Abstract
Linkage maps are widely used to investigate structure, function, and evolution of genomes. In speciation research, maps facilitate the study of the genetic architecture of reproductive isolation by allowing identification of genomic regions underlying reduced fitness of hybrids. Here we present a linkage map for European newts of the Lissotriton vulgaris species complex, constructed using two families of F2 L. montandoni × L. vulgaris hybrids. The map consists of 1146 protein-coding genes on 12 linkage groups, equal to the haploid chromosome number, with a total length of 1484 cM (1.29 cM per marker). It is notably shorter than two other maps available for salamanders, but the differences in map length are consistent with cytogenetic estimates of the number of chiasmata per chromosomal arm. Thus, large salamander genomes do not necessarily translate into long linkage maps, as previously suggested. Consequently, salamanders are an excellent model to study evolutionary consequences of recombination rate variation in taxa with large genomes and a similar number of chromosomes. A complex pattern of transmission ratio distortion (TRD) was detected: TRD occurred mostly in one family, in one breeding season, and was clustered in two genomic segments. This is consistent with environment-dependent mortality of individuals carrying L. montandoni alleles in these two segments and suggests a role of TRD blocks in reproductive isolation. The reported linkage map will empower studies on the genomic architecture of divergence and interactions between the genomes of hybridizing newts.
Keywords: Lissotriton, linkage map, reproductive isolation, salamander, transmission ratio distortion
Linkage maps are widely used to explore genome structure, function, and evolution. Maps allow identification of loci determining specific phenotypes, such as genomic regions underlying economically important traits (Mao et al. 2015; Royaert et al. 2016), or genetic basis of adaptive ecological traits, such as migration (Hale et al. 2013) or industrial melanism (van’t Hof et al. 2016). In comparative genomics, linkage maps are indispensable to investigate the conservation of chromosomal segments at various evolutionary time scales (Postlethwait et al. 2000; Hollenbeck et al. 2015). Linkage maps also improve quality of genome assemblies and inform research on evolutionary consequences of variation in recombination rate withing the genome (Kawakami et al. 2014; Corbett-Detig et al. 2015).
In speciation research, linkage maps are particularly useful in studies of the genetic basis of reproductive isolation. Linkage analysis and mapping provide a framework for examining both the genomic architecture underlying the reproductive barriers between species or populations and interactions between differentiated genes in hybrid genomes (Rieseberg et al. 2000; Rieseberg and Buerkle 2002). Examples of the former include identification of chromosomal rearrangements that may act as barriers to gene flow (Lowry and Willis 2010; Ostberg et al. 2013; Barb et al. 2014), loci contributing to ecological reproductive isolation (Bradshaw and Schemske 2003), and phenotypic and expression quantitative trait loci (QTL) that differentiate hybridizing species and affect fitness of hybrids (Gagnaire et al. 2013; Lowry et al. 2015; Fruciano et al. 2016). Genic incompatibilities causing low fitness of hybrids can also be mapped in natural populations (Schumer et al. 2014) and in laboratory crosses (Sweigart et al. 2006). Finally, linkage maps facilitate the discovery and characterization of loci subject to adaptive introgression between hybridizing species (Fitzpatrick et al. 2010; Whitney et al. 2015).
In interspecific hybrids, genotype frequencies often deviate from those expected under Mendelian segregation (Jenczewski et al. 1997; Rieseberg et al. 2000; Fishman et al. 2001). Such segregation or transmission ratio distortion (TRD) may be caused by various mechanisms acting both prior to (gametic TRD) and after (zygotic TRD) fertilization [reviewed in Huang et al. (2013)]. The frequency of TRD generally increases with increasing divergence between crossed individuals (Matsubara et al. 2011). Because the TRDs often reflect genetic incompatibilities between differentiated genomes, detection and mapping of TRD loci provide insights into mechanisms of postzygotic reproductive isolation between divergent populations (Hall and Willis 2005) or species (Fishman et al. 2001; Moyle and Graham 2006; Brennan et al. 2014; Liu et al. 2016).
The Carpathian (Lissotriton montandoni) and the smooth (Lissotriton vulgaris) newts are closely related species that hybridize wherever their parapatric ranges meet around the Carpathians (Babik et al. 2003; Zieliński et al. 2014). They form a bimodal hybrid zone, with a large excess of parental genotypes even in the center of the zone, indicative of strong assortative mating of parental species (Babik et al. 2003; Babik and Rafiński 2004). As a consequence, early generation hybrids are rare in nature (Zieliński et al. 2013, 2014). Further, laboratory experiments indicate substantial asymmetric premating isolation, with L. montandoni females being more choosy (Michalak et al. 1997; Michalak and Rafiński 1999). So far there has been no information about the strength and form of postzygotic reproductive isolation between L. montandoni and L. vulgaris. Despite strong prezygotic isolation, there is evidence for extensive interspecific flow of the mitochondrial DNA (Babik et al. 2005; Zieliński et al. 2013) as well as for some nuclear gene flow, which is heterogeneous across the genome (Nadachowska-Brzyska et al. 2012; Zieliński et al. 2014; Stuglik and Babik 2016; Zieliński et al. 2016). The linkage map that we report here will facilitate an understanding of the basis of reproductive isolation between these species. In particular, detection and mapping of loci showing TRD in hybrid families may provide clues about the genetic architecture of reproductive isolation.
The linkage map of Lissotriton newts can also shed light on the possible correlation between physical genome size and length of the linkage map in salamanders. In most taxa, at least a single crossover per chromosomal arm is required for successful meiosis and consequently the total number of chromosomal arms is considered the main determinant of the map length (Sybenga 1996). Accordingly, Pardo-Manuel de Villena and Sapienza (2001) found a strong correlation between the number of chiasmata, map length, and the haploid number of chromosomal arms in mammals. They also surveyed a variety of other taxa (mainly plants, arthropods, and fishes) and confirmed that such correlation is generally observed. This evidence combined with high variability of recombination rate and limited range of genome sizes in mammals indicates the lack of correlation between physical size of the genome and length of linkage maps. However, the two linkage maps available for salamander genera Ambystoma (Smith et al. 2005; Voss et al. 2011) and Notophthalmus (Keinath et al. 2016) are several-fold longer than the expectations based on the number of chromosomal arms. Because salamanders exhibit genomic gigantism (Gregory 2015), a causal relationship between the genome size and map length was suggested (Smith et al. 2005). However, although salamanders share large genome size and most families have similar numbers of chromosomes (Sessions 2008), the number of chiasmata per chromosomal arm varies considerably between taxa (Callan 1966; Herrero and López-Fernández 1986; Zbożeń and Rafiński 1993). Lissotriton newts are at the lower end of this continuum, with ∼1 chiasma per chromosomal arm. Lissotriton genome sizes are similar to that of Ambystoma mexicanum (32 Gb, Keinath et al. 2015): the L. montandoni genome was estimated at 29.1 Gb, while the range for L. vulgaris, which comprises several evolutionary lineages, is 27.7–30.8 Gb (Litvinchuk et al. 2007). If the number of chiasmata predicts the map length in salamanders, we would expect a ∼1200-cM map in Lissotriton. A comparison of the Ambystoma, Lissotriton, and Notophthalmus maps may thus clarify whether the large size of linkage map is universal among salamanders or is an attribute of only some species.
