Abstract
Importance
The association of biomarkers with patient survival after recurrence (SAR) is poorly understood, yet may guide management and treatment.
Objective
To determine the association of DNA mismatch repair (MMR) status and somatic mutations in BRAFV600E or KRAS (exon 2) in the primary tumor with SAR in patients with stage III colon carcinomas treated with adjuvant FOLFOX-based chemotherapy.
Design
Tumor biomarkers were analyzed in relationship to SAR in participants in adjuvant chemotherapy trials.
Intervention
Patients with resected, stage III colon cancers who were randomized to adjuvant FOLFOX ± cetuximab (NCCTG N0147) or FOLFOX ± bevacizumab (NSABP C-08).
Main Outcome Measure(s)
Associations of biomarkers with SAR were analyzed using Cox proportional hazards models adjusted for clinicopathological features and time-to-recurrence. The interaction effect of primary tumor sidedness on the association of biomarkers with SAR was determined.
Results
Among patients with cancer recurrence [N0147 (N=871); C-08 (N= 524)], multivariable analysis revealed that those whose tumors had deficient (d) vs proficient (p) MMR had significantly better SAR (adjusted hazard ratio [HRadj.], 0.70, 95% CI, 0.52 - 0.96, adjusted P [Padj.] <.029). Patients whose tumors harbored mutant BRAFV600E (HRadj., 2.45, 95% CI, 1.85 - 3.25, Padj.<.0001) or mutant KRAS (HRadj., 1.21, 95% CI, 1.00 - 1.47, Padj.=0.052) had worse SAR compared to tumors that had wild-type copies of both genes, although only results for BRAFV600E achieved statistical significance. Significant interactions were found for MMR (Padj.=.029) and KRAS (Padj.=.025) by primary tumor site for SAR. Improved SAR was observed for patients with dMMR tumors of the proximal vs distal colon (HRadj., 0.57, 95% CI, 0.40 - 0.83, Padj. =.003), and worse SAR for mutant KRAS tumors of the distal colon (codon 12: HRadj., 1.76, 95% CI, 1.30 - 2.38, Padj. =.0003; codon 13: HRadj., 1.76, 95% CI, 1.08 - 2.86, Padj. =.022].
Conclusions and Relevance
In patients with recurrence, dMMR was significantly associated with better SAR and this benefit was limited to primary tumors of the proximal colon. Mutations in BRAFV600E were significantly associated with worse SAR, and worse SAR for BRAFV600E or KRAS mutant tumors was more strongly associated with distal cancers. These biomarkers have implications for patient management at recurrence.
Trial Registration
NCCTG NO147, NCT00079274; NSABP C-08, NCT00096278
Introduction
Prognostic biomarkers in patients with tumor recurrence have the potential to influence management and treatment decisions. Approximately 30% of patients with stage III colon carcinoma will experience recurrence of their disease despite adjuvant chemotherapy1. Studies have shown that DNA mismatch repair (MMR) status and mutations in BRAFV600E or KRAS genes can provide prognostic information in patients with stage III disease2. However, the association of biomarkers with survival after recurrence (SAR) remains poorly understood, and studies have been underpowered given the relatively low frequency of these alterations and modest rates of tumor recurrence.
In stage III patients who participated in adjuvant chemotherapy trials, those whose tumors showed dMMR or microsatellite instability (MSI) have generally had better clinical outcomes compared to those with proficient (p) MMR or microsatellite stability3. However, the association of dMMR/MSI with prognosis is less robust in stage III vs stage II disease4, and limited data exist in patients treated with standard adjuvant FOLFOX in contrast to 5-fluorouracil alone5-8. As in patients with metastatic disease9, BRAFV600E mutations have been shown to be significantly associated with poorer survival10-12 with a stronger impact seen for overall survival (OS) compared to disease-free (DFS) or progression-free survival13 (PFS) for reasons that remain unclear. Since BRAFV600E mutations are significantly enriched in sporadic colon cancers with dMMR/MSI (due to epigenetic inactivation of MLH1)14,15, the combined MMR/BRAF variable may be more informative than either alone. In this regard, a new consensus guideline for the molecular testing of colorectal cancer (CRC) recommends that BRAF be analyzed in conjunction with MMR for prognostic stratification. Data for the association of KRAS mutation with clinical outcome have been less consistent than for BRAFV600E 13,16-18. In participants in the North Central Cancer Treatment Group (NCCTG) N0147 and the Pan European Trial Adjuvant Colon Cancer (PETACC)-8 adjuvant trials, stage III colon cancers with mutant vs wild-type (WT) KRAS had poorer DFS rates 16,19.
We studied the association of MMR and mutations in BRAFV600E or KRAS in the primary tumor with SAR in participants in the NCCTG N0147 and the National Surgical Adjuvant Breast and Bowel Project (NSABP) C-08 adjuvant chemotherapy trials. These trials evaluated FOLFOX chemotherapy alone or combined with cetuximab (N0147)20 or bevacizumab (NSAPB C-08)21 where neither antibody significantly improved patient outcome vs FOLFOX alone. We also determined whether the association of biomarkers with SAR depended on primary tumor site within the colon given recent data suggesting prognostic differences by tumor site6,22.
Materials and Methods
The study population consists of patients with stage III colon adenocarcinoma who developed recurrence during participation in phase III adjuvant chemotherapy studies NCCTG N0147[N= 871]20 and NSABP C-08 [N=524]21. We categorized primary tumor site as located proximal to, or at or distal to the splenic flexure. Each trial was approved by the respective Institutional Review Boards (IRB) and by the NCCTG (now part of Alliance for Clinical Trials in Oncology) or NSABP (now part of NRG Oncology). Each participant signed an IRB-approved, protocol-specific informed consent document. Data quality was ensured by review by the Statistics and Data Center of the Alliance or NRG, and by the study chairpersons per established policies.
