Abstract
Microproteins are peptides and small proteins encoded by small open reading frames (smORFs). Newer technologies have led to the recent discovery of hundreds to thousands of new microproteins. The biological functions of a few microproteins have been elucidated, and these microproteins have fundamental roles in biology ranging from limb development to muscle function, highlighting the value of characterizing these molecules. The identification of microprotein–protein interactions (MPIs) has proven to be a successful approach to the functional characterization of these genes; however, traditional immunoprecipitation methods result in the enrichment of nonspecific interactions for microproteins. Here, we test and apply an in situ proximity tagging method that relies on an engineered ascorbate peroxidase 2 (APEX) to elucidate MPIs. The results demonstrate that APEX tagging is superior to traditional immunoprecipitation methods for microproteins. Furthermore, the application of APEX tagging to an uncharacterized microprotein called C11orf98 revealed that this microprotein interacts with nucleolar proteins nucleophosmin and nucleolin, demonstrating the ability of this approach to identify novel hypothesis-generating MPIs.
Bioactive peptides have essential roles in biology. For example, the hormone insulin, which is secreted by β-cells in the pancreas, regulates blood glucose levels.1−3 Most peptide hormones and neuropeptides share a common biosynthetic pathway that produces the mature peptide after limited proteolysis of a longer precursor protein (i.e., a prepropeptide or propeptide).4,5 Outside of some notable exceptions, such as angiotensin, most bioactive peptides were thought to be produced through this secretory pathway, but this view has begun to change as novel peptides, or microproteins, encoded by small open reading frames (smORFs) are steadily being discovered.6,7
smORFs were missed during genome annotations because they are too short for gene-finding algorithms, or the RNAs that encode these smORFs were not known.8,9 Instead, the detection of smORFs and microproteins has relied on the advent and application of novel genomics and proteomics methods.10,11 So far, hundreds to thousands of smORFs and the corresponding microprotein products have been identified from prokaryotic and eukaryotic genomes,12−14 and biological studies in flies and mammals have demonstrated that at least some of these microproteins have fundamental biological functions in development, metabolism, and muscle function.15−17
Because only a few of the discovered smORFs have been characterized, the functional characterization of these microproteins represents a major challenge in the field. A common feature of functional microproteins is that they all seem to partake in protein–protein interactions or, more specifically, microprotein–protein interactions to regulate biology. For example, Magny and colleagues demonstrated that a Drosophila microprotein called sarcolamban (Scl) binds to an endoplasmic reticulum (ER) calcium channel, SERCA, regulating channel function and heart muscle contraction,17 and mammalian homologues of Scl that inhibit or activate SERCA have been identified and provide new insights into musculoskeletal biology.18−20 Other examples, such as a smORF that regulates limb development in flies16,21−23 and a smORF called NoBody that regulates mRNA decapping,24 also operate through microprotein–protein interactions. Consequently, the elucidation of the proteins and protein complexes that associate with microproteins can be used to characterize the functions of microproteins.
Immunoprecipitation of FLAG-tagged microproteins provides a general approach for revealing microprotein-associated proteins. These proteins could be direct interactors of the microprotein or parts of a larger complex that contain the microprotein. For example, the 69-amino acid microprotein modulator of retroviral infection (MRI) associates with Ku70 and Ku80, two essential proteins that mediate cellular repair of double-stranded DNA breaks (i.e., nonhomologous end joining DNA repair).25 The interaction between the MRI microprotein and Ku70/Ku80 suggests that this microprotein is involved in cellular DNA repair, highlighting the utility of defining microprotein-associated proteins as a powerful hypothesis-generating approach.
In addition to Ku70 and Ku80, the immunoprecipitation of MRI microprotein also enriched housekeeping and heat shock proteins. Imaging studies ruled out cytosolic heat shock proteins as bona fide interactors because MRI microprotein localizes to the nucleus where it associates with Ku70 and Ku80.25,26 We believe, however, that MRI may be intrinsically unfolded and that the interaction with heat shock proteins occurs after the cells are lysed during the immunoprecipitation. The identification of many of the same heat shock proteins during the immunoprecipitation of completely unrelated microproteins being studied in our lab (unpublished results) indicates that microproteins might be particularly susceptible to artifacts generated by interactions with heat shock proteins in lysates. Therefore, we needed to find a better approach for identifying microprotein-associated proteins and protein complexes to characterize these novel genes.
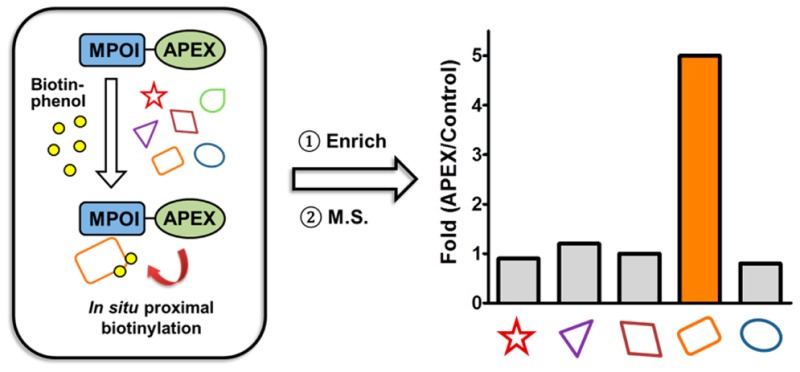
Ting and colleagues developed an ingenious in situ proximity labeling method using an engineered ascorbate peroxidase27 and then optimized the stability and activity of this enzyme through evolution to afford APEX228−31 (the APEX abbreviation refers to the APEX2 protein in the text and figures). In this approach, APEX is fused to a protein of interest. Expression of the APEX fusion protein followed by treatment of cells with hydrogen peroxide (H2O2) in the presence of biotin-phenol covalently labels proteins proximal to the APEX fusion protein with biotin. In this scheme, the H2O2 fuels the catalytic oxidation of biotin-phenol by APEX to generate a highly reactive biotin-phenoxyl radical. The lifetime of the radical is <1 ms that restricts the labeling radius to 20 nm. These biotinylated proteins can then be enriched and analyzed by mass spectrometry, and because proteins adjacent to the APEX fusion protein are preferentially biotinylated, the resulting mass spectrometry data provide a readout of the protein environment around the fusion protein (Figure 1).
