Abstract
Aim:
The aim of this study was to figure the prevalence, phenotypic and genotypic antibiotic resistance (AR) pattern of Staphylococcus aureus isolated from bovine and swine nares.
Materials and Methods:
Colonies with typical morphology on Baird-Parker agar supplemented with egg-yolk tellurite emulsion were selected and biochemically/genotypically identified as S. aureus. These strains were further subjected to epsilometer test for their sensitivity to various clinically important antibiotics and antibiotic susceptibility testing for amoxicillin/clavulanic acid, and double-disk diffusion testing was performed by the standard disc diffusion method following CLSI guidelines. S. aureus strains were also tested for the presence of AR genes, viz., blaZ, mecA, aacA-aphD, erm (ermA, ermB, ermC), tet (efflux genes tetK and tetL, tetM and tetO of the ribosomal protection family), and vanA.
Results:
The nasal cavities of 17 out of 47 randomly selected bovine and 20 out of 28 randomly selected swine were positive for S. aureus, representing the prevalence of 36.2% (95% confidence interval [CI]: 22.5-49.9) and 71.4% (95% CI: 54.7-88.1), respectively. Most of the S. aureus strains showed higher resistance to penicillin (94.6%, minimal inhibitory concentration [MIC] ≥1.5 µg/ml) followed by ciprofloxacin (56.7%, MIC ≥32 µg/ml) and tetracycline (18.9%, MIC ≥32 µg/ml). About 10-15% of the strains were resistant to gentamicin (MIC 16 µg/ml) and oxacillin (MIC 6-8 µg/ml). None of the strains were resistant to vancomycin (MIC 0.25-1.5 µg/ml). In this study, 32.4% strains were resistant to three or more than three antibiotics and prevalence of this multi-drug resistant S. aureus was 45% (95%CI: 26.6-63.4) and 17.6% (95%CI: 6.7-28.5) in swine and bovine nasal samples, respectively. Four strains from pigs were borderline oxacillin-resistant S. aureus MIC 6-8 µg/ml, but none were mecA positive. Two of these strains were β-lactamase hyperproducers. Among the resistance genes blaZ, tetK, tetL, tetM, ermB, and aacA-aphD were found.
Conclusion:
Our results demonstrated the absence of mecA and pvl gene, but the presence of multi-drug resistant S. aureus in the nares of healthy animals which has a potential to spread in a community.
Keywords: antibiotic resistance genes, epsilometer test, livestock nasal swabs, multidrug resistance, Staphylococcus aureus
Introduction
In food animal production, antibiotics are used extensively and are often applied sub-therapeutically for growth promotion and disease prevention in India. This has resulted in the rise of antibiotic-resistant Staphylococcus aureus in food animals and foods of animal origin. S. aureus resistance to oxacillin carrying the mecA gene, now known as methicillin-resistant S. aureus (MRSA), was first reported in the year 1961 and since then reported in hospitals, food, animals, community and the environment [1]. Recently, MRSA strains, consistent with the main livestock-associated (LA)-MRSA type have caused outbreaks in healthcare settings thereby raising the concern. In addition, a new variant of methicillin resistance determinant (mecC) has been described in specific LA-MRSA lineages that are primarily associated with swine, cows, and sheep but again display a broad host range [2].
Sustained efforts are therefore required to analyze antibiotic resistance (AR) patterns of S. aureus strains isolated from different origins, including livestock due to their changing patterns of resistance and its potential to serve as a reservoir of AR genes and MRSA. AR of S. aureus nasal carriage isolates from bovine and swine has not been pursued aggressively, especially in Punjab.
Therefore, the aim of this study was to figure the prevalence, phenotypic and genotypic AR pattern of S. aureus isolated from bovine and swine nares.
Materials and Methods
Ethical approvals
The study was conducted under the supervision of institutional ethical committee.
Sampling
Nasal swabs of 47 apparently healthy bovines and 28 apparently healthy swine were obtained from the Teaching Veterinary Hospital and the pig farm of Guru Angad Dev Veterinary and Animal Sciences University (GADVASU), Ludhiana, Punjab, respectively, during February to April 2014.