In the present study, we report a linkage map obtained using F2 generation L. montandoni × L. vulgaris hybrids from two families. Resequenced fragments of protein-coding genes were used as genetic markers. Specifically, we (i) constructed the first linkage map in Lissotriton newts containing >1000 protein-coding genes—this map constitutes an essential resource for evolutionary studies in this system; (ii) compared the relationship between the Lissotriton map length and physical genome size with the results obtained for other salamander species; and (iii) identified loci showing TRDs in F2 hybrids and examined their distribution through the genome, which provides insights into the genetic basis of reproductive isolation between species.
Materials and Methods
Mapping population
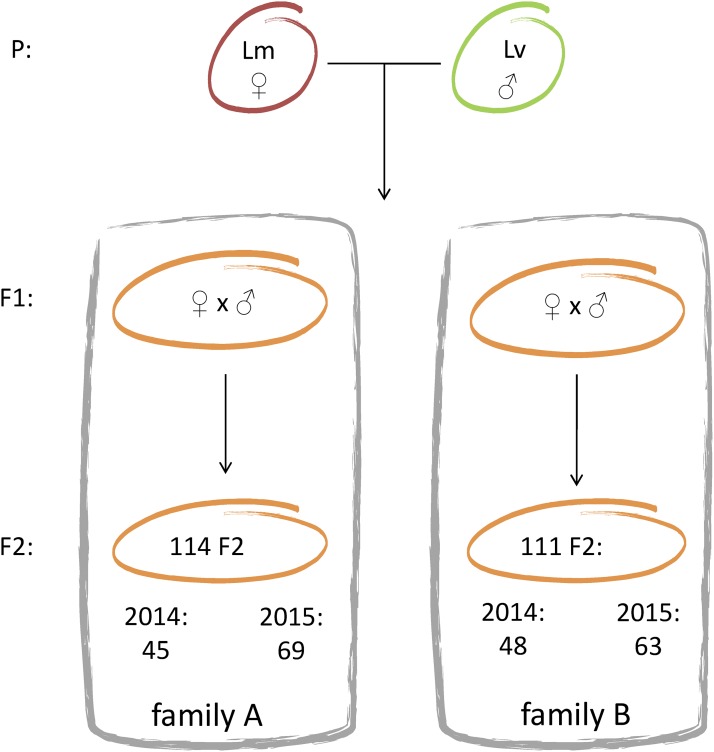
A female L. montandoni and male L. vulgaris were collected from allopatric populations in southern Poland (latitude and longitude of 49.51 N and 20.14 E, and 49.99 N and 19.47 E, respectively) and crossed in the laboratory (generation P). Hybrids of F1 generation that resulted from this cross were reared to maturity and two pairs were used to obtain the F2 hybrids (families A and B). The F2 progeny from these crosses, collected over two consecutive breeding seasons (2014 and 2015), were used for mapping. The F2 larvae from each family were maintained in a separate container and fed ad libitum with Artemia shrimps. The larvae of minimum 1 cm total length were anesthetized in MS222 and preserved in ethanol. We obtained 114 offspring from family A and 111 offspring from family B (Figure 1). DNA was extracted from tail tips of adults (P and F1 generation) and from whole F2 larvae using the Wizard Genomic DNA Purification Kit (Promega). DNA was dissolved in 100 μl of TE buffer.
Figure 1.
Experimental setup. Interspecific mating (generation P) resulting in F1 offspring occurred in the laboratory. Two pairs of F1 were crossed, forming families A and B, which were used to construct the linkage maps. Offspring from both families was collected in two consecutive breeding seasons, 2014 and 2015. Lm, Lissotriton montandoni; Lv, Lissotriton vulgaris.
Molecular markers
Molecular Inversion Probe (MIP) markers (O’Roak et al. 2012; Niedzicka et al. 2016) were designed in Lissotriton transcript sequences (available at http://newtbase.eko.uj.edu.pl). Each MIP amplifies a 112-bp target and all MIPs are assayed simultaneously in a single reaction. Each marker was located in a single exon to allow amplification from genomic DNA. The bioinformatics pipeline used to design markers, laboratory procedures, and sequencing are described in detail in Niedzicka et al. (2016). The MIP markers used to construct the linkage map fell into two groups (Supplemental Material, Table S1 in File S1):
Markers informative in the F2 cross, i.e., MIPs containing single nucleotide polymorphisms (SNPs) homozygous for different alleles in the individuals of generation P (mm × vv). Such markers were identified in previously resequenced transcriptomes of two L. montandoni and two L. vulgaris individuals sampled from the same geographic regions as the individuals of generation P (Stuglik and Babik 2016). Although the sample size of four gene copies per species is small, SNPs diagnostic in such a sample probably have highly differentiated allele frequencies between species. Indeed, generation P individuals were alternative homozygotes in an overwhelming majority of such SNPs. A subset of 205 group 1 markers were reported in Niedzicka et al. (2016).
Markers designed in genes involved in immune response reported by Fijarczyk et al. (2016) and other randomly picked transcripts. Several MIPs were designed for most genes. As these markers were designed without taking into account interspecific differentiation of allele frequencies, their utility for mapping varied.
In the initial screen to filter out poorly amplifying markers and paralogs, all markers were resequenced in parents and 21 F2 offspring from family A. Deviations from the expected Mendelian segregation ratio were checked using the exact multinomial test (EMT) in R package EMT (Menzel 2013). Markers that amplified poorly or contained SNPs with an excess of heterozygotes at a false discovery rate (FDR) of q < 0.01 were removed from further analyses. The remaining MIPs were resequenced in both families. Additional 204 F2 informative (group 1) markers acquired from another project were resequenced only in family A (Table S1 in File S1).
Mapping and SNP calling were performed as described in Niedzicka et al. (2016). Briefly, 2 × 75 bp paired-end or 150 bp single-end Illumina reads were mapped to the reference with Bowtie2. SNP calling was performed using GenomeAnalysisTK (GATK) UnifiedGenotyper (DePristo et al. 2011). Genotypes with coverage <16× or genotype quality <30 phred were considered missing. To estimate genotyping error 29 individuals were amplified and resequenced twice. Genotyping error was expressed as the nonreference discrepancy rate (NRD) in GATK module GenotypeConcordance.
Construction of the linkage map
Mendelian segregation was tested again on the full dataset, and markers containing SNPs with an excess of heterozygotes in either family at the FDR q < 0.01 were removed from both families. These could be paralogs missed in the initial screen due to a limited sample size or inclusion of only a single family. The remaining markers were evaluated in each family separately. Markers with a genotype missing in either parent or with >5% missing data were excluded from the analysis.
Separate linkage maps were constructed for each family. In family A, we used all markers informative in the inbred cross (intercross) with F2 segregation pattern 1:2:1 (P: mm × vv) to extract full information based on a three-generation pedigree (File S2). Markers showing strong TRD in a given family (see below) were excluded from mapping to prevent spurious linkage. Numerous MIPs in family B were excluded for this reason, which reduced the number of available markers. To maximize the number of useful markers in family B, we conducted outbred linkage mapping, for which several marker segregation types are suitable. The suitable markers were identified based on the genotypic information from parents of family B (File S2).
The linkage maps were constructed using the R/onemap package (Margarido et al. 2007). Linkage groups (LGs) were found using the maximum recombination frequency set to 0.55 and a minimum logarithm of odds (LOD) score of 8. The map distances were obtained by applying the Haldane mapping function. The order of markers in each LG was defined using the algorithm RECORD (Mollinari et al. 2009); for eight markers within each LG all possible orders were calculated and compared to each other to choose the best one by the multipoint likelihood in order to create a framework for algorithm operations. The summary statistics (genome length and map coverage) were calculated for the intercross map. The genome length was estimated by method 4 of Chakravarti et al. (1991). Map coverage, expressed as the proportion of the genome within x centimorgan of a mapped marker under the assumption of uniform distribution of markers on the map, was calculated according to Fishman et al. (2001). To check for uniformity of the distribution of markers along the intercross map, a chi-square test was performed against Poisson expectations for markers evenly distributed over 10-cM intervals. A graphical representation of linkage maps was prepared in MapChart (Voorrips 2002) and the consensus map was constructed with MergeMap (Wu et al. 2011).
