Abstract
Many leukemia patients suffer from dysregulation of their immune system, making them more susceptible to infections and leading to general weakening (cachexia). Both adaptive and innate immunity are affected. The fruit fly Drosophila melanogaster has an innate immune system, including cells of the myeloid lineage (hemocytes). To study Drosophila immunity and physiology during leukemia, we established three models by driving expression of a dominant-active version of the Ras oncogene (RasV12) alone or combined with knockdowns of tumor suppressors in Drosophila hemocytes. Our results show that phagocytosis, hemocyte migration to wound sites, wound sealing, and survival upon bacterial infection of leukemic lines are similar to wild type. We find that in all leukemic models the two major immune pathways (Toll and Imd) are dysregulated. Toll–dependent signaling is activated to comparable extents as after wounding wild-type larvae, leading to a proinflammatory status. In contrast, Imd signaling is suppressed. Finally, we notice that adult tissue formation is blocked and degradation of cell masses during metamorphosis of leukemic lines, which is akin to the state of cancer-dependent cachexia. To further analyze the immune competence of leukemic lines, we used a natural infection model that involves insect-pathogenic nematodes. We identified two leukemic lines that were sensitive to nematode infections. Further characterization demonstrates that despite the absence of behavioral abnormalities at the larval stage, leukemic larvae show reduced locomotion in the presence of nematodes. Taken together, this work establishes new Drosophila models to study the physiological, immunological, and behavioral consequences of various forms of leukemia.
Keywords: Ras, oncogene, nematodes, insect immunity, hemocyte, Genetics of Immunity
Immune dysregulation and a general weakness (cachexia or wasting syndrome), including loss of appetite and fatigue, are general features of leukemia (Forconi and Moss 2015). Immune dysregulation affects cells of both the adaptive and the innate branch (Ganan-Gomez et al. 2015; Forconi and Moss 2015). Although the mechanisms for immunosuppression are poorly understood, the leukemic cells may confer part of the suppressive activity themselves. This is due to dysregulation of immune response and to features the leukemic clones share with regulatory B cells (Forconi and Moss 2015). Effects on innate immunity include reduced levels of complement proteins and defects in neutrophils and natural killer cells. In addition, a shift toward nonclassical activation of monocytes to an M2-like phenotype, which is associated with repair and healing functions (Mills and Ley 2014), is observed (Muallem and Hunter 2014; Forconi and Moss 2015). Mutations of the small GTPase, Ras, were shown to occur in several forms of leukemia, similar to other types of tumors (Niemeyer 2014). In general, heterozygosity for mutations of the members in the Ras/Raf/MEK/ERK signaling pathway, collectively called “RASopathies,” predispose to cancer or myeloproliferative disorders during infancy (Niemeyer 2014).
Fewer mutations are sufficient to induce a tumorous or pro-tumorous state in flies compared to mammals (Gateff 1978), which renders Drosophila melanogaster a suitable model to study tumorigenesis (Bangi 2013). Mutants in single genes can affect the brain and the imaginal discs, as well as blood cells (hemocytes), and lead to hyperplastic and neoplastic growth (Brumby and Richardson 2005; Bangi 2013). Drosophila models provide insight into the molecular causes of malignancy and have the potential to identify new therapeutics (Gonzalez 2013). Affecting single genes that are required for hemocyte development can alter their proliferative capacity and lead to a massive increase in hemocyte numbers during larval development (Gateff et al. 1980; Crozatier and Vincent 2011). Starting in the early 2000s, the use of molecular genetics allowed induction of hypertrophic and hyperplastic growth in a tissue-specific and clonal manner (Brumby and Richardson 2003; Pagliarini and Xu 2003). This led to the dissection of cell-autonomous and nonautonomous pathways, which enhanced or limited tumor growth [summarized in Pastor-Pareja and Xu (2013); Parisi et al. 2014; Tipping and Perrimon 2014]. Expression of a dominant-active form of the Ras oncogene (RasV12) and mutations of tumor suppressors such as scribble (scrib) and lethal giant larvae (l(2)gl) turned out to be very efficient at inducing overproliferation in eye discs (Brumby and Richardson 2003; Pagliarini and Xu 2003). In addition to the fly’s short generation cycle, a wide range of molecular tools facilitate whole organism studies of the genes and pathways involved in leukemia progression and modulation (Christofi and Apidianakis 2013; Parisi et al. 2014; Hauling et al. 2014).