Molecular Testing
DNA MMR proteins MLH1, MSH2, and MSH6 were analyzed in FFPE tumor tissues from the N0147 trial as previously described10; MLH1 and MSH2 expression were analyzed in tumors from C-08 as reported23. MMR protein loss was defined as the absence of nuclear staining in tumor cells in the presence of nuclear staining in normal colonic epithelium and lymphocytes. Tumors with loss of an MMR protein were categorized as having deficient (d) MMR, those with intact expression as having proficient (p) MMR. All biomarker assays were interpreted with investigators blinded to patient outcomes.
BRAFV600E and KRAS mutation status were determined using genomic DNA extracted from macrodissected FFPE tumor tissue collected prospectively. In N0147, testing for the BRAF c.1799T>A (V600E) mutation in exon 15 was performed using a multiplex allele-specific PCR–based assay and an automated sequencing technique, as previously described10. KRAS exon 2 mutation status was analyzed using the DxS Mutation Test Kit KR-03/04 (DxS), assessing for seven different mutations in codons 12 and 1316. In N0147, molecular analyses was performed in a Clinical Laboratory Improvement Amendments (CLIA)-compliant laboratory. Mutation profiling of tumor specimens from C-08 was performed using OncoCarta and ColoCarta panel assays, with the running of samples on the MassSpec platform as described previously23.
Statistical Analysis
SAR, defined as the time from recurrence to death due to any cause, was the primary study outcome. Due to the potential for significant confounding, all analyses were based on multivariable models that were adjusted for clinicopathological variables, time-to-recurrence, and biomarkers. The distribution of SAR between patient subgroups by biomarkers was estimated based on direct adjusted survival curves24-26. Since initial results showed significant differences in SAR among the four arms of the two adjuvant chemotherapy trials (p = 0.026), multivariable Cox models (stratified by the four treatment groups) were applied to assess the impact of biomarkers on SAR among patients with recurrence. Models were adjusted for age, sex, performance score, initial T/N stage, histologic grade, time from initial treatment to recurrence, primary tumor site, and biomarkers when applicable. The proportional hazard assumption was confirmed by examination of Schoenfeld residuals plot. Interaction effects of the primary tumor site on the impact of biomarkers on SAR were determined. Subgroup analyses were performed when there were statistically significant interaction effects. Association analyses were performed in patients from the mFOLFOX6 alone treatment arms from both studies due to clinical relevance. Two-sided P values are reported; values < 0.05 were considered statistically significant and were not adjusted for multiple comparisons. Analyses were performed using SAS version 9.4 (SAS Institute Inc., NC).
Results
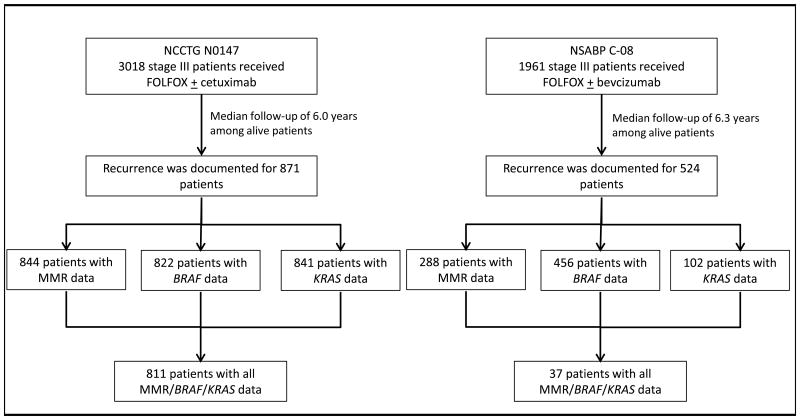
Among the adjuvant trial participants, 3018 patients received mFOLFOX6 ± cetuximab (NCCTG N0147)20 and 1961 patients received mFOLFOX6 ± bevacizumab (NSABP C-08)21. At a median follow-up of 6.0 years (N0147) and 6.3 years (C-08), 871 and 524 patients from each study, respectively, had a documented first recurrence and are included in this report. Among these patients, 848 had complete and available data on MMR and the mutational status of BRAF and KRAS genes (Fig. 1).
Figure 1.
Consort flow diagram of the study population.
Molecular Markers and SAR
The multivariable associations of patient demographics and clinicopathological features, adjusting for biomarkers (MMR, KRAS and BRAF), with SAR are presented in Table 1. Patients with distal tumors had significantly better SAR than did patients with proximal tumors (adjusted hazard ratio [HRadj.], 0.70, 95% confidence interval (CI), 0.58 - 0.84, adjusted P value [Padj.]=.0002). Longer time-to-recurrence (TTR) following primary resection was associated with significantly better SAR (for one year delay, HRadj. 0.79, 95% CI, 0.72 - 0.87, Padj<.0001)[Table 1]. In addition, patient performance score, N stage, and histologic grade were significantly associated with SAR. Among patients who experienced recurrence, those whose tumors showed pMMR (vs dMMR) or had wild-type (WT) KRAS and BRAF (vs either mutated) had significantly longer median TTR (Supplemental Table 1).
Table 1.
Multivariable*$ associations between patient demographics and disease characteristics with survival after recurrence (SAR), adjusting for biomarkers (MMR, KRAS, and BRAF).
