. Then the in vivo fluorescent imaging was visualized at different time points (0.5, 1, 2, and 3 hr) postinjection using the In Vivo IVIS spectrum imaging system (PerkinElmer). At 3 hr postinjection, the mice were sacrificed and all major organs including brains were harvested; the ex vivo imaging of the organs was also carried out.
CCK-8 Assay
The inhibitory effect of the precise oligomer/DNA NPs at different N/P ratios was evaluated in U87 cells by CCK-8 assay. U87 cells were seeded in 96-well plates at a density of 1 × 104 cells/well and allowed to grow for 48 hr. After rinsing once with PBS, cells were treated with different NPs containing 1 μg of DNA or the same amount of free I6P7 peptide in 100 μL of medium. After 24, 48, and 72 hr, 10 μL of CCK-8 reagent was added to each well and incubated for another 1 hr. The absorbance was measured in a microplate reader (absorption: 450 nm). Corresponding cells without any treatment were used as controls, and the assays were performed in quadruplicate.
In Vivo Anti-glioma Efficacy and H&E Staining
The orthotopic glioma-bearing mice were established as described above. Eleven days after surgery, the mice were intravenously administered with saline, I6P7scr-Stp-His/pING4 and I6P7-Stp-His/pING4 (N/P ratio of 36, containing 50 μg of DNA/dose/mouse) on every other day for three times. For the positive control, mice were intragastrically administered with temozolomide at a dose of 50 mg/kg once a day for five days. Two days after administration of all preparations, two nude mice from each group were sacrificed, and all major organs were harvested. Part of the fresh excised tissues was used to analyze ING4 gene expression by western blot. These excised tissues were lysed using radioimmunoprecipitation (RIPA) lysis buffer. Proteins were separated by 12% SDS-PAGE gels and then transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with 5% defatted milk in 0.5% Tris-buffered saline containing Tween 20 (TBST) for 1 hr and then incubated with primary antibodies ING4 and ACTIN overnight at 4°C. After washing with TBST three times, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 30 min at room temperature. Finally, protein expression levels were visualized by using the enhanced chemiluminescence detection system. Another part was fixed with 4% paraformaldehyde for 48 hr, embedded in paraffin wax, and cut into slices of 4 μm thickness, followed by staining with H&E. In addition, frozen sections (10 μm) were stained using TUNEL apoptosis detection kit according to the manufacturer’s instruction. Finally, all sections were observed under a fluorescent microscope. Eight nude mice from each group were continually maintained to monitor survival and measure the body weight every other day.
Preliminary Safety Assessment
Two days after healthy ICR mice were administered with all preparations, whole blood was collected from the orbit (n = 3). Part of the whole blood was directly used for basic blood tests detected by automated hematology cell analyzer. Another part was centrifuged at 3,000 rpm for 5 min, and the serum was collected carefully and then assayed by IL-6 and IL-10 ELISA kit according to the manufacturer’s instruction.
Statistical Analysis
Statistical analysis was performed with GraphPad InStat 3 software. All quantitative results were carried out as mean ± SD. Statistical comparisons were assessed using two-tailed Student’s t test. The survival data were analyzed with a log rank test by GraphPad Prism 5 and presented as Kaplan-Meier plots.
Author Contributions
S.W., U.L., E.W., and R.H. designed the experiments. S.W., S.R., C.L., M.Q., H.J., and Y.D. performed experiments and data analysis. W.L., E.W., and R.H. obtained grant support for the project. All authors contributed to the writing of the manuscript.
Conflicts of Interest
The authors declare no competing financial interest.
Acknowledgments
This work was supported by grants from the Sino-German Research Project (grant GZ995), National Key Basic Research Program of China (973 Program) (grant 2013CB932502), National Natural Science Foundation of China (grant 81573002), and German Research Foundation DFG Cluster of Excellence “Nanosystems Initiative Munich” (NIM).
Footnotes
Supplemental Information includes Supplemental Materials and Methods and six figures and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.04.023.
Contributor Information
Ernst Wagner, Email: ernst.wagner@cup.uni-muenchen.de.
