Abstract
Cataract is the most prevalent leading cause of visual impairment and blindness worldwide. In comparison to congenital cataract, which affects relatively few individuals, age-related cataract is responsible for slightly half of all cases of blindness worldwide. Although significant work has been done, the genetic aspect of age-related cataract is still in its infancy. The current study was performed to analyze the mutations and polymorphisms in the CRYAA, CRYAB, CRYBB1, and GJA8 genes in 40 unrelated age-related cataract patients. Mutational analysis of the above-mentioned genes in 40 cataract cases revealed 14 different substitutions of which 8 variants were novel and 6 were reported SNPs. Two disease-causing mutations, g.44590631G>A (p.R65Q) and g.44592224G>A (p.R119H), were also observed in the CRYAA gene. The disease-causing variants mildly affect the stability, functionality, and localization of crystallin, and, with progressing age, a small change in the microenvironment of the crystallin lens occurs. This change in combination with a mutation may significantly alter the functionality of the crystallin protein, leading to age-related cataract.
Key Words: Cataract, Crystallin, India, Lens, Mutation
Cataract formation is one of the leading causes behind visual impairment and blindness worldwide characterized by opacity of the lens that affects vision [Brian and Taylor, 2001]. Cataracts can be congenital in origin or can develop in advanced age (age-related cataract). According to a WHO report, age-related cataract with reduced vision of 3/60 or worse accounts for about 20 million people (48% of world blindness), and it is predicted to reach 50 million by the year 2020 [Asbell et al., 2005; Murthy et al., 2008]. Both congenital and age-related cataract have been associated with genetic and extrinsic factors [McCarty and Taylor, 2001; Hejtmancik and Kantorow, 2004; Iyengar et al., 2004]. Several factors such as UV-B exposure, certain medications, cigarette smoking, diabetes, gout as well as family history are known to be risk factors in the development of age-related cataract [McCarty and Taylor, 2001; Hejtmancik and Kantorow, 2004]. Cataract can be classified morphologically into nuclear, cortical, posterior subcapsular, and mixed types [Hejtmancik and Kantorow, 2004]. Congenital cataract can appear as an isolated abnormality or as part of a syndrome. There are approximately 44 independent loci and 33 genes reported to be involved in nonsyndromic autosomal dominant congenital cataract and can be divided into subgroups of genes such as crystallin, beaded filament structural protein 1 gene (BFSP1), lens intrinsic membrane proteins, heat shock factor proteins, transferases, and gap junction protein alpha 8 gene (GJA8) [Tsai et al., 2003; Iyengar et al., 2004]. In contrast to congenital cataract, only a few loci and genes have been reported for age-related cataract probably due to its complex inheritance pattern [Shiels and Hejtmancik, 2016]. The first gene variant reported as a risk for age-related cataract was galactokinase-1 (GALK1) in a Japanese cohort [Okano et al., 2001]. Apart from this, several other gene variants were also reported as a risk for age-related cataract, namely polymorphisms in EPH receptor A2 (EPHA2), CRYAA, SLC16A12 (monocarboxylate transporter), GJA8, and crystallin βB2 (CRYBB2) [Shiels et al., 2008; Bhagyalaxmi et al., 2009, 2010; Liu et al., 2011; Zhou et al., 2011; Sundaresan et al., 2012; Abplanalp et al., 2013; Liao et al., 2014]. Some previous studies were also carried out to establish the association of GSTM1, GSTT1, DNA repair genes (WRN, XPD, and XRCC1), HSF4, and the kinesin light chain 1 gene (KLC1) with age-related cataract but resulted into inconsistent or inconclusive association [Sun et al., 2010; Padma et al., 2011; Jiang J et al., 2013; Jiang S et al., 2013; Su et al., 2013; Liao et al., 2015].
Lens crystallin has been known to undergo a wide variety of alterations with age, and many of these alterations are accelerated in the presence of oxidative, osmotic, or other stresses [McCarty and Taylor, 2001; Hejtmancik and Kantorow, 2004]. The alterations include proteolysis, an increase in disulfide bridges, deamidation of asparagine and glutamine residues, racemization of aspartic acid residues, phosphorylation, nonenzymatic glycosylation, and carbamylation [Hejtmancik and Kantorow, 2004]. Some mutations in crystallin cause protein aggregation which lead to congenital cataract; however, some mutations make them susceptible to environmental insults such as light and hyperglycemic or oxidative damage, possibly contributing to age-related cataract [Brian and Taylor, 2001; Congdon et al., 2003; Shiels et al., 2010].