Like all invertebrates, Drosophila lack cells of the lymphoid lineage but contains cells that are akin to the myeloid lineage (Gold and Bruckner 2014; Honti et al. 2014). These are collectively called hemocytes and the most dominant class of hemocytes in Drosophila are the plasmatocytes (Crozatier and Vincent 2011). Plasmatocytes combine the phagocytic function of monocytes/macrophages (Gold and Bruckner 2014) with some of the functions of polymorphonuclear leukocytes (Theopold et al. 2014) and display a surprising degree of plasticity (Anderl et al. 2016). RasV12 has been used to induce overproliferation of plasmatocytes. RasV12-expressing plasmatocytes retained normal phagocytic capacity and showed differential expression of cell-cycle regulators, as well as several immune genes (Asha et al. 2003). Here, in addition to expressing RasV12 in hemocytes alone, we also increased the protumorous phenotype by coexpressing RNAi constructs targeting l(2)gl or scrib. At 25°, development and behavior appear normal for all combinations until the pupal stage. However, at higher temperatures, we observe inhibition of adult tissue formation and degradation of cell masses during metamorphosis, which is similar to cancer-associated cachexia. We show that larval hemocytes retain several of their normal activities, such as phagocytic ability and migration to wound sites, in all three leukemic lines. We also find that expression of the Toll reporter Drosomycin (Drs) is activated in contrast to Imd reporters, which are suppressed. Although we observe only subtle changes in immune competence, the larvae coexpressing RasV12 with a RNAi construct for the tumor suppressors l(2)gl or scrib are more sensitive to infection with entomopathogenic nematodes (EPNs), indicating additional layers of immune regulation. We show that locomotion of scribRNAi; RasV12 larvae is reduced in the presence of nematodes, potentially contributing to the increase in larva sensitivity toward EPNs.
Materials and Methods
Drosophila stocks and genetics
Fly stocks were kept under standard conditions and crosses were performed at both 25° and 29° (see figure legends for each experimental condition). The w; hml (Δ)-GAL4 UAS-eGFP (II) stock was a generous gift from Dan Hultmark. w; hmlΔ-GAL4 UAS-eGFP (II) abbreviated to HFP. BxMS1096-GAL4 and UAS-Ras85DV12 (III) strains were obtained from the Bloomington Drosophila Stock Center, USA. UAS-Ras85DV12 (III) is referred to as RasV12 in the text. In order to monitor hemocytes, UAS-GFP was recombined with HmlΔ-GAL4 on the second chromosome. All RNAi lines for tumor suppressors Scrib and l(2)gl were received from the Vienna Drosophila RNAi Center (Dietzl et al. 2007). UAS-Scrib. RNAi (v105412/KK); UAS-Ras85DV12 and UAS-l(2)gl. RNAi (v109604/KK); UAS-Ras85DV12 lines were generated in the laboratory by combining UAS-Ras85DV12 (III) and RNAi line (II) for tumor suppressor. Abbreviations for control and all leukemia model genotypes are: HFP/w- w; hml (Δ)-GAL4 UAS-eGFP × w1118, HR- HFP/w; UAS-Ras85DV12/+, HRS- HFP/UAS-Scrib.RNAi; UAS-Ras85DV12/+, and HRL-HFP/UAS-l(2)gl.RNAi; UAS-Ras85DV12/+.
Hemocyte preparation and counting
Late third instar (wandering stage) larvae were washed with 25° tap water. Then larvae were quickly dipped in 70% ethanol, followed by submerging in water to remove ethanol. Larvae were gently brushed to release their tissue-resident sessile hemocytes into circulation (Makhijani et al. 2011; Arefin et al. 2015) and placed individually in one well of a 12-well slide (Hendley-Essex 12 multispot slides), where each well contained 30 μl Schneider medium. Anticoagulant phenylthiourea (PTU) was mixed with Schneider medium beforehand to prevent melanization while bleeding. Hemocytes were counted using a hemocytometer. For microscopy, 30 min were allowed to let the hemocytes settle, followed by live imaging.