Rongqin Huang, Email: rqhuang@fudan.edu.cn.
Supplemental Information
References
- 1.Yang H.W., Hua M.Y., Hwang T.L., Lin K.J., Huang C.Y., Tsai R.Y., Ma C.C., Hsu P.H., Wey S.P., Hsu P.W. Non-invasive synergistic treatment of brain tumors by targeted chemotherapeutic delivery and amplified focused ultrasound-hyperthermia using magnetic nanographene oxide. Adv. Mater. 2013;25:3605–3611. doi: 10.1002/adma.201301046. [DOI] [PubMed] [Google Scholar]
- 2.Serwer L.P., James C.D. Challenges in drug delivery to tumors of the central nervous system: an overview of pharmacological and surgical considerations. Adv. Drug Deliv. Rev. 2012;64:590–597. doi: 10.1016/j.addr.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Pardridge W.M. Blood-brain barrier endogenous transporters as therapeutic targets: a new model for small molecule CNS drug discovery. Expert Opin. Ther. Targets. 2015;19:1059–1072. doi: 10.1517/14728222.2015.1042364. [DOI] [PubMed] [Google Scholar]
- 4.Morgan M.A., Parsels L.A., Maybaum J., Lawrence T.S. Improving the efficacy of chemoradiation with targeted agents. Cancer Discov. 2014;4:280–291. doi: 10.1158/2159-8290.CD-13-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nasrollahi F., Varshosaz J., Khodadadi A.A., Lim S., Jahanian-Najafabadi A. Targeted delivery of docetaxel by use of transferrin/poly(allylamine hydrochloride)-functionalized graphene oxide nanocarrier. ACS Appl. Mater. Interfaces. 2016;8:13282–13293. doi: 10.1021/acsami.6b02790. [DOI] [PubMed] [Google Scholar]
- 6.Xie H., Zhu Y., Jiang W., Zhou Q., Yang H., Gu N., Zhang Y., Xu H., Xu H., Yang X. Lactoferrin-conjugated superparamagnetic iron oxide nanoparticles as a specific MRI contrast agent for detection of brain glioma in vivo. Biomaterials. 2011;32:495–502. doi: 10.1016/j.biomaterials.2010.09.024. [DOI] [PubMed] [Google Scholar]
- 7.Yan H., Wang L., Wang J., Weng X., Lei H., Wang X., Jiang L., Zhu J., Lu W., Wei X., Li C. Two-order targeted brain tumor imaging by using an optical/paramagnetic nanoprobe across the blood brain barrier. ACS Nano. 2012;6:410–420. doi: 10.1021/nn203749v. [DOI] [PubMed] [Google Scholar]
- 8.Chen N., Shao C., Qu Y., Li S., Gu W., Zheng T., Ye L., Yu C. Folic acid-conjugated MnO nanoparticles as a T1 contrast agent for magnetic resonance imaging of tiny brain gliomas. ACS Appl. Mater. Interfaces. 2014;6:19850–19857. doi: 10.1021/am505223t. [DOI] [PubMed] [Google Scholar]
- 9.Vallières L., Rivest S. Regulation of the genes encoding interleukin-6, its receptor, and gp130 in the rat brain in response to the immune activator lipopolysaccharide and the proinflammatory cytokine interleukin-1β. J. Neurochem. 1997;69:1668–1683. doi: 10.1046/j.1471-4159.1997.69041668.x. [DOI] [PubMed] [Google Scholar]
- 10.Pan W., Yu C., Hsuchou H., Zhang Y., Kastin A.J. Neuroinflammation facilitates LIF entry into brain: role of TNF. Am. J. Physiol. Cell Physiol. 2008;294:C1436–C1442. doi: 10.1152/ajpcell.00489.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sturzu A., Heckl S. Magnetic resonance imaging of human glioma cells by means of an interleukin-6 receptor-targeted contrast agent. Chem. Biol. Drug Des. 2010;75:369–374. doi: 10.1111/j.1747-0285.2009.00936.x. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka T., Narazaki M., Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014;6:a016295. doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopf M., Baumann H., Freer G., Freudenberg M., Lamers M., Kishimoto T., Zinkernagel R., Bluethmann H., Köhler G. Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature. 1994;368:339–342. doi: 10.1038/368339a0. [DOI] [PubMed] [Google Scholar]
