Abstract
Aim::
This study aimed to detect putative virulence genes in Arcobacter species of animal and human origin.
Materials and Methods::
A total of 41 Arcobacter isolates (16 Arcobacter butzleri, 13 Arcobacter cryaerophilus, and 12 Arcobacter skirrowii) isolated from diverse sources such as fecal swabs of livestock (21), raw foods of animal origin (13), and human stool samples (7) were subjected to a set of six uniplex polymerase chain reaction assays targeting Arcobacter putative virulence genes (ciaB, pldA, tlyA, mviN, cadF, and cj1349).
Results::
All the six virulence genes were detected among all the 16 A. butzleri isolates. Among the 13 A. cryaerophilus isolates, cadF, ciaB, cj1349, mviN, pldA, and tlyA genes were detected in 61.5, 84.6, 76.9, 76.9, 61.5, and 61.5% of isolates, respectively. Among the 12 A. skirrowii isolates, cadF, ciaB, cj1349, mviN, pldA, and tlyA genes were detected in 50.0, 91.6, 83.3, 66.6, 50, and 50% of isolates, respectively.
Conclusion::
Putative virulence genes were detected in majority of the Arcobacter isolates examined. The results signify the potential of Arcobacter species as an emerging foodborne pathogen.
Keywords: Arcobacter, Arcobacter butzleri, Arcobacter cryaerophilus, Arcobacter skirrowii, polymerase chain reaction, virulence genes
Introduction
The genus Arcobacter encompasses a group of Gram-negative, fastidious, nonspore forming, motile, spirally curved rods of the family Campylobacteraceae [1]. Arcobacter species differ from Campylobacter species in their aerotolerance and growth at 15°C [2]. In contrast with the acceptance of Escherichia coli, Salmonella, and Campylobacter species as the main foodborne pathogens, the reports dealing with the association of Arcobacter species are limited. Over the past few years, studies regarding the isolation of Arcobacter species from animals, raw foods of animal origin, and diarrheic humans signify the potential food safety concern associated with them [3-5]. Among several Arcobacter species, Arcobacter butzleri, Arcobacter cryaerophilus, and Arcobacter skirrowii are reported to have veterinary and public health significance [2]. Furthermore, the presence of virulence genes and its cytopathogenic effect on in vitro cell lines resulted in the categorization of A. butzleri as a “serious hazard” to human health by the International Commission on Microbiological Specifications for Foods [6].
Putative virulence determinants such as ciaB (encodes Campylobacter jejuni invasion antigen B that contributes to host cell invasion), mviN (encodes virulence factor, inner membrane protein required for peptidoglycan biosynthesis), pldA (encodes outer membrane phospholipase A associated with lysis of erythrocytes), tlyA (the hemolysin gene), and cadF and cj1349 (encodes fibronectin-binding proteins which promote the binding of bacteria to intestinal cells) were reported to be present in A. butzleri (RM4018) genome [7]. In addition, cadF (Campylobacter adhesion to fibronectin) protein also induces the internalization of bacterial cells by the activation of GTPases [7]. Despite increasing reports of the association of Arcobacter species with livestock and human diseases, the mechanisms of pathogenicity of Arcobacter species are still poorly understood [8,9].
Perusal of the available literature revealed a lack of information on virulence gene profiles of Arcobacter species of animal and human origin in India. The present study was carried out to characterize virulence gene profiles of Arcobacter species isolated from animals, foods of animal origin, and human sources in Andhra Pradesh, India.
Materials and Methods
Ethical approval
This work does not require ethical approval as we have collected fecal swabs after defecation.
Reference strain
The reference strain A. butzleri (ATCC 49616) used in the present study was obtained from the Division of Veterinary Public Health, Indian Veterinary Research Institute, Izatnagar.
