Abstract
The fungal pathogen Fusarium oxysporum f. sp. cubense causes Fusarium wilt, one of the most destructive diseases in banana and plantain cultivars. Pathogenic race 1 attacks the “Gros Michel” banana cultivar, and race 4 is pathogenic to the Cavendish banana cultivar and those cultivars that are susceptible to Foc1. To understand the divergence in gene expression modules between the two races during degradation of the host cell wall, we performed RNA sequencing to compare the genome-wide transcriptional profiles of the two races grown in media containing banana cell wall, pectin, or glucose as the sole carbon source. Overall, the gene expression profiles of Foc1 and Foc4 in response to host cell wall or pectin appeared remarkably different. When grown with host cell wall, a much larger number of genes showed altered levels of expression in Foc4 in comparison with Foc1, including genes encoding carbohydrate-active enzymes (CAZymes) and other virulence-related genes. Additionally, the levels of gene expression were higher in Foc4 than in Foc1 when grown with host cell wall or pectin. Furthermore, a great majority of genes were differentially expressed in a variety-specific manner when induced by host cell wall or pectin. More specific CAZymes and other pathogenesis-related genes were expressed in Foc4 than in Foc1 when grown with host cell wall. The first transcriptome profiles obtained for Foc during degradation of the host cell wall may provide new insights into the mechanism of banana cell wall polysaccharide decomposition and the genetic basis of Foc host specificity.
Keywords: Fusarium oxysporum f. sp. cubense, transcriptome, carbohydrate-active enzymes, cell wall-degrading enzymes, pathogenicity genes
The soil-borne fungus Fusarium oxysporum f. sp. cubense (Foc) causes Fusarium wilt, one of the most important lethal diseases in banana, which has a devastating effect on banana production worldwide (Ploetz 2006, 2015a). According to the susceptibility of specific banana cultivars, Foc can be divided into four races (Stover and Buddenhagen 1986; Stover 1990). Race 1 (Foc1) causes disease in the “Gros Michel” (AAA) and “Silk” (AAB) cultivars (Waite and Stover 1960). Race 2 is the causal agent of this disease in the hybrid triploid Bluggoe (ABB). Race 3 is virulent only to Heliconia spp. and does not affect Musa spp. (Stover 1962). Race 4 (Foc4) infects Cavendish (AAA) cultivars and those cultivars susceptible to races 1 and 2 (Su et al. 1986). In the middle of the past century, Foc1 devastated the Gros Michel cultivar, which was the main commercial banana cultivar grown for the export trade (Stover 1962). This forced the trade to shift to resistant cultivars of the Cavendish variety (Buddenhagen 1990). However, the virulent strain Foc4, to which Cavendish is susceptible, spread rapidly to banana-growing regions and now threatens the global production of Cavendish and other currently popular cultivars (Ploetz 2006; Butler 2013). Regardless of biological, chemical, or cultural measures, no appropriate management strategies for Fusarium wilt are currently available to eliminate this pathogen once plants are infected (Ploetz 2007, 2015b).
Despite the importance and necessity of controlling this disease, the molecular mechanisms of pathogenesis in banana and the genetic basis for host specificity are still poorly understood. Only a few pathogenicity genes involved in Foc4 infection of Cavendish cultivars have been identified thus far, which include the following: Foatf1, encoding a basic leucine zipper (bZIP) transcription factor (TF); FoOCH1, encoding a putative α-1,6-mannosyltransferase; FoSlt2, FoMkk2, and FoBck1, encoding mitogen-activated protein kinases (MAPKs); and four G-protein subunit genes, FGA1, FGA2, FGA3, and FGB1 (Qi et al. 2013; Li et al. 2014; Ding et al. 2015; Guo et al. 2016a,b).
Recently, with the extensive application of next-generation sequencing technologies to plant phytopathogen genomics, increasing numbers of gene sets have been found to be involved in pathogenicity and pathogen–plant interactions with essential roles in plant disease. The genomes of F. oxysporum isolates, including F. oxysporum f. sp. lycopersici (Fol), F. oxysporum f. sp. cubense, F. oxysporum f. sp. ciceris, and F. oxysporum f. sp. melonis, have been sequenced and have been shown to contain a large number of pathogenicity genes and virulence-related genes, including predicted effectors, TFs, transmembrane transporters, and CAZymes, among others (Ma et al. 2010; Guo et al. 2014; van Dam et al. 2016; Williams et al. 2016). A significant finding is that lineage-specific genes on mobile and partly dispensable chromosomes can be transferred intraspecifically and possibly interspecifically, thus constituting determinants of pathogenicity and host range, which have been identified in Fusarium species and other fungal species (Han et al. 2001; Chuma et al. 2003; Coleman et al. 2009; Ma et al. 2010; Schmidt et al. 2016; Williams et al. 2016). Whole-genome sequence analysis of Foc1 and Foc4 has led to the identification of a large number of potential pathogenicity genes. Although the genomes of the two Foc races are highly syntenic with Fol, neither contains the lineage-specific genomic regions that are exclusive to Fol (Guo et al. 2014).
Transcriptomic analysis of phytopathogens is also one of the most effective ways to obtain a full understanding of fungal pathogenesis at the molecular level, potentially allowing the characterization of all actively expressed genes and transcripts at different stages of plant infection or under various conditions. In Fusarium species, transcriptomic studies have mainly focused on the facultative pathogen F. graminearum under a variety of culture conditions (different carbon and nitrogen sources) in vitro and during different stages of the infection of host plants in planta (Guldener et al. 2006; Seong et al. 2008; Gardiner et al. 2009; Lysoe et al. 2011; Carapito et al. 2013; Boedi et al. 2016). Based on these studies, specific subsets of F. graminearum genes are considered to be pathogenicity-related or virulence genes. However, there are few transcriptomic resources available for Foc in the NCBI database. To date, only the transcriptome of Foc during early Cavendish variety infection has been examined. This study indicated that pathogenicity genes encoding MAPK, G-proteins, and a two-component system involved in signaling are activated in Foc4 rather than Foc1 during early Cavendish variety infection (Guo et al. 2014). However, more information about the changes in the expression of pathogenicity-related genes at different critical points in disease establishment and under various growth conditions in vitro, as well as the molecular basis of the difference in virulence between the two races, is still needed.
The plant cell wall is primarily composed of three types of polysaccharides: cellulose, hemicellulose, and pectin. It is the first physical and chemical barrier against pathogen invasion but is also a nutrient source for invading pathogens. Pectin, the most sophisticated polysaccharide in terms of structure, is a highly diverse polymer that contains several specific polysaccharides (Pérez et al. 2000; Caffall and Mohnen 2009). Many pathogenic fungi have the genetic potential to decompose plant cell wall polysaccharides by producing an extensive set of CAZymes (Aro et al. 2005; Coutinho et al. 2009; van den Brink and de Vries 2011). The plant cell wall-degrading enzymes (CWDEs) secreted by fungi are all assigned to CAZymes, which are one type of pathogenicity factor used by pathogens or saprophytes to invade plants (Götesson et al. 2002; Ospina-Giraldo et al. 2003, 2009). The genomes of Foc1 and Foc4 contain large proportions of CAZymes, implying that effective banana cell wall degradation may be important for the virulence of Foc (Guo et al. 2014).
In the current study, to go a step further in the analysis of Foc genes that participate in host cell wall decomposition and contribute to pathogenicity in the banana host, RNA-seq was used to compare the global transcriptional profiles of Foc1 and Foc4 cultured in media containing banana cell wall, pectate, or glucose as the sole carbon source. The aims were as follows: (1) to elucidate the mechanism of the Foc genes involved in the decomposition of different polysaccharides from the primary plant cell wall; (2) to identify differences in gene contents and transcriptional levels between the two Foc races that might account for the variations in the virulence of Foc races during infection of their particular hosts; and (3) to identify Foc pathogenicity genes responsible for decomposition of the host cell wall. This work will provide further transcriptomic information on the mechanisms of Fusarium–plant interactions and contribute to the development of feasible strategies for controlling Foc infection.
Materials and Methods
Fungal strains and growth conditions
The “Brazil” banana cultivar (AAA group, Cavendish) is Foc4-susceptible, but highly resistant to Foc1, while the “Fenjiao” cultivar (ABB group, Pisang Awak) is susceptible to both Foc4 and Foc1. Single-conidium pure cultures of the Foc1 C2 and Foc4 DZ1 strains were isolated from diseased rhizome tissues of the Fenjiao and Brazil cultivars, respectively. Both Foc isolates underwent molecular characterization of Foc and were maintained as microconidial suspensions in 30% (v/v) glycerol at −80° in our laboratory. Foc isolates were periodically inoculated onto their respective host banana cultivars in a growth chamber to confirm their pathogenicity. The Foc4 DZ1 strain was identified as F. oxysporum f. sp. cubense tropical race 4 (Foc TR4). Banana cell walls of the Brazil and Fenjiao cultivars were prepared from greenhouse-grown 6-wk-old banana plants using the method described for maize (Sposato et al. 1995).
Foc isolates (106 conidia) were shaken in synthetic medium (SM) (Garcia Maceira et al. 1997) supplemented with 1% [w/v] glucose at 28° at 120 rpm. After 48 hr of growth, the mycelium was harvested, washed three times in sterile water, and transferred to fresh SM supplemented with different carbon sources: 1% [w/v] glucose, 1% [w/v] citrus pectin (Sigma), or 1% banana cell wall (cell wall from the Brazil banana cultivar for Foc4 or from Fenjiao for Foc1). The mycelia were shaken at 28° at 120 rpm for 24 hr and were then collected using previously sterilized tweezers, washed three times with sterile water treated with diethylpyrocarbonate (DEPC, Sigma), filtered through nitrocellulose membranes (0.22 μm), and flash frozen in liquid nitrogen.
RNA extraction, library preparation, and sequencing of the Foc transcriptome
Total RNA from three independent cultures grown under each condition was extracted separately using the TRIzol Reagent (Invitrogen) following the manufacturer’s instructions, then treated with RNase-free DNase I (TaKaRa, China). One part of total RNA from three independent cultures for each condition were pooled in equal amounts for RNA-seq analysis, and the other part was stored at −80° for further real-time quantitative PCR (qRT-PCR) verification. The quality and quantity of the total RNA was validated using agarose gel electrophoresis and a 2100 Bioanalyzer (Agilent Technologies). A total of 3 μg high-quality RNA (RNA integrity number > 8) per sample was prepared to generate cDNA libraries. Sequencing libraries were generated using the NEBNext Ultra RNA library prep kit for Illumina (New England BioLabs). The cDNA libraries were then sequenced on the Illumina HiSequation 2000 platform using a 100 bp paired-end sequencing strategy at the Novogene Corporation, Beijing, China.
