Abstract
Aim::
This study was conducted to determine the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in cattle and buffalo and to study their antibiotic resistance pattern.
Materials and Methods::
A total of 136 samples (skin and nasal swab) from cattle and buffalo were collected. MRSA was identified by conventional bacterial culture techniques which were further confirmed by amplification of S. aureus-specific 16S rRNA by polymerase chain reaction (PCR). The isolates were further analyzed for the presence of mec A gene by PCR. The antimicrobial susceptibility profiling was performed by disc diffusion method.
Results::
The prevalence of MRSA in the current study was 28.57% and 34.28% in cattle nasal and skin swab, respectively, with an overall prevalence of 31.43% MRSA among cattle. Buffalo nasal and skin sample showed MRSA prevalence of 54.55% and 39.4%, respectively, with 46.9% overall prevalence. PCR could detect mec A gene in 36.4% and 58% MRSA isolates from cattle and buffalo, respectively. Antimicrobial susceptibility test found MRSA resistant to penicillin and oxytetracycline (88% each), cefoxitin (75%), cotrimoxazole (62%), and amoxyclav (50%). 100% sensitivity was observed against ciprofloxacin, amikacin, chloramphenicol, and gentamicin. Three (16.7%) MRSA isolates from buffalo were found resistant to vancomycin.
Conclusion::
Cattle and buffalo were identified as a potential carrier of MRSA in Bihar (India). The isolation of vancomycin-resistant S. aureus (VRSA) in the current study indicates the emergence of VRSA in animal population which may be transmitted to the human beings working in close contact to the animals.
Keywords: antibiogram, bovine, mec A gene, methicillin resistant Staphylococcus aureus
Introduction
Staphylococcus aureus is an opportunistic pathogen of human and animal [1] which usually colonizes anterior nares and causes infection in immunocompromised patients [2]. Alexander Ogston in 1880 first isolated S. aureus and first isolation of penicillin-resistant S. aureus was made in 1942 from clinical cases [3]. It was later on circumvented by the introduction of methicillin and the first methicillin resistant S. aureus (MRSA) appeared in the year 1961 which later on became a serious nosocomial infection worldwide [4]. The organism has developed resistance to methicillin by integration of a 21-67 kbp mobile genetic element, termed as staphylococcal cassette chromosome mec [5] into their genome which harbors the methicillin resistance (mecA) gene and other antibiotic resistance determinants [6,7], leaving vancomycin as the drug of last choice to treat the MRSA infections. This gene encodes for a penicillin-binding protein 2 (PBP2a) expressed in the bacterial cell wall and has a low affinity for β-lactam antibiotics. Thus, this group of antibiotics becomes insensitive to bacteria expressing mecA gene.
Data regarding the prevalence of MRSA in India have been reported from various regions [8-11]. However, no systematic study is available on the prevalence and antimicrobial resistance pattern of MRSA, in Bihar.
Hence, this study was planned for isolation, identification and molecular detection of methicillin resistance in staphylococci and their antibiotic resistance pattern from bovines.
Materials and Methods
Collection of sample
Skin and nasal swab samples (136) from apparently healthy cattle and buffalo were collected from Institutional Livestock Farm Complex, Bihar Veterinary College (BVC), Patna, and from the patients coming to teaching veterinary clinical complex of BVC, Patna. The sample comprised skin and anterior nostril swab of cattle (35 each) and buffalo (33 each). The skin samples were collected from the axillary region of the animals. The samples were inoculated in tryptone soya broth (TSB) with 10% sodium chloride salt (TSB-S) (Hi-media) and incubated at 37°C for 24 h. The samples which showed turbidity in TSB-S broth were streaked on mannitol salt agar (Hi-media) with 6 mg/L oxacillin (O-MS agar) and incubated for 24 h at 37°C. Bacterial growth with mannitol fermentation was observed for the presence of round-shaped, typical golden, yellow, or pale color colonies of oxacillin-resistant staphylococci. The presumptive isolates were further confirmed by Gram’s-staining (Hi-media) and catalase test [12]. The test was performed with a loopful culture of isolates mixed with a drop of 3% aqueous solution of hydrogen peroxide observed for the production of gas bubbles by catalase-positive isolates. The oxacillin resistant, mannitol fermenter, Gram-positive cocci in bunch showing positive catalase activity were considered as MRSA isolates. Further these positive isolates were confirmed by S. aureus-specific polymerase chain reaction (PCR) targeting 16S rRNA using the primer sequence as F-5’ GTAGGTGGCCAAGCGTTATCC 3’ and R-5’ CGCACATCAGCGTCAG 3’ [13]. The PCR was performed with 1X PCR buffer (pH-8.3; 15 mM MgCl2), 5 mM of deoxynucleotide triphosphates (dNTPs), 20 pmol of forward and reverse primers, 1 U Taq DNA polymerase, 2 µl of DNA template per reaction and final volume was adjusted to 25 µl with nuclease free water. The cycling conditions were optimized with initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 1 min with the final extension at 72°C for 10 min.