Phagocytosis assay
Late third-instar larvae were bled in PTU containing Schneider medium and incubated in a humid chamber for 10 min at room temperature (RT). A 10-fold excess of Texas Red–conjugated heat-killed Escherichia coli (k-12 strain) was applied to hemocytes and incubated for 20 min at RT. Extracellular fluorescence was quenched by adding trypan blue to a final concentration of 0.2% to the cells just before microscopy (Kurucz et al. 2007). The phagocytic index was calculated as the number of hemocytes displaying phagocytosis divided by the total number of hemocytes and multiplied by the average number of phagocytosed bacteria per hemocyte (Kocks et al. 2005). The control (HFP × w1118, referred to as HFP/w) was set to 100 and the respective comparisons were relative to the control. Approximately 70–100 hemocytes from six independent larvae were analyzed.
Hemocyte migration and wound sealing assay
White prepupae were gently washed and dried with Kleenex tissue paper. Then prepupae were wounded with a needle (125 μm in diameter) at the dorsal side of the posterior end and immediately monitored for hemocyte migration to the wound edge [a modified protocol after Regan et al. (2013)]. Within a few minutes hemocytes started migrating to the wound edge. For the wound-sealing assay, late third-instar larvae were wounded and followed until bleeding stopped. Puparium formation was used as an indication for successful sealing and quantified.
RT-qPCR
Late third-instar larvae were washed and dipped in 70% ethanol followed by rinsing with water. Total RNA extraction was performed as previously described (Arefin et al. 2014). RNA quality and concentration was determined with NanoDrop 2000 spectrophometer (Thermo Scientific). A total of 1000 ng of total RNA was used to convert cDNA using SuperScript III Reverse Transcriptase (Invitrogen) and oligo(dT) (20-mer). RT-qPCR was performed using KAPA PROBE FAST Universal qPCR Master Mix (Kapa Biosystems) and TaqMan Expression Assays (Applied Biosystems) for Drs and Cecropin A1. Diptericin A1 was synthesized by the custom TaqMan Expression Assays (Applied Biosystems) (Arefin et al. 2015; Dantoft et al. 2013). A Rotor-gene Q machine (Qiagen) was used to run qPCR. Each sample was run in triplicate. Relative mRNA expression levels were was analyzed by normalizing to the reference gene rpl32 and compared to the control genotype. The data were then transformed to Log2 fold changes and presented as mean ± SDs of the mean of at least three independent biological replicates. In the uninfected wounded situation, two independent biological samples were analyzed and both show a similar pattern.
Survival analysis after wounding and bacterial infection
Late third-instar larvae were washed with 25° tap water. Afterward, they were dipped in 70% ethanol followed by rinsing with water to remove ethanol. Prior to sticking them on tape onto a glass slide, they were dried with Kleenex (Marque) tissue paper. Larval wounding was performed according to Galko and Krasnow (2004) with a needle (250 μm in diameter). Wounded larvae were immediately submerged either only in PBS (without microbes) or in highly concentrated microbial suspension (Neyen et al. 2014) for 3 min, allowing microbes to enter via wound. Then they were transferred into specially prepared fly vials (the sponge cap used for fly vials were cut into half and put on top of wet tissue paper on the bottom of vial, and the sponge was made wet to ensure humidity before transferring larvae). Survival was monitored twice a day until eclosion. Data were process in the GraphPad 6.0.
Nematode infection
EPNs of the species Heterorhabditis bacteriophora were used to infect Drosophila larvae. Nematode infections were performed according to the protocol described earlier (Arefin et al. 2014; Kucerova et al. 2015). In short, 62–68-hr-old Drosophila larvae were washed in 25° tap water and placed on TORK advanced soft tissue paper. Subsequently, 10 μl of nematode suspension (dose: 25 nematodes/10 μl) was added to each well in a 96-well plate followed by quickly adding individual larvae. The plate was covered with parafilm and incubated for 48 hr at 25°. Physically examining larvae by gently touching with forceps scored mortality. Three groups were infected at a time for a genotype where each group contained 48 larvae. In total three independent biological replicates (48 × 3 × 3 = 432 larvae) were analyzed to determine whether a fly line is sensitive to nematodes.
Microscopy
To select the correct genotype (GFP enabled), a Leica MZ FLIII fluorescence stereomicroscope coupled with a Panasonic DMC-G2 camera was used. Analysis of samples was performed with a Hamamatsu ORCA-ER camera (C4742-95) associated with a Zeiss Axioplan 2 microscope.