Isolation and identification of S. aureus
Isolation of S. aureus from swab samples was performed as per the bacteriological analytical manual [3]. Colonies with typical morphology on Baird-Parker agar supplemented with egg-yolk tellurite emulsion were selected and subjected to Gram-staining and catalase test. Gram and catalase positive isolates were biochemically identified as S. aureus using the HiStaph™ Identification Kit (HiMedia Labs, Mumbai). These isolates were further subjected to coagulase test using rabbit plasma (HiMedia Lab, Mumbai) following manufacturer’s instructions. The S. aureus isolates were purified and maintained in 20% (v/v) glycerol at −20°C till further processing.
Antibiotic susceptibility testing (AST) of selected S. aureus strains
The AST of S. aureus strains was performed by the Epsilometer test. All the selected S. aureus strains were tested for their sensitivity to various antibiotics, viz., oxacillin, penicillin, tetracycline, chloramphenicol, cotrimazole, ceftriaxone, gentamicin, erythromycin, ciprofloxacin, and vancomycin using Ezy MIC™ strip (HiMedia Lab, Mumbai). AST for amoxicillin/clavulanic acid and double- disk diffusion testing (D-test) was performed by the standard disc diffusion method following CLSI guidelines [4].
Identification of antibiotic resistant genes
S. aureus strains were tested for the presence of following AR genes: blaZ, mecA, aacA-aphD, erm (ermA, ermB, ermC), tet (efflux genes tetK and tetL, tetM and tetO of the ribosomal protection family) and vanA, encoding for penicillin, oxacillin, gentamicin, erythromycin, tetracycline and vancomycin resistance, respectively, by amplification of the existing gene using multiplex polymerase chain reaction (PCR). Primers for the respective genes are listed in Table-1 [5-10].
Table-1.
Primers used for detection of antibiotic resistant genes in S. aureus.
| Gene | Oligonucleotide sequence (5`-3`) | Amplicon size | References |
|---|---|---|---|
| aacA-aphD | TAA TCC AAG AGC AAT AAG GGC GCC ACA CTA TCA TAA CCA CTA |
227 | [5] |
| ermA | AAG CGG TAA ACC CCT CTG A TTC GCA AAT CCC TTC TCA AC |
190 | [5] |
| ermB | CTATCTGATTGTTGAAGAAGGATT GTTTACTCTTGGTTTAGGATGAAA |
142 | [6] |
| ermC | AAT CGT CAA TTC CTG CAT GT TAA TCG TGG AAT ACG GGT TTG |
299 | [5] |
| tetK | GTA GCG ACA ATA GGT AAT AGT GTA GTG ACA ATA AAC CTC CTA |
360 | [5] |
| tetM | AGT GGA GCG ATT ACA GAA CAT ATG TCC TGG CGT GTC TA |
158 | [5] |
| tetL | GTCGTTGCGCGCTATATTCC GTGAACGGTAGCCCACCTAA |
696 | [7] |
| tetO | AATGAAGATTCCGACAATTT CTCATGCGTTGTAGTATTCCA |
781 | [7] |
| mecA | AAA ATC GAT GGT AAA GGT TGG C AGT TCT GCA GTA CCG GAT TTG C |
532 | [5] |
| blaZ | ACT TCA ACA CCT GCT GCT TTC TGA CCA CTT TTA TCA GCA ACC |
173 | [6] |
| vanA | ATGAATAGAATAAAAGTTGC TCACCCCTTTAACGCTAATA |
1032 | [8] |
| 16SrDNA (Staphylococcus genus specific) | CAG CTC GTG TCG TGA GAT GT AAT CAT TTG TCC CAC CTT CG |
420 | [5] |
| Coa | ATA GAG ATG CTG GTA CAG G GCT TCC GAT TGT TCG ATG C |
547 (550-875) | [9] |
| Nuc (Species specific) | GCGATTGATGGTGATACGGTT AGCCAAGCCTTGACGAACTAAAGC |
279 | [10] |
S. aureus=Staphylococcus aureus
Antibiotic resistant genes from strains were amplified, purified and sequenced (Invitrogen) for use as a control. Sequence comparisons were performed using the BLAST online program (http://www.ncbi.nlm.- nih.gov/BLAST/), and respective sequences were submitted to NCBI. S. aureus strain ATCC 33591 and ATCC 33592 were used as MRSA (mecA +ve) and MSSA (mecA −ve) positive control, respectively. KU872013, KP834338/KP834339, KP658721, KP658723, KP886833, KT454736, KT454737, S. aureus strains were used as positive control for genes blaZ, aacA-aphD, tetK, tetL, tetM, ermB, ermC, respectively.