Lens crystallins form a target for accumulated damage over the lifetime of an individual, and αA-crystallin's association with age-related cataract has already been established. Therefore, for the present study both α-crystallin genes were selected, i.e., CRYAA and CRYAB encoding αA- and αB-crystallins, respectively, as both proteins share 57% sequence similarity and similar phosphorylation patterns. CRYBB1 is a major subunit of the β-crystallins (9% of the total soluble crystallin), which plays an important role in protein aggregation and orientation. Studies also revealed that the loss of the terminal arm of CRYBB1 can hamper dimerization of the β-crystallins, which causes cataract [Yang et al., 2008]. GJA8 and CRYBB2 have already been reported as susceptible genes for age-related cataracts. In the light of the above-mentioned facts, 4 genes – CRYAA, CRYAB, CRYBB 1, and GJA8 – were selected to screen age-related cataract patients from a northern Indian cohort. The study revealed 14 variants of which 2 were suspected to be pathogenic.
Materials and Methods
A total of 40 patients, ≥40 years of age, with late-onset cataract were recruited from the eastern Uttar Pradesh and western Bihar states of India. Only patients with primary cataract who had visual impairment due to cataract were included in the study. Patients with secondary cataract, i.e., acquired cataract, due to trauma and toxins as well as complicated cataract, due to inflammatory and degenerative ocular diseases, were excluded from the study. In addition, patients with associated conditions such as diabetes, hypertension, myopia, glaucoma, and patients under medications known to be associated with development of cataract (e.g., steroids) were not considered. Controls were also selected from the same population by ruling out the possibility of having cataract, diabetes or any other eye defect in the individuals and their families.
Diagnosis of different types of cataract was done based on the slit-lamp examination following the Lens Opacities Classification System III (LOCS-III). The LOCS-III grading of all patients are shown in Table 1.
Table 1.
Grading of age-related cataract patients according to LOCS-III grading system
| Patient ID | Age, years | Sex | LOCS-III grading |
|---|---|---|---|
| P1 | 58 | M | N3 |
| P2 | 54 | F | P3N2 |
| P3 | 62 | F | N3 |
| P4 | 40 | M | P2N2 |
| P5 | 48 | M | C2N1 |
| P6 | 55 | M | N2C2 |
| P7 | 62 | F | P3N3 |
| P8 | 60 | M | N3 |
| P9 | 65 | M | P2C3 |
| P10 | 50 | M | C3 |
| P11 | 50 | F | P2C1 |
| P12 | 50 | F | P3 |
| P13 | 60 | F | N3 |
| P14 | 60 | M | N2C1 |
| P15 | 70 | F | P2N3C2 |
| P16 | 40 | F | P2N1 |
| P17 | 92 | M | N4 |
| P18 | 40 | M | N3C2 |
| P19 | 74 | M | P2N3C1 |
| P20 | 66 | F | P3N3 |
| P21 | 65 | F | P3C3 |
| P22 | 70 | M | N4 |
| P23 | 70 | F | N3C2 |
| P24 | 42 | M | P2N2 |
| P25 | 45 | M | P3N1 |
| P26 | 65 | F | C2N1 |
| P27 | 70 | F | C3N3 |
| P28 | 50 | M | P2N2 |
| P29 | 52 | M | C2N2 |
| P30 | 75 | F | N3 |
| P31 | 80 | M | P4 |
| P32 | 70 | M | N5 |
| P33 | 58 | F | P2C3 |
| P34 | 47 | F | P1N2 |
| P35 | 60 | F | N3 |
| P36 | 65 | F | C2N2 |
| P37 | 65 | F | N2 |
| P38 | 54 | M | C3N2 |
| P39 | 40 | F | P2N2 |
| P40 | 46 | F | C2N2 |
LOCS III, Lens Opacities Classification System III.