Larval locomotion behavior by FIMTrack
An agarose gel was poured as a crawling surface at a concentration of 0, 8% in deionized water, which produced a solid gel (thickness of 2 mm; see Kunc et al. (2017)). To improve illumination properties, the gel was kept moist by tap water. A salt barrier was poured to prevent larvae from escaping the experimental area by adding 5 M NaCl in 2.5% agarose gel. The gels were surrounded by wet tissue paper to prevent drying out. For image acquisition, a Basler A601f camera was used. FIMTrack v2 Windows (X86) software was downloaded from http://fim.uni-muenster.de/. Data obtained from the software were analyzed in Prism 6 (GraphPad).
Statistics
ANOVA followed by Tukey’s multiple comparison test, Student’s t-test (unpaired, two sided), Fisher’s LSD test, and Log-rank test on GraphPad 6.0 (survival analysis) were used to determine statistical significance. Data represent means of SD (Figure 1, Figure 2, Figure 3, Figure 4, Figure 5, and Figure 6) and SEM (Figure 7); ** P < 0.01, * P < 0.05.
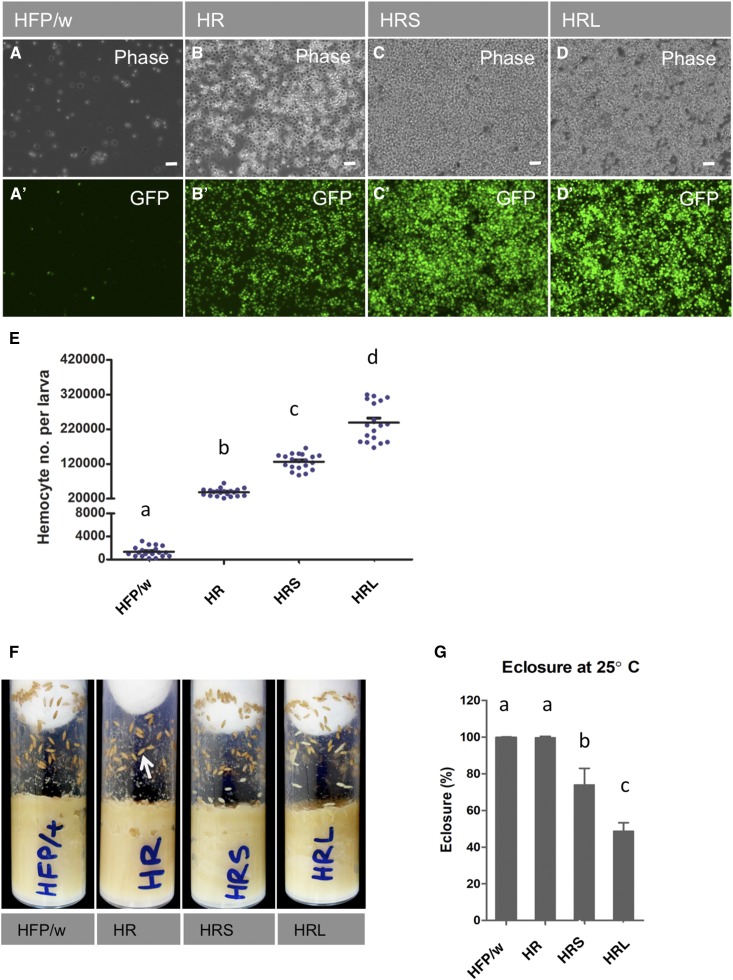
Figure 1.
Establishing Drosophila leukemia by expressing RasV12 alone and in combination with knockdown of tumor suppressors in hemocytes. (A) Larval hemocytes from control crosses expressing eGFP (w; hml (Δ)-GAL4 UAS-eGFP × w1118, which is abbreviated to HFP/w). (B–D) The number of hemocytes increases drastically when a dominant active form of Ras (UAS-RasV12) alone (HR) or in combination with RNAi lines of tumor suppressors scribble (scrib, HRS) or lethal giant larvae [(l(2)gl, HRL] is expressed using the same driver as in (A). (A–D) show phase contrast exposures while (A’–D’) show the corresponding images under fluorescence, revealing eGFP-expressing hemocytes. (E) Quantification of late third-instar larval hemocytes for the genotypes shown in (A–D). (F) Leukemia lines are viable in the prepupal stage (12 hr of metamorphosis) at 25°. No sign of larval lethality is seen in the leukemic lines. White arrow indicates the normal prepupa. (G) Quantification of eclosion rates at 25° for all lines. Expression of RasV12 alone in hemocytes (HR) does not interfere with eclosion rates at 25°; however, reduced eclosion is observed with HRS and HRL. ANOVA followed by Tukey’s multiple comparison test was performed. The same and different letters above columns on the graphs indicate nonsignificance [a to a for instance, see (G)] and significance [a to b for example, see (E) and (G)], respectively. Data represent the means with SD; P < 0.0001 and P < 0.01 was found for (E) and (G), respectively. Bar in (A–D), 50 μm.