The confirmed isolates of MRSA were further screened for the presence of mecA gene by PCR. The PCR assay was standardized for amplification of mecA gene by forward (5’ GTA GAA ATG ACT GAA CGT CCG ATAA 3’) and reverse (5’ CCAATTCC ACATTGT TTCG GTC TAA 3’) primer [14] with some modifications. The PCR reaction mixture was prepared in 25 µl reaction volume each containing 2.5 µl ×10 PCR buffer (pH-8.3; 15 mM MgCl2), 0.5 µl of dNTP mixture (10 mM each), 2 µl (10 pmol/µl) of forward and reverse primers, 1 µl (1 unit) Taq DNA polymerase, 5 µl of bacterial lysate and final volume was adjusted with nuclease free water. The bacterial lysate was prepared by boiling and snap chilling [15] from overnight grown culture in TSB. The cycling conditions used were initial denaturation at 94°C for 5 min, followed by 32 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 1 min and extension at 72°C for 1 min with the final extension phase at 72°C for 10 min.
The amplified products were analyzed by agarose gel electrophoresis using 1.5% LE agarose (Hi media, India) on tris acetate-ethylenediaminetetraacetic acid buffer containing ethidium bromide (0.5 µg/ml). Gels were visualized and photographed under gel documentation system (Biorad).
Antibiogram study of MRSA isolates was performed by disc diffusion method [16] using a group of antibiotics, viz., ciprofloxacin (5 µg), amoxiclav (30 µg), ofloxacin (5 µg), amikacin (30 µg), cefoxitin (30 µg), ceftriaxone (30 µg), chloramphenicol (30 µg), clindamycin (2 µg), penicillin (10 µg), gentamicin (10 µg), oxytetracycline (30 µg), tetracycline (30 µg), cotrimoxazole (25 µg), and vancomycin (30 µg). The interpretation of results was made according to CLSI guidelines [9].
Results
A total of 136 nasal and skin swab samples were processed in this study. The prevalence of MRSA, out of 136 nasal and skin swab samples studied, is shown in Table-1. 22 and 31 MRSA isolates were collected from cattle and buffalo respectively, based on biochemical confirmation and amplification of S. aureus-specific 16S rRNA (228 bp) gene (Table-1). The prevalence of MRSA was 28.57% and 34.28% in cattle nasal and skin swabs, respectively, with an overall prevalence of 31.43% among cattle, whereas buffalo nasal and skin showed 54.55% and 39.4%, respectively, with an overall prevalence of 46.9% MRSA among buffalo (Table-1).
Table-1.
Prevalence of MRSA among cattle and buffalo.
| Sample type | n=136 | % MRSA detected by mannitol fermentation and 16S rRNA amplification (n) | % mecA positive MRSA (n) |
|---|---|---|---|
| Cattle | |||
| Nose swab | 35 | 28.57 (10) | 50.00 (05) |
| Skin swab | 35 | 34.28 (12) | 25.00 (03) |
| Sub total | 70 | 31.43 (22) | 36.36 (08) |
| Buffalo | |||
| Nose swab | 33 | 54.55 (18) | 50.00 (09) |
| Skin swab | 33 | 39.39 (13) | 69.23 (09) |
| Sub total | 66 | 46.97 (31) | 58.06 (18) |
N: Total number of samples; n: number of samples in each column. MRSA=Methicillin resistant Staphylococcus aureus
PCR amplification of the mecA gene has been used for specific and rapid identification of MRSA among the oxacillin-resistant S. aureus isolates. The test detected mecA gene (Figure-1) in 58.06% MRSA isolates from buffalo, whereas only 36.4% of MRSA isolates from cattle were found positive for mecA gene. The frequency of detection of mecA gene in cattle nostril and skin swab was 50% and 25%, respectively, wherein it was 50% and 69.23% in buffalo nostril and skin swab, respectively (Table-1).
Figure-1.