Figure 1.
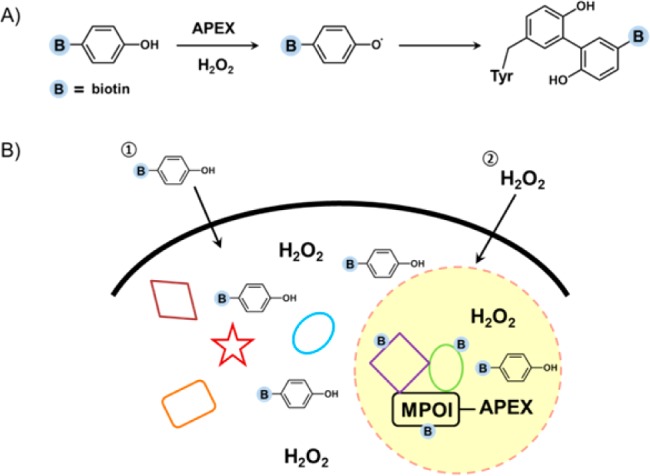
Schematic illustration of the identification of microprotein-associated proteins by APEX tagging. (A) Biotin-phenol is an APEX substrate, and the phenol is converted to a phenoxyl radical by APEX upon H2O2 treatment. The highly reactive phenoxyl radical forms covalent bonds with nearby aromatic residues such as tyrosine. (B) APEX tagging can be applied to the identification of microprotein-associated proteins by fusing the microprotein of interest (MPOI) to APEX. Cells expressing the MPOI–APEX fusion protein are pretreated with biotin-phenol followed by the addition of H2O2 to initiate biotin labeling. The hyper-reactivity of the biotin-phenoxyl radical results in a short half-life to favor labeling of nearby proteins, and any biotinylated proteins are considered to be near the microprotein. The biotinylated proteins are identified by streptavidin enrichment and proteomics.
Using this method, Ting and co-workers comprehensively mapped proteins to distinct intracellular compartments such as mitochondrial intermembrane space and mitochondrial matrix.27,32 APEX fusion proteins have also been used to identify the proteome at junctions between the plasma membrane (PM) and endoplasmic reticulum (ER), demonstrating the generality of this approach.33 We hypothesized that APEX–microprotein fusion proteins could solve the background issues with identifying microprotein-associated proteins and protein complexes because the interactions take place in the context of a living cell, not a lysate. Here, we demonstrate that APEX–microprotein fusion proteins provide a superior approach to identifying microprotein–protein interactions by comparing MRI–FLAG immunoprecipitation to an MRI–APEX experiment. Then we demonstrate that APEX–microprotein fusion proteins can be used to identify novel proteins that interact with the microprotein and protein complexes using a microprotein from the C11orf98 gene. These results highlight the value of APEX tagging for discovering microprotein-associated proteins.
Experimental Procedures
Biotin-Phenol Labeling in Live Cells
Biotin-phenol labeling in live cells was performed according to previously published protocols.29 Briefly, constructs harboring microprotein–APEX fusion proteins or the APEX control were transiently transfected into HEK293T cells using Lipofectamine 2000. Twenty-four hours post-transfection, cell culture medium was changed to fresh growth medium containing 500 μM biotin-tyramide (CDX-B0270, Adipogen). After incubation at 37 °C for 30 min, H2O2 was added to each plate at a final concentration of 1 mM, and the plates were gently agitated for 1 min. Cells were then washed three times with a quenching solution [5 mM Trolox, 10 mM sodium azide, and 10 mM sodium ascorbate in phosphate-buffered saline (PBS)], and the pellet was collected by centrifugation at 1000g for 5 min.
Western Blot and Proteomic Analysis of Biotin-Phenol Labeling
Cell pellets were lysed on ice for 20 min in RIPA buffer (Thermo catalog no. 89901) supplemented with a Roche complete protease inhibitor cocktail tablet and 1 mM phenylmethanesulfonyl fluoride (PMSF) followed by centrifugation at 20000g for 20 min at 4 °C to remove cell debris. Cell lysates were added to prewashed streptavidin agarose resin (Thermo catalog no. 20359), rotated at 4 °C for 4 h, and then washed three times with TBST and 0.5% (v/v) sodium dodecyl sulfate (SDS). Bound proteins were eluted with 2× SDS loading buffer and analyzed by Western blotting. For proteomics, eluted samples were precipitated with trichloroacetic acid (TCA, MP Biomedicals catalog no. 196057) overnight at 4 °C. Dried pellets were dissolved in 8 M urea, reduced with 5 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP, Thermo catalog no. 20491), and alkylated with 10 mM iodoacetamide (Sigma I1149). Proteins were then digested overnight at 37 °C with trypsin (Promega V5111). The reaction was quenched with formic acid at a final concentration of 5% (v/v). Digested samples were analyzed on a Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer.
Co-Immunoprecipitation
FLAG-tagged microprotein constructs [or the empty pcDNA3.1(+) vector] were transfected into a 10 cm dish of HEK293T cells using Lipofectamine 2000 according to the manufacturer’s protocol. Forty-eight hours post-transfection, cells were harvested and lysed in RIPA buffer (Thermo catalog no. 89901) supplemented with a Roche complete protease inhibitor cocktail tablet and 1 mM PMSF. Cells were lysed on ice for 20 min followed by centrifugation at 20000g for 20 min at 4 °C to remove cell debris. Cell lysates were added to prewashed mouse IgG agarose beads (Sigma catalog no. A0919) and rotated at 4 °C for 1 h. The supernatants were collected and added to prewashed anti-FLAG M2 Affinity Gel (Sigma, catalog no. A2220). The suspensions were rotated at 4 °C overnight and washed four times with TBST. Bound proteins were eluted with 3× FLAG peptide (Sigma, catalog no. F4799) at 4 °C for 1 h. The eluents were then separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and analyzed by Western blotting using the indicated antibodies.