Bacterial isolates
A total of 41 Arcobacter isolates recovered from diverse sources such as fecal swabs of livestock (21), raw foods of animal origin (13), and human stool samples (7) were used in this study. The identification of each isolate was carried out using the following tests: Gram-staining (Gram-negative, short ‘S’ shaped rods), dark-field microscopy (corkscrew motility), oxidase (positive), catalase (positive), nitrate reduction (positive), and hippurate hydrolysis (negative) [10]. Further, all the 41 isolates were confirmed at genus level as Arcobacter by genus-specific polymerase chain reaction (PCR) targeting 16S rRNA gene [11] and at species level as A. butzleri (16), A. cryaerophilus (13), and A. skirrowii (12) by multiplex PCR targeting 16S and 23S rDNA [12]. Arcobacter isolates from fecal swabs of livestock include those from pigs (8), chicken (6), turkey (2), cattle (2), sheep (2), and duck (1). Arcobacter isolates from raw foods of animal origin include those from chicken (5), pork (4), milk (2), and mutton (2). Arcobacter isolates from human stool samples include those from farm workers of pig/poultry (3), veterinary students (2), and diarrheic humans (2). Whole-cell DNA was extracted by boiling and snap chilling method [4]. The absorbance of the DNA at wavelengths 260 and 280 nm was measured using Nanodrop (Thermo Scientific, USA).
Detection of putative virulence genes by PCR
All the 41 Arcobacter isolates were subjected to a set of six uniplex PCR assays for the detection of six putative virulence genes (ciaB, pldA, tlyA, mviN, cadF, and cj1349) [13]. Primer sequences, expected amplicon sizes, and virulence gene details are listed in Table-1. A preliminary gradient PCR was conducted with DNA of A. butzleri (ATCC 49616) which carried all the six putative virulence genes to assess the correct annealing temperature. With annealing temperatures lower than 56.0°C (for ciaB, cj1349, and mviN genes) and 58.0°C (for cadF, pldA, and tlyA genes), lower molecular weight products were detected in addition to expected amplicons. Thus, annealing temperatures of 56.0°C (for amplification of ciaB, cj1349, and mviN genes) and 58.0°C (for cadF, pldA, and tlyA genes) were optimized for future PCR reactions. All the six PCR assays were carried out in Eppendorf thermal cycler (USA) with a heated lid.
Table-1.
Oligonucleotide primers used for detection of Arcobacter putative virulence genes.
| Primer/Target gene | Virulence factor | Nucleotide sequence (5’ 3’) | Amplicon size (bp) |
|---|---|---|---|
| cadF | Fibronectin-binding proteins | TTACTCCTACACCGTAGT | 283 |
| AAACTATGCTAACGCTGGTT | |||
| ciaB | Invasion antigen B | TGGGCAGATGTGGATAGAGCTTGGA | 284 |
| TAGTGCTGGTCGTCCCACATAAAG | |||
| cj1349 | Fibronectin-binding proteins | CCAGAAATCACTGGCTTTTGAG | 659 |
| GGGCATAAGTTAGATGAGGTTCC | |||
| mviN | Virulence factor | TGCACTTGTTGCAAAACGGTG | 294 |
| TGCTGATGGAGCTTTTACGCAAGC | |||
| pldA | Phospholipase A | TTGACGAGACAATAAGTGCAGC | 293 |
| CGTCTTTATCTTTGCTTTCAGGGA | |||
| tlyA | Hemolysin | CAAAGTCGAAACAAAGCGACTG | 230 |
| TCCACCAGTGCTACTTCCTATA |
The PCR assays for ciaB, cj1349, and mviN genes were optimized in 25 µl reaction mixture containing 2 µl of DNA template; Taq buffer (10×) - 2.50 μl; dNTP mix (10 mM) - 1 μl; MgCl2 (25 mM) - 0.75 μl; forward primer (10 pmol/μl) - 1.50 μl; reverse primer (10 pmol/μl) - 1.50 μl; Taq DNA polymerase (1 U/μl) - 1 μl; and nuclease free water - 14.75 μl, under the following standardized cycling conditions: initial denaturation at 94°C for 5 min, 30 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 45 s, elongation at 72°C for 1 min, and final elongation at 72°C for 10 min and hold at 4°C.