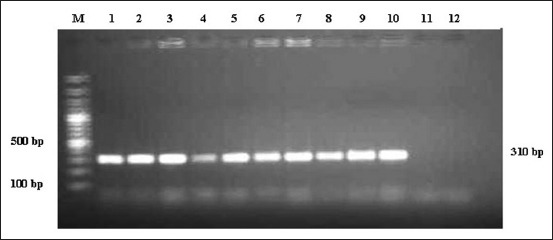
Polymerase chain reaction amplification of mecA gene of methicillin resistant Staphylococcus aureus. M: Gene ruler 100 bp plus DNA ladder, L1: Positive control, L2-L10: Positive amplicons of methicillin resistant Staphylococcus aureus, L11: Isolate with no amplicon, L12: Negative control.
Antimicrobial susceptibility test was performed for all the 53 MRSA isolates of cattle and buffalo origin. It revealed that 88% MRSA isolates of cattle origin, having mecA gene were resistant to penicillin and oxytetracycline, whereas 75% were resistant to cefoxitin followed by cotrimoxazole (62%) and amoxyclav (50%) whereas no resistance was observed against ciprofloxacin, amikacin, chloramphenicol, gentamicin, and vancomycin (Figure-2a). Similar pattern of antimicrobial resistance for penicillin, cotrimoxazole, cefoxitin, ciprofloxacin, amikacin and chloramphenicol was observed in mecA positive isolates of buffalo origin. The finding also revealed resistance against vancomycin, in 16.7% of MRSA isolates from buffalo (Figure-2b).
Figure-2.
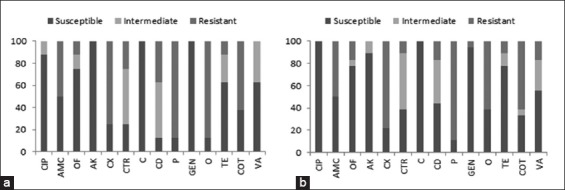
Antimicrobial susceptibility test result for methicillin resistant Staphylococcus aureus from cattle (a) and buffalo (b). CIP: Ciprofloxacin, AMC: Amoxiclav, OF: Ofloxacin, AK: Amikacin, CX: Cefoxitin, CTR: Ceftriaxone, C: Chloramphenicol, CD: Clindamycin, P: Penicillin, GEN: Gentamicin, O: Oxytetracycline, TE: Tetracycline, COT: Cotrimoxazole and VA: Vancomycin.
Discussion
The findings of this study revealed that prevalence of MRSA in buffalo (46.9%) was higher in comparison to cattle (31.43%) which corroborate with earlier findings on isolation of MRSA from nasal or skin samples of cattle [17,18]. However, no reports are available on isolation of MRSA from buffalo nasal and skin samples. Although low prevalence (0.7%) of MRSA harboring in nose and skin of pigs have been reported from Thailand [19]. The difference in the sample size and geographical variations may be the causes of this discrepancy in the prevalence of MRSA. The presence of mecA gene which encodes a modified penicillin-binding protein (PBP), i.e., PBP2a is a useful molecular marker of β-lactam resistance in Staphylococci [20,21]. Hence, PCR amplification of the mecA gene have been used in the present study for specific identification of MRSA among the oxacillin-resistant S. aureus isolates [22,23]. The study revealed a total 36.36% and 58.06% of MRSA isolates carrying mecA gene from cattle and buffalo, respectively. Similar findings of mecA gene detection in cattle nasal carriage have been reported by Alzohairy [24]. In contrast, a lower prevalence of 0.3% and 1% of MRSA has been reported in farm cattle and calves [25]. The lower percentage of mecA gene amplification by PCR among MRSA isolates in the current study may be due to penicillinase activity of S. aureus [26] or due to the presence of new mecA gene homologue, mecALGA251 called mecC gene in the isolates [27] which could be responsible for MRSA isolates.
The antibiogram study revealed resistance to penicillin and oxytetracycline (88%) in MRSA isolates of cattle, whereas 75% were resistant to cefoxitin, (62%) cotrimoxazole and (50%) amoxyclav. No resistance was observed against ciprofloxacin, amikacin, chloramphenicol, gentamicin and vancomycin. Similar pattern of antimicrobial resistance for penicillin, cotrimoxazole, cefoxitin, ciprofloxacin, amikacin, and chloramphenicol was observed in mecA positive isolates of buffalo origin. It revealed that 88% MRSA isolates of cattle origin, having mecA gene were resistant to penicillin and oxytetracycline whereas 75% were resistant to cefoxitin followed by cotrimoxazole (62%) and amoxyclav (50%) whereas no resistance was observed against ciprofloxacin, amikacin, chloramphenicol, gentamicin, and vancomycin (Figure-2a). This antimicrobial resistance pattern was found similar to that of buffalo isolates. The finding also revealed resistance against vancomycin, in 16.7% of MRSA isolates from buffalo. Kumar et al. [8], Vishnupriya et al. [9], and Chandrasekaran et al. [10] have the similar findings of antimicrobial resistance against a group of antibiotics in MRSA.