Reciprocal Immunoprecipitation
HEK293T cells were cotransfected with C11orf98-FLAG and NPM1-HA (empty vector as a control). Lysates from both samples were incubated with mouse anti-HA agarose beads (Sigma, catalog no. A2095) to immunoprecipitate HA-tagged NPM1. Alternatively, lysates from HEK293T cells co-expressing C11orf98-FLAG and NPM1-HA were incubated with either mouse IgG beads (Sigma, catalog no. A0919) or mouse anti-HA agarose beads. After being washed three times with TBST, bound proteins were eluted with HA peptide (Sigma, catalog no. I2149) at 4 °C for 1 h. The eluents were then separated by SDS–PAGE and analyzed by Western blotting using the indicated antibodies.
Immunofluorescence and Confocal Imaging
HeLa cells were seeded onto a coverslip (Fisher Scientific, catalog no. 12-541-B) in a six-well plate, which was pretreated with 50 μg/mL poly-l-lysine (Sigma, catalog no. P1399). The next day, cells were cotransfected with 1 μg of NPM1-HA and 1 μg of C11orf98-FLAG using Lipofectamine 2000. Forty-eight hours post-transfection, cells were fixed with 4% paraformaldehyde (Polysciences, Inc., catalog no. 18814) and permeabilized with 0.1% saponin (Alfa Aesar, catalog no. A18820). After being incubated with 4% BSA in PBS for 1 h at room temperature, cells were stained with primary antibodies (rabbit anti-FLAG and mouse anti-HA) at a 1:1000 dilution overnight at 4 °C. Then cells were washed three times with PBS, followed by incubation with secondary antibodies (goat anti-mouse Alexa Fluor 647 and goat anti-rabbit Alexa Fluor 488, 1:500 in PBS) for 1 h at room temperature. Nuclei were counterstained with Hoechst 33258 (Sigma, catalog no. 94403; 1:2000 in PBS). After three PBS washes, the coverslip was mounted with Prolong Gold Antifade Mountant (Life Technologies, catalog no. P36930) and submitted for confocal imaging using a Zeiss LSM 710 laser scanning confocal microscope with a 63× oil immersion objective. Images were analyzed with FIJI software.
Results and Discussion
Live Cell Proximity Labeling Using the MRI–APEX Fusion
We used the MRI microprotein to optimize the elucidation of microprotein-associated proteins. As mentioned, MRI interacts with the Ku70/Ku80 heterodimer, and therefore, our readout would be the addition of biotin to these proteins. Our construct consists of an APEX–myc fusion at the C-terminus of an N-terminal FLAG-tagged MRI microprotein (MRI–APEX), and APEX was used as a control in these experiments (Figure 2A). We were concerned that the addition of the much larger APEX protein to MRI might interfere with Ku70/Ku80 binding and used the FLAG tag to test whether the complex was intact. Transient transfection of HEK293T cells with the MRI–APEX fusion protein, followed by FLAG immunoprecipitation, enriched Ku70 and Ku80 and confirmed that the MRI–APEX fusion protein still binds to the Ku70/Ku80 heterodimer (Figure 2B and Figure S1).
Figure 2.
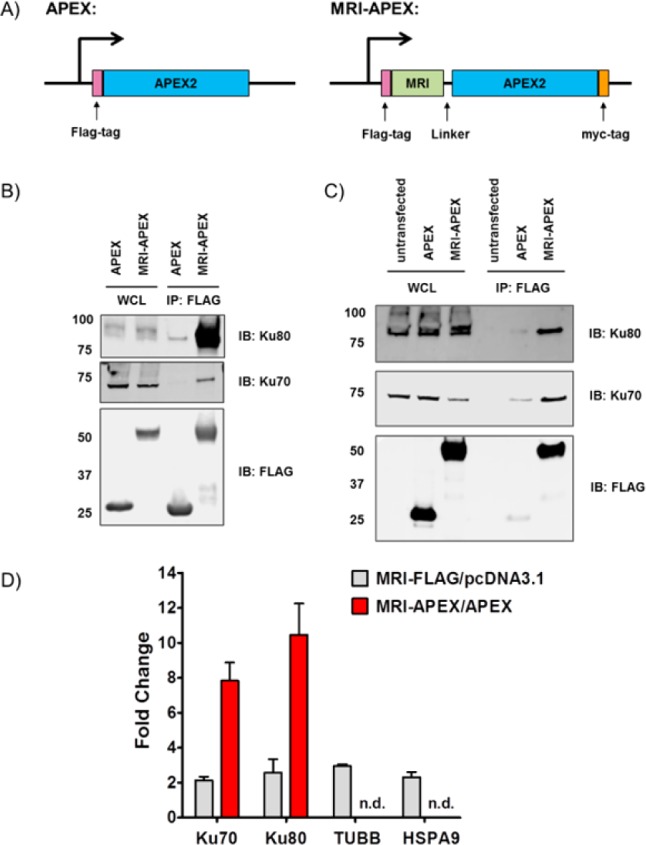
Identification of MRI microprotein–protein interactions in live cells by APEX tagging. (A) Schematic illustration of APEX and MRI–APEX constructs. (B) Anti-FLAG immunoprecipitation of HEK293T cells expressing APEX or the MRI–APEX fusion. Eluted proteins were separated by SDS–PAGE and visualized by Western blotting using the indicated antibodies (WCL, whole cell lysate). (C) Western blot of the MRI–APEX fusion labeling proteome indicating that the Ku70/Ku80 complex is selectively biotinylated by the MRI–APEX fusion. (D) Spectral count analysis indicated that APEX labeling has an improved fold change and a lower background compared to those of FLAG IP. Error bars represent the standard error of the mean of triplicate biological tests.