Transcriptome assembly and analysis
Clean reads were obtained for the six libraries by removing reads containing adapters, poly-Ns, and low-quality bases from the raw reads. The sequencing quality of the clean reads was calculated at the Q20, Q30, GC-content, and sequence duplication levels. De novo transcriptome assembly was performed with Trinity (Grabherr et al. 2011). After sequence assembly, unigenes with the expected length and size were generated from the longest transcripts at each locus.
All unigenes in the Foc transcriptome were aligned to protein databases with a priority order of the NCBI nonredundant protein (Nr) database, Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG), and the EuKaryotic Orthologous Groups (KOG) database via BLASTX alignment (E-value ≤ 1 × 10−5) (Altschul et al. 1997). The coding sequences (CDSs) and corresponding amino acid sequences of the unigenes were extracted from the BLAST analysis results. For the unigenes with no hits in BLAST, ESTScan software was used to predict their CDSs and corresponding amino acid sequences (Iseli et al. 1999). The protein families of all unigenes were annotated through Pfam analysis (Finn et al. 2014). Gene ontology (GO) annotations according to the biological process, cellular component, and molecular function ontologies were assigned for each unigene using the Blast2GO program (Conesa et al. 2005). The nucleotide sequences of all unigenes were aligned to the NCBI nucleotide sequences (Nt) database with the BLAST algorithm (E-value ≤ 1 × 10−5). CAZymes were identified and classified using the CAZymes Analysis Toolkit (Park et al. 2010), applying a cut-off E-value ≤ 1 × 10−5, and through CAZyme annotation (dbCAN) (Yin et al. 2012). Foc pathogenicity genes were predicted using Blastp (E-value ≤ 1 × 10−5), against protein sequences in the pathogen–host interaction database (Winnenburg et al. 2006).
Gene expression profiling analyses
To estimate the RNA transcriptional abundance for each gene, the clean reads from six Foc libraries were mapped back to the reference-assembled transcriptome using RSEM (Li and Dewey 2011). Gene expression levels were calculated as FPKM values (expected fragments per kilobase of transcript per million fragments sequenced) using the program Cufflinks (Trapnell et al. 2010). The read counts from six libraries were first adjusted using the edgeR program package through one scaling-normalized factor (Robinson et al. 2010). Then, the differential expression levels of each unigene were analyzed using the DESeq R package by employing a negative binomial distributions model (Anders and Huber 2010; Wang et al. 2010). Differentially expressed genes (DEGs) were identified using a q-value (an adjusted p-value) ≤ 0.005 and a |log2(fold change)| ≥ 1 as the threshold (Storey and Tibshirani 2003). To further characterize the biological functions and metabolic pathways of DEGs, GO enrichment analyses were performed using Goseq (Young et al. 2010), and KEGG pathway enrichment analysis was carried out using KOBAS (Mao et al. 2005), with a cut-off p-value ≤ 0.05.
qRT-PCR verification
For validation of the RNA-seq results, 15 genes with different expression levels and diverse functions were randomly selected for qRT-PCR analysis. The total RNA samples (pretreated with DNase I) were the same samples obtained from three independent cultures for each condition but were not pooled, as for RNA-seq. Total RNA was reverse transcribed in a 20 μl reaction mixture using the PrimeScript RT Master Mix Kit (TaKaRa, China). The qRT-PCR was performed using a Thermal Cycler Dice TP900 (Takara, Japan) in combination with the SYBR Premix Ex Taq II Kit (TaKaRa, China) according to the manufacturer’s protocol. The gene-specific primers designed for qRT-PCR analysis are listed in Supplemental Material, Table S10 in File S2. The qRT-PCR conditions were as follows: initial denaturation at 95° for 30 sec, followed by 40 cycles of 95° for 5 sec, and 60° for 30 sec. All amplification reactions were run in triplicate with three biological replicates. The expression levels of target genes in response to growth in the presence of host cell wall or pectin were normalized to the constitutively expressed β-tubulin gene (reference gene), and were calibrated compared with the levels recorded during growth in glucose culture (set as 1) using the 2−ΔΔCt method (Livak and Schmittgen 2001).
Data availability
Strains of Foc used in this study are available upon request. The raw RNA-seq reads from the six samples are available at the NCBI Sequence Read Archive database under the accessions SRA486974. Supplemental figures and supplemental tables contain detailed information of all supplemental files to support this study.
Results
RNA sequencing and de novo assembly of the Foc transcriptome
To obtain a global view of the Foc1 and Foc4 transcriptomes during host cell wall degradation, RNA-seq was performed on Foc1 and Foc4 cultures containing banana cell wall, pectin, and glucose as the sole carbon source, and the gene expression profiles of each strain were compared. Six cDNA libraries prepared from the pooled total RNA extracted from two Foc isolates grown under three carbon conditions were paired-end sequenced on the Illumina HiSequation 2000 platform. The sequencing results for the six libraries (designated G_Foc1, G_Foc4, P_Foc1, P_Foc4, FCW_Foc1, and BCW_Foc4) are summarized in Table 1. After filtering out repetitive, low-complexity, and low-quality reads, the clean reads were assembled into 38,950 transcripts with an average length of 1937 bp and an N50 length of 3235 bp. The longest transcript for each locus was defined as the unigene, resulting in 23,417 unigenes with an average length of 1419 bp and an N50 length of 2542 bp (Table 2). The size distribution of these transcripts and unigenes is shown in Figure S1 in File S1.
Table 1. The six Foc libraries quality of RNA-seq.
| Samplea | Raw Reads | Clean Reads | Clean Bases | Error (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|
| G_Foc1 | 75163928 | 70921444 | 7.10G | 0.035 | 97.39 | 91.33 | 52.35 |
| G_Foc4 | 67448538 | 62516786 | 6.26G | 0.035 | 97.39 | 91.33 | 52.55 |
| P_Foc1 | 63999398 | 60148382 | 6.02G | 0.035 | 97.37 | 91.28 | 52.79 |
| P_Foc4 | 69432346 | 65433836 | 6.54G | 0.035 | 97.36 | 91.22 | 52.66 |
| FCW_Foc1 | 79810356 | 75082316 | 7.50G | 0.040 | 97.30 | 91.05 | 53.24 |
| BCW_Foc4 | 58060848 | 55168204 | 5.52G | 0.040 | 97.33 | 91.19 | 53.21 |
G, glucose; P, pectin; FCW, cell wall of Fengjiao cultivar; BCW, cell wall of Brazil cultivar.
Table 2. Length distribution of assembled transcripts and unigenes.
| Transcript | Unigene | |
|---|---|---|
| Total number of transcripts or unigenes | 38,950 | 23,417 |
| Number of transcripts or unigenes (200–500 bp) | 10,082 | 8,810 |
| Number of transcripts or unigenes (500–1000 bp) | 5,945 | 3,971 |
| Number of transcripts or unigenes (1000–2000 bp) | 9,003 | 5,045 |
| Number of transcripts or unigenes (> 2000 kbp) | 13,920 | 5,591 |
| Total length (bp) | 75,459,645 | 33,233,628 |
| Min length (bp) | 201 | 201 |
| Max length (bp) | 24,163 | 24,163 |
| Mean length (bp) | 1,937 | 1,419 |
| N50 (bp)a | 3,235 | 2,542 |
| N90 (bp)a | 1,010 | 605 |
The N50 size is computed by sorting all transcripts from largest to smallest and by determining the minimum set of transcripts whose sizes total 50% of the entire transcript and unigene was the same; N90 was counted in a similar way.
Functional gene annotation
The functional annotation of all 23,417 assembled unigenes was mainly based on BLAST homology searches against seven public protein databases. A total of 20,879 unigenes were annotated in at least one database, accounting for 89.16% of the unigenes (Table S1 in File S2). Among these unigenes, 15,883 (67.82%) showed high homology with known proteins in the Nr database, and 8212 unigenes (35.06%) were annotated in the Swiss-Prot database.
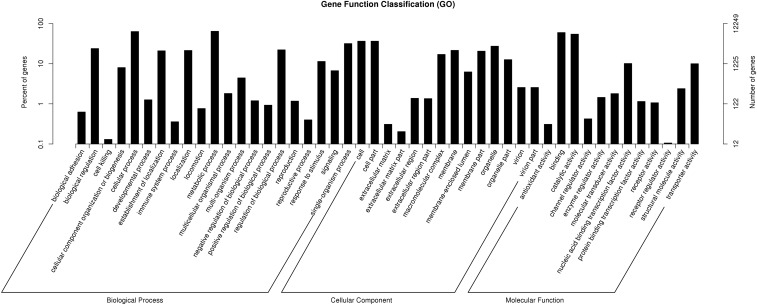
GO assignments were utilized to predict the functions of Foc unigenes by classifying them according to biological processes. A total of 12,249 unigenes (52.3%) were assigned to at least one GO term and were categorized into 54 functional groups in three categories (biological process, cellular component, and molecular function) (Figure 1 and Table S2 in File S2). Under biological processes, metabolic process and cellular process were the most prominent GO terms. Within the molecular function ontology, genes associated with the “binding,” “catalytic,” “nucleic acid binding transcription factor activity,” and “transporter activity” groups were the most abundant. These results indicate that Foc grows rapidly and exhibits extensive metabolic activity under each of the three tested carbon source conditions.
Figure 1.
Gene Ontology (GO) categories assigned to the Foc unigenes. The right y-axis indicates the number of genes in a category. The left y-axis indicates the percentage of a specific category of genes in that main category.
Using the KOG database to further annotation, a total of 5411 unigenes (23.1%) were clustered into 25 KOG categories (Figure S2 in File S1). Among these categories, the majority of genes were associated with “general functional prediction only” (1142 unigenes, 21.1%), “post-translational modification, protein turnover, and chaperones” (479 unigenes, 8.9%), and “secondary metabolite biosynthesis, transport, and catabolism” (458 unigenes, 8.5%).