The isolation of genomic DNA from S. aureus strains was done using HiPurA™ bacterial genomic DNA purification kit (HiMedia Lab, Mumbai). Each strain was subjected to a separate multiplex PCR assays for a detection of each group (gp) of genes: gp1 (16S rDNA-genus specific, nuc-species specific, mecA), gp2 (tetK, tetL, tetM and tetO), gp3 (ermA, ermB, ermC and aacA-aphD), and gp4 (coa and blaZ). Separate PCR was run for vanA and pvl gene.
The PCR amplification was carried out in a total reaction volume of 25 µl containing 0.4 mM deoxynucleotide triphosphates, 4 mM MgCl2 (Promega, USA), 10 pmol/µl of each primer set containing forward and reverse primers, 1 U Taq DNA polymerase (Promega, USA), 0.01-0.2 µg template and sterilized nuclease free water was added to make up the reaction volume of 25 µl.
The cycling conditions of multiplex PCR for gp1, gp2, and gp3 and of single PCR for vanA gene were as per methodology of Strommenger et al. [5] (with little modifications) and Saha et al. [8], respectively. However, cycling condition for coa and blaZ (gp 4) included an initial denaturation of DNA at 94°C for 45 s, followed by 30 cycles of denaturation at 94°C 20 s, annealing at 55°C for 15 s and extension at 70°C for 15 s, followed by a final extension of 2 min at 72°C and hold at 4°C. The cycling condition for pvl includes 94°C for 1 min, 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min followed by a final extension at 72°C for 5 min.
All the PCR amplicons were visualized using an ultraviolet light box after electrophoresis on a 1.5% agarose gel containing 0.5 µg/ml ethidium bromide.
Statistical analysis
Microsoft excel was used for statistical analysis. The variables were compared using a Chi-squared or Fisher’s exact test, as appropriate. Differences were considered significant when the p<0.05. Dendrogram based on antibiogram was composed using the unweighted paired group method with Bionumeric software V07.
Results
The nasal cavities of 17 out of 47 randomly selected bovine and 20 out of 28 randomly selected swine were positive for S. aureus, representing the prevalence of 36.2% (95% confidence interval [CI]: 22.5-49.9) and 71.4% (95% CI: 54.7-88.1), respectively. Furthermore, all the S. aureus strains were coagulase positive except one swine nasal swab (1S1). One strain from each sample was screened against antibiotics that are of clinical importance and percentage of strains classified as susceptible, intermediate or resistant were used to summarize resistance percentage overall and disaggregated by species type (Figure-1).
Figure-1.
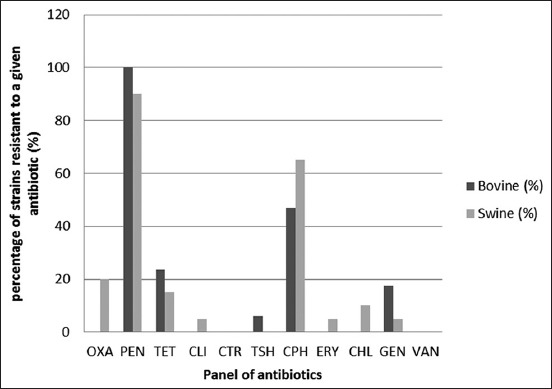
Percentage of Staphylococcus aureus strains resistant to different antibiotics.
Phenotypic resistance profile
Most of the S. aureus strains showed higher resistance to penicillin (94.6%, MIC ≥1.5 µg/ml) followed by ciprofloxacin (56.7%, MIC ≥32 µg/ml) and tetracycline (18.9%, MIC ≥32 µg/ml). About 10-15% of the strains were resistant to gentamicin (MIC 16 µg/ml) and oxacillin (MIC 6-8 µg/ml). However, 21.6% of S. aureus strains showed intermediate resistance to chloramphenicol (MIC 16 µg/ml). Similarly, for erythromycin and ceftriaxone, most of the strains were intermediate resistant with MIC 1-3 µg/ml, 54.05% and MIC 16-32 µg/ml, 21.6%, respectively. Vancomycin MIC values for S. aureus strains were in the susceptible range (0.25-1.5 µg/ml). Around 16 unique resistance profiles (taking five antibiotics as cutoff value) were found (Figure-2).