TRDs and epistatic interactions
For each family, deviations from the Mendelian segregation ratio were tested within each year with the EMT followed by the FDR correction. For each marker showing TRD at FDR q < 0.01, we followed Leppälä et al. (2013) to check whether data supported the gametic or zygotic causes of TRD. We calculated the likelihoods of three models: (i) a null model assuming Mendelian segregation, (ii) a gametic model assuming that all TRD was caused by departures of gamete frequencies from those expected under Mendelian segregation, and (iii) a zygotic model assuming that differential survival of diploid genotypes contributed to the observed TRD. The models were compared using likelihood ratio tests, with P < 0.001 considered significant (Leppälä et al. 2013).
To test for possible epistatic interactions between pairs of unlinked markers, contingency tables with the observed two-locus genotype counts were calculated in Genepop (Rousset 2008). The chi-square test was then used to compare the observed counts with those expected, assuming independence between markers. This test was performed for all pairs of markers located on different LGs.
Data availability
File S2 contains genotypes for each individual.
Results
Linkage map
To evaluate the amplification efficiency and Mendelian inheritance of markers, 1741 MIPs were genotyped in 23 individuals (parents and 21 offspring) from family A. After excluding markers that amplified poorly or showed signs of paralogy, 1166 MIP markers were resequenced in both families (Figure 1 and Table S1 in File S1). At this stage, an additional 31 markers were excluded due to an excess of heterozygotes in either family, suggesting paralogy. The estimated genotyping error (NRD) was 0.008. An additional 204 MIPs were resequenced only in family A (Table S1 in File S1). If more than one MIP per gene was resequenced, the MIP containing the most informative SNP was used for mapping.
Separate linkage maps were constructed for each family. The map for family A was based on the three-generation pedigree (intercross map) with one type of markers (P: mm × vv). After excluding two markers with unstable position within a LG, 1076 markers were used and all were assigned to LGs (Table S2 in File S1). We obtained 13 LGs, one more than the haploid number of Lissotriton chromosomes (n = 12). Reduction of the LOD score threshold from eight to six did not cause any LGs to merge. The predicted genome length is 1404 cM (Table 1) and 99.9 and 95.3% of the genome was estimated to be within 5 and 2 cM of the mapped markers, respectively. The markers were not uniformly distributed on the map ( = 80.7, P = 1.3 × 10−10).
Table 1. Summary of the linkage maps.
| Map A | Map B | Consensus Map | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LG | Length (cM) | N Markers | N Unique Mapped | Marker Interval (cM) | Length (cM) | N Markers | N Unique Mapped | Marker Interval (cM) | Length (cM) | N Markers | Marker Interval (cM) |
| 1 | 139.51 | 135 | 53 | 1.04 | 165.55 | 114 | 41 | 1.47 | 181.28 | 148 | 1.23 |
| 2 | 140.62 | 131 | 55 | 1.08 | 107.83 | 111 | 48 | 0.98 | 160.22 | 140 | 1.15 |
| 3 | 141.06 | 132 | 53 | 1.08 | 145.77 | 119 | 48 | 1.24 | 157.32 | 138 | 1.15 |
| 4 | 140.13 | 119 | 56 | 1.19 | 55.76 | 46 | 26 | 1.24 | 142.38 | 120 | 1.20 |
| 5 | 134.02 | 85 | 31 | 1.59 | 87.19 | 74 | 29 | 1.19 | 132.07 | 88 | 1.52 |
| 6 | 108.62 | 73 | 34 | 1.51 | 108.74 | 61 | 29 | 1.81 | 116.23 | 79 | 1.49 |
| 7 | 102.45 | 81 | 35 | 1.28 | 105.35 | 59 | 26 | 1.82 | 112.33 | 81 | 1.40 |
| 8 | 97.04 | 52 | 29 | 1.90 | 79.36 | 41 | 23 | 1.98 | 101.90 | 56 | 1.85 |
| 9 | 48.35 | 43 | 27 | 1.15 | 82.12 | 56 | 25 | 1.49 | 100.63 | 61 | 1.68 |
| 10 | 106.05 | 88 | 36 | 1.22 | 55.02 | 28 | 16 | 2.04 | 100.35 | 89 | 1.14 |
| 11 | 102.75 | 54 | 30 | 1.94 | 73.08 | 48 | 18 | 1.55 | 96.23 | 63 | 1.55 |
| 12 | 78.73 | 78 | 37 | 1.02 | 71.34 | 65 | 32 | 0.94 | 82.88 | 83 | 1.01 |
| 13 | 21.88 | 5 | 5 | 5.47 | |||||||
| Total | 1361.21 | 1076 | 481 | 1137.11 | 822 | 361 | 1483.82 | 1146 | |||
Map A: intercross map constructed for the family A (see Results). Map B: outcross map constructed for the family B (see Results). Consensus map was constructed by merging maps A and B. Linkage groups (LGs) are numbered according to the length on the consensus map.
Numerous highly distorted markers (HDMs) were excluded from mapping in family B. To maximize the number of useful markers in this family, we conducted outbred linkage mapping, allowing for several types of segregation. The map was constructed with 822 markers, 13 LGs were obtained and one marker remained unlinked. Reduction of the LOD score threshold from eight to seven linked two LGs together, reducing their final number to 12, consistent with the haploid Lissotriton chromosome count (Table S2 in File S1). Groups 9 and 13 from the family A map were linked together as LG9 in the family B map. The map length is 1137 cM (Table 1). Because distorted markers clustered in two blocks on LG4 and LG10, removal of these markers resulted in shortening these LGs in family B (Figure S1). The overall difference in the length of corresponding LGs between the two maps was not significant (Wilcoxon test V = 60, P = 0.11).
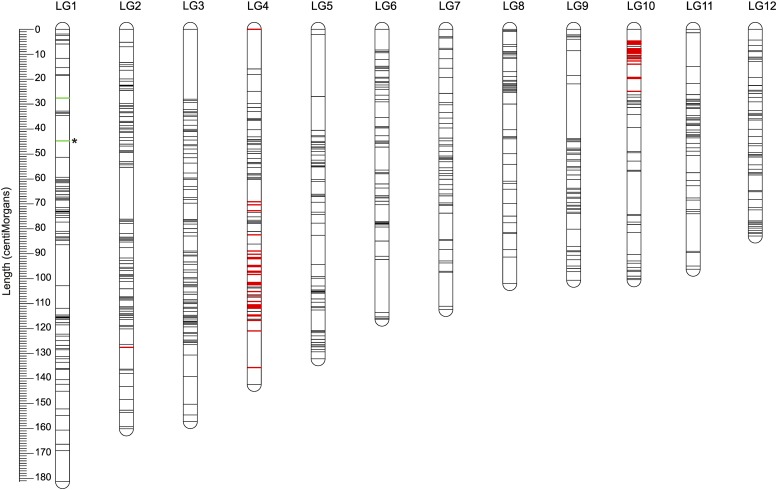
To integrate information from all markers, a consensus map was constructed by merging maps from both families. The consensus linkage map is 1484 cM and contains a total of 1146 markers on 12 LGs, i.e., on average one marker per 1.29 cM (Figure 2 and Table 1). Detailed characteristics of LGs and the order of markers on the consensus map are provided in Table S3 in File S1.