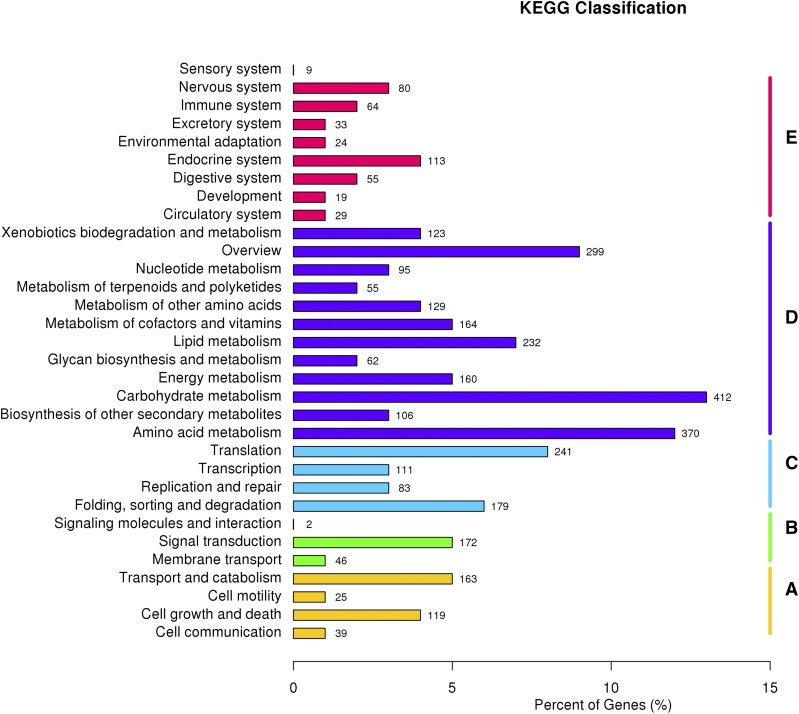
To determine the biological functions and interactions of Foc unigenes in the three carbon source cultures, a total of 3194 unigenes were assigned to the 259 KEGG biochemical pathways (Figure 2). Most of the unigenes were related to the category “metabolism,” with pathways “carbohydrate metabolism” (412 unigenes) and “amino acid metabolism” (370 unigenes) being the most representative (Figure 2). KEGG pathways belonging to “carbohydrate metabolism,” such as “starch and sucrose metabolism” and “pentose and glucuronate interconversions,” were more likely to be involved in banana cell wall polysaccharide degradation by Foc (Table S3 in File S2).
Figure 2.
Pathways assignment based on the Kyoto Encyclopedia of Genes and Genomes (KEGG). A = Cellular Processes; B = Environmental Information Processing; C = Genetic Information Processing; D = Metabolism; and E = Organismal Systems.
CAZymes present in the Foc transcriptome
CAZymes secreted by fungal pathogens can break down plant cell walls; therefore, they are important in establishing infection and in accessing nutrients during growth. CAZymes are assigned to CAZy families of glycoside hydrolases (GHs), carbohydrate esterases (CEs), polysaccharide lyases (PLs), glycosyl transferases (GTs), auxiliary activities (AAs), and carbohydrate-binding modules (CBMs). In this study, a total of 2105 CAZyme modules were identified among 1970 unigenes using the CAZymes Analysis Toolkit and dbCAN (Tables S4 and S5 in File S2). These unigenes were sorted into superfamilies, with GH being the most abundant, followed by GT, CE, CBM, AA, and PL (Table S4 in File S2). Based on the CAZyme modules for plant cell wall degradation, a total of 393 unigenes were identified as putative CWDEs, including 28 GHs, 3 CEs, 3 CBMs, 4 PLs, and 1 AA family (Table S6 in File S2). According to the substrates degraded by CWDEs, these putative CWDEs were categorized into 33 classes of enzymes (Table S6 in File S2). Unigenes encoding endo- and exopolygalacturonase were the most abundant, followed by β-1,4-xylosidase and β-1,4-glucosidase. These results indicate that Foc produces a large arsenal of plant CWDEs during degradation of host cell wall polysaccharides, with pectinase being the most abundant, followed by hemicellulase and cellulase.
Pathogenicity-associated genes in the Foc transcriptome
The PHI database contains expert information on experimentally verified pathogenicity, virulence, lethal, and effector genes from fungal and oomycete pathogens that infect animal, plant, and fungal hosts. The annotation results showed that 1202 unigenes were characterized as known genes proven to affect the outcome of pathogen–host interactions in various pathogenic fungi. In addition, 432 unigenes homologous to PHI genes associated with loss of pathogenicity (86), reduced virulence (340), and pathogenic effectors (6) were considered to be pathogenicity determinants for Foc (Table S7 in File S2). These unigenes encoded effectors, G-proteins and G-protein-coupled receptors, signaling protein, TFs, CWDEs, cutinases, transporters, cytochrome P450s, polyketide synthases, and chitin synthases, among others (Table S8 in File S2). GO analysis revealed that Foc genes homologous to PHI genes fell into a variety of metabolic processes, thereby highlighting the important roles of these processes in pathogenicity (Figure S3 in File S1).
DEGs induced by host cell wall or pectin and qRT-PCR verification
Having generated a Foc reference transcriptome, our next goal was to identify genes displaying significant changes in expression when Foc was grown with host cell wall polysaccharides. For each library, all clean reads were mapped back to the Foc reference transcriptome. The percentage of clean reads from each library that could be uniquely mapped ranged from 90.67 to 91.75%, thus providing good coverage of the transcript profiles (Table S9 in File S2). Six Foc libraries showed similar FPKM density distributions (Figure S4 in File S1). To identify expression patterns of similarity across six Foc samples, the principal component analysis and Pearson correlation analysis of scaled expression profiles revealed six distinct clusters when Foc1 and Foc4 grew on cultures containing three different carbon sources, suggesting that the gene expression profiles induced by host cell wall were significantly different as compared to those induced by pectin or glucose (Figure S5 in File S1). The major cause for this result was that the host cell wall is composed of different types of polysaccharides, including cellulose, hemicellulose, and pectin, which could induce more genes expression in Foc than pectin or glucose. Besides, the gene expression profiles of Foc4 were also significantly different from those of Foc1 induced by the same carbon source.
DEGs were analyzed via pairwise comparisons of P_Foc4 vs. G_Foc4, P_Foc1 vs. G_Foc1, P_Foc4 vs. P_Foc1, FCW_Foc1 vs. G_Foc1, BCW_Foc4 vs. G_Foc4, and BCW_Foc4 vs. FCW_Foc1. To verify the RNA-seq results, 15 unigenes were randomly selected for qRT-PCR analysis. These genes, which were involved in carbohydrate metabolism, signaling, and transport, or were genes of unknown function, were either upregulated, downregulated, or unaffected in the two races. Seventy-nine (87.7%) of the 90 qRT-PCR results agreed with the changes in transcript levels detected through RNA-seq (r = 0.89, p-value ≤ 10−5) (Figure S6 in File S1 and Table S10 in File S2). Thus, the RNA-seq data were considered reliable for the identification of DEGs induced by host cell wall and pectin in this study.
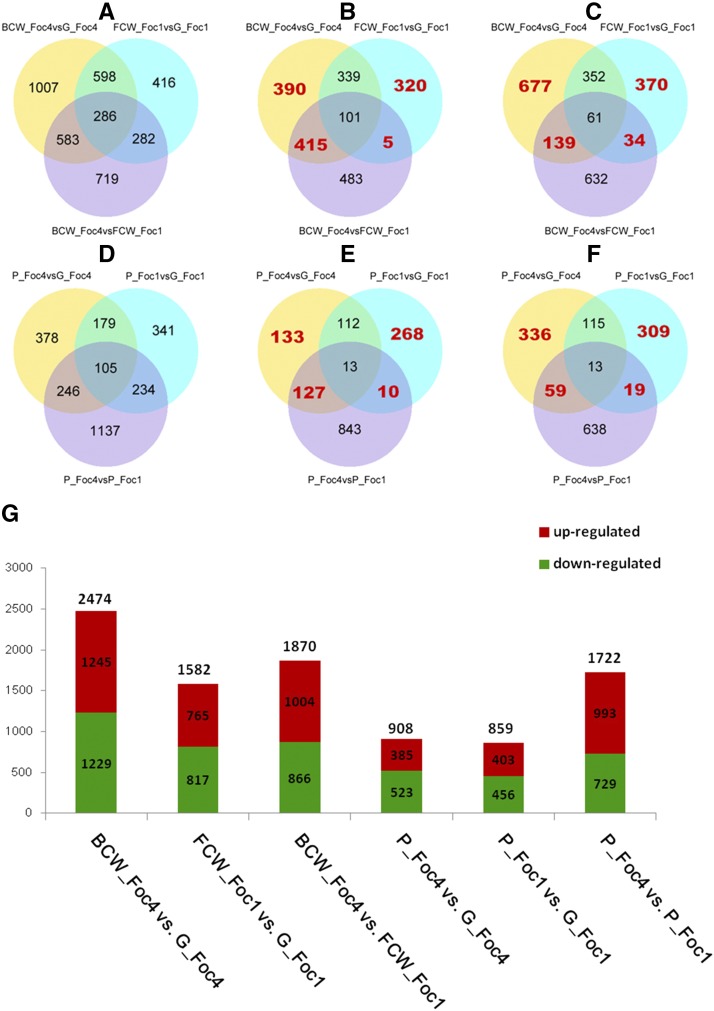
The global gene expression profiles of each comparison are shown in Figure 3. Overall, a much larger number of genes showed altered expression levels in Foc4 (2474 DEGs) grown in the presence of host cell wall than in Foc1 (1582 DEGs). A slightly greater number of genes showed altered expression levels in Foc4 (908 DEGs) grown in the presence of pectin than in Foc1 (859 DEGs) (Figure 3). Additionally, the intensity of gene expression was different between the two races grown in the presence of host cell wall or pectin, which could be found by comparing BCW_Foc4 vs. FCW_Foc1 or P_Foc4 vs. P_Foc1. Compared with downregulated DEGs, more upregulated DEGs were found when comparing BCW_Foc4 vs. FCW_Foc1 or P_Foc4 vs. P_Foc1, which indicated that total mRNA expression in Foc4 was higher than in Foc1 (Figure 3). These results implied that Foc4 degrades host cell wall polysaccharides by changing the expression of more genes than Foc1. GO enrichment analyses showed that a large number of DEGs were significantly enriched in a variety of metabolic processes (Tables S11 and S12 in File S2). KEGG pathway enrichment analysis revealed that the specific metabolism pathways were significantly enriched among the DEGs regulated by Foc during degradation of host cell wall polysaccharides (Tables S13 and S14 in File S2).