Next, we tested whether Ku70 and Ku80 are biotinylated after treatment of MRI–APEX fusion-expressing HEK293T cells with biotin-phenol and H2O2. After H2O2 treatment, cells were lysed and biotinylated proteins were enriched from the lysate using streptavidin beads. Western blotting of the biotin pull down revealed that Ku70 and Ku80 were biotinylated in cells transfected with the MRI–APEX fusion but not the APEX control or untransfected samples (Figure 2C), indicating successful labeling of known MRI-associated proteins. Furthermore, total cell lysates from the MRI–APEX fusion and APEX are biotinylated to the same extent, showing that the increase in the level of Ku70 and Ku80 biotinylation is due to their proximity to MRI and not due to a difference in biotinylation activity between the two samples (Figures S2 and S3).
To determine whether the APEX approach provided data that were superior to those from FLAG immunoprecipitation, we performed a shotgun proteomics experiment and analyzed the enrichment of Ku70 and Ku80 (Tables S1 and S2). The APEX conditions provided a dramatically improved fold enrichment for Ku70 and Ku80 (Figure 2D). In the FLAG sample, where the pull down occurs in cellular lysates, we find an ∼2-fold enrichment for Ku70 and Ku80 (MRI–FLAG fusion vs pcDNA3.1-transfected samples), but in situ proximity labeling with the MRI–APEX fusion resulted in an 8–10-fold increase for the same proteins (MRI–APEX fusion- vs APEX-transfected samples). This superior enrichment is due to the significantly lower background of these proteins under the APEX conditions. Also, we analyzed the enrichment of tubulin (TUBB) and heat shock 70 kDa protein 9 (HSPA9). These two proteins were enriched in our previous studies.25 FLAG pull down enriched these proteins ∼2-fold, but they were not detected (n.d.) in the MRI–APEX samples. The observed enrichment of TUBB and HSPA9 by the MRI–FLAG fusion highlights the problem with immunoprecipitations from cellular lysates. The FLAG immunoprecipitation, which disrupts cellular localization due to cell lysis, indicates a protein interaction between MRI and TUBB and HSPA9. By contrast, the MRI–APEX fusion experiment that takes place in an intact cell shows no enrichment of TUBB or HSPA9. Because the cytosolic TUBB or HSPA9 proteins should not interact with nuclear MRI microprotein, it follows that APEX provides a more accurate picture of cellular microprotein–protein interactions by avoiding interactions that occur only upon cell lysis. The power of this approach clearly demonstrates the benefit of using APEX tagging to identify microprotein-associated proteins while weakening nonspecific interactions and improving fold enrichment for target proteins.
APEX Fusions of C11orf98 to Discover Interactors
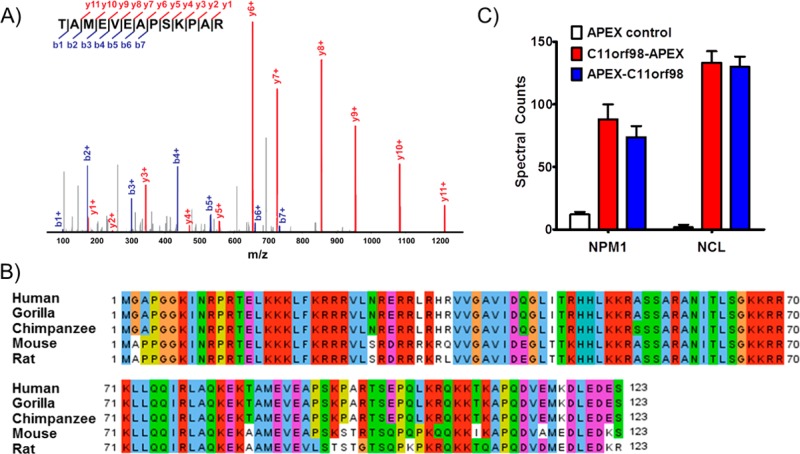
We sought to apply the APEX labeling approach to identify interaction partners for uncharacterized microprotein interactions. We chose an uncharacterized 123-amino acid microprotein encoded by the C11orf98 smORF. A tryptic peptide from this microprotein was detected in HEK293T cells by shotgun mass spectrometry (Figure 3A). The C11orf98 transcript is identified as a validated protein-coding gene in RefSeq, but this gene remains uncharacterized. The C11orf98 microprotein is highly conserved between humans and mice, suggesting a potential function (Figure 3B).
Figure 3.
Identification of C11orf98 microprotein-associated proteins. (A) MS/MS spectrum of the unique C11orf98 tryptic peptide, with detected fragment ions marked in blue (b-ions) and red (y-ions). (B) A multiple-sequence alignment of the C11orf98 microprotein indicates that it is highly conserved in mammals. (C) Semiquantitative proteomics by spectral counting revealed that NPM1 and NCL were enriched ∼7- and ∼80-fold, respectively, in both N- and C-terminal APEX fusion samples compared to APEX control samples. Error bars represent the standard deviation of triplicate biological tests.