| Biomarkers | N of patients (%) | HR | 95% CI | P-value |
|---|---|---|---|---|
| Age, 10 year increase | 832 | 1.06 | 0.98-1.15 | 0.17 |
|
| ||||
| Sex | ||||
| Female | 387 (46.5%) | 0.90 | 0.76-1.06 | 0.21 |
| Male | 445 (53.5%) | Ref | ||
|
| ||||
| Performance Score | ||||
| 0 | 629 (75.6%) | Ref | ||
| 1 | 198 (22.7%) | 1.23 | 1.01-1.49 | 0.037 |
| 2 | 5 (0.6%) | 7.97 | 3.19-19.88 | <.0001 |
|
| ||||
| T-stage | ||||
| T1/2 | 47 (5.6%) | Ref | ||
| T3 | 631 (75.8%) | 1.32 | 0.88-1.99 | 0.1794 |
| T4 | 154 (18.5%) | 1.41 | 0.91-2.19 | 0.1264 |
|
| ||||
| N-stage | ||||
| N1 | 339 (40.7%) | Ref | ||
| N2 | 493 (59.3%) | 1.39 | 1.17-1.66 | 0.0002 |
|
| ||||
| Primary tumor site | ||||
| Distal | 384 (46.2%) | 0.70 | 0.58-0.84 | 0.0002 |
| Proximal | 448 (53.8%) | Ref | ||
|
| ||||
| Histologic grade | ||||
| Low grade (1-2) | 587 (70.6%) | Ref | ||
| High grade (3/4/anaplastic) | 245 (29.4%) | 1.40 | 1.17-1.68 | 0.0003 |
|
| ||||
| Time-to-recurrence, 1 year increase | 832 | 0.79 | 0.72-0.87 | <.0001 |
Abbreviations: HR, hazard ratio; CI, confidence interval.
Multivariable model in 832 patients includes complete data on all covariates (age, sex, performance score, T-stage, N-stage, primary tumor site, histologic grade, time-to-recurrence, MMR, KRAS, and BRAF. The HR, 95% CI and p-value associated with MMR, KRAS, BRAF are presented in Table 2.
Stratified Cox models with four treatment arms as individual strata.
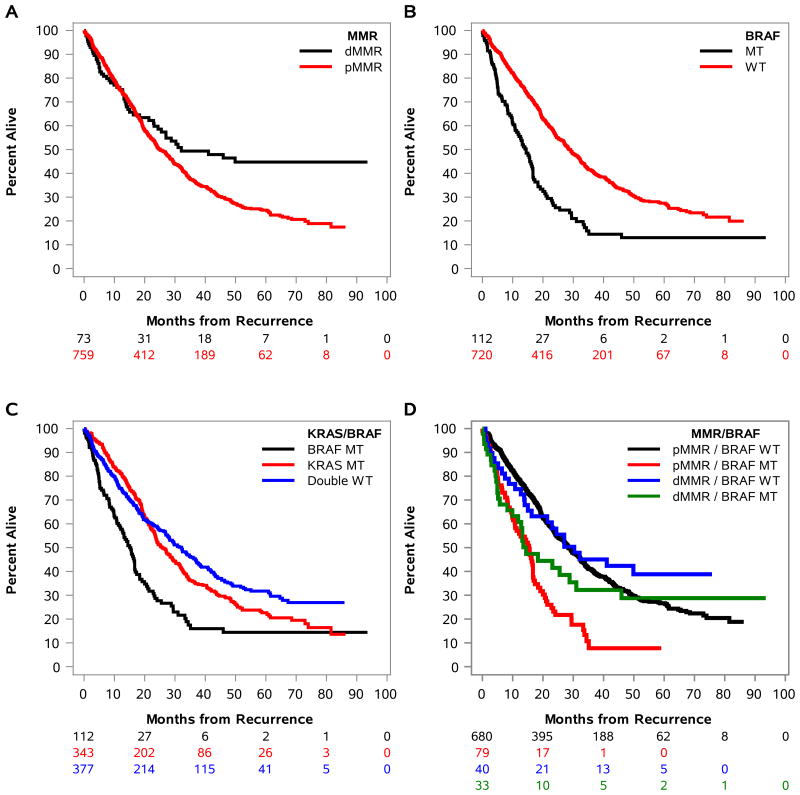
Multivariable associations of molecular markers with SAR are shown in Table 2. After adjustment for covariates including TTR after primary treatment, patients with dMMR vs pMMR tumors had significantly better SAR (HRadj., 0.70, 95% CI, 0.52 - 0.96, Padj=.028) [Fig. 2A, Table 2]. Patients with BRAFV600E mutant tumors had significantly worse SAR compared to those whose tumors had wild-type (WT) BRAF (HRadj., 2.45, 95% CI, 1.85 - 3.25, Padj.<.0001) [Fig. 2B,C, Table 2]. Given that MMR status and BRAFV600E are strongly associated, we analyzed MMR/BRAF as a combined variable. Patients whose tumors had dMMR or pMMR plus mutant BRAFV600E had similarly poor adjusted median SAR times of 14.5 (HRadj., 1.52, 95% CI, 0.99- 2.34, Padj =.058) and 15.4 months (HRadj., 2.64, 95% CI, 1.96 - 3.57, Padj.<.0001), respectively, and shorter SAR compared to pMMR/WT BRAF (referent) [Table 2, Fig. 2D]. In contrast, patients whose tumors had dMMR or pMMR with WT BRAF showed better SAR with 30.3 and 28.4 month adjusted median SAR, respectively, and there was no statistical difference between these two groups (Table 2, Fig. 2D). Within the subset of dMMR tumors, we observed that those with BRAF mutations had significantly poorer SAR compared to those with WT BRAF (HRadj., 2.70, 95% CI, 1.23 – 5.93, Padj= .0136)[Table 2]. Patients whose tumors harbored KRAS exon 2 mutations had shorter SAR compared to those whose tumors were WT for KRAS and BRAF (25.9 vs 32.1 months; p=.052)[Table 2]. When KRAS was analyzed by codon 12 or 13 mutations vs WT KRAS, the associations did not reach statistical significance (Table 2). Patients whose tumors had both WT BRAF and WT KRAS had the longest SAR (adjusted median of 32.1 months) of all groups that was significantly improved compared to patients whose tumors had BRAFV600E mutation (15 months; p<.0001) [Table 2].