Figure 3.
Number of differentially expressed genes (DEGs) in cells grown with host cell wall polysaccharides. The number of DEGs that were up- and downregulated in each comparison are shown in the Venn diagrams (A–F) and histogram (G). The number of DEGs (A), including upregulated (B) and downregulated DEGs (C), derived from comparing cells grown in the presence of host cell wall. The number of DEGs (D), including upregulated (E) and downregulated DEGs (F), derived from comparing cells grown in the presence of pectin. The numbers of specific DEGs regulated by Foc1 or Foc4 were shown in the nonoverlapping regions (red numbers). The DEGs were identified by applying a threshold of q-value ≤ 0.005 and an absolute value of |log2(fold-change)| ≥ 1.
DEGs specifically found in Foc grown in the presence of host cell wall or pectin
The great majority of Foc genes are differentially expressed in a variety-specific manner during decomposition of the host cell wall. The numbers of specific DEGs up- and downregulated by Foc1 or Foc4 during the degradation of the host cell wall or pectin were shown in the nonoverlapping regions (red numbers) of the Venn diagram (Figure 3, B, C, E, and F and Table S15 in File S2). Compared with Foc1 grown with host cell wall, many more DEGs were specifically regulated in Foc4 (805 upregulated and 816 downregulated specific DEGs); more than twice as many DEGs were specifically regulated in Foc4 than in Foc1 (325 upregulated and 404 downregulated specific DEGs) (Figure 3, B and C and Table S15 in File S2). In contrast, the number of DEGs that were specifically regulated in Foc4 (260 upregulated and 395 downregulated specific DEGs) was greater than in Foc1 (278 upregulated and 328 downregulated specific DEGs) grown with pectin (Figure 3, D and F and Table S15 in File S2). Additionally, the gene expression levels of the specific DEGs in Foc4 were higher than in Foc1 (Figure 3, B, C, E, and F). More specific DEGs were upregulated in the comparison of BCW_Foc4 vs. FCW_Foc1 or P_Foc4 vs. P_Foc1.
Additionally, the annotation of specific DEGs showed that Foc4 specifically expresses more pathogenesis-related genes than Foc1 during degradation of host cell wall polysaccharides, which may account for the variation in the virulence of these races during infection of their particular hosts (Table 3). The number of specific DEGs related to fungal pathogenicity identified in the comparison of BCW_Foc4 vs. G_Foc4 (439 DEGs) was almost twice as great as in the comparison of FCW_Foc1 vs. G_Foc1 (219 DEGs) (Table 3). In particular, more specific DEGs annotated as G-protein, chitin synthase, ATP-binding cassette (ABC) transporter, zinc finger protein, bZIP TF, serine/threonine kinase, sucrose nonfermenting (SNF) protein kinase, F-box protein, peroxidase, and polyketide synthase were specifically upregulated by Foc4 than by Foc1 during the degradation of the host cell wall. Furthermore, ∼26.6% of the Foc4-specific DEGs and 18.2% of the Foc1-specific DEGs induced by growth in the presence of host cell wall were unannotated. In addition, the number of specific DEGs related to pathogenicity in the comparison of P_Foc4 vs. G_Foc4 (142 DEGs) was slightly lower than for P_Foc1 vs. G_Foc1 (165 DEGs) (Table 3). However, more DEGs annotated as a G-protein, chitin synthase, ABC transporter, or bZIP TF were specifically upregulated by Foc4 than by Foc1 during the degradation of pectin. Additionally, ∼15.4% of the Foc4-specific DEGs and 14.0% of the Foc1-specific DEGs induced by growth with pectin were unannotated.
Table 3. The number of DEGs specifically regulated by Foc4 and Foc1 during decomposition of host cell wall polysaccharides, which were known to associated with fungal pathogenicity.
| Gene Function | BCW_Foc4vsG_Foc4 | FCW_Foc1vsG_Foc1 | P_Foc4vsG_Foc4 | P_Foc1vsG_Foc1 | ||||
|---|---|---|---|---|---|---|---|---|
| Upregulation | Downregulation | Upregulation | Downregulation | Upregulation | Downregulation | Upregulation | Downregulation | |
| Effector | 1 | 2 | 0 | 0 | 0 | 1 | 0 | 0 |
| SIX protein | 0 | 3 | 0 | 1 | 0 | 2 | 0 | 0 |
| G-protein | 35 | 13 | 2 | 6 | 5 | 3 | 2 | 8 |
| G-β | 28 | 5 | 1 | 2 | 5 | 1 | 0 | 2 |
| G-protein α subunit | 2 | 2 | 0 | 0 | 0 | 1 | 0 | 1 |
| G-protein β/γ-subunit complex binding | 2 | 2 | 0 | 0 | 0 | 1 | 1 | 1 |
| RagA G protein | 5 | 3 | 0 | 0 | 0 | 1 | 1 | 1 |
| G-protein-coupled receptor | 4 | 6 | 0 | 4 | 0 | 0 | 0 | 5 |
| Chitin synthase | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cutinase | 0 | 2 | 0 | 3 | 0 | 1 | 0 | 3 |
| Cutinase transcription factor | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| MFS transporter | 18 | 25 | 35 | 20 | 16 | 12 | 9 | 21 |
| ABC transporter | 17 | 15 | 1 | 7 | 1 | 5 | 3 | 7 |
| Zinc finger protein | 46 | 34 | 14 | 19 | 4 | 25 | 9 | 12 |
| Zn(II)2Cys6-type transcription factor | 13 | 38 | 16 | 10 | 6 | 12 | 5 | 28 |
| bZIP transcription factor | 5 | 6 | 4 | 3 | 0 | 5 | 1 | 0 |
| Serine/threonine kinase | 23 | 21 | 7 | 9 | 1 | 8 | 5 | 4 |
| Two-component system | 1 | 0 | 2 | 0 | 0 | 0 | 1 | 0 |
| Response regulator | 2 | 1 | 2 | 2 | 0 | 1 | 2 | 1 |
| Histidine kinase | 2 | 4 | 2 | 2 | 1 | 1 | 3 | 0 |
| MAPK/MAPKK/MAPKKK | 0 | 4 | 0 | 1 | 0 | 0 | 1 | 0 |
| SNF protein kinase | 6 | 1 | 0 | 1 | 0 | 0 | 2 | 1 |
| Other protein kinase | 43 | 24 | 8 | 21 | 7 | 12 | 9 | 11 |
| F-box protein | 8 | 1 | 1 | 3 | 1 | 4 | 1 | 5 |
| Peroxidase | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Polyketide synthase | 3 | 4 | 1 | 1 | 1 | 4 | 1 | 1 |
| Cytochrome P450 | 2 | 13 | 7 | 4 | 0 | 3 | 1 | 8 |
| No function | 77 | 137 | 46 | 87 | 38 | 64 | 58 | 27 |
BCW, cell wall of Brazil cultivar; FCW, cell wall of Fengjiao cultivar; P, pectin; SIX, secreted in xylem; RagA, Ras-related GTP-binding protein A; MFS, major facilitator superfamily; ABC, ATP-binding cassette; bZIP, basic leucine zipper; MAPK, mitogen-activated protein kinase; SNF, sucrose nonfermenting.
Furthermore, 111 genes were specifically differentially expressed in BCW_Foc4 vs. G_Foc4 but not expressed in FCW_Foc1 vs. G_Foc1, including six DEGs that were upregulated and 105 DEGs that were downregulated (Table S16 in File S2). Additionally, 45 genes were specifically differentially expressed in P_Foc4 vs. G_Foc4 but not expressed in P_Foc1 vs. G_Foc1, including 9 DEGs that were upregulated and 36 DEGs that were downregulated (Table S17 in File S2). These genes may play essential roles as host-specific virulence genes when Foc4 invades Brazil banana cultivars. Among these DEGs, the genes known to be associated with fungal pathogenicity were all downregulated by Foc4 during the degradation of host cell wall polysaccharides (Table S18 in File S2). But the DEGs that were specifically upregulated in Foc4 grown in the presence of host cell wall or pectin were all genes of unknown function, which may need to be further characterized.
DEGs encoding CWDEs induced by growth with host cell wall or pectin
The number of DEGs encoding putative CAZymes in each comparison and their expression profiles are shown in Figure S7 in File S1. Compared with Foc1, more DEGs encoding putative CAZymes were assigned to GHs, GTs, and CEs in Foc4 grown with host cell wall (Table S19 in File S2). During the degradation of host cell wall polysaccharides, the expression pattern of CAZyme genes assigned to each CAZy family in Foc4 was obviously different than in Foc1 (Figure S7G in File S1). Additionally, many genes encoding putative CAZymes were differentially expressed in a variety-specific manner when the two races degraded host cell wall polysaccharides. More specific DEGs encoding CAZyme were found in Foc4 than in Foc1 grown in the presence of host cell wall (Figure S7, A–F in File S1).
According to the analysis of DEGs encoding CWDE in Foc grown with host cell wall polysaccharides, the DEGs encoding hemicellulases and pectinases were the most abundant, followed by cellulases (Table S20 in File S2). Interestingly, the CWDE genes of Foc1 were more highly expressed during degradation of the host cell wall (Figure S8G in File S1). On the other hand, some genes involved in cellulose, hemicellulose, pectin, inulin, and starch degradation were specifically differentially expressed in the two races during the degradation of host cell wall polysaccharides (Figure S8, A–F in File S1). These results indicated that hemicellulases and pectinases produced by Foc might play a leading role in the degradation of the host cell wall, and that Foc1 may be better able to decompose banana cell wall than Foc4.