The PCR assays for cadF, pldA, and tlyA genes were optimized in 25 µl reaction mixture containing 2 µl of DNA template; Taq buffer (10×) - 2.50 μl; dNTP mix (10 mM) - 1 μl; MgCl2 (25 mM) - 1.5 μl; forward primer (10 pmol/μl) - 1.0 μl; reverse primer (10 pmol/μl) - 1.0 μl; Taq DNA polymerase (1 U/μl) - 1 μl; and nuclease free water - 15.0 μl, under the following standardized thermal cycling conditions: initial denaturation at 94°C for 5 min, 30 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 45 s, elongation at 72°C for 1 min, and final elongation at 72°C for 10 min and hold at 4°C.
Results and Discussion
Putative virulence genes were detected in majority of screened Arcobacter isolates. The ciaB gene was predominant gene detected (92.6%, 38/41 isolates), followed by cj1349 (87.8%, 36/41), mviN (82.9%, 34/41), cadF (73.1%, 30/41), pldA (73.1%, 30/41), and tlyA (73.1%, 30/41) (Figure-1 and Table-2). All the 16 A. butzleri isolates carried all six putative virulence genes. None of the 13 A. cryaerophilus and 12 A. skirrowii isolates possessed all the six virulence genes. The present results were in accordance with the previous studies from Berlin [9] and Belgium [13] where 100% prevalence of these putative virulence genes in A. butzleri strains was reported.
Figure-1.
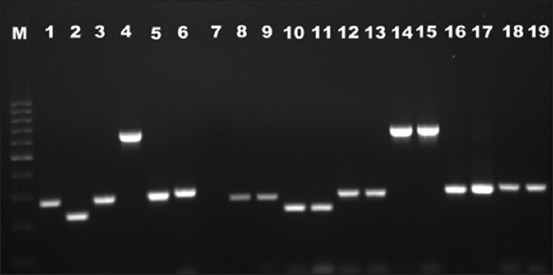
Gel photograph of polymerase chain reaction assays targeting Arcobacter putative virulence genes; Lane M: Molecular weight marker (100 bp); L1-6: Positive control of Arcobacter butzleri (ATCC 49616) carrying all 6 putative virulence genes, i.e., cadF (283 bp), tlyA (230 bp), ciaB (284 bp), cj1349 (659 bp), pldA (293 bp), and mviN (294 bp); L7: Negative control; L8: A. butzleri isolate with cadF (283 bp) gene; L9: Arcobacter cryaerophilus isolate with cadF (283 bp) gene; L10: A. butzleri isolate with tlyA (230 bp) gene; L11: Arcobacter skirrowii isolate with tlyA (230 bp) gene; L12: A. butzleri isolate with ciaB (284 bp) gene; L13: A. cryaerophilus isolate with ciaB (284 bp) gene; L14: A. butzleri isolate with cj1349 (659 bp) gene; L15: A. skirrowii isolate with cj1349 (659 bp) gene; L16: A. butzleri isolate with pldA (293 bp) gene; L17: A. cryaerophilus isolate with pldA (293 bp) gene; L18: A. butzleri isolate with mviN (294 bp) gene; L19: A. skirrowii isolate with mviN (294 bp) gene.
Table-2.
Putative virulence-associated genes detected in Arcobacter isolates using PCR.