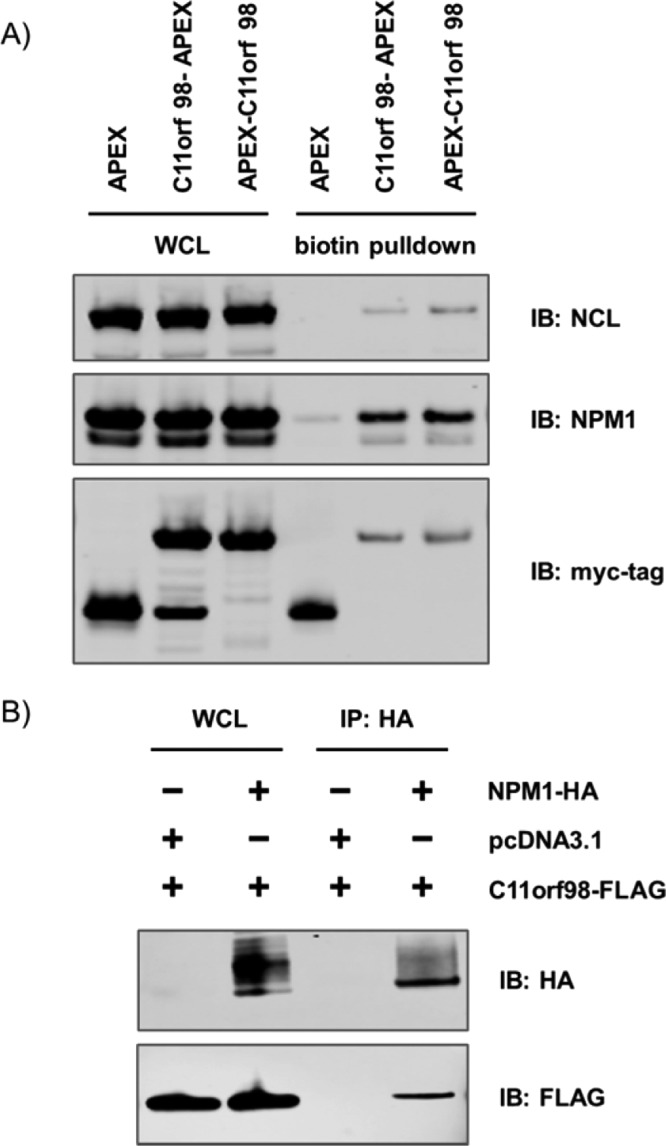
We expressed an N-terminal APEX-tagged C11orf98 (APEX–C11orf98) or a C-terminal APEX-tagged C11orf98 (C11orf98–APEX) fusion protein in HEK293T cells to characterize this microprotein (Figure S4). Cells were then treated with biotin-phenol and H2O2, followed by harvesting of the cells and cell lysis. Biotinylated proteins were isolated from each sample by streptavidin beads and analyzed by proteomics. Unfused APEX was used as the control in these experiments.
Candidate C11orf98-interacting proteins were identified by filtering the proteomics data for proteins with a spectral count of >5, a >2-fold increase versus the control sample, and a p value of <0.05. This analysis resulted in 112 proteins for the APEX–C11orf98 fusion and 137 proteins for the C11orf98–APEX fusion, with 99 proteins present in both data sets (Tables S3 and S4). The strong overlap between the APEX–C11orf98 and C11orf98–APEX data sets provides additional confidence in the reliability of these data.
The majority of proteins enriched by APEX–C11orf98 and C11orf98–APEX fusions are reported to have a nuclear localization according to Human Protein Atlas [97 of 112 for the APEX–C11orf98 fusion and 115 of 137 for C11orf98 (Figure S5)]. Two of the most robust C11orf98-associated proteins are nucleolin (NCL) and nucleophosmin (NPM1) (Figure 3C). NCL had the largest number of spectral counts in both the APEX–C11orf98 and C11orf98–APEX data sets, and an ∼80-fold increase with respect to the control. NPM1, an interaction partner of NCL, was also found among the top hits with a strong signal and an ∼7-fold increase in the APEX–C11orf98 and C11orf98–APEX data sets. Given the high spectral counts and robust fold changes for NCL and NPM1, and the established interaction between NCL and NPM1,34,35 our data indicate that C11orf98 is associated with an NCL and NPM1 complex. These criteria led us to focus on characterizing the C11orf98–NPM1–NCL interactions, but we could not completely rule out the possibility that C11orf98 may also interact with other proteins in the list, which might end up being important in the biology.
Validation of Interactions of the C11orf98 Microprotein with NPM1 and NCL
To validate the association among NPM1, NCL, and the C11orf98 microprotein, we repeated the APEX–C11orf98 and C11orf98–APEX experiments and performed Western blots using NPM1 and NCL specific antibodies. Consistent with the proteomics data, we observed the enrichment of NPM1 and NCL after streptavidin enrichment of lysates from the APEX–C11orf98 and C11orf98–APEX samples. NPM1 and NCL were not enriched in the control sample with an unfused APEX (Figure 4A and Figure S6). In addition, immunoprecipitation of a FLAG-tagged C11orf98 microprotein also enriched the NPM1 and NCL, which demonstrated that the interactions among C11orf98, NPM1, and NCL are mediated by C11orf98 and are not unique to the C11orf98–APEX fusions (Figures S7 and S8).
Figure 4.
Validation of the interaction of the C11orf98 microprotein with NPM1 and NCL. (A) Western blot of the C11orf98–APEX labeling proteome with anti-NCL, anti-NPM1, and anti-myc tag antibodies. (B) Reciprocal anti-HA immunoprecipitation of NPM1-HA from HEK293T cells co-expressing the C11orf98–FLAG fusion, with cells expressing the C11orf98–FLAG fusion alone as a control. Eluted proteins were analyzed by Western blotting.
Furthermore, we validated the association of the C11orf98 microprotein with NPM1 by a reciprocal immunoprecipitation experiment. Cells co-expressed a FLAG-tagged C11orf98 microprotein and HA-tagged NPM1. As a control, cells were co-expressed with FLAG-tagged C11orf98 microprotein and an empty vector. Lysates were prepared and immunoprecipitated with an anti-HA antibody against HA-tagged NPM1. Western blot analysis of the eluates using anti-HA and anti-FLAG antibodies revealed that HA-tagged NPM1 enriched FLAG-tagged C11orf98, further supporting an interaction between these proteins (Figure 4B and Figure S9). Control experiments using a control IgG did not enrich FLAG-tagged C11orf98, which provided additional evidence of an interaction between NPM1 and the C11orf98 microprotein (Figures S10 and S11). Moreover, Gygi’s group and Mann’s group independently published a recent proteome-wide protein interaction database, and the data are available to be searched.36,37 We downloaded MS raw files for NPM1 IP from Mann’s data, and both NPM1 and NCL IPs from Gygi’s Bioplex database. Reanalyzing this data using the human Uniprot proteome appended with the C11orf98 microprotein revealed that C11orf98 is detected with multiple peptide and spectral counts (Figure S12). This experiment demonstrates that C11orf98 interacts with NPM1 and NCL even when it is not overexpressed.