DEGs related to pathogenicity induced by growth with host cell wall or pectin
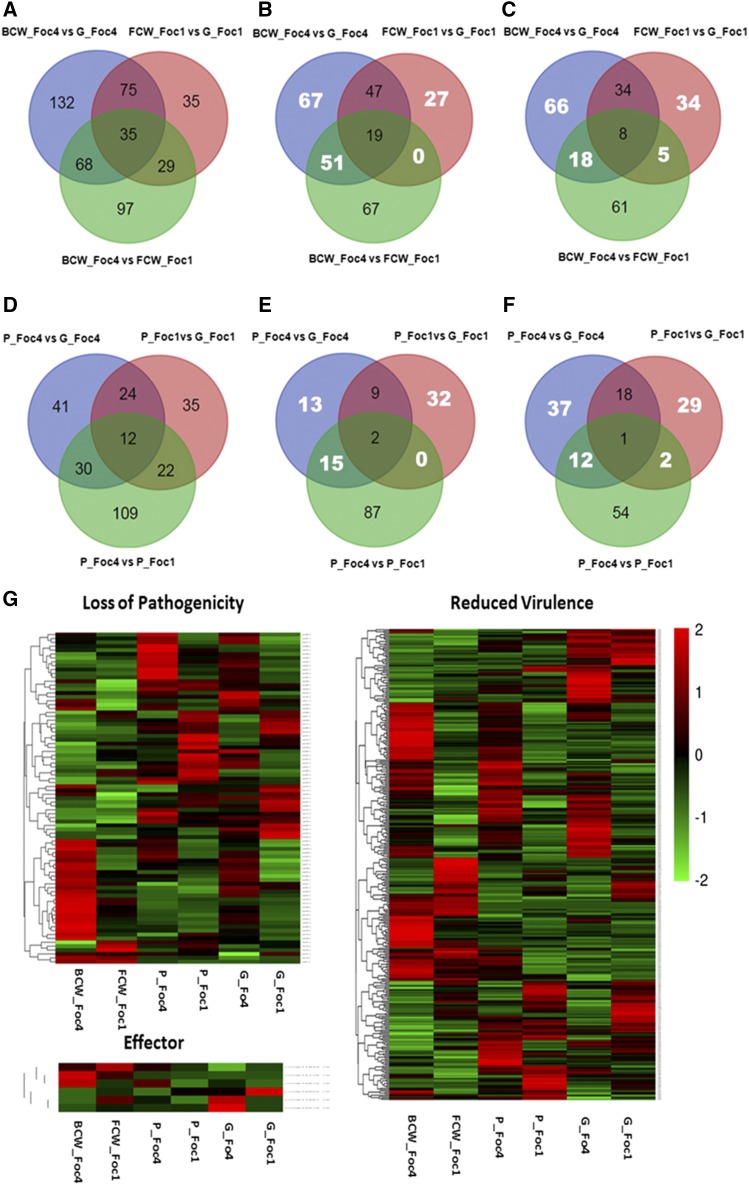
The expression profiles of genes related to host–pathogen interactions in various pathogenic fungi and the number of these DEGs identified in each comparison are summarized in Figure 4. Overall, Foc4 expressed more PHI genes than Foc1 when degrading the host cell wall. The gene expression levels of these DEGs were higher in Foc4 than in Foc1 (Figure 4G). In addition, many more DEGs homologous to PHI genes were specifically regulated in Foc4 than in Foc1 when grown in the presence of host cell wall or pectin (Figure 4, A–F).
Figure 4.
The expression profiles of genes related to host–pathogen interactions induced by host cell polysaccharides. The number of differentially expressed genes (DEGs) related to host–pathogen interactions showed in Venn diagram form (A–F). The number of DEGs (A), including upregulated (B) and downregulated DEGs (C), derived from comparisons induced by host cell wall. The number of DEGs (D), including upregulated (E) and downregulated DEGs (F), derived from comparisons induced by pectin. The numbers of specific DEGs homologous to pathogen–host interaction (PHI) genes regulated by Foc1 or Foc4 were shown in the nonoverlapping regions (white numbers). Expression profiles of putative pathogenicity genes homologous to genes associated with loss of pathogenicity, reduction of virulence, and pathogenic effectors in the PHI database were present in Heatmaps (G). Heatmaps represent fragments per kilobase of transcript per million fragments sequenced (FPKM) values of a unigene in each library.
The genes associated with loss of pathogenicity and reduced virulence were more highly expressed in Foc4 than in Foc1 grown in the presence of host cell wall or pectin (Figure 4G). Furthermore, more DEGs associated with loss of pathogenicity and reduced virulence were specifically differentially expressed in Foc4 than in Foc1 grown in the presence of host cell wall (Figures S9 and S10 in File S1). Among the pathogenicity genes associated with loss of pathogenicity, DEGs encoding F-box (PHI: 490), signaling cascade (PHI: 182), and adenosine triphosphate (ATP) citrate lyase (PHI: 2386) proteins were specifically upregulated in Foc4 grown in the presence of host cell wall, while DEGs related to AF-toxin biosynthesis (PHI: 508), malate synthase (PHI: 2267 and PHI: 365,) and glyoxal oxidase (PHI: 352) were specifically upregulated in Foc1 grown in the presence of host cell wall. On the other hand, among the pathogenicity genes associated with loss of pathogenicity, DEGs encoding ATP citrate lyase (PHI: 2386) were specifically upregulated in Foc4 grown with pectin, while DEGs encoding phospholipase C (PHI: 2106), transcription activator (PHI: 481), and trehalose-6-phosphate synthase (PHI: 322, PHI: 2172, and PHI: 1064) were specifically upregulated in Foc1. Among the pathogenicity genes associated with reduced virulence, genes encoding pectin methylesterase (PME) (PHI: 1028 and PHI: 278), small secreted protein (PHI: 860), and apoptosis (PHI: 2334) proteins were found only among the DEGs that were specifically upregulated in Foc4 grown with host cell wall, while DEGs encoding cytochrome C peroxidase precursor (PHI: 854), hydrophobin (PHI: 458 and PHI: 487), and δ-aminolevulinic acid synthase (PHI: 2248) proteins were specifically upregulated in Foc1. In addition, more TFs orthologous to PHI genes involved in reduced virulence were differentially expressed in Foc4 than in Foc1 when grown in the presence of host cell wall. Of the pathogenicity genes involved in reduced virulence, DEGs related to alcohol oxidase (PHI: 199) and Ca2+ pump (PHI: 2095) proteins were specifically upregulated in Foc4 grown with pectin, while DEGs encoding phospholipase C (PHI: 2106) and a protein kinase (PHI: 1190) were specifically upregulated in Foc1 (Tables S8 and S21 in File S2). These results revealed that different pathogenicity genes are activated when two races decompose their particular host’s cell wall, and Foc4 exhibits a broader expression profile of pathogenicity genes than Foc1, which may also account for the variation in their virulence.
Discussion
Pectinases and hemicellulases may contribute greatly to the ability of Foc to decompose the host cell wall
A critical phase of pathogen infection in plants is penetration of the plant cell wall. Depending on the distinct host preference, pathogenic fungi require different CAZymes to decompose the plant cell wall. Genome analyses show that the content and distribution of CAZymes differ among pathogenic fungi. The genomes of Botrytis cinerea and Sclerotinia sclerotiorum, which tend to infect flowers and fruits rich in pectin, contain a larger number of CAZymes involved in pectin and hemicellulose decomposition than those involved in the decomposition of cellulose (Amselem et al. 2011). In contrast, the genomes of Phaeosphaeria nodorum and Pyrenophora teres f. teres, and Magnaporthe oryzae, whose hosts cell walls contain less pectin, exhibit higher proportions of CAZymes involved in cellulose and hemicellulose decomposition than of those involved in pectin decomposition (Amselem et al. 2011). Although the genome of Foc has been published, CWDEs remain uncharacterized in Foc (Guo et al. 2014). Analysis of the Foc transcriptome during early Cavendish variety infection showed that the two examined Foc isolates possess different CAZymes for attacking Brazil banana, and PL genes were mainly induced in Foc after infection for 48 hr (Guo et al. 2014). In this study, we annotated 393 CAZymes associated with plant cell wall degradation in the Foc transcriptome, among which a large proportion are involved in pectin and hemicellulose decomposition. Furthermore, we focused on the gene expression profiles of CWDEs during the degradation of host cell wall polysaccharides, which showed that Foc genes encoding pectinase and hemicellulase were strongly induced by the presence of host cell wall and pectin (Table S20 in File S2). Thus, our results suggest that the expression of genes encoding CWDEs in Foc during infection could be properly stimulated under in vitro growth conditions in the presence of host cell wall. In previous studies, the activity of polygalacturonases (PGs) and pectate lyases has been detected in the culture supernatant when Foc is grown in media containing pectin, which is also in agreement with our results (Dong and Wang 2010, 2015; Dong et al. 2011).
Although there was no obvious difference in the quantity of CWDE DEGs induced by host cell wall polysaccharides, the significantly different expression patterns of CWDE genes in the two races showed that Foc1 may be better able to decompose the host cell wall than Foc4 (Figure S8 in File S1). Plants can not only recognize cell wall-derived molecules following triggering of the plant defense response but also produce PG-inhibiting proteins to inhibit PG secreted by pathogens (Prade et al. 1999; Maulik et al. 2012; Zhang et al. 2014; Benedetti et al. 2015). The lower expression levels of CWDE genes in Foc4 are presumably associated with avoidance of the plant immune response. However, compared with Foc1 grown in the presence of host cell wall polysaccharides, a larger number of CAZymes genes were specifically regulated by Foc4, indicating that Foc4 exhibits a greater adaptive ability for different host nutrients.
Additionally, CWDEs are very important determinants of fungal pathogenicity. The roles of pectinases and hemicellulases in virulence have been investigated in several phytopathogens (Ten et al. 1998; Garcia-Maceira et al. 2000; Kars et al. 2005; Brito et al. 2006). In the present study, a DEG encoding PME orthologous to Bcpme1 (PHI: 278) was specifically upregulated by Foc4 during degradation of the host cell wall. Disruption of Bcpme1 in a B. cinerea strain led to a reduction of virulence in apple, grapevine, and Arabidopsis thaliana (Valettecollet et al. 2003).
Genes involved in signaling may play important roles in Foc4 virulence
Protein kinases are responsible for the reversible phosphorylation of proteins, which play a major part in the regulation of fungal growth, developmental processes, and responses to environmental stimuli (Cohen 2000). The deletion of protein kinase genes results in loss of pathogenicity or low virulence of pathogenic fungi, as observed for the SNF protein kinase gene FgSNF1 and the protein kinase gene FgSsn3 in F. graminearum, as well as three MAPK genes (FoSlt2, FoMkk2, and FoBck) in Foc4 (Yu et al. 2014; Ding et al. 2015; Cao et al. 2016). In this study, GO molecular function analysis revealed that a large number of putative pathogenic factors that exhibit kinase activity were induced by growth in the presence of host cell wall polysaccharides. This result suggests that protein kinases may affect the transcription levels of CWDEs (Figure S3 in File S1). Mutation of the MAPK gene fmk1 in Fol resulted in loss of pathogenicity in tomato plants and a serious reduction in the expression levels of pl1, encoding PL (Di Pietro et al. 2001). Additionally, more DEGs encoding protein kinases, including serine/threonine kinases and SNF kinases, were upregulated specifically in Foc4 than in Foc1 grown in the presence of host cell wall. Two DEGs that were specifically upregulated in Foc4 grown with host cell wall were orthologous to the Gin4-like protein kinase gene (PHI: 1184) and the Dbf2/Dbf20 protein kinase gene (PHI: 1187). Mutants with deletions of these genes showed a reduction of pathogenicity in F. graminearum (Wang et al. 2011).