Figure 2.
The consensus linkage map. The consensus map contains 1146 markers and has a length of 1484 cM. Red shows highly distorted markers (HDMs) with a deficit of L. montandoni allele; green shows HDMs with a deficit of L. vulgaris allele. Only a single HDM (asterisk) was detected in family A, the remaining HDM were detected only in family B.
TRD and epistatic interactions
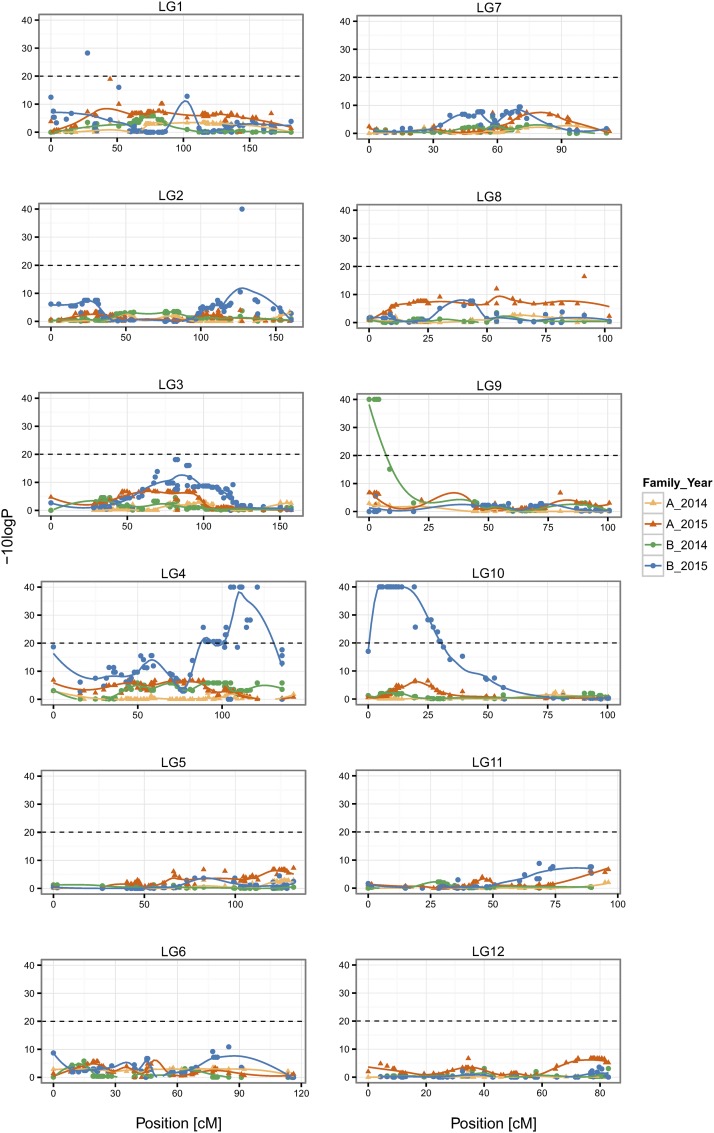
Deviations from Mendelian segregation at the nominal P < 0.05 were widespread: 23% (264) and 26% (240) of markers were distorted in families A and B, respectively. Only 42 deviating markers were common to both families, which suggests that many significant results may represent false positives. The families differed drastically in the number of HDMs (FDR q < 0.01): we found only one HDM in family A and as many as 95 in family B (HDMs were not shared between families). In all HDMs, one type of homozygote was underrepresented, and in 95% it was the mm homozygote. In order to further investigate the causes of the difference between families, EMT was also calculated within years (Table 2). The results show that the difference between families in the number of HDM was mostly due to data from 2015: markers highly distorted in that year did not show much distortion in 2014 (Figure 3 and Table 2). The distribution of HDMs within the genome was analyzed using the consensus map. Most HDMs clustered in two separate blocks on LG4 and LG10 (Figure 3). All markers in these blocks had deficit of mm homozygotes. The gametic causes of TRD were inferred for most markers (86), while zygotic or both gametic and zygotic models were supported for eight and two markers, respectively.
Table 2. Transmission ratio distortions (TRDs) in families and breeding seasons (2014 and 2015).
| Family | P | 2014 | 2015 | 2014 and 2015 Combined |
|---|---|---|---|---|
| A | Nominal < 0.05 | 89 | 223 | 264 |
| B | Nominal < 0.05 | 86 | 240 | 240 |
| A | FDR < 0.01 | 0 | 0 | 1 |
| B | FDR < 0.01 | 4 | 88 | 95 |
Deviations from the expected Mendelian segregation ratio were evaluated with the exact multinomial test (EMT). Both the nominal and false discovery rate (FDR) corrected P values are shown. Note that the more numerous significant results at the nominal P < 0.05 may partially reflect larger sample sizes (Figure 1) and thus higher statistical power in season 2015.
Figure 3.
Transmission ratio distortions (TRDs) shown on the consensus map. Departures of genotype counts from Mendelian expectations were tested with the Exact Multinomial Test and the nominal P are given as −10logP, the dashed line indicates the nominal P of 0.01. Polynomial regression trend lines are shown.
To detect epistatic interactions, we tested the statistical independence of two-marker genotypes within families for all pairs of markers located on different LGs. Numerous tests had the nominal P < 10−4 (184 and 11 in families A and B, respectively), but no test remained significant even at an FDR of 0.1. We note that interpretation of a standard FDR is unclear here, because of the partial nonindependence of tests due to linkage between markers. This, together with the very large number of tests performed (exceeding 0.5 million in family A), makes our tests for epistasis highly conservative. Therefore we briefly describe interactions with P < 10−4 as possible cases of epistasis (Table S4 in File S1). Only a single interaction was common to both families; as both markers were heterozygous in the L. vulgaris grandparent, this result is difficult to interpret in the context of reproductive isolation. In family A, almost all tests with P < 10−4 involved two blocks of markers, on LG6 and LG12, characterized by a strong deficit or lack of both mm:mm and vv:mm genotypes, which again is difficult to interpret in the context of reproductive isolation. In family B, nine out of 11 tests with P < 10−4 represent two possible epistatic interactions between a single HDM (deficit of v alleles) on LG1, and two separate regions on LG6. Interestingly, these appear as negative epistatic interactions of the Dobzhansky–Muller (DM) type, with vv:mm genotypes missing and vv:mv underrepresented.
Discussion
In this study a gene-based linkage map of newts from the L. vulgaris species complex was constructed using F2 L. montandoni × L. vulgaris hybrid progeny from two families. The map contains 1146 genes in 12 LGs, equal to the haploid number of Lissotriton chromosomes, and has excellent genomic coverage but is notably shorter than two other available salamander linkage maps. Because progeny from each family was collected over two consecutive breeding seasons, we were able to detect a complex pattern of segregation distortion in the hybrid genomes, providing some insight into the nature of reproductive isolation between the parental species.