C11orf98 Localizes to the Nucleolus
NPM1 and NCL localize to the nucleolus, an organelle within the nucleus that is the site of ribosome biogenesis and newly emerging functions in protein regulation using noncoding RNAs.38,39 The high percentage of nucleolar proteins enriched by C11orf98 indicates that this microprotein should also be localized to the nucleolus. Confocal imaging of overexpressed FLAG-tagged C11orf98 in HeLa cells validated this hypothesis by revealing a nucleolar localization for this microprotein (Figure 5). Furthermore, the FLAG-tagged C11orf98 microprotein overlaps entirely with the HA-tagged NPM1 in the nucleoli, providing additional evidence that NPM1 and the C11orf98 microprotein are likely to interact with each other.
Figure 5.
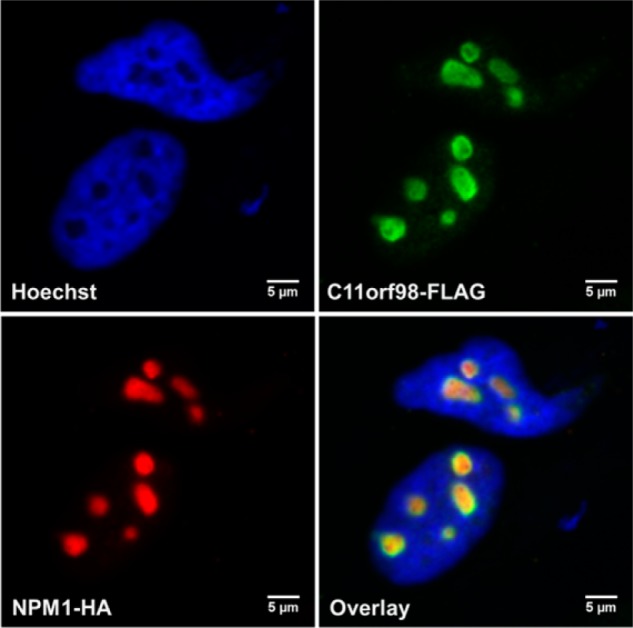
Colocalization of the C11orf98 microprotein and NPM1 in the cell nucleolus. HeLa cells were transfected with C11orf98-FLAG and NPM1-HA, fixed, and stained with anti-FLAG and anti-HA antibodies to visualize C11orf98 and NPM1 proteins. Nuclei were stained with Hoechst. Scale bars are 5 μm.
Functional studies have revealed that both the NCL and NPM1 proteins are multifunctional, with roles in ribosome biogenesis, cell cycle and apoptosis, transcriptional regulation, and DNA replication and repair.40,41 As a putative interaction partner of NPM1 and NCL, we hypothesize that the C11orf98 microprotein will likely have a role in at least some of these processes. These data highlight the value of MPIs in generating novel hypotheses that can lead to the functional characterization of microproteins.
Conclusion
Here, we demonstrate the value of APEX for identifying MPIs. Unlike traditional approaches that include immunoprecipitation from lysates, APEX captures endogenous interactions in the context of a living cell. This feature is particularly useful for microproteins because we have observed that microproteins undergoing immunoprecipitation enrich many nonspecific interactors, which are not found in the APEX experiments. We suspect that microproteins are becoming unstructured during FLAG-based immunoprecipitations, leading to more nonspecific interactions. Furthermore, the reduction in background afforded by the APEX technology also leads to a greater fold increase for the MRI binding partners Ku70 and Ku80, which makes it much easier to identify bona fide microprotein-interacting partners.
Indeed, the application of APEX tagging to the C11orf98 microprotein led to the discovery that this microprotein interacts with NCL and NPM1, and several other nucleolar proteins. NCL and NPM1 are multifunctional proteins found in the nucleolus where they participate in the synthesis and maturation of ribosomes. NCL and NPM1 are also reported to interact with each other. Mutations and amplifications of NPM1 have been linked to many cancers, including acute myelogenous leukemia, though the exact mechanism for this connection is still being worked out.42,43 Thus, the identification of an MPI among the C11orf98 microprotein, NCL, and NPM1 is of fundamental and clinical interest and will lead to new testable hypotheses about the function of the C11orf98 microprotein. Furthermore, the apparent improvement in the APEX data supports the application of APEX to the remaining uncharacterized microproteins.
Acknowledgments
We thank members of Saghatelian’s lab for fruitful discussions. We are grateful for generous help from Dr. Uri Manor at the Waitt Advanced Biophotonics Core Facility of the Salk Institute. We thank Jamie Simon, Dr. Gordon Louie, and Prof. Joseph Noel for their help with graphics. Imaging work in this study was supported by the Waitt Advanced Biophotonics Core Facility of the Salk Institute with funding from NIH-NCI CCSG Grant P30 014195, NINDS Neuroscience Core Grant NS072031, and the Waitt Foundation. We also are thankful for Cancer Center Support Grant P30 CA014195, which was used to purchase and provide access to the MGC collection.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.7b00265.
Q.C. is a postdoctoral fellow funded by the George E. Hewitt Foundation for medical research. This study was supported by the National Institutes of Health with a National Cancer Institute Cancer Center Support Grant P30 (CA014195 MASS core, A.S.), R01 (GM102491, A.S.), P41 (P41 GM103533, J.R.Y.) and R01 (MH67880, J.R.Y.). Additional support was provided by the Leona M. and Harry B. Helmsley Charitable Trust grant (2012-PG-MED002 to A.S.), and Dr. Frederick Paulsen Chair/Ferring Pharmaceuticals (A.S.).
The authors declare no competing financial interest.