Among signal transduction pathways, fungal TFs can regulate diverse biological processes. Zinc finger, bZIP, and Zn(II)2CyS6 TFs are considered to be pathogenicity-associated genes in some pathogenic fungi (Imazaki et al. 2007; Heimel et al. 2010; Chung et al. 2013; Qi et al. 2013; Shantappa et al. 2013; Yue et al. 2015). In this study, a set of TFs related to fungal pathogenicity was found to be activated during the degradation of host cell wall polysaccharides (Figure S3 in File S1). Additionally, more DEGs encoding zinc finger and bZIP TFs were upregulated specifically in Foc4 than in Foc1 grown with host cell wall. Nine DEGs specifically upregulated by Foc4 grown with host cell wall were similar to TFs (PHI: 1440, PHI: 1606, PHI: 1354, PHI: 1325, PHI: 1529, PHI: 1422, PHI: 1933, PHI: 1915, and PHI: 1556), including bZIP and zinc finger TFs. Mutations in these transcription factors result in multiple defects in virulence, growth, and toxin production in F. graminearum (Son et al. 2011).
Heterotrimeric GTP-binding proteins (G-proteins), consisting of Gα, Gβ, and Gγ subunits, are essential signaling components that mediate various cellular responses to environmental stimuli in eukaryotic organisms (Simon and Gautam 1991). In plant pathogenic fungi, G-proteins are indispensable for growth, asexual and sexual development, and virulence (Li et al. 2006). Mutations in G-protein genes generated in some fungi, such as Cryphonectria parasitica, M. grisea, B. cinerea, and F. oxysporum f. sp. cucumerinum, result in loss or reduction of pathogenicity (Choi et al. 1995; Liu and Dean 1997; Gronover et al. 2001; Jain et al. 2002). Compared with Foc1, Foc4 may activate FGA1-mediated G protein signaling during early Cavendish variety infection (Guo et al. 2014). In this study, DEGs including two G-protein α subunits, two G-protein β subunits, and four G-protein-coupled receptors were specifically upregulated in Foc4, while none of these genes were specifically regulated in Foc1 (Table 3). These results imply that some Foc4-specific G-protein α and β subunit-mediated signaling pathways may be activated during the degradation of the host cell wall compared with what occurs in Foc1.
In addition, F-box proteins, which are considered fungal pathogenic factors, mediate the ubiquitination of proteins targeted for degradation by the proteasome and play roles in signal transduction and regulation of diverse cell functions (Jonkers and Rep 2009; Jonkers et al. 2011; Guo et al. 2015). In this study, one DEG that was specifically upregulated in Foc4 grown with host cell wall was similar to Frp1 (PHI: 490), which encodes an F-box protein, and a mutation in this gene in Fol results in loss of pathogenicity in tomato (Duyvesteijn et al. 2005).
Thus, more genes involved in signaling are specifically expressed in Foc4 than in Foc1, suggesting that Foc4 may be better able to adapt to host environments.
Genes involved in fungal nutrition and transportation may play important roles in Foc4 virulence
The most obvious explanation for the interaction of fungi with host plants is to achieve more efficient nutrient acquisition, and fatty acids are one of the most basic nutritional elements. Mutations in peroxisome genes in the glyoxylate pathway, which is involved in fatty acid oxidation, lead to a loss of pathogenicity (Kimura et al. 2001). Peroxidase also protects pathogenic fungi against the oxidative stress generated by the host plant during infection (Tanabe et al. 2010; Singh et al. 2012). In our study, a DEG encoding peroxidase was upregulated specifically in Foc4 grown with host cell wall, while none of these genes were affected in Foc1 (Table 4).
Phytopathogenic fungi are champions of transmembrane transporters, which mediate the selective uptake and efflux of nutrients, metabolites, toxic compounds, and ions from their host environments. Therefore, transporters play a role in the acquisition of nutrients, pathogenesis, and fungicide sensitivity and resistance (Diallinas 2016). The ABC superfamily plays a role in protecting pathogenic fungi against plant defense compounds at an advanced phase of disease, which has been characterized in various phytopathogenic fungi (Stergiopoulos et al. 2002, 2003; Sun et al. 2006; Gupta and Chattoo 2008). The major facilitator superfamily (MFS) transporters are largely known as secondary active carriers that play an important role in antifungal drug resistance and pathogen–host interactions (Costa et al. 2014). In our study, more DEGs for ABC transporters were upregulated specifically in Foc4 than in Foc1 grown with host cell wall (Table 4). When grown with host cell wall, one of the DEGs that was specifically upregulated in Foc4 was orthologous to the MFS transporter-like gene CTB4 (PHI: 737 and PHI: 2329). A Cercospora nicotianae CTB4 mutant displays a drastic reduction of cercosporin toxin accumulation and fungal virulence (Choquer et al. 2007).
During degradation of the host cell wall, genes involved in fungal nutrition and transportation may be required for Foc4 virulence against plant defense.
Effectors, chitin synthase, and polyketide synthase may play a role in Foc4 infection
Effectors are small molecular weight secreted proteins produced by pathogenic fungi at various stages of infection that can alter host cell metabolism, inhibit or stimulate effector-triggered immune responses, and facilitate infection (Ellis et al. 2009; Wit et al. 2009). Chitin synthases contribute to the structural integrity of the fungal cell wall and are important for fungal growth, development, and pathogenicity (Werner et al. 2007). Polyketide synthases, which are involved in the biosynthesis of fungal toxins, have been shown to participate in the regulation of fungal virulence (Takano et al. 1995; Tsai et al. 1998; Bohnert et al. 2004). In our study, slightly more DEGs encoding these pathogenic factors were specifically upregulated in Foc4 than in Foc1 grown with host cell wall (Table 4). Under induction by host cell wall, one of the DEGs that was specifically upregulated by Foc4 was orthologous to MSP1 (PHI: 860), which encodes a small secreted protein required for the virulence of M. grisea (Jeong et al. 2007). Another DEG that was specifically upregulated in Foc4 is orthologous to CgCHSIII (PHI: 1055), encoding chitin synthase, which is expressed during pathogenic development but does not affect pathogenicity of Colletotrichum graminicola (Werner et al. 2007). Overall, these genes may be activated by growth in the presence of host cell wall and may also play important roles in Foc4 virulence at different stages of infection.
Conclusions
A comprehensive transcriptome resource for Foc was generated in this study, and the gene expression profiles of fungi grown in the presence of host cell wall polysaccharides were characterized using RNA-seq. Comparative transcriptome analysis allowed the identification of a set of DEGs when Foc decomposed host cell wall polysaccharides, revealing the molecular mechanisms of pathogenesis in banana and the genetic basis of host specificity in Foc. In this study, a large number of Foc4- and Foc1-specific regulated genes induced when fungi grew with host cell wall polysaccharide were first characterized, including unknown genes. In particular, the results indicated that genes that are specifically regulated by Foc4, especially genes involved in penetration of the host cell wall, signaling, and transportation, may have a significant impact on virulence. Other pathogenicity factors also attracted our attention. Functional analysis of these Foc genes needs to be carried out to discover new pathogenicity-related or virulence genes.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.042226/-/DC1.
Acknowledgments
We are grateful for the technical support for Illumina sequencing and initial data analysis that we received from the Novogene Corporation at Beijing, China. This work was financially supported by the National Natural Science Foundation of China (30971888) and National Public Welfare Sectors (Agriculture) Special Research of China (grant no. 200903049-05).