Figure-2.
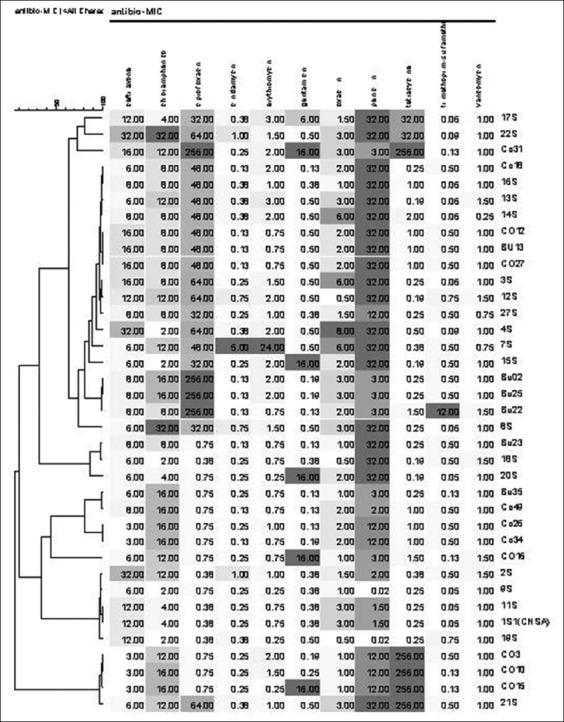
Dendrogram based on minimal inhibitory concentration antibiogram of 37 Staphylococcus aureus strains. The dendrogram was obtained using the unweighted paired group method with bionumeric software V07. The color indicates the susceptibility gradient with Dark-resistant, gray-intermediate resistant and white or light gray-susceptible. The top left horizontal scale indicates the number of antibiotics 1-11.
In this study, 32.4% strains were resistant to three or more than three antibiotics, and they were designated as multidrug resistance (MDR). The prevalence of MDR S. aureus was 45% (95% CI: 26.6-63.4) and 17.6% (95% CI: 6.7-28.5) in swine and bovine nasal samples, respectively.
Relationship between AR phenotype and genotype
Penicillin
One S. aureus strain (Bu35) was negative for blaZ gene, but resistant to penicillin with MIC 3 µg/ml and was intermediately resistant to chloramphenicol (Figure-2 and Table-2).
Table-2.
Correlation between the phenotypic and genotypic AR to penicillin, oxacillin and gentamicin in S. aureus.
| Gene | Number of strains | Number of strains tested by E strip (MIC µg/ml) | ||
|---|---|---|---|---|
| Sensitive | Intermediate | Resistant | ||
| blaZ | Penicillin MIC≤0.12 | Penicillin MIC 0.12-0.25 | Penicillin MIC≥0.25 | |
| Positive | 34 | 0 | 0 | 34 |
| Negative | 3 | 2 | 1 | |
| mecA | Oxacillin MIC≤2 | Oxacillin MIC 2-4 | Oxacillin MIC≥4 | |
| Positive | 0 | 0 | 0 | 0 |
| Negative | 37 | 33 | 0 | 4 |
| aacA-aphD | Gentamicin MIC≤4 | Gentamicin MIC 8 | Gentamicin MIC≥16 | |
| Positive | 15 | 10 | 0 | 5 |
| Negative | 22 | 22 | 0 | 0 |
S. aureus=Staphylococcus aureus, MIC=Minimal inhibitory concentration, AR=Antibiotic resistance
Oxacillin
All S. aureus strains were mecA negative. However, four S. aureus strains that were phenotypically resistant to oxacillin but genotypically mecA negative showed borderline resistance with MIC ranges from 6 to 8 µg/ml (Table-2). Out of these four S. aureus strains, S7 and S14 were β-lactamases hyperproducers (Table-3).
Table-3.