The L. montandoni × L. vulgaris linkage map is three to four times shorter than maps available for two other salamander genera, Ambystoma (Ambystomatidae) and Notophthalmus (Salamandridae), even though these genera have similar number of chromosomes as Lissotriton (Table 3). The difference in map length has two plausible explanations: actual differences in the rate of meiotic recombination between salamander taxa or reduced recombination in F1 Lissotriton hybrids. The former explanation is supported by ample cytogenetic evidence. All Lissotriton species have 12 chromosome pairs and ∼1 chiasma per chromosomal arm in meiosis (Wickbom 1945; Watson and Callan 1963; Barsacchi et al. 1970; Herrero and López-Fernández 1986; Zbożeń and Rafiński 1993), which predicts a map length of ∼1200 cM, slightly shorter than reported here. However, the number of chiasmata in Lissotriton is at the lower end of the scale observed in salamanders (Table 3), which typically have more than one chiasma per chromosome arm (Wickbom 1945; Callan and Spurway 1951), even in excess of four in A. mexicanum (Callan 1966). Crossover rates are more variable among salamanders than among mammals, in which a maximum of 2.5 chiasmata per chromosome arm were reported (Dumont 2017). A comparison within Ambystoma revealed two times fewer chiasmata in Ambystoma macrodactylum than in A. mexicanum (Kezer et al. 1980). The recombination rate in salamanders can thus evolve relatively quickly despite conservative karyotypes, consistent with theory and data from other systems (True et al. 1996; Dumont et al. 2011; Smukowski and Noor 2011).
Table 3. Comparison of the number of chromosomes, number of chiasmata per meiotic cell, and expected and observed map lengths in ambystomatid and salamandrid salamanders.
| Expected Map Length (cM) Based on | ||||||
|---|---|---|---|---|---|---|
| Species | No. of Chr | No. of Arms | No. of Chias | No. of Arms | No. of Chias | Observed Map Length (cM) |
| Ambystomatidae | ||||||
| Ambystoma macrodactylum | 14 | 28a | 53a | 1400 | 2650 | NA |
| Ambystoma mexicanumb | 14 | 28 | 113 | 1400 | 5650 | 4200c |
| Salamandridae | ||||||
| Ichthyosaura alpestris | 12 | 24d | 32.3e | 1200 | 1615 | NA |
| Lissotriton boscai | 12 | 24d | 21.7e | 1200 | 1085 | NA |
| Lissotriton helveticus | 12 | 24d | 22.2e | 1200 | 1110 | NA |
| Lissotriton montandoni | 12 | 24d | 23.3f | 1200 | 1165 | 1484g |
| Lissotriton vulgaris | 12 | 24d | 24.2h | 1200 | 1210 | |
| Notophthalmus viridescens | 11 | 22d | NA | 1100 | NA | 6162i |
| Pleurodeles waltl | 12 | 24d | 28.1j | 1200 | 1405 | NA |
| Salamandra salamandra | 12 | 24d | 36.8k | 1200 | 1840 | NA |
| Triturus carnifex | 12 | 24d | 31.4l | 1200 | 1570 | NA |
| Triturus cristatus | 12 | 24d | 37.4l | 1200 | 1870 | NA |
| Triturus karelinii | 12 | 24d | 40.8l | 1200 | 2040 | NA |
| Triturus marmoratus | 12 | 24d | 25.7e | 1200 | 1285 | NA |
Expected map length based on the number of chromosomal arms assumes a single crossover per arm. No. of chr, haploid number of chromosomes; No. of arms, number of chromosomal arms; No. of chias, average number of chiasmata per meiotic cell; NA, not available.
All salamandrids are characterized by biarmed chromosomes (Sessions 2008).
This study.
L. vulgaris meridionalis (Barsacchi et al. 1970).
Another potential explanation for the shorter map in Lissotriton compared to other salamanders is underestimation of map length due to reduced recombination in F1 hybrids. This phenomenon has been reported in many systems, including Triturus newts (Callan and Spurway 1951). However, in other cases interspecific maps are not shorter than intraspecific maps (Beukeboom et al. 2010) and can even be longer (Woram et al. 2004). Strong reduction of recombination in L. montandoni × L. vulgaris hybrids is unlikely for two reasons. First, the karyotypes of the parental species are very similar indicating a lack of major chromosomal rearrangements (Zbożeń and Rafiński 1993; Zbożeń 1997). Second, in hybrid males, meiotic recombination assessed by cytogenetic methods is similar to males of pure parental species (Zbożeń 1997). Because males are heterogametic in Lissotriton, in accordance with Haldane’s rule they are more likely to experience meiotic problems. Evidence to the contrary suggests a lack of strong reduction of meiotic recombination in hybrids of either sex. Reduced recombination in hybrids is thus unlikely the sole explanation of the observed map length. It cannot be however dismissed completely as an additional factor. Therefore, it would be interesting to compare the length of the map reported here with that of intraspecific and backcross maps. Such comparisons would also check for the presence of smaller scale rearrangements between genomes of parental species, not detectable with cytogenetic methods.
Our results, together with extensive cytogenetic data accumulated over decades, show that the large genome size shared by salamander species does not automatically translate into long linkage maps. Such a relationship was hypothesized previously based on the Ambystoma map size (Smith et al. 2005). The several-fold differences in map length among salamanders make this group well suited to study the evolutionary consequences of recombination rate variation between taxa characterized by large genomes and similar numbers of chromosomes.
Numerous markers showed TRD in F2 hybrids. Distortions occurred mostly in one family, in one breeding season, and clustered in two blocks on LG4 and 10. Almost all HDMs, including all markers in the two blocks had a deficit of the mm genotype, and also mv heterozygotes were often underrepresented. Apparent TRD can result from various methodological artifacts, such as genotyping errors or extensive missing data. Such major artifacts are unlikely in our study because of the high quality of the MIP markers, highly repeatable genotyping, and the fact that TRD occurred mostly in a single family and a single season. Therefore, we are convinced that the observed TRDs reflect biological reality.
True TRD may arise before (gametic) or after (zygotic) fertilization and both types can have multiple causes [reviewed in Huang et al. (2013)]. In principle, one can use the observed genotype counts to distinguish between gametic and zygotic processes by comparing the likelihoods of respective models (Leppälä et al. 2013). Unfortunately this approach is sometimes inconclusive. For example, under a realistic scenario of viability selection against a partially dominant allele, the zygotic model has only slightly better fit than the gametic model even though all distortion is due to differential survival after fertilization. This observation is important because, although the comparison of models point to mostly prezygotic causes of TRD, their occurrence in only one breeding season argues against this explanation. The gametic causes of distortion cannot be dismissed completely, as environment- or age-dependent examples of meiotic drive are known (Orr and Irving 2005; Lindholm et al. 2016). However, additional observations speak against such possibility because the parents of the two families were siblings of the same age, and they shared an environment as juveniles and then as adults were kept separately only during the breeding season. Although we used only a single cross direction, we can rule out cytonuclear interactions as a major cause of TRD because L. montandoni alleles were underrepresented in most distorted markers, while all hybrids carried L. montandoni mitochondrial DNA. Hence, in our opinion, the most likely explanation of the observed TRDs is environment-dependent mortality of certain genotypes, and little difference in fit between gametic and zygotic models suggests partial dominance of L. montandoni alleles. The environment-dependent mortality may have been sex-dependent, but we were not able to evaluate this possibility because the sex of the larvae could not be determined and sex-linked molecular markers are not available. The F2 larvae from each family were raised in the same laboratory and at the same time but in separate aquaria. Although larvae were given the same food (Artemia hatchlings, ad libitum), we did not tightly control important environmental conditions, such as larval density, oxygen concentration, amount of substrate, or decaying organic matter at the bottom of aquaria. Embryonal and larval mortality during development were not recorded either, because young newt larvae are small and their bodies decompose quickly. Larvae from both families were preserved at the same time and stage, so increased mortality of some developmental stages is not a plausible explanation for the observed differences in TRDs. Therefore, it is likely that environmental conditions differed between families and if some genotypes had low survival only under stressful conditions, differences in the patterns of TRDs between families and/or years would ensue.