- 14.Kudo M., Jono H., Shinriki S., Yano S., Nakamura H., Makino K., Hide T., Muta D., Ueda M., Ota K. Antitumor effect of humanized anti-interleukin-6 receptor antibody (tocilizumab) on glioma cell proliferation. Laboratory investigation. J. Neurosurg. 2009;111:219–225. doi: 10.3171/2008.12.JNS081284. [DOI] [PubMed] [Google Scholar]
- 15.Kronfol Z., Remick D.G. Cytokines and the brain: implications for clinical psychiatry. Am. J. Psychiatry. 2000;157:683–694. doi: 10.1176/appi.ajp.157.5.683. [DOI] [PubMed] [Google Scholar]
- 16.Su J.L., Lai K.P., Chen C.A., Yang C.Y., Chen P.S., Chang C.C., Chou C.H., Hu C.L., Kuo M.L., Hsieh C.Y., Wei L.H. A novel peptide specifically binding to interleukin-6 receptor (gp80) inhibits angiogenesis and tumor growth. Cancer Res. 2005;65:4827–4835. doi: 10.1158/0008-5472.CAN-05-0188. [DOI] [PubMed] [Google Scholar]
- 17.Gao H., Pan S., Yang Z., Cao S., Chen C., Jiang X., Shen S., Pang Z., Hu Y. A cascade targeting strategy for brain neuroglial cells employing nanoparticles modified with angiopep-2 peptide and EGFP-EGF1 protein. Biomaterials. 2011;32:8669–8675. doi: 10.1016/j.biomaterials.2011.07.069. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y.C., Chiang C.F., Chen L.F., Liang P.C., Hsieh W.Y., Lin W.L. Polymersomes conjugated with des-octanoyl ghrelin and folate as a BBB-penetrating cancer cell-targeting delivery system. Biomaterials. 2014;35:4066–4081. doi: 10.1016/j.biomaterials.2014.01.042. [DOI] [PubMed] [Google Scholar]
- 19.Stock K., Kumar J., Synowitz M., Petrosino S., Imperatore R., Smith E.S., Wend P., Purfürst B., Nuber U.A., Gurok U. Neural precursor cells induce cell death of high-grade astrocytomas through stimulation of TRPV1. Nat. Med. 2012;18:1232–1238. doi: 10.1038/nm.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lächelt U., Kos P., Mickler F.M., Herrmann A., Salcher E.E., Rödl W., Badgujar N., Bräuchle C., Wagner E. Fine-tuning of proton sponges by precise diaminoethanes and histidines in pDNA polyplexes. Nanomedicine (Lond.) 2014;10:35–44. doi: 10.1016/j.nano.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Lo Y.L., Chou H.L., Liao Z.X., Huang S.J., Ke J.H., Liu Y.S., Chiu C.C., Wang L.F. Chondroitin sulfate-polyethylenimine copolymer-coated superparamagnetic iron oxide nanoparticles as an efficient magneto-gene carrier for microRNA-encoding plasmid DNA delivery. Nanoscale. 2015;7:8554–8565. doi: 10.1039/c5nr01404b. [DOI] [PubMed] [Google Scholar]
- 22.Kievit F.M., Veiseh O., Fang C., Bhattarai N., Lee D., Ellenbogen R.G., Zhang M. Chlorotoxin labeled magnetic nanovectors for targeted gene delivery to glioma. ACS Nano. 2010;4:4587–4594. doi: 10.1021/nn1008512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein P.M., Müller K., Gutmann C., Kos P., Krhac Levacic A., Edinger D., Höhn M., Leroux J.C., Gauthier M.A., Wagner E. Twin disulfides as opportunity for improving stability and transfection efficiency of oligoaminoethane polyplexes. J. Control. Release. 2015;205:109–119. doi: 10.1016/j.jconrel.2014.12.035. [DOI] [PubMed] [Google Scholar]