Table 2.
| Biomarkers | Events/N | Adjusted median time in months (95% CI)1,2 | HR | 95% CI | P-value | Interaction P-value with tumor Site | Interaction P-value with treatment |
|---|---|---|---|---|---|---|---|
| MMR | 0.029 | 0.0026 | |||||
| dMMR | 54/73 | 32.1 (24.4-NR) | 0.70 | 0.52-0.96 | 0.028 | ||
| pMMR | 521/759 | 25.1 (23.3-28.3) | Ref | ||||
|
| |||||||
| KRAS | 0.025 | 0.23 | |||||
| Codon 12 MT | 201/275 | 23.8 (21.2-27.2) | 1.20 | 0.98-1.47 | 0.076 | ||
| Codon 13 MT | 55/68 | 27.2 (22.0-33.8) | 1.24 | 0.91-1.67 | 0.17 | ||
| WT | 319/489 | 28.1 (24.1-32.3) | Ref | ||||
|
| |||||||
| BRAF | 0.056 | 0.57 | |||||
| MT | 102/112 | 14.5 (10.8-16.7) | 2.45 | 1.85-3.25 | <.0001 | ||
| WT | 473/720 | 28.7 (26.8-32.1) | Ref | ||||
|
| |||||||
| KRAS/BRAF | 0.0005 | 0.67 | |||||
| BRAF MT | 102/112 | 15.0 (12.1-16.9) | 2.45 | 1.85-3.25 | <.0001 | ||
| KRAS MT | 256/343 | 25.9 (23.5-29.4) | 1.21 | 1.00-1.47 | 0.052 | ||
| Both WT | 217/377 | 32.1 (27.6-37.1) | Ref | ||||
|
| |||||||
| MMR/BRAF | 0.076 | 0.016 | |||||
| MT BRAF dMMR | 28/33 | 14.5 (11.8-45.9) | 1.52 | 0.99-2.34 | 0.058 | ||
| WT BRAF dMMR | 26/40 | 30.3 (21.4-NR) | 0.85 | 0.56-1.28 | 0.43 | ||
| MT BRAF pMMR | 74/79 | 15.4 (10.8-16.7) | 2.64 | 1.96-3.57 | <.0001 | ||
| WT BRAF pMMR | 447/680 | 28.4 (26.2-31.9) | Ref | ||||
| BRAF in dMMR patients | |||||||
| MT BRAF | 28/33 | 2.70 | 1.23-5.93 | 0.0136 | |||
| WT BRAF | 26/40 | Ref | |||||
Abbreviations: HR, hazard ratio; CI, confidence interval; MT, mutant; WT, wild-type; NR, not reached
Adjusting for age, sex, performance score, T/N stage, primary tumor site, histologic grade, biomarkers (when applicable), and time-to-recurrence
Stratified Cox models with four treatment arms as individual strata.
Based on direct adjusted survival curves from Cox model
Median not calculated for BRAF in dMMR patients due to small sample size (model convergence failed)
Figure 2.
In patients with stage III colon carcinoma treated with FOLFOX-containing adjuvant therapy, direct adjusted plots of survival after recurrence (SAR) are shown by DNA mismatch repair (MMR) status (A), mutated vs wild-type BRAF (B) or KRAS (C), and the combined variables of KRAS/BRAF (D), or MMR/BRAF (E). MUT: mutant; WT: wild-type. MMR status: deficient (d) or proficient (p).
Analysis by Primary Tumor Site
Based on statistically significant interactions between biomarkers and primary tumor site for SAR (Table 2), we separately examined the associations between biomarkers and SAR among patients with proximal or distal tumors (Table 3, Suppl. Fig. 1). After adjustment for covariates, patients with dMMR tumors of the proximal but not the distal colon had significantly better SAR [HR adj., 0.57, 95% CI, 0.40 - 0.83, Padj.=.0028] (Table 3, Suppl. Fig. 1A), interaction p =.029 (Table 2). Patients with BRAFV600E mutated tumors had significantly shorter SAR for both proximal (HR adj., 1.90, 95% CI 1.37 – 2.64,, Padj. = 0.0001; Suppl. Fig. 1D) and distal cancers (HRadj., 5.84, 95% CI 3.27 – 10.43,Padj. < 0.0001) versus those whose tumors had WT BRAF (Suppl. Fig. 1B) or WT BRAF/WT KRAS (Suppl. Fig. 1C)[Table 3], although the interaction between BRAF and primary tumor site for SAR did not achieve significance (Padj. = 0.056) [Table 2]. A significant interaction was observed for KRAS mutations (codon 12, 13) [P adj. =.025] and the combined KRAS/BRAF variable (p=.0005) with primary tumor site for SAR (Table 2). Compared to tumors with WT KRAS, patients whose tumors harbored KRAS mutations at codon 12 (HR adj., 1.76, 95% CI, 1.30 - 2.38, Padj. =0.0003) or codon 13 (HR adj., 1.76, 95% CI, 1.08 - 2.86, Padj. = 0.022) each had significantly worse SAR among distal, but not proximal cancers (Table 3, Suppl. Fig. 1C). For the combined MMR/BRAF variable, the adjusted median SAR was shorter for patients with dMMR and mutant BRAFV600E tumors of the distal vs proximal colon (5.7 vs 14.5 months). Furthermore, dMMR and mutant BRAFV600E tumors in the distal colon had significantly shorter SAR than did patients with pMMR and WT BRAF tumors (HRadj., 9.38, 95% CI, 3.23 - 27.28, Padj. < 0.0001; Table 3, Suppl. Fig. 1D).
Table 3.