Several mechanisms may underlie the environment-dependent mortality of certain genotypes: (i) inbreeding depression, (ii) low survival of hybrids due to genetic incompatibilities between the parental genomes revealed only in some environmental conditions, and (iii) low survival under particular conditions of hybrid genotypes carrying L. montandoni alleles at specific genomic locations. Regarding inbreeding depression, the parents of families A and B may have differed by chance in the load of recessive deleterious alleles. These would be exposed in the F2 generation, leading to differences in TRDs between families. We deem this explanation unlikely, because in most distorted markers heterozygotes were also underrepresented, implying partial codominance of deleterious alleles, which is not compatible with inbreeding depression. The two remaining mechanisms are plausible and both may contribute to reproductive isolation. Increased mortality of some hybrid genotypes is customarily attributed to genetic incompatibilities between parental populations or species, which are often environment-dependent (Bordenstein and Drapeau 2001; Willett and Burton 2003; Hou et al. 2015). If these incompatibilities take the form of negative epistatic interactions of the DM type, as commonly observed (Coyne and Orr 2004; Corbett-Detig et al. 2013), statistical associations are expected between genotypes at unlinked loci. In family B, we detected possible DM type interactions at the nominal P < 10−4, which may suggest environment-dependent incompatibilities between the parental genomes; however, none were significant after the correction for multiple tests. Blocks of distorted markers on LG4 and LG10 were not involved in any such interaction, and consequently, there is no support for environment-dependent DM type incompatibilities as the causes of TRD in these regions. However, because of the very large number of tests, samples sizes would need to be much larger to achieve good statistical power. Further, complex epistasis between TRD blocks and multiple other genomic regions, which may be even more difficult to detect, cannot be excluded [reviewed in Fraïsse et al. (2014)]. An alternative explanation not evoking genetic incompatibilities is poor survival under some environmental conditions of larvae carrying L. montandoni alleles in these genomic regions. The species differ ecologically (Zavadil et al. 2003; Schmidtler and Franzen 2004) and differences in survival of their larvae in some or even most environmental conditions would not be surprising. The TRD blocks could then even mark genomic regions involved in prezygotic habitat isolation between the parental species.
Our experiment was not designed to distinguish between various isolating barriers. Hence, although we ruled out some explanations of TRD, unambiguous links between TRD and mechanisms of reproductive isolation could not be established. Fortunately, tools and experimental designs to detect and quantify components of reproductive isolation are available (Rundle and Whitlock 2001; Nosil 2012). Common garden experiments comparing survival of pure L. montandoni, L. vulgaris, hybrid, and backcross larvae under various conditions in combination with genotyping of mapped markers would be especially helpful in this respect. Regardless of the exact mechanisms, there is little doubt that TRD blocks play a role in reproductive isolation. If so, reduced interspecific introgression of these genomic segments is expected, and this prediction can be tested in L. montandoni × L. vulgaris hybrid zones by comparing the width and location of allele frequency clines in TRD segments to the genomic average. The map reported here is an essential tool to study genomic architecture of divergence between species and interactions between their genomes in natural hybrid zones. Another important direction of research enabled by the linkage map is elucidation of the genetic basis of phenotypic and ecological differences between species by QTL analysis.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.041178/-/DC1.
Acknowledgments
We thank A. Kret, A. Osikowski, and G. Pabijan for help in newt maintenance and breeding; M. Stuglik for assistance in bioinformatics analyses; and M. Pabijan for valuable comments. This work was funded by the Polish National Science Centre (2012/04/A/NZ8/00662 to W.B.) and the Jagiellonian University (DS/WBiNoZ/INoS/762/16).
Footnotes
Communicating editor: F. F. Pardo Manuel de Villena
Literature Cited
- Babik W., Rafiński J., 2004. Relationship between morphometric and genetic variation in pure and hybrid populations of the smooth and Montandon’s newt (Triturus vulgaris and T. montandoni). J. Zool. (Lond.) 262: 135–143. [Google Scholar]
- Babik W., Szymura J. M., Rafiński J., 2003. Nuclear markers, mitochondrial DNA and male secondary sexual traits variation in a newt hybrid zone (Triturus vulgaris × T. montandoni). Mol. Ecol. 12: 1913–1930. [DOI] [PubMed] [Google Scholar]
- Babik W., Branicki W., Crnobrnja-Isailovic J., Cogalniceanu D., Sas I., et al. , 2005. Phylogeography of two European newt species - discordance between mtDNA and morphology. Mol. Ecol. 14: 2475–2491. [DOI] [PubMed] [Google Scholar]
- Barb J. G., Bowers J. E., Renaut S., Rey J. I., Knapp S. J., et al. , 2014. Chromosomal evolution and patterns of introgression in Helianthus. Genetics 197: 969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsacchi G., Bussotti L., Mancino G., 1970. The maps of the lampbrush chromosomes of Triturus (Amphibia urodela). Chromosoma 31: 255–279. [DOI] [PubMed] [Google Scholar]
- Beukeboom L., Niehuis O., Pannebakker B., Koevoets T., Gibson J., et al. , 2010. A comparison of recombination frequencies in intraspecific vs. interspecific mapping populations of Nasonia. Heredity 104: 302–309. [DOI] [PubMed] [Google Scholar]
- Bordenstein S., Drapeau M., 2001. Genotype‐by‐environment interaction and the Dobzhansky–Muller model of postzygotic isolation. J. Evol. Biol. 14: 490–501. [Google Scholar]
- Bradshaw H., Schemske D. W., 2003. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature 426: 176–178. [DOI] [PubMed] [Google Scholar]
- Brennan A. C., Hiscock S. J., Abbott R. J., 2014. Interspecific crossing and genetic mapping reveal intrinsic genomic incompatibility between two Senecio species that form a hybrid zone on Mount Etna, Sicily. Heredity 113: 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan H. G., 1966. Chromosomes and nucleoli of the axolotl, Ambystoma mexicanum. J. Cell Sci. 1: 85–108. [DOI] [PubMed] [Google Scholar]
- Callan H. G., Spurway H., 1951. A study of meiosis in interracial hybrids of the newt, Triturus cristatus. J. Genet. 50: 235–249. [DOI] [PubMed] [Google Scholar]
- Chakravarti A., Lasher L. K., Reefer J. E., 1991. A maximum likelihood method for estimating genome length using genetic linkage data. Genetics 128: 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig R. B., Zhou J., Clark A. G., Hartl D. L., Ayroles J. F., 2013. Genetic incompatibilities are widespread within species. Nature 504: 135–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig R. B., Hartl D. L., Sackton T. B., 2015. Natural selection constrains neutral diversity across a wide range of species. PLoS Biol. 13: e1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 2004. Speciation, Sinauer, Sunderland, MA. [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont B. L., 2017. Variation and evolution of the meiotic requirement for crossing over in mammals. Genetics 205: 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont B. L., White M. A., Steffy B., Wiltshire T., Payseur B. A., 2011. Extensive recombination rate variation in the house mouse species complex inferred from genetic linkage maps. Genome Res. 21: 114–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijarczyk A., Dudek K., Babik W., 2016. Selective landscapes in newt immune genes inferred from patterns of nucleotide variation. Genome Biol. Evol. 8: 3417–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L., Kelly A. J., Morgan E., Willis J. H., 2001. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics 159: 1701–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick B. M., Johnson J. R., Kump D. K., Smith J. J., Voss S. R., et al. , 2010. Rapid spread of invasive genes into a threatened native species. Proc. Natl. Acad. Sci. USA 107: 3606–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraïsse C., Elderfield J., Welch J., 2014. The genetics of speciation: are complex incompatibilities easier to evolve? J. Evol. Biol. 27: 688–699. [DOI] [PubMed] [Google Scholar]
- Fruciano C., Franchini P., Kovacova V., Elmer K. R., Henning F., et al. , 2016. Genetic linkage of distinct adaptive traits in sympatrically speciating crater lake cichlid fish. Nat. Commun. 7: 12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnaire P. A., Normandeau E., Pavey S. A., Bernatchez L., 2013. Mapping phenotypic, expression and transmission ratio distortion QTL using RAD markers in the Lake Whitefish (Coregonus clupeaformis). Mol. Ecol. 22: 3036–3048. [DOI] [PubMed] [Google Scholar]
- Gregory, T. R., 2015 Animal Genome Size Database. Available at: http://www.genomesize.com. Accessed: December 8, 2016.