| Species and source | Number of strains examined | Number of strains generating specific gene amplicon | |||||
|---|---|---|---|---|---|---|---|
| cadF | ciaB | cj1349 | mviN | pldA | tlyA | ||
| A. butzleri | |||||||
| Poultry feces | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Pig feces | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Cattle feces | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Chicken meat | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Pork | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Milk | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Veterinary students | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Farm workers | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Diarrheic humans | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Total (%) | 16 | 16 (100) | 16 (100) | 16 (100) | 16 (100) | 16 (100) | 16 (100) |
| A. cryaerophilus | |||||||
| Poultry feces | 2 | 1 | 2 | 2 | 2 | 1 | 1 |
| Pig feces | 3 | 1 | 2 | 2 | 2 | 2 | 2 |
| Cattle feces | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| Chicken meat | 3 | 2 | 3 | 3 | 2 | 2 | 2 |
| Pork | 3 | 2 | 2 | 1 | 2 | 2 | 2 |
| Milk | 1 | 1 | 1 | 1 | 1 | 0 | 1 |
| Total (%) | 13 | 8 (61.5) | 11 (84.6) | 10 (76.9) | 10 (76.9) | 8 (61.5) | 8 (61.5) |
| A. skirrowii | |||||||
| Poultry feces | 5 | 3 | 5 | 4 | 4 | 2 | 3 |
| Pig feces | 3 | 1 | 3 | 2 | 1 | 2 | 1 |
| Sheep feces | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Mutton | 2 | 1 | 1 | 2 | 2 | 1 | 1 |
| Total (%) | 12 | 6 (50) | 11 (91.6) | 10 (83.3) | 8 (66.6) | 6 (50) | 6 (50) |
| Grand total (%) | 41 | 30 (73.1) | 38 (92.6) | 36 (87.8) | 34 (82.9) | 30 (73.1) | 30 (73.1) |
PCR=Polymerase chain reaction, A. butzleri=Arcobacter butzleri, A. cryaerophilus=Arcobacter cryaerophilus, A. skirrowii=Arcobacter skirrowii
Among the 13 A. cryaerophilus isolates, ciaB (84.6%, 11/13 isolates), cj1349 (76.9%, 10/13), and mviN (76.9%, 10/13) genes were detected frequently, followed by cadF (61.5%, 8/13), pldA (61.5%, 8/13), and tlyA (61.5%, 8/13) genes. In a study from Chile, putative virulence genes such as ciaB (100%), mviN (100%), tlyA (66.7%), pldA (33.3%), and cj1349 (14.3%) were detected in Arcobacter species, and cadF gene was reported to be not detected in Arcobacter species [14]. Among 12 A. skirrowii isolates, ciaB (91.6%, 11/12 isolates), cj1349 (83.3%, 10/12), and mviN (66.6%, 8/12) genes were frequently detected, followed by cadF (50%, 6/12), pldA (50%, 6/12), and tlyA (50%, 6/12) genes. In a study from Chile, putative virulence genes such as ciaB (93.8%), mviN (81.3%), tlyA (68.8%), cj1349 (31.3%), pldA (25%), and cadF (12.5%) genes were reported to be detected in Arcobacter species [14]. Predominance of putative virulence genes in A. butzleri was noticed compared to A. cryaerophilus and A. skirrowii, as evidenced by detection of all the six putative virulence genes in all the 16 A. butzleri isolates, whereas none of the 13 A. cryaerophilus and 12 A. skirrowii isolates carried all the six virulence genes, which was in agreement with the previous studies [14,15]. This may indicate possibility of differential pathogenic behavior of Arcobacter species or higher genomic heterogeneity [13] or bias from the use of A. butzleri ATCC 49616 genome sequence only in designing the virulence gene primers [8].
In a study from Belgium [13], three Arcobacter type strains (i.e., A. butzleri LMG 10828, A. cryaerophilus LMG 10210, and A. skirrowii LMG 6621) were reported to carry nine putative Campylobacter virulence genes (cadF, ciaB, cj1349, hecA, hecB, irgA, mviN, pldA, and tlyA). In addition, no significant difference was noticed in cadF gene detected among different strains of Arcobacter isolated from humans, cattle, horses, sheep, dogs, and chicken [13]. However, the role of these putative virulence determinants or Campylobacter homologs in the pathogenicity Arcobacter species was still contradictory [9,16]. Arcobacter species were reported to have ability to adhere and invade the intestinal epithelial cells of host and induce inflammatory responses [17]. Arcobacter infection was reported to be associated with leak flux type of watery diarrhea resulting from epithelial barrier dysfunction [18], and adhesion, invasion and toxin production could be the mechanisms of Arcobacter pathogenicity [9,19].