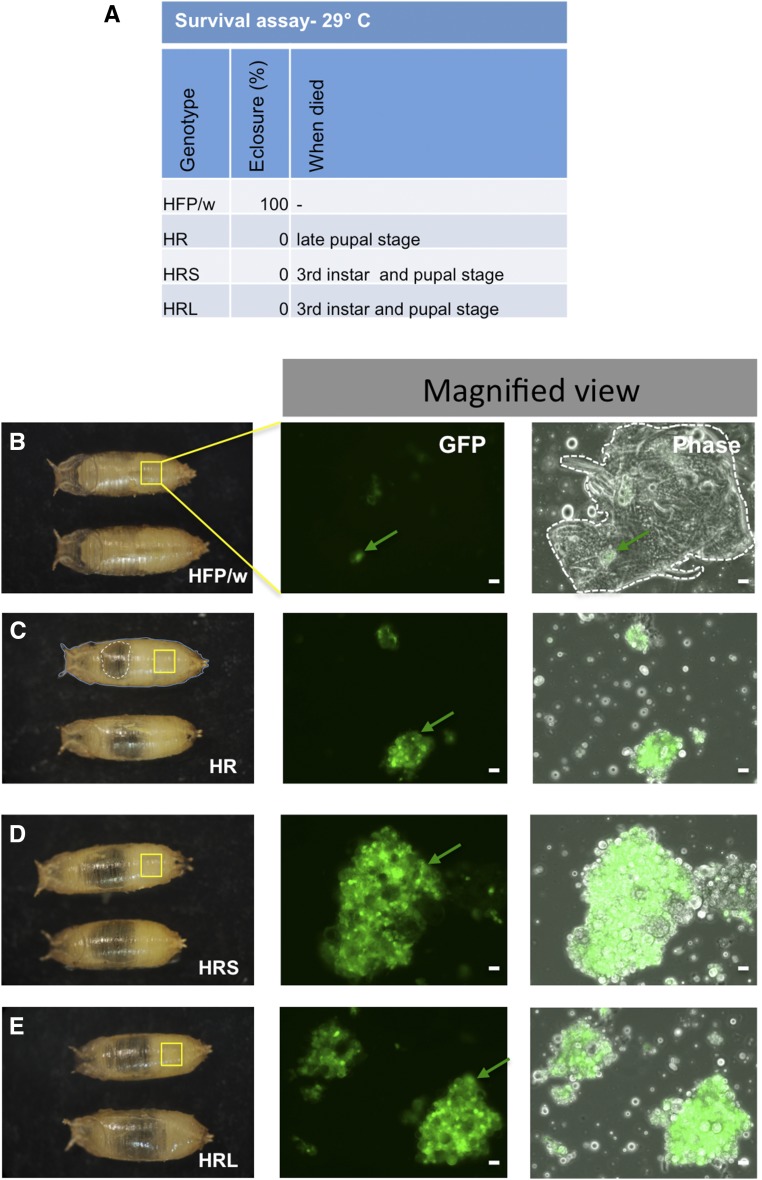
Figure 2.
Leukemic lines at higher temperatures die and contain hemocyte aggregates at the pupal stage. (A) At 29°, HR failed to eclose and larval/pupal mortality was observed for HRS and HRL. (B) Control HFP/w pupae. Opening of pupal cage at 46 hr after puparium formation (APF) at 29° showed fewer hemocytes and formation of adult structures (outlined in phase contrast). (C–E) Opening of pupae from leukemic lines showed clusters of hemocytes and an absence of adult structures. The dashed line in (C) outlines the vacuole-like structure (perhaps due to drying out). Green arrows point toward GFP-positive hemocytes. Scale bar, 20 μm.