Adjusted associations between biomarkers and survival after recurrence (SAR) among patients by primary tumor site*$
| Proximal tumor | Distal tumor | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Biomarkers | Events/N | Adjusted median time in months (95% CI)1 | HR | 95% CI | P-value | Events/N | Adjusted median time in months (95% CI)1 | HR | 95% CI | P-value |
| MMR | ||||||||||
| dMMR | 41/55 | 30.3 (16.3-NR) | 0.57 | 0.40-0.83 | 0.0028 | 13/18 | 27.0 (24.4-NR) | 1.26 | 0.69-2.28 | 0.45 |
| pMMR | 308/393 | 19.0 (17.2-20.6) | Ref | 213/366 | 36.6 (33.3-43.3) | Ref | ||||
|
| ||||||||||
| KRAS | ||||||||||
| Codon 12 MT | 120/162 | 21.8 (19.7-25.3) | 0.85 | 0.65-1.12 | 0.26 | 81/113 | 27.0 (23.2-34.4) | 1.76 | 1.30-2.38 | 0.0003 |
| Codon 13 MT | 35/43 | 21.9 (16.6-30.2) | 0.91 | 0.61-1.36 | 0.65 | 20/25 | 33.8 (24.4-48.1) | 1.76 | 1.08-2.86 | 0.022 |
| WT | 194/243 | 16.7 (14.5-19.4) | Ref | 125/246 | 43.3 (37.1-55.1) | Ref | ||||
|
| ||||||||||
| BRAF | ||||||||||
| MT | 85/94 | 13.2 (9.9-16.7) | 1.90 | 1.37-2.64 | 0.0001 | 17/18 | 11.3 (6.5-23.4) | 5.84 | 3.27-10.43 | <.0001 |
| WT | 264/354 | 21.8 (19.3-23.8) | Ref | 209/366 | 39.2 (33.9-44.2) | Ref | ||||
|
| ||||||||||
| KRAS/BRAF | ||||||||||
| BRAF MT | 85/94 | 12.8 (9.9-16.0) | 1.90 | 1.37-2.64 | 0.0001 | 17/18 | 16.7 (6.6-25.5) | 5.84 | 3.27-10.43 | <.0001 |
| KRAS MT | 155/205 | 23.3 (21.1-27.2) | 0.87 | 0.67-1.12 | 0.28 | 101/138 | 29.1 (24.5-36.3) | 1.76 | 1.32-2.34 | <.0001 |
| Both WT | 109/149 | 19.1 (14.7-26.8) | Ref | 108/228 | 45.4 (38.5-61.4) | Ref | ||||
|
| ||||||||||
| MMR/BRAF | ||||||||||
| MT BRAF dMMR | 24/29 | 14.5 (11.8-NR) | 1.08 | 0.66-1.75 | 0.76 | 4/4 | 5.7 (2.8-NR) | 9.38 | 3.23-27.28 | <.0001 |
| WT BRAF dMMR | 17/26 | 30.3 (16.3-NR) | 0.59 | 0.35-0.99 | 0.047 | 9/14 | 27.0 (24.4-NR) | 1.13 | 0.56-2.29 | 0.73 |
| MT BRAF pMMR | 61/65 | 15.0 (9.9-16.7) | 1.92 | 1.35-2.73 | 0.0003 | 13/14 | 14.5 (6.6-NR) | 5.38 | 2.81-10.31 | <.0001 |
| WT BRAF pMMR | 247/328 | 20.7 (19.0-23.5) | Ref | 200/352 | 39.2 (34.4-44.2) | Ref | ||||
Abbreviations: HR, hazard ratio; CI, confidence interval; MT, mutant; WT, wild-type; NR, not reached
Adjusting for age, sex, performance score, T/N stage, tumor site, histologic grade, biomarkers (when applicable), and time-to-recurrence
Stratified Cox models with four arms as individual stratums.
Based on direct adjusted survival curves from Cox model
Analysis by Study Treatment Arm
A statistically significant interaction was observed between the study treatment arm and MMR status (Padj. =.0026), and for the combined variable of MMR/BRAF (Padj. =.016) for SAR [Table 2]. The significantly favorable impact of dMMR on SAR shown in multivariable analysis was evident in the FOLFOX arms from both adjuvant trials (HR adj., 0.50, 95% CI, 0.31 - 0.81, Padj. = 0.0043, but was not observed in the FOLFOX + cetuximab arm of the N0147 trial (HR adj., 1.19, 95% CI, 0.78 - 1.82, Padj. = 0.43) [Suppl. Table 2]. An association of mutant BRAFV600E with significantly poorer SAR was observed in patients treated with FOLFOX alone or combined with cetuximab. However, patients whose cancers were dMMR and mutant BRAFV600E showed significantly poorer SAR when cetuximab was added to FOLFOX (HR adj., 2.95, 95% CI, 1.64 - 5.32, Padj. = 0.0003), but not in patients whose tumors were treated with FOLFOX alone (HR adj., 1.03, 95% CI, 0.52 - 2.01, Padj. = 0.94) [Suppl. Table 2]. A similar effect was observed for tumors with KRAS codon 12 mutations whereby their SAR was worse than in patients with WT KRAS tumors when treated with FOLFOX plus cetuximab, but not FOLFOX alone (Suppl. Table 2). Among patients with KRAS WT tumors, no differences in SAR were observed within proximal or distal primary tumors by treatment arm.
Discussion
We determined the impact of biomarkers on SAR in stage III colon cancer patients who participated in two large adjuvant chemotherapy trials of FOLFOX-containing therapy. In the overall cohort, patients whose tumors had mutant BRAF had significantly worse SAR with a 14.2 month decrease in adjusted median survival time compared to WT BRAF tumors. This result can explain, at least in part, prior data showing that mutant BRAFV600E was more strongly associated with OS compared to DFS or relapse-free survival in the N01476 and Pan European Trial Adjuvant Colon Cancer (PETACC)-3 adjuvant chemotherapy trials, respectively13. Furthermore, these findings suggest that the impact of BRAFV600E mutation on tumor aggressiveness is enhanced at the time of tumor recurrence since recurrence of these tumors led to accelerated patient mortality. In this regard, patients whose tumors harbored BRAFV600E mutations had a ∼3-fold increase in early peritoneal metastases compared to those patients whose tumors showed WT BRAF in the stage III N0147 cohort27. These data are consistent with other reports showing adverse outcome 28 and significantly higher rates of peritoneal and distant lymph node metastases among BRAFV600E mutant metastatic CRCs9.