- 24.Maier K., Wagner E. Acid-labile traceless click linker for protein transduction. J. Am. Chem. Soc. 2012;134:10169–10173. doi: 10.1021/ja302705v. [DOI] [PubMed] [Google Scholar]
- 25.Kos P., Lächelt U., Herrmann A., Mickler F.M., Döblinger M., He D., Krhač Levačić A., Morys S., Bräuchle C., Wagner E. Histidine-rich stabilized polyplexes for cMet-directed tumor-targeted gene transfer. Nanoscale. 2015;7:5350–5362. doi: 10.1039/c4nr06556e. [DOI] [PubMed] [Google Scholar]
- 26.Yao H., Wang K., Wang Y., Wang S., Li J., Lou J., Ye L., Yan X., Lu W., Huang R. Enhanced blood-brain barrier penetration and glioma therapy mediated by a new peptide modified gene delivery system. Biomaterials. 2015;37:345–352. doi: 10.1016/j.biomaterials.2014.10.034. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Y., Su C., Zhai H., Tian Y., Sheng W., Miao J., Yang J. Synergistic antitumor effect of adenovirus-mediated hING4 gene therapy and (125)I radiation therapy on pancreatic cancer. Cancer Lett. 2012;316:211–218. doi: 10.1016/j.canlet.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y., Yang J., Sheng W., Xie Y., Liu J. Adenovirus-mediated ING4/PTEN double tumor suppressor gene co-transfer modified by RGD enhances antitumor activity in human nasopharyngeal carcinoma cells. Int. J. Oncol. 2015;46:1295–1303. doi: 10.3892/ijo.2015.2822. [DOI] [PubMed] [Google Scholar]
- 29.Irvine J.D., Takahashi L., Lockhart K., Cheong J., Tolan J.W., Selick H.E., Grove J.R. MDCK (Madin-Darby canine kidney) cells: a tool for membrane permeability screening. J. Pharm. Sci. 1999;88:28–33. doi: 10.1021/js9803205. [DOI] [PubMed] [Google Scholar]
- 30.Schaffert D., Badgujar N., Wagner E. Novel Fmoc-polyamino acids for solid-phase synthesis of defined polyamidoamines. Org. Lett. 2011;13:1586–1589. doi: 10.1021/ol200381z. [DOI] [PubMed] [Google Scholar]
- 31.Schaffert D., Troiber C., Salcher E.E., Fröhlich T., Martin I., Badgujar N., Dohmen C., Edinger D., Kläger R., Maiwald G. Solid-phase synthesis of sequence-defined T-, i-, and U-shape polymers for pDNA and siRNA delivery. Angew. Chem. Int. Ed. Engl. 2011;50:8986–8989. doi: 10.1002/anie.201102165. [DOI] [PubMed] [Google Scholar]
- 32.Brunner S., Sauer T., Carotta S., Cotten M., Saltik M., Wagner E. Cell cycle dependence of gene transfer by lipoplex, polyplex and recombinant adenovirus. Gene Ther. 2000;7:401–407. doi: 10.1038/sj.gt.3301102. [DOI] [PubMed] [Google Scholar]
- 33.Baish J.W., Gazit Y., Berk D.A., Nozue M., Baxter L.T., Jain R.K. Role of tumor vascular architecture in nutrient and drug delivery: an invasion percolation-based network model. Microvasc. Res. 1996;51:327–346. doi: 10.1006/mvre.1996.0031. [DOI] [PubMed] [Google Scholar]
- 34.Garkavtsev I., Kozin S.V., Chernova O., Xu L., Winkler F., Brown E., Barnett G.H., Jain R.K. The candidate tumour suppressor protein ING4 regulates brain tumour growth and angiogenesis. Nature. 2004;428:328–332. doi: 10.1038/nature02329. [DOI] [PubMed] [Google Scholar]
- 35.Xu S., Olenyuk B.Z., Okamoto C.T., Hamm-Alvarez S.F. Targeting receptor-mediated endocytotic pathways with nanoparticles: rationale and advances. Adv. Drug Deliv. Rev. 2013;65:121–138. doi: 10.1016/j.addr.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.