Supplementary Material
References
- Wilcox G. (2005) Insulin and insulin resistance. Clin. Biochem. Rev. (Ultimo, Aust.) 26, 19–39. [PMC free article] [PubMed] [Google Scholar]
- Andrali S. S.; Sampley M. L.; Vanderford N. L.; Ozcan S. (2008) Glucose regulation of insulin gene expression in pancreatic beta-cells. Biochem. J. 415, 1–10. 10.1042/BJ20081029. [DOI] [PubMed] [Google Scholar]
- Saltiel A. R.; Kahn C. R. (2001) Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414, 799–806. 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- Hook V.; Funkelstein L.; Lu D.; Bark S.; Wegrzyn J.; Hwang S. R. (2008) Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu. Rev. Pharmacol. Toxicol. 48, 393–423. 10.1146/annurev.pharmtox.48.113006.094812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokfelt T.; Broberger C.; Xu Z. Q. D.; Sergeyev V.; Ubink R.; Diez M. (2000) Neuropeptides - an overview. Neuropharmacology 39, 1337–1356. 10.1016/S0028-3908(00)00010-1. [DOI] [PubMed] [Google Scholar]
- Saghatelian A.; Couso J. P. (2015) Discovery and characterization of smORF-encoded bioactive polypeptides. Nat. Chem. Biol. 11, 909–916. 10.1038/nchembio.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. J.; Rothnagel J. A. (2014) Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 15, 193–204. 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- Chu Q.; Ma J.; Saghatelian A. (2015) Identification and characterization of sORF-encoded polypeptides. Crit. Rev. Biochem. Mol. Biol. 50, 134–141. 10.3109/10409238.2015.1016215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble W. L.; Keegan K. P.; D’Souza M.; Wilke A.; Wilkening J.; Gilbert J.; Meyer F. (2012) Short-read reading-frame predictors are not created equal: sequence error causes loss of signal. BMC Bioinf. 13, 183. 10.1186/1471-2105-13-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspden J. L.; Eyre-Walker Y. C.; Phillips R. J.; Amin U.; Mumtaz M. A.; Brocard M.; Couso J. P. (2014) Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. eLife 3, e03528. 10.7554/eLife.03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavoff S. A.; Mitchell A. J.; Schwaid A. G.; Cabili M. N.; Ma J.; Levin J. Z.; Karger A. D.; Budnik B. A.; Rinn J. L.; Saghatelian A. (2013) Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 9, 59–64. 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladoukakis E.; Pereira V.; Magny E. G.; Eyre-Walker A.; Couso J. P. (2011) Hundreds of putatively functional small open reading frames in Drosophila. Genome Biol. 12, R118. 10.1186/gb-2011-12-11-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagoub D.; Tay A. P.; Chen Z.; Hamey J. J.; Cai C.; Chia S. Z.; Hart-Smith G.; Wilkins M. R. (2015) Proteogenomic Discovery of a Small, Novel Protein in Yeast Reveals a Strategy for the Detection of Unannotated Short Open Reading Frames. J. Proteome Res. 14, 5038–5047. 10.1021/acs.jproteome.5b00734. [DOI] [PubMed] [Google Scholar]
- Vanderperre B.; Lucier J. F.; Bissonnette C.; Motard J.; Tremblay G.; Vanderperre S.; Wisztorski M.; Salzet M.; Boisvert F. M.; Roucou X. (2013) Direct Detection of Alternative Open Reading Frames Translation Products in Human Significantly Expands the Proteome. PLoS One 8, e70698. 10.1371/journal.pone.0070698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.; Zeng J.; Drew B. G.; Sallam T.; Martin-Montalvo A.; Wan J.; Kim S. J.; Mehta H.; Hevener A. L.; de Cabo R.; Cohen P. (2015) The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 21, 443–454. 10.1016/j.cmet.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T.; Hashimoto Y.; Kato K.; Inagaki S.; Hayashi S.; Kageyama Y. (2007) Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat. Cell Biol. 9, 660–665. 10.1038/ncb1595. [DOI] [PubMed] [Google Scholar]
- Magny E. G.; Pueyo J. I.; Pearl F. M.; Cespedes M. A.; Niven J. E.; Bishop S. A.; Couso J. P. (2013) Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 341, 1116–1120. 10.1126/science.1238802. [DOI] [PubMed] [Google Scholar]
- Anderson D. M.; Anderson K. M.; Chang C.-L.; Makarewich C. A.; Nelson B. R.; McAnally J. R.; Kasaragod P.; Shelton J. M.; Liou J.; Bassel-Duby R.; Olson E. N. (2015) A Micropeptide Encoded by a Putative Long Noncoding RNA Regulates Muscle Performance. Cell 160, 595–606. 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B. R.; Makarewich C. A.; Anderson D. M.; Winders B. R.; Troupes C. D.; Wu F.; Reese A. L.; McAnally J. R.; Chen X.; Kavalali E. T.; Cannon S. C.; Houser S. R.; Bassel-Duby R.; Olson E. N. (2016) A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271–275. 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. M.; Makarewich C. A.; Anderson K. M.; Shelton J. M.; Bezprozvannaya S.; Bassel-Duby R.; Olson E. N. (2016) Widespread control of calcium signaling by a family of SERCA-inhibiting micropeptides. Sci. Signaling 9, ra119. 10.1126/scisignal.aaj1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pueyo J. I.; Couso J. P. (2008) The 11-aminoacid long Tarsal-less peptides trigger a cell signal in Drosophila leg development. Dev. Biol. 324, 192. 10.1016/j.ydbio.2008.08.025. [DOI] [PubMed] [Google Scholar]
- Galindo M. I.; Pueyo J. I.; Fouix S.; Bishop S. A.; Couso J. P. (2007) Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 5, e106. 10.1371/journal.pbio.0050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanet J.; Benrabah E.; Li T.; Pelissier-Monier A.; Chanut-Delalande H.; Ronsin B.; Bellen H. J.; Payre F.; Plaza S. (2015) Pri sORF peptides induce selective proteasome-mediated protein processing. Science 349, 1356–1358. 10.1126/science.aac5677. [DOI] [PubMed] [Google Scholar]