Among patients with dMMR tumors, we found that their adjusted median SAR was 7 months longer than patients with pMMR tumors indicating a clinically significant survival advantage for this patient subset. This finding is consistent with the longer recurrence-free interval (i.e, TTR) observed for dMMR vs pMMR tumors in the overall study cohort. Importantly, the analysis was adjusted for covariates that included BRAF mutation status, TTR, and primary tumor site which were the variables whose inclusion in the multivariable model had the greatest impact on SAR in dMMR tumors. The longer SAR for patients with dMMR tumors may be explained, in part, by the increase in recurrence rates at regional vs distant sites, such as the liver, that was observed in the N0147 cohort27. Among patients whose tumors had mutant KRAS, a trend was seen toward poorer SAR that did not reach statistical significance for codon 12 or 13 mutations.
Sporadic colon cancers with dMMR are highly enriched with BRAFV600E mutations5,14, and a forthcoming consensus guideline recommends that BRAFV600E mutation testing be done in conjunction with MMR analysis for prognostic stratification. A similarly poor SAR was observed for patients with BRAFV600E mutant dMMR or pMMR cancers with 14.5 and 15.4 month adjusted median SAR, respectively. In contrast, patients whose tumors had WT BRAF showed significantly better SAR with 30.3 (for dMMR) and 28.4 (for pMMR) month adjusted median SAR, respectively. Therefore, the mutational status of BRAF is an important determinant of SAR that confers adverse outcome in patients with both dMMR and pMMR cancers. In a pooled analysis of stage II and III patients from the NSABP C-07 and C-08 adjuvant studies where dMMR was associated with a lower rate of tumor recurrence, a trend toward worse SAR was seen for patients with dMMR colon cancers, although the analysis was not adjusted for BRAF 23. The authors, however, postulated that the association of dMMR with shorter SAR was due to mutant BRAFV600E since patients with BRAFV600E mutant tumors had significantly shorter SAR23. In another study of patients with stage I-IV colorectal cancers, transcriptomic data were used to categorize tumors into four consensus molecular subtypes (CMS). The CMS1 subtype was enriched for tumors with MSI-H and BRAFV600E mutations, and patients with these tumors had a poorer SAR compared to the other three subtypes (CMS I-III) by univariate analysis29. However, the study data used to generate CMS were not adjusted for BRAF (or KRAS) status nor for TTR which was strongly associated with SAR as shown in our dataset.
We observed a statistically significant interaction between biomarkers (MMR, KRAS) and primary tumor site for SAR. The significant association of dMMR with better SAR was limited to cancers of the proximal vs distal colon. While not prognostic overall, analysis of KRAS mutations by primary tumor site revealed a significantly shorter SAR for patients with distal but not proximal cancers. This finding for SAR is consistent with TTR data from the N0147 cohort where the association of KRAS mutations with TTR and OS was stronger in patients with distal cancers6. Conversely and relevant to anti-EGFR therapy, patient tumors with WT KRAS alleles had significantly better SAR for distal vs proximal cancers. However, stage III patients with WT KRAS tumors treated with FOLFOX + cetuximab vs FOLFOX had similar SAR irrespective of tumor site. In patients with metastatic CRC, a recent report suggests that distal cancers respond more favorably to cetuximab than do proximal tumors (CALGB 80405)30. Patients whose tumors harbored mutations in BRAFV600E had significantly poorer SAR independent of primary site, yet the association was stronger for distal tumors. Of note, an association between primary tumor site and SAR was also been seen in stage III colon cancer patients treated with non oxaliplatin-containing chemotherapy in the PETACC-3 study31. Factors not studied in our report that may contribute to observed differences in prognosis by tumor site include epigenetic16 and/or other genomic31 alterations that may be embryologically influenced since the origin of the proximal colon is from the midgut and distal colon from the hindgut. In addition, microbial composition or metabolites may be relevant factors. Analysis of the associations between biomarkers and SAR by study treatment arm revealed that the better SAR for patients with dMMR tumors seen among FOLFOX-treated patients did not extend to those who also received cetuximab for reasons that are unclear. Due to the modest number of patients with complete biomarker data in C-08, results for SAR from the FOLFOX + bevacizumab study arm are not reported.
Strengths of our study include the two clinical trial cohorts receiving standard adjuvant FOLFOX-based chemotherapy with mature recurrence and survival data. All molecular analyses were performed on prospectively collected biospecimens. Our study findings are relevant to clinical practice in that National Comprehensive Cancer Network (NCCN) guidelines and a forthcoming consensus guideline recommend testing of all newly diagnosed CRCs for expanded RAS and BRAFV600E mutations in combination with MMR/MSI for prognostic stratification and identification of Lynch Syndrome patients. Study limitations include the fact that biomarkers were analyzed in only a subset of the C-08 cohort and that KRAS testing was limited to exon 2. However, a recent study found that clinicopathologic features, survival outcomes, and gene expression profiles were similar between patients whose CRC harbored KRAS codon 12/13 mutations and those with KRAS 61/146 or NRAS mutations32. Analysis of biomarkers by tumor site for SAR resulted in some small patient subsets for which cautious interpretation of the data is warranted. Lastly, no data were available on patient treatment after tumor recurrence for which we cannot exclude an impact on SAR.
In conclusion, the association of dMMR with more favorable SAR suggests that some of these patients may be candidates for an aggressive surgical approach at recurrence. Furthermore, therapy with an immune checkpoint inhibitor is a new therapeutic option in patients with metastatic dMMR/MSI CRCs where impressive tumor responses and extended PFS were observed33. In patients with both dMMR and pMMR tumors, BRAFV600E mutations were associated with significantly poorer SAR indicating the need for novel therapies in this subset33,34. The significant interactions of MMR and KRAS mutation status with SAR by primary tumor site indicates that these biomarkers should be interpreted in this context. Taken together, these data have important implications for stage III colon cancer patients at the time of tumor recurrence where they can be utilized to inform clinical decision-making.