Blood samples and clinical photographs were collected from each patient after receiving written informed consent. Genomic DNA was isolated from 3 to 5 mL peripheral blood according to standard protocol. All the exons as well as exon-intron boundaries of the CRYAA, CRYBB1, CRYAB, and GJA8 genes were PCR amplified and directly resequenced using the genetic analyzer-3130 (Applied Biosystems, Carlsbad, CA, USA) following the manufacturer's protocol. Primer sequences used in this study were taken from Guo et al. [2012]. Two disease-causing variants (g.44590631G>A; p.R65Q and g.44592224G>A; p.R119H) were screened in 160 control individuals from the same population using ARMS PCR. The primer sets used to screen g.44590631G>A were 5′-GGCAGGTGACCGAAGCATC-3′ (forward primer) and 5′-GAAGGCATGGTGCAGGTG-3′ (reverse primer) along with either wild-type allele-specific primer, 5′-CACGTTTGGATTTCAGGTTCG-3′, or mutant allele-specific primer 5′-CACGTTTGGATTTCAGGTTCA-3′. Primer sets used to screen g.44592224G> A were 5′-GCAGCTTCTCTGGCATGG-3′ (forward primer) and 5′-GGGAAGCAAAGGAAGACAGA-3′ (reverse primer) along with either wild-type allele-specific primer, 5′-AGTTCCACCGCCGCTACCG-3′, or mutant allele-specific primer 5′-AGTTCCACCGCCGCTACCA-3′.
Sequencing data were analyzed by several in silico analysis tools as mentioned below. The effect of each SNP was predicted by in silico analysis using MutationTaster and other prediction tools such as SIFT, Panther, SNAP, Polyphen-1, Polyphen-2, PhD-SNP, MAPP, and PredictSNP. The Exome Variant Server (http://evs.gs.washington.edu/EVS/), dbSNP (www.ncbi.nlm.nih.gov/SNP), 1000 Genomes (http://www.internationalgenome.org/), and ExAC (http://exac.broadinstitute.org/) were used to determine the frequency of sequence alteration in the population.
The comparison of the protein sequence with the genomic DNA sequence was performed using GeneWise (http://www.ebi.ac.uk/Tools/psa/genewise/) to reveal the altered amino acid due to nucleotide substitution. In silico nucleotide to protein translation (comparing normal and mutant protein) was done using the online tool EMBOSS Sixpack (http://www.ebi.ac.uk/Tools/st/emboss_sixpack./). The tertiary structure of protein was downloaded from the Protein Data Bank (http://www.pdb.org) and visualized on the standalone tool DeepView – Swiss-PdbViewer (http://spdbv.vital-it.ch/).
Results and Discussion
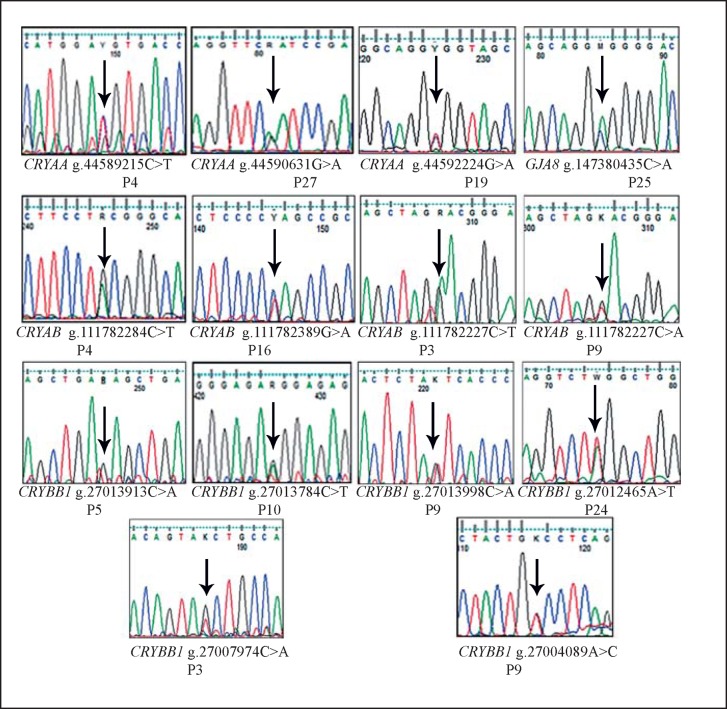
Resequencing of the CRYAA, CRYBB1, CRYAB, and GJA8 genes identified 14 single nucleotide substitutions of which 6 were reported and 8 were novel (Fig. 1; Table 2). In CRYAA, 3 single nucleotide substitutions were identified, of which 2 were predicted to be disease causing (g.44590631G>A; p.R65Q in patient P27 and g.44592224G>A; p.R119H in patient P19) by MutationTaster as well as other prediction tools. These 2 disease-causing variants were not found in 160 unrelated control individuals from the same population.
Fig. 1.
Electropherograms of all 14 variants observed during screening of the CRYAA, CRYBB1, CRYAB, and GJA8 genes in cataract patients.
Table 2.