Conclusion
Food safety needs a thorough investigation of virulence properties of potentially emerging pathogenic bacteria in animals and foods of animal origin. The presence of Arcobacter strains carrying virulence markers in animals, humans and foods of animal origin have not yet been a subject of investigation in India. The present study reporting the presence of six virulence-associated genes of three emerging Arcobacter species isolated from animal and human sources adds to the significance of Arcobacter as an emerging foodborne pathogen with zoonotic potential. Further studies should focus on examining the interactions of Arcobacter species virulence factors with eukaryotic cells.
Authors’ Contributions
MSS is the student worked for M.V.Sc thesis. SRT as a major guide and BKC and VSK as minor guides designed and supervised the research work. NMS collected the samples and executed the isolation. MSS was involved in the molecular characterization. The manuscript was drafted and revised by MSS and NMS under the guidance of SRT, BKC, and VSK. All authors read and approved the final manuscript.
Acknowledgments
The authors would like to thank Sri Venkateswara Veterinary University, Tirupati, Andhra Pradesh, for providing necessary funds (grant number 2370/BG/B1/2016, dated 13-04-2016 of the Comptroller, SVVU, Tirupati to the department of Veterinary Public Health and Epidemiology, NTR C.V.Sc., Gannavaram) and facilities to carry out the research work.
Competing Interests
The authors declare that they have no competing financial interests.
References
- 1.Vandamme P, Vancanneyt M, Pot B, Mels L, Hoste B, Dewettinck D, Vlaes L, Van den Borre C, Higgins R, Hommez J, Kersters K. Polyphasic taxonomic study of the emended genus Arcobacter with Arcobacter butzleri comb. nov Arcobacter skirrowii sp. nov, an aerotolerant bacterium isolated from veterinary specimens. Int. J. Syst. Evol. Microbiol. 1992;42(3):344–356. doi: 10.1099/00207713-42-3-344. [DOI] [PubMed] [Google Scholar]
- 2.Collado L, Figueras M.J. Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin. Microbiol. Rev. 2011;24(1):174–192. doi: 10.1128/CMR.00034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patyal A, Rathore R.S, Mohan H.V, Dhama K, Kumar A. Prevalence of Arcobacter spp. in humans, animals and foods of animal origin including sea food from India. Transbound. Emerg. Dis. 2011;58(5):402–410. doi: 10.1111/j.1865-1682.2011.01221.x. [DOI] [PubMed] [Google Scholar]
- 4.Ramees T.P, Rathore R.S, Bagalkot P.S, Mohan H.V, Kumar A, Dhama K. Detection of Arcobacter butzleri and Arcobacter cryaerophilus in clinical samples of humans and foods of animal origin by cultural and multiplex PCR based methods. Asian J. Anim. Vet. Adv. 2014;9:243–252. [Google Scholar]
- 5.Mohan H.V, Rathore R.S, Dhama K, Ramees T.P, Patyal A, Bagalkot P.S, Wani M.Y, Bhilegaonkar K.N, Kumar A. Prevalence of Arcobacter spp. In humans, animals and foods of animal origin in India based on cultural isolation, antibiogram, PCR and multiplex PCR detection. Asian J. Anim. Vet. Adv. 2014;9(8):452–466. [Google Scholar]
- 6.ICMSF. 2002 in foods. 7. Microbiological testing in food safety management. International Commission on Microbiological Specifications for Foods. New York: Kluwer Academic Plenum Publishers; 2002. [Google Scholar]