- D’Lima N. G.; Ma J.; Winkler L.; Chu Q.; Loh K. H.; Corpuz E. O.; Budnik B. A.; Lykke-Andersen J.; Saghatelian A.; Slavoff S. A. (2017) A human microprotein that interacts with the mRNA decapping complex. Nat. Chem. Biol. 13, 174–180. 10.1038/nchembio.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavoff S. A.; Heo J.; Budnik B. A.; Hanakahi L. A.; Saghatelian A. (2014) A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining. J. Biol. Chem. 289, 10950–10957. 10.1074/jbc.C113.533968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy G. J.; Rulten S. L.; Arribas-Bosacoma R.; Davidson K.; Kozik Z.; Oliver A. W.; Pearl L. H.; Caldecott K. W. (2016) The Ku-binding motif is a conserved module for recruitment and stimulation of non-homologous end-joining proteins. Nat. Commun. 7, 11242. 10.1038/ncomms11242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee H. W.; Zou P.; Udeshi N. D.; Martell J. D.; Mootha V. K.; Carr S. A.; Ting A. Y. (2013) Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331. 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S. S.; Martell J. D.; Kamer K. J.; Deerinck T. J.; Ellisman M. H.; Mootha V. K.; Ting A. Y. (2015) Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54. 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V.; Udeshi N. D.; Lam S. S.; Loh K. H.; Cox K. J.; Pedram K.; Carr S. A.; Ting A. Y. (2016) Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 11, 456–475. 10.1038/nprot.2016.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.; Udeshi N. D.; Deerinck T. J.; Svinkina T.; Ellisman M. H.; Carr S. A.; Ting A. Y. (2017) Proximity Biotinylation as a Method for Mapping Proteins Associated with mtDNA in Living Cells. Cell Chem. Biol. 24, 404–414. 10.1016/j.chembiol.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K. H.; Stawski P. S.; Draycott A. S.; Udeshi N. D.; Lehrman E. K.; Wilton D. K.; Svinkina T.; Deerinck T. J.; Ellisman M. H.; Stevens B.; Carr S. A.; Ting A. Y. (2016) Proteomic Analysis of Unbounded Cellular Compartments: Synaptic Clefts. Cell 166, 1295–1307. 10.1016/j.cell.2016.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V.; Zou P.; Rhee H. W.; Udeshi N. D.; Cracan V.; Svinkina T.; Carr S. A.; Mootha V. K.; Ting A. Y. (2014) Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol. Cell 55, 332–341. 10.1016/j.molcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing J.; He L.; Sun A. M.; Quintana A.; Ding Y. H.; Ma G. L.; Tan P.; Liang X. W.; Zheng X. L.; Chen L. Y.; Shi X. D.; Zhang S. L.; Zhong L.; Huang Y.; Dong M. Q.; Walker C. L.; Hogan P. G.; Wang Y. J.; Zhou Y. B. (2015) Proteomic mapping of ER-PM junctions identifies STIMATE as a regulator of Ca2+ influx. Nat. Cell Biol. 17, 1339–1347. 10.1038/ncb3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin E. L.; Ting L.; Bruckner R. J.; Gebreab F.; Gygi M. P.; Szpyt J.; Tam S.; Zarraga G.; Colby G.; Baltier K.; Dong R.; Guarani V.; Vaites L. P.; Ordureau A.; Rad R.; Erickson B. K.; Wühr M.; Chick J.; Zhai B.; Kolippakkam D.; Mintseris J.; Obar R. A.; Harris T.; Artavanis-Tsakonas S.; Sowa M. E.; De Camilli P.; Paulo J. A.; Harper J. W.; Gygi S. P. (2015) The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 162, 425–440. 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. P.; Busch R. K.; Valdez B. C.; Busch H. (1996) C23 interacts with B23, a putative nucleolar-localization-signal-binding protein. Eur. J. Biochem. 237, 153–158. 10.1111/j.1432-1033.1996.0153n.x. [DOI] [PubMed] [Google Scholar]
- Huttlin E. L.; Ting L.; Bruckner R. J.; Gebreab F.; Gygi M. P.; Szpyt J.; Tam S.; Zarraga G.; Colby G.; Baltier K.; Dong R.; Guarani V.; Vaites L. P.; Ordureau A.; Rad R.; Erickson B. K.; Wuhr M.; Chick J.; Zhai B.; Kolippakkam D.; Mintseris J.; Obar R. A.; Harris T.; Artavanis-Tsakonas S.; Sowa M. E.; De Camilli P.; Paulo J. A.; Harper J. W.; Gygi S. P. (2015) The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 162, 425–440. 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein M. Y.; Hubner N. C.; Poser I.; Cox J.; Nagaraj N.; Toyoda Y.; Gak I. A.; Weisswange I.; Mansfeld J.; Buchholz F.; Hyman A. A.; Mann M. (2015) A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723. 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- Boisvert F.-M.; van Koningsbruggen S.; Navascues J.; Lamond A. I. (2007) The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585. 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Matera A. G.; Terns R. M.; Terns M. P. (2007) Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8, 209–220. 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Colombo E.; Alcalay M.; Pelicci P. G. (2011) Nucleophosmin and its complex network: a possible therapeutic target in hematological diseases. Oncogene 30, 2595–2609. 10.1038/onc.2010.646. [DOI] [PubMed] [Google Scholar]
- Scott D. D.; Oeffinger M. (2016) Nucleolin and nucleophosmin: nucleolar proteins with multiple functions in DNA repair. Biochem. Cell Biol. 94, 419–432. 10.1139/bcb-2016-0068. [DOI] [PubMed] [Google Scholar]
- Grisendi S.; Mecucci C.; Falini B.; Pandolfi P. P. (2006) Nucleophosmin and cancer. Nat. Rev. Cancer 6, 493–505. 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- Box J. K.; Paquet N.; Adams M. N.; Boucher D.; Bolderson E.; O’Byrne K. J.; Richard D. J. (2016) Nucleophosmin: from structure and function to disease development. BMC Mol. Biol. 17, 19. 10.1186/s12867-016-0073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.