Single nucleotide substitutions observed in age-related cataract patients while screening of CRYAA, CRYAB, CRYBB1, and GJA8 genes
| No. | Patient ID | Gene | Genomic coordinate | Amino acid change | Database status | In silico prediction |
|---|---|---|---|---|---|---|
| 1 | P4, P6, P7, P11, P12, P13, P14, P19, P25, P29, P38, P40 | CRYAA | g.44589215C>T | p.D2D | reported, rs872331 | polymorphism |
| 2 | P27 | CRYAA | g.44590631G>A | p.R65Q | reported, rs199640007 | disease causing |
| 3 | P19 | CRYAA | g.44592224G>A | p.R119H | novel | disease causing |
| 4 | P4, P13, P20, P34 | CRYAB | g.111782284C>T | p.L55L | reported, rs2228387 | polymorphism |
| 5 | P16, P29, P32 | CRYAB | g.111782389G>A | p.P20P | reported, rs4252582 | polymorphism |
| 6 | P2, P3, P5 | CRYAB | g.111782227C>T | intron | novel | polymorphism |
| 7 | P9, P10, P12, P15, P17, P18, P21, P22, P23, P24, P30, P31 | CRYAB | g.111782227C>A | intron | novel | polymorphism |
| 8 | P5 | CRYBB1 | g.27013913C>A | intron | novel | polymorphism |
| 9 | P10 | CRYBB1 | g.27013784C>T | intron | reported, rs77926469 | polymorphism |
| 10 | P9 | CRYBB1 | g.27013998C>A | 5′UTR | novel | polymorphism |
| 11 | P11, P24, P25, P26, P13, P15, P40 | CRYBB1 | g.27012465A>T | intron | novel | polymorphism |
| 12 | P3 | CRYBB1 | g.27007974C>A | intron | novel | polymorphism |
| 13 | P9 | CRYBB1 | g.27004089A>C | intron | reported, rs5761626 | polymorphism |
| 14 | P25 | GJA8 | g.147380435C>A | p.A118E | novel | polymorphism |
Clinical History and in silico Analysis of Mutational Effects
CRYAA, g.44590631G>A (p.R65Q, rs199640007)
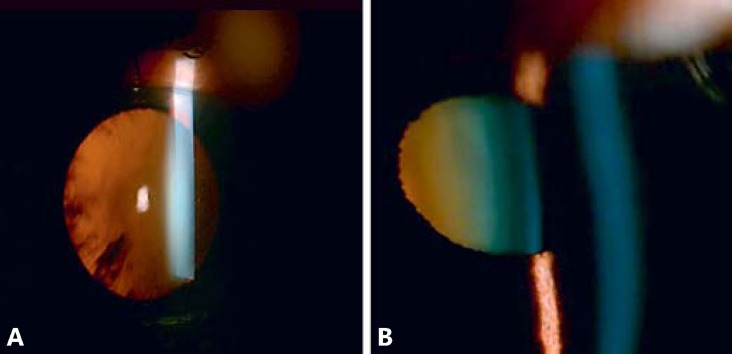
The 70-year-old female patient (P27) presented with bilateral cataract (LOCS III grading C3N3; Fig. 2A). The age of onset was 55 years as stated by the patient. Her mother was also affected by late-onset cataract, but her 2 brothers and their children were not affected. She has 2 children, 42 and 45 years old, but they were not available for testing.
Fig. 2.
Clinical photographs of cataract patients. A Slit-lamp image of patient P27 showing C3N3 cataract by LOCS-III grading. B Slit-lamp image of patient P19 with P2N3C1 cataract.