Supplementary Material
Suppl. Figure 1. Direct adjusted plots of survival after recurrence (SAR) for molecular biomarkers by anatomic site of the primary tumor. Proximal (left side) vs distal (right side) tumor site for status of DNA mismatch repair (MMR) (A), BRAF (B), and combined variables of KRAS/BRAF (C), or MMR/BRAF (D). MUT: mutant; WT: wild-type. MMR status: deficient (d) or proficient (p).
Acknowledgments
The authors with to acknowledge the patient accrual contributions of the following individuals: Daniel M. Anderson, M.D. (Department of Hematology, Oncology, and Transplantation, University of Minnesota, Minneapolis, MN), Anthony F. Shields, M.D. (Karmanos Cancer Institute, Wayne State University, Detroit, MI), Morton S. Kahlenberg, M.D. (Surgical Oncology Associates of South Teas, San Antonio, TX), Emily Chan, M.D. (Division of Hematology/Oncology, Vanderbilt University, Nashville, TN), and Sharlene Gill, M.D. (Division of Medical Oncology, University of British Columbia, Vancouver, BC, Canada).
Research support: This work was supported by a National Cancer Institute Senior Scientist Award (grant number K05CA142885; to F.A. Sinicrope). The study was also supported, in part, by grants from the National Cancer Institute to the North Central Cancer Treatment Group (NCCTG; grant number U10CA025224); the NCCTG Biospecimen Resource (grant number U24CA114740); the Alliance for Clinical Trials in Oncology (grant number U10CA-031946); the Alliance Statistics and Data Center (grant number U10CA-033601); the National Surgical Adjuvant Breast and Bowel Project (NSABP) (grant numbers U10CA-180868, UG1CA-189867,U24CA-196067, U10CA-180822), and a grant from the Pennsylvania Department of Health (which specifically disclaims responsibility for any analyses, interpretations or conclusions). The study was also supported, in part, by unrestricted support from Sanofi U.S., Pfizer, Inc. and Imclone Systems, Inc.
Footnotes
The corresponding author had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. The authors report no conflicts of interest with the content of this manuscript.
Contributor Information
Frank A. Sinicrope, Departments of Medicine and Oncology, Mayo Clinic, Rochester, MN
Qian Shi, Biomedical Statistics and Informatics, Mayo Clinic, Rochester, MN
Carmen J. Allegra, Division of Hematology and Oncology, University of Florida, Gainesville, FL
Thomas C. Smyrk, Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN
Stephen N. Thibodeau, Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN
Richard M. Goldberg, Ohio State University Comprehensive Cancer Center, Columbus OH
Jeffrey P. Meyers, Biomedical Statistics and Informatics, Mayo Clinic, Rochester, MN
Kay L. Pogue-Geile, Pathology Laboratory, National Surgical Adjuvant Breast and Bowel Project (NSABP)/NRG Oncology, Pittsburgh, PA
Greg Yothers, NRG Oncology and Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA
Daniel J. Sargent, Biomedical Statistics and Informatics, Mayo Clinic, Rochester, MN
Steven R. Alberts, Department of Oncology, Mayo Clinic, Rochester, MN
References
- 1.Shi Q, Andre T, Grothey A, et al. Comparison of outcomes after fluorouracil-based adjuvant therapy for stages II and III colon cancer between 1978 to 1995 and 1996 to 2007: evidence of stage migration from the ACCENT database. J Clin Oncol. 2013 Oct 10;31(29):3656–3663. doi: 10.1200/JCO.2013.49.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaanan A, Bachet JB, Andre T, Sinicrope FA. Prognostic impact of deficient DNA mismatch repair and mutations in KRAS, and BRAFV600E in patients with lymph node-positive colon cancer. Curr Colorectal Cancer Rep. 2014 Sep 1;10(3):346–353. doi: 10.1007/s11888-014-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawakami H, Zaanan A, Sinicrope FA. Microsatellite instability testing and its role in the management of colorectal cancer. Curr Treat Options Oncol. 2015 Jul;16(7):30. doi: 10.1007/s11864-015-0348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klingbiel D, Saridaki Z, Roth AD, Bosman FT, Delorenzi M, Tejpar S. Prognosis of stage II and III colon cancer treated with adjuvant 5-fluorouracil or FOLFIRI in relation to microsatellite status: results of the PETACC-3 trial. Ann Oncol. 2015 Jan;26(1):126–132. doi: 10.1093/annonc/mdu499. [DOI] [PubMed] [Google Scholar]
- 5.Sinicrope FA, Shi Q, Smyrk TC, et al. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology. 2015 Jan;148(1):88–99. doi: 10.1053/j.gastro.2014.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinicrope FA, Mahoney MR, Yoon HH, et al. Analysis of molecular markers by anatomic tumor site in stage III colon carcinomas from adjuvant chemotherapy Trial NCCTG N0147 (Alliance) Clin Cancer Res. 2015 Dec 1;21(23):5294–5304. doi: 10.1158/1078-0432.CCR-15-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gavin PG, Paik S, Yothers G, Pogue-Geile KL. Colon cancer mutation: prognosis/prediction--response. Clin Cancer Res. 2013 Mar 1;19(5):1301. doi: 10.1158/1078-0432.CCR-13-0020. [DOI] [PubMed] [Google Scholar]
- 8.Andre T, de Gramont A, Vernerey D, et al. Adjuvant fluorouracil, leucovorin, and oxaliplatin in stage II to III colon cancer: Updated 10-year survival and outcomes according to BRAF mutation and mismatch repair status of the MOSAIC Study. J Clin Oncol. 2015 Dec 10;33(35):4176–4187. doi: 10.1200/JCO.2015.63.4238. [DOI] [PubMed] [Google Scholar]
- 9.Tran B, Kopetz S, Tie J, et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer. 2011 Oct 15;117(20):4623–4632. doi: 10.1002/cncr.26086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinicrope FA, Mahoney MR, Smyrk TC, et al. Prognostic impact of deficient DNA mismatch repair in patients with stage III colon cancer from a randomized trial of FOLFOX-based adjuvant chemotherapy. J Clin Oncol. 2013 Oct 10;31(29):3664–3672. doi: 10.1200/JCO.2013.48.9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farina-Sarasqueta A, van Lijnschoten G, Moerland E, et al. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol. 2010 Dec;21(12):2396–2402. doi: 10.1093/annonc/mdq258. [DOI] [PubMed] [Google Scholar]