Characteristics of isolates resistant to oxacillin but negative for the mecA gene by PCR.
| Source | Strains | Oxacillin MIC (µg/ml) | Ceftriaxone MIC (µg/ml) | Amoxyclave inhibition zone* (mm) | blaZ (PCR detection) | β-lactamase hyper producers |
|---|---|---|---|---|---|---|
| Swine nasal swab | S3 | 6 | 16 | 14 | + | - |
| Swine nasal swab | S4 | 8 | 32 | 15 | + | - |
| Swine nasal swab | S7 | 6 | 6 | 40 | + | + |
| Swine nasal swab | S14 | 6 | 8 | 35 | + | + |
The diameter of the inhibition zone with amoxyclave (amoxicillin-clavulanic acid: 20 and 10 µg, respectively) if exceeded 20 mm then strains were considered β-lactamases hyperproducers. Ceftriaxone MIC≤8 µg/ml - susceptible, MIC=1632 µg/ml-intermediate resistant and MIC≥64 µg/ml. MIC=Minimal inhibitory concentration, PCR=Polymerase chain reaction
Tetracycline
The majority of the S. aureus strains in this study either carried tetK or tetL whereas one strain contained both tetK and tetM gene (Table-4). tetL was found only among swine nasal S. aureus strains, whereas tetK was common among bovine nasal S. aureus strains. This is agreeing with Kadlec et al. [11] and Ugwu et al. [12]. Furthermore, among S. aureus strains that were carrying the gene, 63.6% showed resistance to tetracycline with higher MIC values (32->256 µg/ml).
Table-4.
Correlation between the phenotypic and genotypic AR to tetracycline in S. aureus.
| tet presence | Number of strains tested by E strip (MIC µg/ml) | |||||
|---|---|---|---|---|---|---|
| tetK | tetL | tetM | Sensitive (≤4) | Intermediate (8) | Resistant (≥16) | |
| Positive | 5 | 6 | 1* | 4 | 0 | 7 |
| Negative | 26 | 26 | 0 | |||
1 samples (Co31) possess both tetK and tetM. MIC=Minimal inhibitory concentration, AR=Antibiotic resistance, S. aureus=Staphylococcus aureus
Erythromycin
Most of S. aureus strains were genotypically negative but phenotypically intermediate resistant with MIC 1-3 µg/ml (Table-5). In this study, all strains considered resistant or intermediate to erythromycin in vitro tested for inducible clindamycin resistance (D-test). None of the strains showed inducible macrolide, lincosamides, and streptogramins Type B (MLSB) phenotype (Ery+/Cli−, D+), but one strain (7S) showed a constitutive MLSB phenotype (Erm+/Cli+, D−) and positive for ermB gene.
Table-5.
Relationship between the phenotypic and genotypic AR to erythromycin S. aureus.
| erm presence | Number of strains tested by E strip (MIC µg/ml) | ||||
|---|---|---|---|---|---|
| ermB | ermA/ermC | Sensitive (≤0.5) | Intermediate (1-4) | Resistant (≥8) | |
| Positive | 5 | 0 | 0 | 4 | 1 |
| Negative | 32 | 16 | 16 | 0 | |
MIC=Minimal inhibitory concentration, AR=Antibiotic resistance, S. aureus=Staphylococcus aureus
Gentamicin
All S. aureus strains resistant to gentamicin with MIC ≥16 µg/ml showed the presence of aacA-aphD gene, and that constitutes only 33.3% of total strains positive for this gene (Table-2).
Discussion
Penicillin, ciprofloxacin, and tetracycline are highly used in veterinary medicine and as feed additive in India [13]. This may be a reason for the increase in resistance to these antibiotics. Though low, but intermediate resistance to ceftriaxone is also a concern because ceftriaxone (third generation of cephalosporin) used to treat animals and humans especially children [14]. These results corroborate similar studies in pig, cattle and clinical isolates [15,16] and obtain in other countries [17,18] which reflects the predominant use of these drugs in pig and dairy cattle husbandry worldwide.
There was no significant difference between the percentage of AR among the bovine and swine nasal S. aureus strains (Fisher’s exact test; p=0.478), but higher prevalence of MDR S. aureus was observed in swine as compared to bovine. Thereby, our data show that bovine and swine nares are often contaminated with MDR S. aureus, which is the public health concern because direct contact with livestock colonized with resistant bacteria is the most documented routes of resistance transmission from the animals into human populations [19].