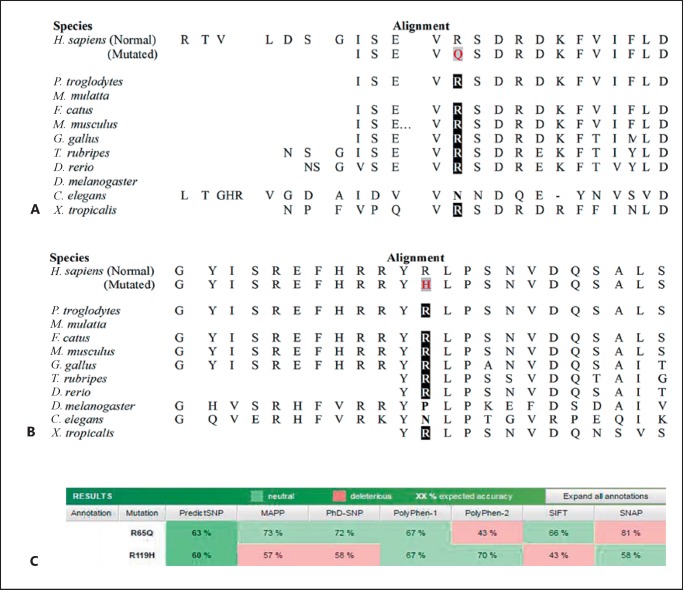
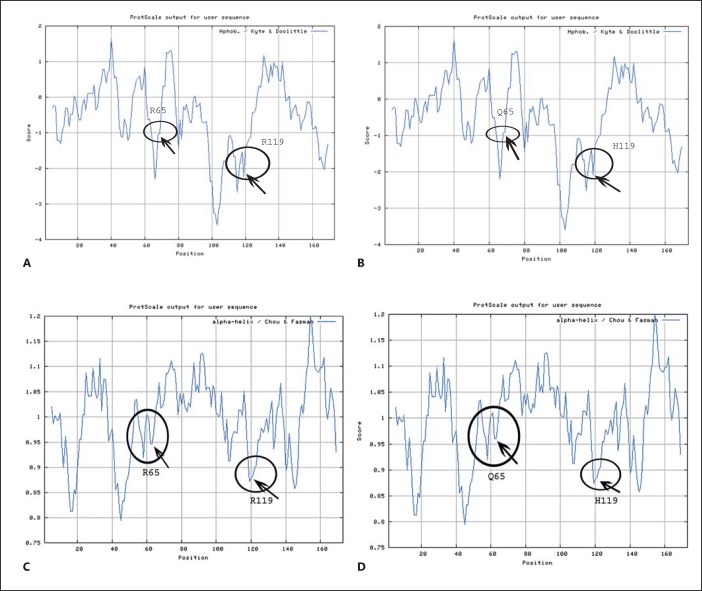
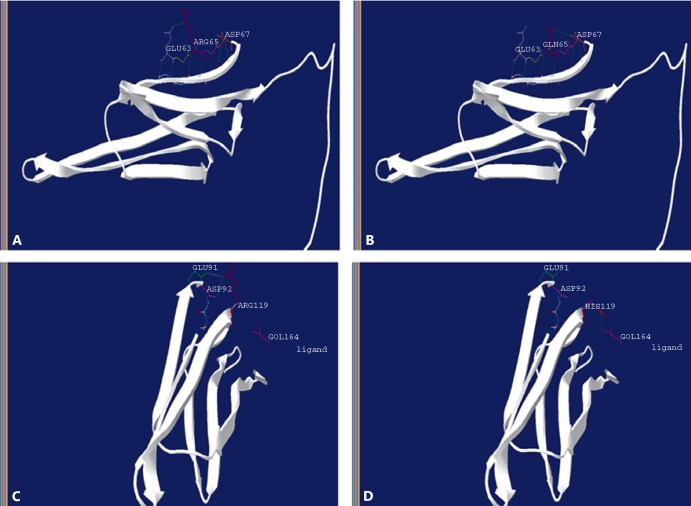
CRYAA screening detected the nonsynonymous variant g.44590631G>A. The allele frequency of the variant analyzed from 1000 Genomes and ExAC revealed that the frequency of the mutant allele A was very low (0.0006 and 0.0005453, respectively) and was present only in the South Asian population. Phylogenetic analysis of the variant across species revealed that the arginine at position 65 was evolutionarily conserved (Fig. 3A, C). Other properties of the protein such as hydrophobicity and helix formation were measured using ProtScale, which revealed a mild alteration in the protein microenvironment due to mutation. This mildly altered microenvironment might have a severe effect later in life due to change in the lens microenvironment that happens with ageing (Fig. 4). Furthermore, the tertiary structure of CRYAA was predicted by ModBase to study the effect of amino acid alteration on its protein structure. The amino acid substitution of R with Q results in a break of the hydrogen bond (H-bond) Glu63 and formation of a new H-bond with Asp67 in the CRYAA protein as predicted in silico (Fig. 5A, B).
Fig. 3.
A The multiple sequence alignment of the p.R65Q mutant protein reveals the conservation of arginine at this position throughout evolution. B Multiple sequence alignment of the p.R119H mutant CRYAA protein reveals the conservation of R119 across species. C In silico prediction of both variants using prediction tools: SIFT, SNAP, Polyphen-1, Polyphen-2, PhD-SNP, MAPP, and PredictSNP.
Fig. 4.