The current finding also revealed resistance against vancomycin, in 16.7% of MRSA isolates from buffalo which is considered as the drug of choice for treatment of MRSA infections [28]. Although a low level of resistance against vancomycin has been reported earlier [28], only a few intermediate susceptibility but no resistance was against vancomycin has been reported [11]. Hence, the isolation of vancomycin-resistant S. aureus (VRSA) in the current study indicates the emergence of VRSA in animal population which may have a serious implication in human infection.
Conclusion
The present finding concludes that cattle and buffalo may act as a potential carrier of MRSA which may act as a risk factor for human’s infection who are in direct contact with live animals [29].
Authors’ Contributions
PK was the mentor and project leader. AK and Anjay were responsible for project design and performed most of the work. PK and MK were responsible for experimental work and support. All authors read and approved the final manuscript.
Acknowledgments
Authors wish to acknowledge the Honbl’e Vice Chancellor, BAU, Sabour and Dean BVC, Patna, for providing all the necessary funds (BAU Communication No. 175/2016), technical support and infrastructure for conducting the study.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Vanderhaeghen W, Herman K, Haesebrouck F, Butaye P. Methicillin resistant Staphylococcus aureus(MRSA) in food production animals. Epdemiol. Infect. 2010;138:606–625. doi: 10.1017/S0950268809991567. [DOI] [PubMed] [Google Scholar]
- 2.Wertheim H.F.L, Lelles D.C, Vos M.C, van Leeuwen W, van Belkum A, Verbrugh H.A, Nouwen J.L. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005;5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 3.Sangappa S, Thiagarajan P. Methicillin resistant S. Aureus:Resistance gene and their regulation. Int. J. Pharm. Pharm. Sci. 2012;4:658–667. [Google Scholar]
- 4.Anon. 2001 Antimicrobial Resistance Surveillance System (EARSS). Annual Report. Bilthoven: National Institute of Public Health and the Environment; 2001. [Google Scholar]
- 5.Rohrer S, Bischoff M, Rossi J, Berger-Bachi B. 2003 of methicillin resistance. In: Fluit A.C, Schmitz F.J, editors. MRSA:Current Perspectives. Norfolk, England: Caister Academic Press; 2003. p. 31.p. 53. [Google Scholar]
- 6.Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2001;45:1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma X.X, Ito T, Tiensasitorn C, Jamklang M, Chongtrakool P, Vavra S.B, Daum R.S, Hiramatsu K. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 2002;46:1147–1152. doi: 10.1128/AAC.46.4.1147-1152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar R, Yadav B.R, Singh R.S. Antibiotic resistance and pathogenicity factors in Staphylococcus aureus isolated from mastitic sahiwal cattle. J. Biosci. 2011;36:175–188. doi: 10.1007/s12038-011-9004-6. [DOI] [PubMed] [Google Scholar]
- 9.Vishnupriya S, Antony P.X, Mukhopadhyay H.K, Pillai R.M, Thanislass J, Srinivas V.M, Kumar S.R. Methicillin resistant staphylococci associated with bovine mastitis and their zoonotic importance. Vet. World. 2014;7:422–427. [Google Scholar]
- 10.Chandrasekaran D, Venkatesan P, Tirumurugaan K.G, Gowri B, Subapriya S, Thirunavukkarasu S. Sub-acute mastitis associated with methicillin resistant Staphylococcus aureus in a cow:A case report. J. Adv. Vet. Anim. Res. 2014;1:235–237. [Google Scholar]
- 11.Sharma L, Verma A.K, Kumar A, Rahat A, Neha Nigam R. Incidence and pattern of antibiotic resistance of Staphylococcus aureus isolated from clinical and subclinical mastitis in cattle and buffaloes. Asia. J. Anim. Sci. 2015;9:100–109. [Google Scholar]
- 12.Agarwal R.K, Bhilegaonkar K.N, Singh D.K, Kumar A, Rathore R.S. In:Laboratory Manual for the Isolation and Identification of Food Borne Pathogens. 1st ed. Izzatnagar: IVRI; 2003. pp. 38–39. [Google Scholar]