- 7.Miller W.G, Parker C.T, Rubenfield M, Mendz G.L, Wosten M.M, Ussery D.W, Stolz J.F, Binnewies T.T, Hallin P.F, Wang G, Malek J.A. 2007 complete genome sequence and analysis of the epsilon proteobacterium Arcobacter butzleri. PLoS One. 2007;2(12):e1358. doi: 10.1371/journal.pone.0001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levican A, Alkeskas A, Günter C, Forsythe S.J, Figueras M.J. Adherence to and invasion of human intestinal cells by Arcobacter species and their virulence genotypes. Appl. Environ. Microbiol. 2013;79(16):4951–4957. doi: 10.1128/AEM.01073-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karadas G, Sharbati S, Hanel I, Messelhauber U, Glocker E, Alter T, Golz G. 2013 of virulence genes, adhesion and invasion of Arcobacter butzleri. J. Appl. Microbiol. 2013;115(2):583–590. doi: 10.1111/jam.12245. [DOI] [PubMed] [Google Scholar]
- 10.Vandamme P, Dewhirst F.E, Paster B.J, On S.L.W, Brenner D.J, Kreig N.P, Staley J.T, Garrity G.M Genus II Arcobacter. In:Bergey’s Manual of Systematic Bacteriology. New York, USA: Springer; 2005. [Google Scholar]
- 11.Harmon K.M, Wesley I.V. Identification of Arcobacter isolates by PCR. Lett. Appl. Microbiol. 1996;23(4):241–244. doi: 10.1111/j.1472-765x.1996.tb00074.x. [DOI] [PubMed] [Google Scholar]
- 12.Houf K, Tutenel A, de Zutter L, van Hoof J, Vandamme P. Development of a multiplex PCR assay for the simultaneous detection and identification of Arcobacter butzleri, Arcobacter cryaerophilus and Arcobacter skirrowii. FEMS Microbiol. Lett. 2000;193(1):89–94. doi: 10.1111/j.1574-6968.2000.tb09407.x. [DOI] [PubMed] [Google Scholar]
- 13.Douidah L, de Zutter L, Bare J, de Vos P, Vandamme P, Vandenberg O, van den Abeele A.M, Houf K. Occurrence of putative virulence genes in Arcobacter species isolated from humans and animals. J. Clin. Microbiol. 2012;50(3):735–741. doi: 10.1128/JCM.05872-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collado L, Jara R, Vasquez N, Telsaint C. Antimicrobial resistance and virulence genes of Arcobacter isolates recovered from edible bivalve molluscs. Food Control. 2014;46:508–512. [Google Scholar]
- 15.Tabatabaei M, Aski H.S, Shayegh H, Khoshbakht R. Occurrence of six virulence-associated genes in Arcobacter species isolated from various sources in Shiraz, Southern Iran. Microb. Pathog. 2014;66:1–4. doi: 10.1016/j.micpath.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira S, Fraqueza M.J, Queiroz J.A, Domingues F.C, Oleastro M. Genetic diversity, antibiotic resistance and biofilm-forming ability of Arcobacter butzleri isolated from poultry and environment from a Portuguese slaughterhouse. Int. J. Food Microbiol. 2013;162(1):82–88. doi: 10.1016/j.ijfoodmicro.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Ho H.T, Lipman L.J, Hendriks H.G, Tooten P.C, Ultee T, Gaastra W. Interaction of Arcobacter spp. with human and porcine intestinal epithelial cells. FEMS Immunol. Med. Microbiol. 2007;50(1):51–58. doi: 10.1111/j.1574-695X.2007.00230.x. [DOI] [PubMed] [Google Scholar]
- 18.Bucker R, Troeger H, Kleer J, Fromm M, Schulzke J.D. Arcobacter butzleri induces barrier dysfunction in intestinal HT-29/B6 cells. J. Infect. Dis. 2009;200(5):756–764. doi: 10.1086/600868. [DOI] [PubMed] [Google Scholar]
- 19.Etonsi M.A. 2013 on Arcobacter species Their Isolation and Pathogenicity. Scotland: Ph.D Thesis Submitted to Heriot-Watt University; 2013. [Google Scholar]