Hydrophobicity profile of wild-type and mutant (p.R65Q and p.R119H) CRYAA proteins are shown predicted by the ProtScale program at the ExPASy server. A The circles represent the hydrophobicity around R65 and R119 in the wild-type protein. B The circles represent the hydrophobicity around Q65 and H119 in the mutant protein showing a mild shift in hydrophobicity compared to the wild type. C Secondary structure formation (alpha helix in this case) property of wild-type and mutant protein predicted by ProtScale program using Chau-Fasman algorithm. D Alpha helix-forming property of each amino acid in the wild-type protein.
Fig. 5.
The tertiary structure of the CRYAA protein predicted by ModBase and visualized on the standalone tool DeepView – Swiss-PdbViewer is shown. A The wild-type CRYAA protein with R65 hydrogen (H)-bonded with Glu63. B The mutant Q65 CRYAA protein formation of a new H-bond occurs with Asp67; the previous H-bonding with Glu63 was diminished. C The wild-type CRYAA protein with R119 has 2 H-bonds with Glu91 and Asp92. D The mutant CRYAA protein with H119 results in a breakage of both H-bonds with Glu91 and Asp92 and creates an additional bond with a ligand.
CRYAA, g.44592224G>A (p.R119H)
The 74-year-old male patient (P19) was clinically diagnosed with bilateral cataract (LOCS III grading P2N3C1; Fig. 2B). The age of onset was 58 years as stated by the patient. He has no family history of cataract, and his 3 children were also unaffected. This could be a case of sporadic age-related cataract, although his children could manifest cataract later in life regarding they are all younger than 50 years.
The nonsynonymous novel single nucleotide substitution in exon 3 of the CRYAA gene causes p.R119H in the patient. Arginine at position 119 was also almost conserved across species except in a few (Fig. 3B, C). The analysis of alteration in hydrophobicity and helix-forming properties of the protein due to this mutation revealed minor changes (Fig. 4). Despite this, in silico analysis of the tertiary structure of the protein revealed major changes in the intramolecular H-bonding pattern due to the p.R119H shift. Wild-type R119H bonded with Glu91 and Asp92, while mutant H119 devastated both H-bonding and created an additional bond with a ligand (Fig. 5C, D). The p.R119H not only disturbed intramolecular H-bonding, but also interfered with the protein-ligand interaction which may create a disturbance in the functionality of the protein. An independent study on congenital nuclear cataract by Li et al. [2010] revealed a heterozygous p.R116C mutation, while Zhang et al. [2011] revealed another mutation, p.R116H, in the CRYAA gene as a potential cause for congenital anterior polar cataract. Although the identified p.R119H variant lies very close to these reported mutations and did not exert its effect immediately, it is probable that it will generate a similar abnormal microenvironment in advanced age, leading to age-related cataract [Pang et al., 2010].
α-Crystallins are molecular chaperones thought to allow the lens to tolerate aging-induced deterioration of the lens proteins. Besides the chaperone's activity, its other functions include remodeling and protection of the cytoskeleton, inhibition of apoptosis, and enhancing the resistance of cells to stress [Andley, 2007]. The fact that the 2 alterations, p.R65Q and p.R119H, probably will interfere with the protein stability and solubility may lead to the development of age-related cataract, not evident in early age of the patients.
The SNP g.44589215C>T (rs872331) was one of the most commonly (in 12 patients) observed variations in patients from the present study as well as in other reported populations (Table 3). In CRYBB1, 6 different substitutions were identified in either UTR or intronic regions, which are predicted to be polymorphic. In the CRYAB gene, 4 different substitutions, g.111782284C>T (rs2228387), g.111782389G>A (rs4252582), g.111782227C>T and g.111782227C>A, were identified. The frequency of the 2 CRYAB variants, i.e., rs2228387 and rs4252582, were very low in various reported populations. In the future, it will be of interest to understand the role of the reported variants (rs4252582 and rs2228387) and novel variants (GJA8; g.147380435C>A) in age-related cataract because the current study did not cover the case-control association study.
Table 3.
Minor allele frequency of nonsynonymous variants in different populations reported on ExAC browser
| Population | rs872331 | rs199640007 | rs2228387 | rs4252582 |
|---|---|---|---|---|
| South Asian | 0.3605 | 0.0005453 | 0.0315 | 0.04843 |
| African | 0.7359 | 0 | 0.003963 | 0 |
| East Asian | 0.1993 | 0 | 0.0002774 | 0 |
| European (Finnish) | 0.5787 | 0 | 0.02085 | 0.0003249 |
| European (non-Finnish) | 0.608 | 0 | 0.02323 | 0.0001922 |
| Latino | 0.4638 | 0 | 0.005371 | 0.00164 |
| Other | 0.5509 | 0 | 0.0176 | 0.009091 |
| Total minor allele frequency | 0.5404 | 7.46e-05 | 0.01935 | 0.008093 |
Here, we reported 2 disease-causing mutations and 12 polymorphic variants in 4 genes in age-related cataract cases from a north Indian cohort.