There was a statistically significant relationship (p<0.05) between phenotypes and genotype resistance pattern in S. aureus strains subjected to the antibiotic penicillin, tetracycline, gentamicin, and erythromycin. However, not all the penicillin-resistant S. aureus strains exhibited genotypic resistance to penicillin (Table-2), corresponding well with the findings of the previous studies [20,21]. This may be because the phenotypic resistance has been caused by point mutations, biofilm formation or antibiotic tolerance [22]. Thereby, our results suggest that blaZ may play a major role but cannot be used alone as an indicator for penicillin resistance.
Borderline oxacillin-resistant S. aureus (BORSA) has been another relatively frequently observed phenotype among S. aureus strains. These strains are cefoxitin/ceftriaxone susceptible and do not carry the mecA or mecC genes, but are shown oxacillin resistance MIC between 1 and 8 µg/ml [23]. In this study, All S. aureus strains were negative for mecA gene and four of those strains were BORSA. Such pattern may be because of hyperproduction of β-lactamases, production of normal penicillin-binding protein 2 with altered binding capacity or variant of mecA gene (mecC) [24]. To exclude the possibility of hyperproduction of β-lactamase, amoxyclave antibiotic disk diffusion test was performed [6]. Two of those strains were found β-lactamases hyperproducers and other two were not (Table-3). Martineau et al. [6] also reported exist of isolates that were phenotypically oxacillin resistant but negative for mecA gene. Likewise Pereira et al. [25] reported 38% of S. aureus strains phenotypically resistant to oxacillin but only 0.68% of those strains showed the presence of mecA gene. In conclusion, our result showed the prevalence of BORSA that is of public health concern because the incidences of BORSA has been reported among human clinical strains [26] and have also been detected in food of animal origin, cattle and pigs [27].
Farmers and veterinarians harbored tetracycline resistant strains more frequently than people without contact with livestock especially pigs [28]. Our result reported the presence of tetK, tetL and tetM genes in bovine and swine nares and is of public health concern because the tetK and tetM genes are on mobile genetic element, such as small plasmids and conjugative transposons of S. aureus [7] that can spread and cause treatment failure both in veterinary and human medicine.
A form of acquired simultaneous resistance to MLSB in clinical isolates is due to evolutionary variants of erm genes [11]. The strains in the current study only had ermB gene. This is in accordance with the study that too reported ermB gene in animal strains [6]. Most of the strains were intermediate resistant to erythromycin but erm gene negative. This intermediate resistance to erythromycin can be possibly associated with mechanism not yet characterized in staphylococcus or may be due to another gene such as msrA and mphC––corroborating with past research [6,12].
In this study, most of the strains were aacA-aphD positive but phenotypically sensitive to gentamicin (Table-2). This is the issue of concern from a clinical prospective as the susceptible strains harboring but not expressing an AR gene is considered as potentially resistant to that antibiotic because there are the experiments that strongly suggest that susceptible strains harboring a resistance gene have the potential to develop resistance on in vivo selection of the appropriate antimicrobial agent [6]. None of the strain was pvl positive.
Conclusion
Hitherto there is no direct evidence that the food can contribute to the community-associated-MRSA but the direct contact with colonized animals, especially swine contact now appears to be a significant risk factor for colonization with MDR S. aureus and MRSA (LA-MRSA), thereby explains the relevance of this type of studies. To our knowledge, this is the first short study in the Punjab examines the prevalence and characteristics of MDR S. aureus and screening for MRSA in bovine and swine nares. Of particular concern in this study was the small sample size, but clearly, shows the requirement of further study at the farm and retail levels involving large sample size over time at different location to better assess the presence of MDR S. aureus and MRSA in livestock and the risk to livestock handlers and consumers. Although all strains were susceptible to vancomycin and negative for mecA and pvl gene, the S. aureus strains have demonstrated resistance to more than two antibiotics. Our result showed the prevalence of BORSA that is of public health concern because the incidences of BORSA have been reported in hospitals. We also observed that most strains presented the genes of resistance, although these were not being expressed, yet demonstrating the future potential for these strains to become resistant to the evaluated antimicrobial agents.
Authors’ Contributions
The study was designed by RS and JPSG. Laboratory work was done by AS. SK and AS prepared manuscript while as RS and AS analyzed data. All authors read and approved the final manuscript.