- Hale M. C., Thrower F. P., Berntson E. A., Miller M. R., Nichols K. M., 2013. Evaluating adaptive divergence between migratory and nonmigratory ecotypes of a salmonid fish, Oncorhynchus mykiss. G3 3: 1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. C., Willis J. H., 2005. Transmission ratio distortion in intraspecific hybrids of Mimulus guttatus. Genetics 170: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero P., López-Fernández C., 1986. The meiotic system of Iberian species of the genus Triturus (Amphibia: Caudata). Caryologia 39: 385–395. [Google Scholar]
- Hollenbeck C. M., Portnoy D. S., Gold J. R., 2015. A genetic linkage map of red drum (Sciaenops ocellatus) and comparison of chromosomal syntenies with four other fish species. Aquaculture 435: 265–274. [Google Scholar]
- Hou J., Friedrich A., Gounot J.-S., Schacherer J., 2015. Comprehensive survey of condition-specific reproductive isolation reveals genetic incompatibility in yeast. Nat. Commun. 6: 7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. O., Labbe A., Infante-Rivard C., 2013. Transmission ratio distortion: review of concept and implications for genetic association studies. Hum. Genet. 132: 245–263. [DOI] [PubMed] [Google Scholar]
- Jenczewski E., Gherardi M., Bonnin I., Prosperi J.-M., Olivieri I., et al. , 1997. Insight on segregation distortions in two intraspecific crosses between annual species of Medicago (Leguminosae). Theor. Appl. Genet. 94: 682–691. [Google Scholar]
- Kawakami T., Smeds L., Backström N., Husby A., Qvarnström A., et al. , 2014. A high‐density linkage map enables a second‐generation collared flycatcher genome assembly and reveals the patterns of avian recombination rate variation and chromosomal evolution. Mol. Ecol. 23: 4035–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath M. C., Timoshevskiy V. A., Timoshevskaya N. Y., Tsonis P. A., Voss S. R., et al. , 2015. Initial characterization of the large genome of the salamander Ambystoma mexicanum using shotgun and laser capture chromosome sequencing. Sci. Rep. 5: 16413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath M. C., Voss S. R., Tsonis P. A., Smith J. J., 2016. A linkage map for the newt Notophthalmus viridescens: insights in vertebrate genome and chromosome evolution. Dev. Biol. DOI: 10.1016/j.ydbio.2016.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kezer J., León P. E., Sessions S. K., 1980. Structural differentiation of the meiotic and mitotic chromosomes of the salamander, Ambystoma macrodactylum. Chromosoma 81: 177–197. [Google Scholar]
- Leppälä J., Bokma F., Savolainen O., 2013. Investigating incipient speciation in Arabidopsis lyrata from patterns of transmission ratio distortion. Genetics 194: 697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm A. K., Dyer K. A., Firman R. C., Fishman L., Forstmeier W., et al. , 2016. The ecology and evolutionary dynamics of meiotic drive. Trends Ecol. Evol. 31: 315–326. [DOI] [PubMed] [Google Scholar]
- Litvinchuk S. N., Rosanov J. M., Borkin L. J., 2007. Correlations of geographic distribution and temperature of embryonic development with the nuclear DNA content in the Salamandridae (Urodela, Amphibia). Genome 50: 333–342. [DOI] [PubMed] [Google Scholar]
- Liu S., Li Y., Qin Z., Geng X., Bao L., et al. , 2016. High‐density interspecific genetic linkage mapping provides insights into genomic incompatibility between channel catfish and blue catfish. Anim. Genet. 47: 81–90. [DOI] [PubMed] [Google Scholar]
- Lowry D. B., Willis J. H., 2010. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 8: e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry D. B., Hernandez K., Taylor S. H., Meyer E., Logan T. L., et al. , 2015. The genetics of divergence and reproductive isolation between ecotypes of Panicum hallii. New Phytol. 205: 402–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancino G., Barsacchi G., Nardi I., 1969. The lampbrush chromosomes of Salamandra salamandra (L.) (Amphibia Urodela). Chromosoma 26: 365–387. [DOI] [PubMed] [Google Scholar]
- Mao X., Kadri N. K., Thomasen J. R., De Koning D., Sahana G., et al. , 2015. Fine mapping of a calving QTL on Bos taurus autosome 18 in Holstein cattle. J. Anim. Breed. Genet. 133: 207–218. [DOI] [PubMed] [Google Scholar]
- Margarido G., Souza A., Garcia A., 2007. OneMap: software for genetic mapping in outcrossing species. Hereditas 144: 78–79. [DOI] [PubMed] [Google Scholar]
- Matsubara K., Ebana K., Mizubayashi T., Itoh S., Ando T., et al. , 2011. Relationship between transmission ratio distortion and genetic divergence in intraspecific rice crosses. Mol. Genet. Genomics 286: 307–319. [DOI] [PubMed] [Google Scholar]
- Menzel, U., 2013 EMT: Exact Multinomial Test: Goodness-of-Fit Test for Discrete Multivariate Data. Available at: https://cran.r-project.org/web/packages/EMT/. Accessed: May 15, 2017.