Statement of Ethics
The study was approved by the institutional ethical committee of the Institute of Medical Sciences, Banaras Hindu University, Varanasi, India.
Disclosure Statement
The authors have no conflicts of interest to declare.
Acknowledgments
The authors are grateful to the patients for their participation in this study. We would like to thank Dr. Samir for his help during sample collection, Ms. Pooja Chauhan and Mr. Chandra Bhan Singh for their support during experimental work, and Dr. V.P. Singh for his support. The Council of Scientific and Industrial Research (CSIR), New Delhi, is highly acknowledged for providing a Junior Research and Senior Research Fellowship to R.P. The authors also thank the anonymous reviewers for their critical reading and suggestions to improve the manuscript.
References
- Abplanalp J, Laczko E, Philp NJ, Neidhardt J, Zuercher J, et al. The cataract and glucosuria associated monocarboxylate transporter MCT12 is a new creatine transporter. Hum Mol Genet. 2013;22:3218–3226. doi: 10.1093/hmg/ddt175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andley UP. Crystallins in the eye: function and pathology. Prog Retin Eye Res. 2007;26:78–98. doi: 10.1016/j.preteyeres.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Asbell PA, Dualan I, Mindel J, Brocks D, Ahmad M, Epstein S. Age-related cataract. Lancet. 2005;365:599–609. doi: 10.1016/S0140-6736(05)17911-2. [DOI] [PubMed] [Google Scholar]
- Bhagyalaxmi SG, Srinivas P, Barton KA, Kumar KR, Vidyavathi M, et al. A novel mutation (F71L) in αA-crystallin with defective chaperone-like function associated with age-related cataract. Biochimica Biophysica Acta. 2009;1792:974–981. doi: 10.1016/j.bbadis.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagyalaxmi SG, Padma T, Reddy GB, Reddy KR. Association of G>A transition in exon-1 of alpha crystallin gene in age-related cataracts. Oman J Ophthalmol. 2010;3:7–12. doi: 10.4103/0974-620X.60014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian G, Taylor H. Cataract blindness-challenges for the 21st century. Bull World Health Organ. 2001;79:249–256. [PMC free article] [PubMed] [Google Scholar]
- Congdon NG, Friedman DS, Lietman T. Important causes of visual impairment in the world today. JAMA. 2003;290:2057–2060. doi: 10.1001/jama.290.15.2057. [DOI] [PubMed] [Google Scholar]
- Guo Y, Su D, Li Q, Yang Z, Ma Z, et al. A nonsense mutation of CRYGC associated with autosomal dominant congenital nuclear cataracts and microcornea in a Chinese pedigree. Mol Vis. 2012;18:1874–1880. [PMC free article] [PubMed] [Google Scholar]
- Hejtmancik JF, Kantorow M. Molecular genetics of age-related cataract. Exp Eye Res. 2004;79:3–9. doi: 10.1016/j.exer.2004.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar SK, Klein BE, Klein R, Jun G, Schick JH, et al. Identification of a major locus for age-related cortical cataract on chromosome 6p12-q12 in the Beaver Dam Eye study. Proc Natl Acad Sci USA. 2004;101:14485–14490. doi: 10.1073/pnas.0400778101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Zhou J, Yao Y, Zhu R, Liang C, et al. Copy number variations of DNA repair genes and the age-related cataract: Jiangsu Eye Study. Invest Ophthalmol Vis Sci. 2013;54:932–938. doi: 10.1167/iovs.12-10948. [DOI] [PubMed] [Google Scholar]
- Jiang S, Hu N, Zhou J, Zhang J, Gao R, et al. Polymorphisms of the WRN gene and DNA damage of peripheral lymphocytes in age-related cataract in a Han Chinese population. Age (Dordr) 2013;35:2435–2444. doi: 10.1007/s11357-013-9512-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FF, Yang M, Ma X, Zhang Q, Zhang M, et al. Autosomal dominant congenital nuclear cataracts caused by a CRYAA gene mutation. Curr Eye Res. 2010;35:492–498. doi: 10.3109/02713681003624901. [DOI] [PubMed] [Google Scholar]