Author contributions: S.Q. and Z.W. conceived and designed the experiments; S.Q. and C.J. performed the experiments; S.Q., Z.W., and Y.L. analyzed the data; S.Q. and Z.W. wrote the paper. All authors have read and approved the final version of the manuscript.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Altschul S. F., Madden T. L., Schaeffer A. A., Zhang J., Zhang Z., et al. , 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amselem J., Cuomo C. A., van Kan J. A., Viaud M., Benito E. P., et al. , 2011. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7: e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W., 2010. Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro N., Pakula T., Penttilä M., 2005. Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol. Rev. 29: 719–739. [DOI] [PubMed] [Google Scholar]
- Benedetti M., Pontiggia D., Raggi S., Cheng Z. Y., Scaloni F., et al. , 2015. Plant immunity triggered by engineered in vivo release of oligogalacturonides, damage-associated molecular patterns. Proc. Natl. Acad. Sci. USA 112: 5533–5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedi S., Berger H., Sieber C., Munsterkotter M., Maloku I., et al. , 2016. Comparison of Fusarium graminearum transcriptomes on living or dead wheat differentiates substrate-responsive and defense-responsive genes. Front. Microbiol. 7: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert H. U., Fudal I., Dioh W., Tharreau D., Notteghem J. L., et al. , 2004. A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 16: 2499–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito N., Espino J. J., González C., 2006. The endo-β-1,4-xylanase xyn11A is required for virulence in Botrytis cinerea. Mol. Plant Microbe Interact. 19: 25–32. [DOI] [PubMed] [Google Scholar]
- Buddenhagen I. W., 1990. Banana breeding and Fusarium wilt, pp. 107–113 in Twenty Contributors Present Reports and Research on the Disease, Fusarium Wilt of Banana, edited by Ploetz R. C. APS Press, St. Paul, MN. [Google Scholar]
- Butler D., 2013. Fungus threatens top banana. Nature 504: 195–196. [DOI] [PubMed] [Google Scholar]
- Caffall K. H., Mohnen D., 2009. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344: 1879–1900. [DOI] [PubMed] [Google Scholar]
- Cao S., Zhang S., Hao C., Liu H., Xu J. R., et al. , 2016. FgSsn3 kinase, a component of the mediator complex, is important for sexual reproduction and pathogenesis in Fusarium graminearum. Sci. Rep. 6: 22333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapito R., Vorwerk S., Jeltsch J. M., Phalip V., 2013. Genome-wide transcriptional responses of Fusarium graminearum to plant cell wall substrates. FEMS Microbiol. Lett. 340: 129–134. [DOI] [PubMed] [Google Scholar]
- Choi G. H., Chen B., Nuss D. L., 1995. Virus-mediated or transgenic suppression of a G-protein alpha subunit and attenuation of fungal virulence. Proc. Natl. Acad. Sci. USA 92: 305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquer M., Lee M. H., Bau H. J., Chung K. R., 2007. Deletion of a MFS transporter-like gene in Cercospora nicotianae reduces cercosporin toxin accumulation and fungal virulence. FEBS Lett. 581: 489–494. [DOI] [PubMed] [Google Scholar]
- Chuma I., Tosa Y., Taga M., Nakayashiki H., Mayama S., 2003. Meiotic behavior of a supernumerary chromosome in Magnaporthe oryzae. Curr. Genet. 43: 191–198. [DOI] [PubMed] [Google Scholar]
- Chung H., Choi J., Park S. Y., Jeon J., Lee Y. H., 2013. Two conidiation-related Zn(II)2Cys6 transcription factor genes in the rice blast fungus. Fungal Genet. Biol. 61: 133–141. [DOI] [PubMed] [Google Scholar]
- Cohen P., 2000. The regulation of protein function by multisite phosphorylation-a 25 year update. Trends Biochem. Sci. 25: 596–601. [DOI] [PubMed] [Google Scholar]
- Coleman J. J., Rounsley S. D., Rodriguezcarres M., Kuo A., Wasmann C. C., et al. , 2009. The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet. 5: 1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M., et al. , 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- Costa C., Dias P. J., Sa-Correia I., Teixeira M. C., 2014. MFS multidrug transporters in pathogenic fungi: do they have real clinical impact? Front. Physiol. 5: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho P. M., Andersen M. R., Kolenova K., vanKuyk P. A., Benoit I., et al. , 2009. Post-genomic insights into the plant polysaccharide degradation potential of Aspergillus nidulans and comparison to Aspergillus niger and Aspergillus oryzae. Fungal Genet. Biol. 46: S161–S169. [DOI] [PubMed] [Google Scholar]
- Di Pietro A., Garcia-Maceira F. I., Meglecz E., Roncero M. I. G., 2001. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39: 1140–1152. [PubMed] [Google Scholar]
- Diallinas G., 2016. Dissection of transporter function: from genetics to structure. Trends Genet. 32: 576–590. [DOI] [PubMed] [Google Scholar]
- Ding Z., Li M., Sun F., Xi P., Sun L., et al. , 2015. Mitogen-activated protein kinases are associated with the regulation of physiological traits and virulence in Fusarium oxysporum f. sp. cubense. PLoS One 10: e0122634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Wang Z., 2010. Isolation and characterization of an exopolygalacturonase from Fusarium oxysporum f.sp. cubense race 1 and race 4. BMC Biochem. 12: 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Wang Z., 2015. Isolation and heterologous expression of a polygalacturonase produced by Fusarium oxysporum f. sp. cubense race 1 and 4. Int. J. Mol. Sci. 16: 7595–7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z. Y., Yang Y., Wang Z. Z., Qin S. W., Jiang Y. X., et al. , 2011. Isolation, expression and comparison of a pectate lyase produced by Fusarium oxysporum f. sp. cubense race 1 and race 4. Afr. J. Biotechnol. 9: 8984–8990. [Google Scholar]
- Duyvesteijn R. G. E., van Wijk R., Boer Y., Rep M., Cornelissen B. J. C., et al. , 2005. Frp1 is a Fusarium oxysporum F-box protein required for pathogenicity on tomato. Mol. Microbiol. 57: 1051–1063. [DOI] [PubMed] [Google Scholar]
- Ellis J. G., Rafiqi M., Gan P., Chakrabarti A., Dodds P. N., 2009. Recent progress in discovery and functional analysis of effector proteins of fungal and oomycete plant pathogens. Curr. Opin. Plant Biol. 12: 399–405. [DOI] [PubMed] [Google Scholar]
- Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., et al. , 2014. Pfam: the protein families database. Nucleic Acids Res. 42: D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Maceira F. I., Di Pietro A., Roncero M. I. G., 2000. Cloning and disruption of pgx4 encoding an in planta expressed exopolygalacturonase from Fusarium oxysporum. Mol. Plant Microbe Interact. 13: 359–365. [DOI] [PubMed] [Google Scholar]
- Garcia Maceira F. I., Di Pietro A., Roncero M. I. G., 1997. Purification and characterization of a novel exopolygalacturonase from Fusarium oxysporum f.sp. lycopersici. FEMS Microbiol. Lett. 154: 37–43. [DOI] [PubMed] [Google Scholar]
- Gardiner D. M., Kazan K., Manners J. M., 2009. Nutrient profiling reveals potent inducers of trichothecene biosynthesis in Fusarium graminearum. Fungal Genet. Biol. 46: 604–613. [DOI] [PubMed] [Google Scholar]
- Götesson A., Marshall J. S., Jones D. A., Hardham A. R., 2002. Characterization and evolutionary analysis of a large polygalacturonase gene family in the oomycete plant pathogen Phytophthora cinnamomi. Mol. Plant Microbe Interact. 15: 907–921. [DOI] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., et al. , 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronover C. S., Kasulke D., Tudzynski P., Tudzynski B., 2001. The role of G protein alpha subunits in the infection process of the gray mold fungus Botrytis cinerea. Mol. Plant Microbe Interact. 14: 1293–1302. [DOI] [PubMed] [Google Scholar]
- Guldener U., Seong K. Y., Boddu J., Cho S. H., Trail F., et al. , 2006. Development of a Fusarium graminearum Affymetrix GeneChip for profiling fungal gene expression in vitro and in planta. Fungal Genet. Biol. 43: 316–325. [DOI] [PubMed] [Google Scholar]
- Guo L., Han L., Yang L., Zeng H., Fan D., et al. , 2014. Genome and transcriptome analysis of the fungal pathogen Fusarium oxysporum f. sp. cubense causing banana vascular wilt disease. PLoS One 9: e95543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Yang L., Liang C., Wang J., Liu L., et al. , 2016a The G-protein subunits FGA2 and FGB1 play distinct roles in development and pathogenicity in the banana fungal pathogen Fusarium oxysporum f. sp. cubense. Physiol. Mol. Plant Pathol. 93: 29–38. [Google Scholar]
- Guo L., Yang Y., Yang L., Wang F., Wang G., et al. , 2016b Functional analysis of the G-protein α subunits FGA1 and FGA3 in the banana pathogen Fusarium oxysporum f. sp. cubense. Physiol. Mol. Plant Pathol. 94: 75–82. [Google Scholar]
- Guo M., Gao F., Zhu X. L., Nie X., Pan Y. M., et al. , 2015. MoGrr1, a novel F-box protein, is involved in conidiogenesis and cell wall integrity and is critical for the full virulence of Magnaporthe oryzae. Appl. Microbiol. Biotechnol. 99: 8075–8088. [DOI] [PubMed] [Google Scholar]
- Gupta A., Chattoo B. B., 2008. Functional analysis of a novel ABC transporter ABC4 from Magnaporthe grisea. FEMS Microbiol. Lett. 278: 22–28. [DOI] [PubMed] [Google Scholar]
- Han Y., Liu X., Benny U., Kistler H. C., Vanetten H. D., 2001. Genes determining pathogenicity to pea are clustered on a supernumerary chromosome in the fungal plant pathogen Nectria haematococca. Plant J. 25: 305–314. [DOI] [PubMed] [Google Scholar]
- Heimel K., Scherer M., Vranes M., Wahl R., Pothiratana C., et al. , 2010. The transcription factor Rbf1 is the master regulator for b-mating type controlled pathogenic development in Ustilago maydis. PLoS Pathog. 6: 761–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imazaki I., Kurahashi M., Iida Y., Tsuge T., 2007. Fow2, a Zn(II)2Cys6-type transcription regulator, controls plant infection of the vascular wilt fungus Fusarium oxysporum. Mol. Microbiol. 63: 737–753. [DOI] [PubMed] [Google Scholar]
- Iseli C., Jongeneel C. V., Bucher P., 1999. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 138–148. [PubMed] [Google Scholar]
- Jain S., Akiyama K., Mae K., Ohguchi T., Takata R., 2002. Targeted disruption of a G protein α subunit gene results in reduced pathogenicity in Fusarium oxysporum. Curr. Genet. 41: 407–413. [DOI] [PubMed] [Google Scholar]
- Jeong J. S., Mitchell T. K., Dean R. A., 2007. The Magnaporthe grisea snodprot1 homolog, MSP1, is required for virulence. FEMS Microbiol. Lett. 273: 157–165. [DOI] [PubMed] [Google Scholar]
- Jonkers W., Rep M., 2009. Lessons from fungal F-box proteins. Eukaryot. Cell 8: 677–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers W., van Kan J. A., Tijm P., Lee Y. W., Tudzynski P., et al. , 2011. The FRP1 F-box gene has different functions in sexuality, pathogenicity and metabolism in three fungal pathogens. Mol. Plant Pathol. 12: 548–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kars I., McCalman M., Wagemakers L., Van Kan J. A. L., 2005. Functional analysis of Botrytis cinerea pectin methylesterase genes by PCR-based targeted mutagenesis: Bcpme1 and Bcpme2 are dispensable for virulence of strain B05.10. Mol. Plant Pathol. 6: 641–652. [DOI] [PubMed] [Google Scholar]