Acknowledgments
The study was performed as part of the M.V.Sc. research work and was funded by the School of Public Health and Zoonoses, GADVASU, Ludhiana, Punjab, India.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Ogata K, Narimatsu H, Suzuki M, Higuchi W, Yamamoto T, Taniguchi H. 2012 distributed meat as a potential vehicle for community-acquired methicillin-resistant Staphylococcus aureus. Appl. Environ. Microbiol. 2012;78:2797–2802. doi: 10.1128/AEM.07470-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larsen R.A. 2014 Online presentation on the Emergence and Spread of MRSA CC398. Event:Molecular typing methods for pathogens, Lyon, france. PGEC lecture Diagnostic Bacteriology and General Microbiology. 2014. [Accessed on 24-08-2016]. https://www.escmid.org/escmid_publications/escmid_elibrary/?q=Larsen&id=2173&L=0&x=0&y=0 .
- 3.Bennett R.W, Lancette G.A. Staphylococcus aureus In:FDA Bacteriological analytical manual. 8th ed. 12. Gaithersburg, MD: AOAC International; 2001. [Google Scholar]
- 4.Clinical and Laboratory Standards Institute 2011. 2011 for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically:Approved Standard. Vol. 31. Wayne, PA: Clinical and Laboratory Standards Institute; 2011. pp. M100–S21. [Google Scholar]
- 5.Strommenger B, Kettlitz C, Werner G, Witte W. 2003 PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J. Clin. Microbiol. 2003;41(9):4089–4094. doi: 10.1128/JCM.41.9.4089-4094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martineau F, Picard J.F, Lansac N, Menard C, Roy H.P, Ouellette M, Bergeron G.M. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2002;44(2):231–238. doi: 10.1128/aac.44.2.231-238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huys G, D’Haene K, Van Eldere J, von Holy A, Swings J. Molecular diversity and characterization of tetracycline-resistant Staphylococcus aureus isolates from a poultry processing plant. Appl. Environ. Microbiol. 2005;71:574–579. doi: 10.1128/AEM.71.1.574-579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saha B, Singh AK, Ghosh A, Bal M. Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia) J. Med. Microbiol. 2008;57(1):72–79. doi: 10.1099/jmm.0.47144-0. [DOI] [PubMed] [Google Scholar]
- 9.Hookey J.V, Richardson J.F, Cookson B.D. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J. Clin. Microbiol. 1998;36(4):1083–1089. doi: 10.1128/jcm.36.4.1083-1089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brakstad O.G, Aasbakk K, Maeland J.A. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 1992;30:1654–1660. doi: 10.1128/jcm.30.7.1654-1660.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadlec K, Febler T.A, Hauschild T, Schwarz S. Novel and uncommon antimicrobial resistance genes in livestock associated methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2012;18:745–755. doi: 10.1111/j.1469-0691.2012.03842.x. [DOI] [PubMed] [Google Scholar]
- 12.Ugwu C.C, Gomez-Sanz E, Agbo C.I, Torres C, Brazilian F.K. Characterization of mannitol-fermenting methicillin-resistant Staphylococci isolated from pigs in Nigeria. J. Microbiol. 2015;46(3):885–892. doi: 10.1590/S1517-838246320140644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Centre for Science and Environment (CSE) 2014 Latest Study by CSE’s Pollution Monitoring Lab Finds Antibiotic Residues in Chicken. Food Safety and Toxins. 2014. [Accessed on 14-08-2016]. Available from: http://www.cseindia.org/node/5487 .