- Michalak P., Rafiński J., 1999. Sexual isolation between two newt species, Triturus vulgaris and T. montandoni (Amphibia, Urodela, Salamandridae). Biol. J. Linn. Soc. Lond. 67: 343–352. [Google Scholar]
- Michalak P., Grzesik J., Rafiński J., 1997. Tests for sexual incompatibility between two newt species, Triturus vulgaris and Triturus montandoni: no-choice mating design. Evolution 51: 2045–2050. [DOI] [PubMed] [Google Scholar]
- Mollinari M., Margarido G., Vencovsky R., Garcia A., 2009. Evaluation of algorithms used to order markers on genetic maps. Heredity 103: 494–502. [DOI] [PubMed] [Google Scholar]
- Moyle L. C., Graham E. B., 2006. Genome-wide associations between hybrid sterility QTL and marker transmission ratio distortion. Mol. Biol. Evol. 23: 973–980. [DOI] [PubMed] [Google Scholar]
- Nadachowska-Brzyska K., Zielinski P., Radwan J., Babik W., 2012. Interspecific hybridization increases MHC class II diversity in two sister species of newts. Mol. Ecol. 21: 887–906. [DOI] [PubMed] [Google Scholar]
- Niedzicka M., Fijarczyk A., Dudek K., Stuglik M., Babik W., 2016. Molecular Inversion Probes for targeted resequencing in non-model organisms. Sci. Rep. 6: 24051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P., 2012. Ecological Speciation. Oxford University Press, Oxford. [Google Scholar]
- O’Roak B. J., Vives L., Fu W., Egertson J. D., Stanaway I. B., et al. , 2012. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338: 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., Irving S., 2005. Segregation distortion in hybrids between the Bogota and USA subspecies of Drosophila pseudoobscura. Genetics 169: 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostberg C. O., Hauser L., Pritchard V. L., Garza J. C., Naish K. A., 2013. Chromosome rearrangements, recombination suppression, and limited segregation distortion in hybrids between Yellowstone cutthroat trout (Oncorhynchus clarkii bouvieri) and rainbow trout (O. mykiss). BMC Genomics 14: 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F., Sapienza C., 2001. Recombination is proportional to the number of chromosome arms in mammals. Mamm. Genome 12: 318–322. [DOI] [PubMed] [Google Scholar]
- Postlethwait J. H., Woods I. G., Ngo-Hazelett P., Yan Y.-L., Kelly P. D., et al. , 2000. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 10: 1890–1902. [DOI] [PubMed] [Google Scholar]
- Rieseberg L. H., Buerkle C. A., 2002. Genetic mapping in hybrid zones. Am. Nat. 159: S36–S50. [DOI] [PubMed] [Google Scholar]
- Rieseberg L. H., Baird S. J., Gardner K. A., 2000. Hybridization, introgression, and linkage evolution. Plant Mol. Biol. 42: 205–224. [PubMed] [Google Scholar]
- Rousset F., 2008. GENEPOP’007: a complete re‐implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour. 8: 103–106. [DOI] [PubMed] [Google Scholar]
- Royaert S., Jansen J., da Silva D. V., de Jesus Branco S. M., Livingstone D. S., et al. , 2016. Identification of candidate genes involved in Witches’ broom disease resistance in a segregating mapping population of Theobroma cacao L. in Brazil. BMC Genomics 17: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundle H. D., Whitlock M. C., 2001. A genetic interpretation of ecologically dependent isolation. Evolution 55: 198–201. [DOI] [PubMed] [Google Scholar]
- Schmidtler J., Franzen M., 2004. Triturus vulgaris (Linnaeus, 1758)–Teichmolch, pp. 847–967 in Handbuch der Reptilien und Amphibien Europas, edited by Bohme W. Aula-Verlag, Wiebelsheim. [Google Scholar]
- Schumer M., Cui R., Powell D. L., Dresner R., Rosenthal G. G., et al. , 2014. High-resolution mapping reveals hundreds of genetic incompatibilities in hybridizing fish species. eLife 3: e02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions S. K., 2008. Evolutionary cytogenetics in salamanders. Chromosome Res. 16: 183–201. [DOI] [PubMed] [Google Scholar]
- Smith J. J., Kump D. K., Walker J. A., Parichy D. M., Voss S. R., 2005. A comprehensive expressed sequence tag linkage map for tiger salamander and Mexican axolotl: enabling gene mapping and comparative genomics in Ambystoma. Genetics 171: 1161–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smukowski C., Noor M., 2011. Recombination rate variation in closely related species. Heredity 107: 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuglik M. T., Babik W., 2016. Genomic heterogeneity of historical gene flow between two species of newts inferred from transcriptome data. Ecol. Evol. 6: 4513–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart A. L., Fishman L., Willis J. H., 2006. A simple genetic incompatibility causes hybrid male sterility in Mimulus. Genetics 172: 2465–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sybenga J., 1996. Recombination and chiasmata: few but intriguing discrepancies. Genome 39: 473–484. [DOI] [PubMed] [Google Scholar]
- True J. R., Mercer J. M., Laurie C. C., 1996. Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics 142: 507–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van’t Hof A. E., Campagne P., Rigden D. J., Yung C. J., Lingley J., et al. , 2016. The industrial melanism mutation in British peppered moths is a transposable element. Nature 534: 102–105. [DOI] [PubMed] [Google Scholar]
- Voorrips R., 2002. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93: 77–78. [DOI] [PubMed] [Google Scholar]
- Voss S. R., Kump D. K., Putta S., Pauly N., Reynolds A., et al. , 2011. Origin of amphibian and avian chromosomes by fission, fusion, and retention of ancestral chromosomes. Genome Res. 21: 1306–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson I., Callan H., 1963. The form of bivalent chromosomes in newt oocytes at first metaphase of meiosis. J. Cell Sci. 3: 281–295. [Google Scholar]
- Whitney K. D., Broman K. W., Kane N. C., Hovick S. M., Randell R. A., et al. , 2015. Quantitative trait locus mapping identifies candidate alleles involved in adaptive introgression and range expansion in a wild sunflower. Mol. Ecol. 24: 2194–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickbom T., 1945. Cytological studies on dipnoi, urodela, anura, and emys. Hereditas 31: 241–346. [DOI] [PubMed] [Google Scholar]
- Willett C. S., Burton R. S., 2003. Environmental influences on epistatic interactions: viabilities of cytochrome c genotypes in interpopulation crosses. Evolution 57: 2286–2292. [DOI] [PubMed] [Google Scholar]
- Woram R., McGowan C., Stout J., Gharbi K., Ferguson M., et al. , 2004. A genetic linkage map for Arctic char (Salvelinus alpinus): evidence for higher recombination rates and segregation distortion in hybrid vs. pure strain mapping parents. Genome 47: 304–315. [DOI] [PubMed] [Google Scholar]
- Wu Y., Close T. J., Lonardi S., 2011. Accurate construction of consensus genetic maps via integer linear programming. IEEE/ACM Trans. Comp. Biol. Bioinf. 8: 381–394. [DOI] [PubMed] [Google Scholar]
- Zavadil V., Pialek J., Dandova R., 2003. Triturus montandoni (Boulenger, 1880) - Karpatenmolch, pp. 657–706 in Handbuch der Reptilen und Amphibien Europas, edited by Bohme W. Aula-Verlag, Wiebelsheim. [Google Scholar]
- Zbożeń, J., 1997 Comparative caryological studies of the smooth (Triturus vulgaris) and Carpathian (T. montandoni) newts (In Polish). Ph.D. Thesis, Jagiellonian University, Kraków. [Google Scholar]
- Zbożeń J., Rafiński J., 1993. A comparative study of mitotic and meiotic chromosomes of the Montandon’s newt, Triturus montandoni (Urodela: Salamandridae) from Poland and Rumania. Genetica 88: 69–77. [Google Scholar]
- Zieliński P., Dudek K., Stuglik M. T., Liana M., Babik W., 2014. Single nucleotide polymorphisms reveal genetic structuring of the Carpathian newt and provide evidence of interspecific gene flow in the nuclear genome. PLoS One 9: e97431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieliński P., Nadachowska‐Brzyska K., Dudek K., Babik W., 2016. Divergence history of the Carpathian and smooth newts modelled in space and time. Mol. Ecol. 25: 3912–3928. [DOI] [PubMed] [Google Scholar]
- Zieliński P., Nadachowska-Brzyska K., Wielstra B., Szkotak R., Covaciu-Marcov S. D., et al. , 2013. No evidence for nuclear introgression despite complete mtDNA replacement in the Carpathian newt (Lissotriton montandoni). Mol. Ecol. 22: 1884–1903. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
File S2 contains genotypes for each individual.