- 12.Ogino S, Shima K, Meyerhardt JA, et al. Predictive and prognostic roles of BRAF mutation in stage III colon cancer: results from intergroup trial CALGB 89803. Clin Cancer Res. 2012 Feb 1;18(3):890–900. doi: 10.1158/1078-0432.CCR-11-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010 Jan 20;28(3):466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 14.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012 Jul 19;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006 Jul;38(7):787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 16.Yoon HH, Tougeron D, Shi Q, et al. KRAS codon 12 and 13 mutations in relation to disease-free survival in BRAF-wild-type stage III colon cancers from an adjuvant chemotherapy trial (N0147 alliance) Clin Cancer Res. 2014 Jun 1;20(11):3033–3043. doi: 10.1158/1078-0432.CCR-13-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchins G, Southward K, Handley K, et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011 Apr 1;29(10):1261–1270. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 18.Imamura Y, Morikawa T, Liao X, et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clin Cancer Res. 2012 Sep 1;18(17):4753–4763. doi: 10.1158/1078-0432.CCR-11-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taieb J, Tabernero J, Mini E, et al. Oxaliplatin, fluorouracil, and leucovorin with or without cetuximab in patients with resected stage III colon cancer (PETACC-8): an open-label, randomised phase 3 trial. Lancet Oncol. 2014 Jul;15(8):862–873. doi: 10.1016/S1470-2045(14)70227-X. [DOI] [PubMed] [Google Scholar]
- 20.Alberts SR, Sargent DJ, Nair S, et al. Effect of oxaliplatin, fluorouracil, and leucovorin with or without cetuximab on survival among patients with resected stage III colon cancer: a randomized trial. JAMA. 2012 Apr 4;307(13):1383–1393. doi: 10.1001/jama.2012.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allegra CJ, Yothers G, O'Connell MJ, et al. Bevacizumab in stage II-III colon cancer: 5-year update of the National Surgical Adjuvant Breast and Bowel Project C-08 trial. J Clin Oncol. 2013 Jan 20;31(3):359–364. doi: 10.1200/JCO.2012.44.4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popovici V, Budinska E, Bosman FT, Tejpar S, Roth AD, Delorenzi M. Context-dependent interpretation of the prognostic value of BRAF and KRAS mutations in colorectal cancer. BMC Cancer. 2013;13:439. doi: 10.1186/1471-2407-13-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gavin PG, Colangelo LH, Fumagalli D, et al. Mutation profiling and microsatellite instability in stage II and III colon cancer: an assessment of their prognostic and oxaliplatin predictive value. Clin Cancer Res. 2012 Dec 1;18(23):6531–6541. doi: 10.1158/1078-0432.CCR-12-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makuch RW. Adjusted survival curve estimation using covariates. J Chronic Dis. 1982;35(6):437–443. doi: 10.1016/0021-9681(82)90058-3. [DOI] [PubMed] [Google Scholar]
- 25.Gail MH, Byar DP. Variance calculations for direct adjusted survival curves, with applications to testing for no treatment effect. Biometrical J. 1986;28(5):487–599. [Google Scholar]
- 26.Zhang X, Loberiza FR, Klein JP, Zhang MJ. A SAS macro for estimation of direct adjusted survival curves based on a stratified Cox regression model. Comput Methods Programs Biomed. 2007 Nov;88(2):95–101. doi: 10.1016/j.cmpb.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Wilcox RE, Shi Q, Sinicrope FA, et al. Influence of molecular alterations on site-specific (ss) time to recurrence (TTR) following adjuvant therapy in resected colon cancer (CC) (Alliance Trial N0147) J Clin Oncol. 2015 May 20;33(15) [Google Scholar]
- 28.Richman SD, Seymour MT, Chambers P, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009 Dec 10;27(35):5931–5937. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- 29.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015 Nov;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venook AP, Niedzwiecki D, Innocenti F, et al. Impact of primary tumor location on overall survival (OS) and progression-free survival (PFS) in patients (pts) with metastatic colorectal cancer (mCRC): analysis of CALGB/SWOG 80405 (Alliance) (abstract) J Clin Oncol. 2016;34(Suppl) Abstract 3504. [Google Scholar]
- 31.Missiaglia E, Jacobs B, D'Ario G, et al. Distal and proximal colon cancers differ in terms of molecular, pathological and clinical features. Ann Surg Oncol. 2014 Jul 23; doi: 10.1093/annonc/mdu275. [DOI] [PubMed] [Google Scholar]
- 32.Morris VK, Lucas FA, Overman MJ, et al. Clinicopathologic characteristics and gene expression analyses of non-KRAS 12/13, RAS-mutated metastatic colorectal cancer. Ann Oncol. 2014 Oct;25(10):2008–2014. doi: 10.1093/annonc/mdu252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015 Jun 25;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corcoran RB, Atreya CE, Falchook GS, et al. Combined BRAF and MEK inhibition with dabrafenib and trametinib in BRAF V600-mutant colorectal cancer. J Clin Oncol. 2015 Dec 1;33(34):4023–4031. doi: 10.1200/JCO.2015.63.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Figure 1. Direct adjusted plots of survival after recurrence (SAR) for molecular biomarkers by anatomic site of the primary tumor. Proximal (left side) vs distal (right side) tumor site for status of DNA mismatch repair (MMR) (A), BRAF (B), and combined variables of KRAS/BRAF (C), or MMR/BRAF (D). MUT: mutant; WT: wild-type. MMR status: deficient (d) or proficient (p).