- 13.Monday S.R, Bohach G.A. Use of multiplex PCR to detect classical and newly described pathogenic toxin gene in staphylococcal isolates. J. Clin. Microbiol. 1999;37:3411–3414. doi: 10.1128/jcm.37.10.3411-3414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braoios A, Fluminhan J.A, Pizzolitto A.C. Multiplex PCR use for Staphylococcus aureus identification and oxacillin and mupirocin resistance evaluation. J. Basic Appl. Pharm. Sci. 2009;30:303–307. [Google Scholar]
- 15.Kaushik P, Anjay Kumari S, Bharti S.K, Dayal S. Isolation and prevalence of Salmonella from chicken meat and cattle milk collected from local 30:Markets of Patna, India. Vet. World. 2014;7:62–65. [Google Scholar]
- 16.Wayne P.A. 2002 Standards of Antimicrobial Susceptibility:National Committee for Clinical Laboratory Standards (NCCLS) NCCLS Approved Standards. 2002:100–159. [Google Scholar]
- 17.Garipcin M, Seker E. Nasal carriage of methicillin-resistant Staphylococcus aureus in cattle and farm workers in Turkey. Vet. Arch. 2015;85:117–129. [Google Scholar]
- 18.Inegol E, Turkyilmaz S. Determination of SCC mec types in methicillin resistant staphylococci isolated from cows and farm workers. Ankara. Univ. Vet. Fak. Derg. 2012;59:89–93. [Google Scholar]
- 19.Patchnee P, Tadee P, Arjkumpa O, Love D, Chanachai K, Alter T, Hinjoy S, Tharavichitkul P. Occurrence and characterization of livestock associated methicillin resistant Staphylococcus aureus in pig industries of Northern Thailand. J. Vet. Sci. 2014;15:529–536. doi: 10.4142/jvs.2014.15.4.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulligan M.E, Murray K.A, Standiford H.C, John J.F, Kauffmann C.A, Yu V.L. Methicillin-resistant Staphylococcus aureus:A consensus review of the microbiology, pathogenesis, and epidemiology with implications for prevention and management. Ame. J. Med. 1993;94:313–328. doi: 10.1016/0002-9343(93)90063-u. [DOI] [PubMed] [Google Scholar]
- 21.Pinho M.G, Filipe S.R, de Lencastre H, Tomasz A. Complementation of the essential peptidoglycan trans-peptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J. Bacteriol. 2001;183:6525–6531. doi: 10.1128/JB.183.22.6525-6531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi S.M, Kim S, Kim C, Lee D, Choi J, Yoo J, Kang J, Shin W, Kang W. Multiplex PCR for the detection of genes encoding aminoglycoside modifying enzymes and methicillin resistance among Staphylococcus species. J. Korean Med. Sci. 2003;18:631–636. doi: 10.3346/jkms.2003.18.5.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalhor H, Shariati L, Validi M, Tabatabaiefar M.A, Nafisi M.R. Comparison of agar screen and duplex-PCR methods in determination of methicillin-resistant Staphylococcus aureus(MRSA) strains isolated from nasal carriage. Afr. J. Microbiol. Res. 2012;6:3722–3726. [Google Scholar]
- 24.Alzohairy M.A. Colonization and antibiotic susceptibility pattern of methicillin resistance Staphylococcus aureus(MRSA) among farm animals in Saudi Arabia. J. Bacteriol. Res. 2011;3:63–68. [Google Scholar]
- 25.Huber H, Koller S, Giezendanner N, Stephan R, Zweifel C. Prevalence and characteristics of methicillin-resistant Staphylococcus aureus in humans in contact with farm animals, in livestock, and in food of animal origin, Switzerland, 2009. Eur. Surv. 2010;15:7–10. [PubMed] [Google Scholar]
- 26.Lowy F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 27.Peterson G.K, Larsen J, Harrison E.M, Larsen A.R, Morgan F.J, Peacock S.J, Parkhill J, Zadoks R.N, Holmes M.A. First detection of livestock-associated meticillin resistant Staphylococcus aureus CC398 in bulk tank milk in the United Kingdom. Eur. Surv. 2012;17:20337. [PMC free article] [PubMed] [Google Scholar]
- 28.Assadullah S, Kakru D.K, Thoker M.A, Bhat F.A, Hussain W, Shah A. Emergence of low level vancomycin resistance in MRSA. Indian J. Med. Microbiol. 2003;21:196–198. [PubMed] [Google Scholar]
- 29.Beninati C, Reich F, Muscolino D, Giarratana F, Panebianco A, Klein G, Atanassova V. ESBL-producing bacteria and MRSA isolated from poultry and Turkey products imported from Italy. Czech. J. Food Sci. 2015;33:97–102. [Google Scholar]