- Kimura A., Takano Y., Furusawa I., Okuno T., 2001. Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. Plant Cell 13: 1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C. N., 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wright S. J., Krystofova S., Park G., Borkovich K. A., 2006. Heterotrimeric G protein signaling in filamentous fungi. Annu. Rev. Microbiol. 61: 423–452. [DOI] [PubMed] [Google Scholar]
- Li M. H., Xie X. L., Lin X. F., Shi J. X., Ding Z. J., et al. , 2014. Functional characterization of the gene FoOCH1 encoding a putative α-1,6-mannosyltransferase in Fusarium oxysporum f. sp. cubense. Fungal Genet. Biol. 65: 1–13. [DOI] [PubMed] [Google Scholar]
- Liu S., Dean R. A., 1997. G protein α subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol. Plant Microbe Interact. 10: 1075–1086. [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Method. Methods. 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lysoe E., Seong K. Y., Kistler H. C., 2011. The transcriptome of Fusarium graminearum during the infection of wheat. Mol. Plant Microbe Interact. 24: 995–1000. [DOI] [PubMed] [Google Scholar]
- Ma L. J., van der Does H. C., Borkovich K. A., Coleman J. J., Daboussi M. J., et al. , 2010. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X. Z., Cai T., Olyarchuk J. G., Wei L. P., 2005. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21: 3787–3793. [DOI] [PubMed] [Google Scholar]
- Maulik A., Sarkar A. I., Devi S., Basu S., 2012. Polygalacturonase-inhibiting proteins - leucine-rich repeat proteins in plant defence. Plant Biol. 14: 22–30. [DOI] [PubMed] [Google Scholar]
- Ospina-Giraldo M. D., Mullins E., Kang S., 2003. Loss of function of the Fusarium oxysporum SNF1 gene reduces virulence on cabbage and Arabidopsis. Curr. Genet. 44: 49–57. [DOI] [PubMed] [Google Scholar]
- Ospina-Giraldo M. D., Griffith J. G., Laird E. W., Mingora C., 2009. The CAZyome of Phytophthora spp.: a comprehensive analysis of the gene complement coding for carbohydrate-active enzymes in species of the genus Phytophthora. BMC Genomics 11: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B. H., Karpinets T. V., Syed M. H., Leuze M. R., Uberbacher E. C., 2010. CAZymes Analysis Toolkit (CAT): web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology 20: 1574–1584. [DOI] [PubMed] [Google Scholar]
- Pérez S., Mazeau K., Penhoat C. H. D., 2000. The three-dimensional structures of the pectic polysaccharides. Plant Physiol. Biochem. 38: 37–55. [Google Scholar]
- Ploetz R. C., 2006. Fusarium wilt of banana is caused by several pathogens referred to as Fusarium oxysporum f. sp cubense. Phytopathology 96: 653–656. [DOI] [PubMed] [Google Scholar]
- Ploetz R. C., 2007. Diseases of tropical perennial crops: challenging problems in diverse environments. Plant Dis. 91: 644–663. [DOI] [PubMed] [Google Scholar]
- Ploetz R. C., 2015a Fusarium wilt of banana. Phytopathology 105: 1512–1521. [DOI] [PubMed] [Google Scholar]
- Ploetz R. C., 2015b Management of Fusarium wilt of banana: a review with special reference to tropical race 4. Crop Prot. 73: 7–15. [Google Scholar]
- Prade R. A., Zhan D., Ayoubi P., Mort A. J., 1999. Pectins, pectinases and plant-microbe interactions. Biotechnol. Genet. Eng. Rev. 16: 361–391. [DOI] [PubMed] [Google Scholar]
- Qi X., Guo L., Yang L., Huang J., 2013. Foatf1, a bZIP transcription factor of Fusarium oxysporum f. sp. cubense, is involved in pathogenesis by regulating the oxidative stress responses of Cavendish banana (Musa spp.). Physiol. Mol. Plant Pathol. 84: 76–85. [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S. M., Lukasiewicz J., Farrer R., van Dam P., Bertoldo C., et al. , 2016. Comparative genomics of Fusarium oxysporum f. sp. melonis reveals the secreted protein recognized by the Fom-2 resistance gene in melon. New Phytol. 209: 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong K. Y., Zhao X., Xu J. R., Guldener U., Kistler H. C., 2008. Conidial germination in the filamentous fungus Fusarium graminearum. Fungal Genet. Biol. 45: 389–399. [DOI] [PubMed] [Google Scholar]
- Shantappa S., Dhingra S., Hernándezortiz P., Espeso E. A., Calvo A. M., 2013. Role of the zinc finger transcription factor SltA in morphogenesis and sterigmatocystin biosynthesis in the fungus Aspergillus nidulans. PLoS One 8: 697–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M. I., Gautam N., 1991. Diversity of G proteins in signal transduction. Science 252: 802–808. [DOI] [PubMed] [Google Scholar]
- Singh S., Brausstromeyer S. A., Timpner C., Valerius O., Von T. A., et al. , 2012. The plant host Brassica napus induces in the pathogen Verticillium longisporum the expression of functional catalase peroxidase which is required for the late phase of disease. Mol. Plant Microbe Interact. 25: 569–581. [DOI] [PubMed] [Google Scholar]
- Son H., Seo Y. S., Min K., Park A. R., Lee J., et al. , 2011. A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum. PLoS Pathog. 7: e1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sposato P., Ahn J.-H., Walton J. D., 1995. Characterization and disruption of a gene in the maize pathogen Cochliobolus carbonum encoding a cellulase lacking a cellulose binding domain and hinge region. Mol. Plant Microbe Interact. 8: 602–609. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos I., Zwiers L. H., Waard M. A. D., 2002. Secretion of natural and synthetic toxic compounds from filamentous fungi by membrane transporters of the ATP-binding cassette and major facilitator superfamily. Eur. J. Plant Pathol. 108: 719–734. [Google Scholar]
- Stergiopoulos I., Zwiers L., De Waard M., 2003. The ABC transporter MgAtr4 is a virulence factor of Mycosphaerella graminicola that affects colonization of substomatal cavities in wheat leaves. Mol. Plant Microbe Interact. 16: 689–698. [DOI] [PubMed] [Google Scholar]
- Storey J. D., Tibshirani R., 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover R. H., 1962. Fusarial Wilt (Panama Disease) of Bananas and Other Musa Species Commonwealth Mycological Institute, Surrey, England. [Google Scholar]
- Stover R. H., 1990. Fusarium wilt of banana: some history and current status of the disease, pp. 1–7 in Twenty Contributors Present Reports and Research on the Disease, Fusarium Wilt of Banana, edited by Ploetz R. C. APS Press, St. Paul, MN. [Google Scholar]
- Stover R. H., Buddenhagen I. W., 1986. Banana breeding: polyploidy, disease resistance and productivity. Fruits 41: 175–191. [Google Scholar]
- Su H., Hwang S. C., Ko W. H., 1986. Fusarial wilt of Cavendish in Taiwan. Plant Dis. 70: 814–818. [Google Scholar]
- Sun C. B., Suresh A., Deng Y. Z., Naqvi N. I., 2006. A multidrug resistance transporter in Magnaporthe is required for host penetration and for survival during oxidative stress. Plant Cell 18: 3686–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano Y., Kubo Y., Shimizu K., Mise K., Okuno T., et al. , 1995. Structural analysis of PKS1, a polyketide synthase gene involved in melanin biosynthesis in Colletotrichum lagenarium. Mol. Gen. Genet. 249: 162–167. [DOI] [PubMed] [Google Scholar]
- Tanabe S., Ishiiminami N., Saitoh K., Otake Y., Kaku H., et al. , 2010. The role of catalase-peroxidase secreted by Magnaporthe oryzae during early infection of rice cells. Mol. Plant Microbe Interact. 24: 163–171. [DOI] [PubMed] [Google Scholar]
- Ten H. A., Mulder W., Visser J., van Kan J. A., 1998. The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol. Plant Microbe Interact. 11: 1009–1016. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H. F., Chang Y. C., Washburn R. G., Wheeler M. H., Kwon-Chung K. J., 1998. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J. Bacteriol. 180: 3031–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valettecollet O., Cimerman A., Reignault P., Levis C., Boccara M., 2003. Disruption of Botrytis cinerea pectin methylesterase gene Bcpme1 reduces virulence on several host plants. Mol. Plant Microbe Interact. 16: 360–367. [DOI] [PubMed] [Google Scholar]
- van Dam P., Fokkens L., Schmidt S. M., Linmans J. H., Kistler H. C., et al. , 2016. Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ. Microbiol. 18: 4087–4102. [DOI] [PubMed] [Google Scholar]
- van den Brink J., de Vries R. P., 2011. Fungal enzyme sets for plant polysaccharide degradation. Appl. Microbiol. Biotechnol. 91: 1477–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite B. H., Stover R. H., 1960. Studies on Fusarium wilt of bananas. VI. Variability and the cultivar concept in Fusarium oxysporum f. cúbense. Can. J. Bot. 38: 985–994. [Google Scholar]
- Wang C. F., Zhang S. J., Hou R., Zhao Z. T., Zheng Q., et al. , 2011. Functional analysis of the kinome of the wheat scab fungus Fusarium graminearum. PLoS Pathog. 7: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Feng Z., Wang X., Wang X., Zhang X., 2010. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26: 136–138. [DOI] [PubMed] [Google Scholar]
- Werner S., Sugui J. A., Steinberg G., Deising H. B., 2007. A chitin synthase with a myosin-like motor domain is essential for hyphal growth, appressorium differentiation, and pathogenicity of the maize anthracnose fungus Colletotrichum graminicola. Mol. Plant Microbe Interact. 20: 1555–1567. [DOI] [PubMed] [Google Scholar]
- Williams A. H., Sharma M., Thatcher L. F., Azam S., Hane J. K., et al. , 2016. Comparative genomics and prediction of conditionally dispensable sequences in legume-infecting Fusarium oxysporum formae speciales facilitates identification of candidate effectors. BMC Genomics 17: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnenburg R., Baldwin T. K., Urban M., Rawlings C., Koehler J., et al. , 2006. PHI-base: a new database for pathogen host interactions. Nucleic Acids Res. 34: D459–D464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wit P. J. G. M. D., Mehrabi R., Burg H. A. V. D., Stergiopoulos I., 2009. Fungal effector proteins: past, present and future. Mol. Plant Pathol. 10: 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y. B., Mao X. Z., Yang J. C., Chen X., Mao F. L., et al. , 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40: W445–W451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. D., Wakefield M. J., Smyth G. K., Oshlack A., 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Son H., Park A. R., Lee S. H., Choi G. J., et al. , 2014. Functional characterization of sucrose non-fermenting 1 protein kinase complex genes in the ascomycete Fusarium graminearum. Curr. Genet. 60: 35–47. [DOI] [PubMed] [Google Scholar]
- Yue X. F., Que Y. W., Xu L., Deng S. Z., Peng Y., et al. , 2015. ZNF1 encodes a putative C2H2 zinc finger protein essential for appressorium differentiation by the rice blast fungus Magnaporthe oryzae. Mol. Plant Microbe Interact. 29: 22–35. [DOI] [PubMed] [Google Scholar]
- Zhang L., Kars I., Essenstam B., Liebrand T. W., Wagemakers L., et al. , 2014. Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the Arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 164: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains of Foc used in this study are available upon request. The raw RNA-seq reads from the six samples are available at the NCBI Sequence Read Archive database under the accessions SRA486974. Supplemental figures and supplemental tables contain detailed information of all supplemental files to support this study.