- Liao J, Su X, Chen P, Wang X, Xu L, et al. Meta-analysis of genome-wide association studies in multiethnic Asians identifies two loci for age-related nuclear cataract. Hum Mol Genet. 2014;23:6119–6128. doi: 10.1093/hmg/ddu315. [DOI] [PubMed] [Google Scholar]
- Liao RF, Ye MJ, Liu CY, Ye DQ. An updated meta-analysis: risk conferred by glutathione S-transferases (GSTM1 and GSTT1) polymorphisms to age-related cataract. J Ophthalmol. 2015;2015:103950. doi: 10.1155/2015/103950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ke M, Yan M, Guo S, Mothobi ME, et al. Association between gap junction protein-alpha 8 polymorphisms and age-related cataract. Mol Biol Rep. 2011;38:1301–1307. doi: 10.1007/s11033-010-0230-z. [DOI] [PubMed] [Google Scholar]
- McCarty CA, Taylor HR. The genetics of cataract. Invest Ophthalmol Vis Sci. 2001;42:1677–1678. [PubMed] [Google Scholar]
- Murthy G, Gupta SK, John N, Vashist P. Current status of cataract blindness and Vision 2020: the right to sight initiative in India. Indian J Ophthalmol. 2008;56:489–494. doi: 10.4103/0301-4738.42774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano Y, Asada M, Fujimoto A, Ohtake A, Murayama K, et al. A genetic factor for age-related cataract: identification and characterization of a novel galactokinase variant, “Osaka,” in Asians. Am J Hum Genet. 2001;68:1036–1042. doi: 10.1086/319512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padma G, Mamata M, Reddy KR, Padma T. Polymorphisms in two DNA repair genes (XPD and XRCC1) – association with age-related cataracts. Mol Vis. 2011;17:127–133. [PMC free article] [PubMed] [Google Scholar]
- Pang M, Su JT, Feng S, Tang ZW, Gu F, et al. Effects of congenital cataract mutation R116H on αA-crystallin structure, function and stability. Biochim Biophys Acta. 2010;1804:948–956. doi: 10.1016/j.bbapap.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Shiels A, Hejtmancik JF. Mutations and mechanisms in congenital and age-related cataracts. Exp Eye Res. 2016 doi: 10.1016/j.exer.2016.06.011. S0014-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels A, Bennett TM, Knopf HL, Maraini G, Li A, et al. The EPHA2 gene is associated with cataracts linked to chromosome 1p. Mol Vis. 2008;14:2042–2055. [PMC free article] [PubMed] [Google Scholar]
- Shiels A, Bennett TM, Hejtmancik JF. Cat-Map: putting cataract on the map. Mol Vis. 2010;16:2007–2015. [PMC free article] [PubMed] [Google Scholar]
- Su S, Yao Y, Zhu R, Liang C, Jiang S, et al. The associations between single nucleotide polymorphisms of DNA repair genes, DNA damage, and age-related cataract: Jiangsu Eye Study. Invest Ophthalmol Vis Sci. 2013;54:1201–1207. doi: 10.1167/iovs.12-10940. [DOI] [PubMed] [Google Scholar]
- Sun L, Xi B, Yu L, Gao XC, Shi DJ, et al. Association of glutathione S-transferases polymorphisms (GSTM1 and GSTT1) with senile cataract: a meta-analysis. Invest Ophthalmol Vis Sci. 2010;51:6381–6386. doi: 10.1167/iovs.10-5815. [DOI] [PubMed] [Google Scholar]
- Sundaresan P, Ravindran RD, Vashist P, Shanker A, Nitsch D, et al. EPHA2 polymorphisms and age-related cataract in India. PLoS One. 2012;7 doi: 10.1371/journal.pone.0033001. e33001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SY, Cheng CY, Hsu WM, Su TP, Liu JH, Chou P. Association between visual impairment and depression in the elderly. J Formos Med Assoc. 2003;102:86–90. [PubMed] [Google Scholar]
- Yang J, Zhu Y, Gu F, He X, Cao Z, et al. A novel nonsense mutation in CRYBB1 associated with autosomal dominant congenital cataract. Mol Vis. 2008;14:727–731. [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang Y, Liu P, Cao W, Tang X, Su S. Congenital anterior polar cataract associated with a missense mutation in the human alpha crystallin gene CRYAA. Mol Vis. 2011;17:2693–2697. [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Wang B, Hu S, Zhang C, Ma X, Qi Y. Genetic variations in GJA3 GJA8 LIM2, and age-related cataract in the Chinese population: a mutation screening study. Mole Vis. 2011;17:621–626. [PMC free article] [PubMed] [Google Scholar]