- 14.Frye J.G, Fedorka-Cray P.J. Prevalence, distribution and characterization of ceftiofur resistance in Salmonella enterica isolated from animals in the USA from 1999 to 2003. Int. J. Antimicrob. Agents. 2007;30:134–142. doi: 10.1016/j.ijantimicag.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 15.Duran N, Ozer B, Duran G.G, Onlen Y, Demir C. Antibiotic resistance genes and susceptibility patterns in Staphylococci. Indian J. Med. Res. 2012;135:389–396. [PMC free article] [PubMed] [Google Scholar]
- 16.Morcillo A, Castro B, Rodríguez-Álvarez C, Abreu R, Aguirre-Jaime A, Arias A. Descriptive analysis of antibiotic-resistant patterns of methicillin-resistant Staphylococcus aureus(MRSA) st398 isolated from healthy swine. Int. J. Environ. Res. Publ. Health. 2015;12:611–622. doi: 10.3390/ijerph120100611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Neeling A.J, van den Broek M.J, Spalburg E.C, van Santen-Verheuvel M.G, Dam-Deisz W.D, Boshuizen H.C, van de Giessen A.W, van Duijkeren E, Huijsdens X.W. High prevalence of methicillin resistant Staphylococcus aureus in pigs. Vet. Microbiol. 2007;122:366–372. doi: 10.1016/j.vetmic.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 18.Zhang C, Song L, Chen H, Liu Y, Qin Y, Ning Y. Antimicrobial susceptibility and molecular subtypes of Staphylococcus aureus isolated from pig tonsils and cow’s milk in China. Can. J. Vet. Res. 2012;76:268–274. [PMC free article] [PubMed] [Google Scholar]
- 19.Schmithausen R.M, Schulze-Geisthoevel S.V, Stemmer F, El-Jade M, Reif M, Hack S, Meilaender A, Montabauer G, Fimmers R, Parcina M, Hoerauf A, Exner M. Analysis of transmission of MRSA and ESBL-E among pigs and farm personnel. PloS One. 2015;10:e0138173. doi: 10.1371/journal.pone.0138173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frey Y, Rodriguez J.P, Thomann A, Schwendener S, Perreten V. Genetic characterization of antimicrobial resistance in coagulase-negative Staphylococci from bovine mastitis milk. J. Dairy Sci. 2013;96:2247–2257. doi: 10.3168/jds.2012-6091. [DOI] [PubMed] [Google Scholar]
- 21.Yang F, Wang Q, Wang X, Wang L, Xiao M, Li X, Luo J, Zhang S, Li H. Prevalence of blaZ gene and other virulence genes in penicillin-resistant Staphylococcus aureus isolated from bovine mastitis cases in Gansu, China. Turk. J. Vet. Anim. Sci. 2015;39:634–636. [Google Scholar]
- 22.Croes S, Deurenberg R.H, Boumans M.L.L, Beisser P.S, Neef C, Stobberingh E.E. Staphylococcus aureus biofilm formation at the physiologic glucose concentration depends on the S. aureus lineage. BMC Microbiol. 2009;9:229. doi: 10.1186/1471-2180-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shore A.C, Coleman D.C. Staphylococcal cassette chromosome mec:Recent advances and new insights. Int. J. Med. Microbiol. 2013;303:350–359. doi: 10.1016/j.ijmm.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Laurent F, Chardon H, Haenni M, Bes M, Reverdy M.E, Madec J.Y, Lagier E, Vandenesch F, Tristan A. MRSA harboring mec A variant gene mec C in France. Emerg. Infect. Dis. 2012;18:1465–1467. doi: 10.3201/eid1809.111920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pereira V, Lopes C, Castro A, Silva J, Gibbs P, Teixeira P. Characterization for enterotoxin production, virulence factors, and antibiotic susceptibility of Staphylococcus aureus isolates from various foods in Portugal. Food Microbiol. 2009;26:278–282. doi: 10.1016/j.fm.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Balslev U, Bremmelgaard A, Svejgaard E, Havstreym J, Westh H. An outbreak of borderline oxacillin-resistant Staphylococcus aureus(BORSA) in a dermatological unit. Microb. Drug Resist. 2005;11:78–81. doi: 10.1089/mdr.2005.11.78. [DOI] [PubMed] [Google Scholar]
- 27.Krupa P, Bystron J, Podkowik M, Empel J, Mroczkowska A, Bania J. Population Structure and Oxacillin Resistance of Staphylococcus aureus from Pigs and Pork Meat in South-West of Poland. Bio. Med. Res Int. 2015;2015:1–9. doi: 10.1155/2015/141475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oppliger A, Moreillon P, Charrière N, Giddey M, Morisset D, Sakwinska O. Antimicrobial resistance of Staphylococcus aureus strains acquired by pig farmers from pigs. Appl. Environ. Microbiol. 2012;78:8010–8014. doi: 10.1128/AEM.01902-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