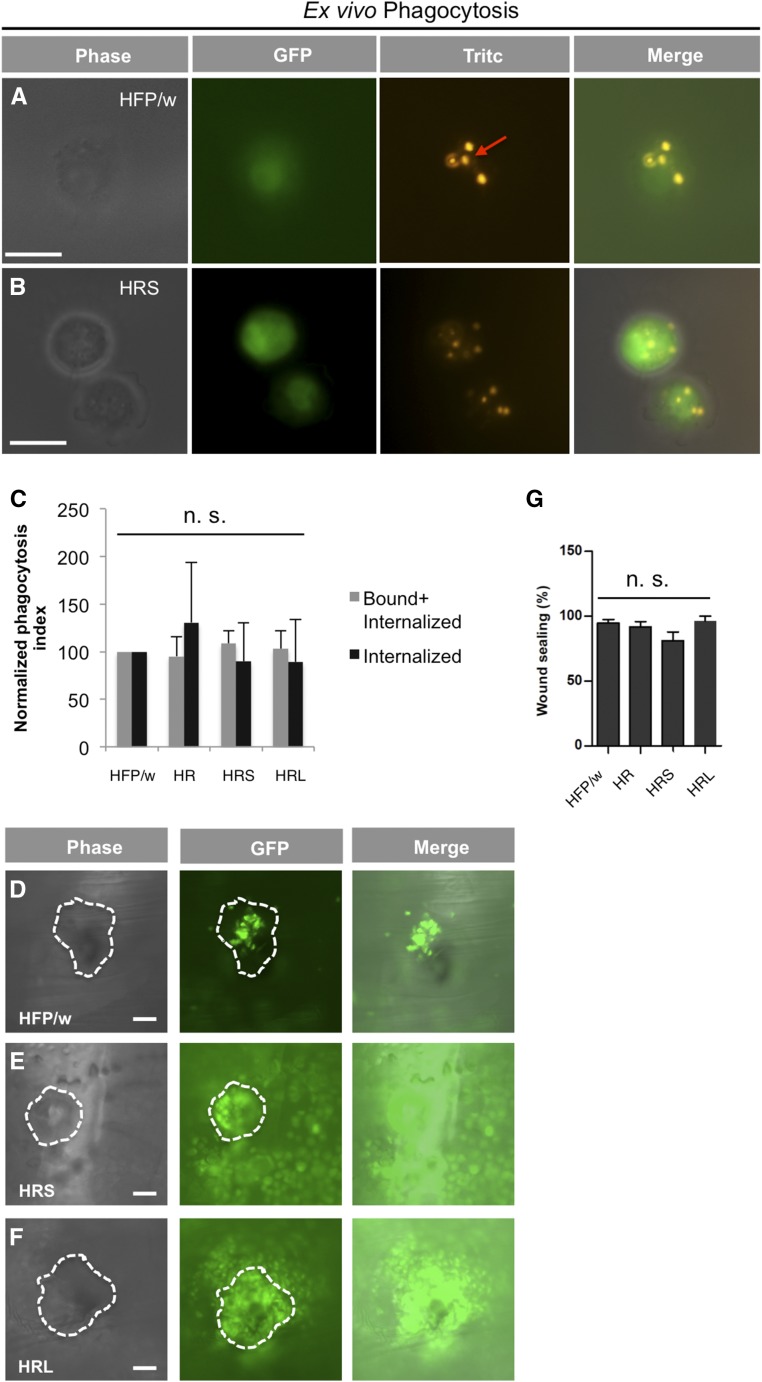
Figure 3.
Phagocytosis, hemocyte recruitment to wounds, and wound sealing are not impaired in leukemic lines. (A and B) Ex-vivo phagocytosis of Texas Red–conjugated E. coli (K-12 strain). A representative image of the HFP/w and HRS-leukemia lines is shown after phagocytosis. The red arrow indicates phagocytosed E. coli. (C) Quantification of the phagocytosis index. All leukemic lines show wild-type levels of phagocytic capacity. (D–F) Hemocyte migration to the wound edge in leukemic situation is comparable to wild type. Dashed line in phase-contrast and the GFP channel encircle the wound edges. (E and F) The increased number of hemocytes in the leukemic lines leads to a more blurry appearance in both GFP and merge channels. (G) Wounds in leukemic lines are sealed equally efficiently as in wild type. Bar in (A) and (B) is 10 μm, and in (D–F) is 50 μm. Data represent means of SD; Student’s t-test on the normalized data was performed in (C). ANOVA followed by Tukey’s multiple comparison test was performed in (G). n.s., not significant.
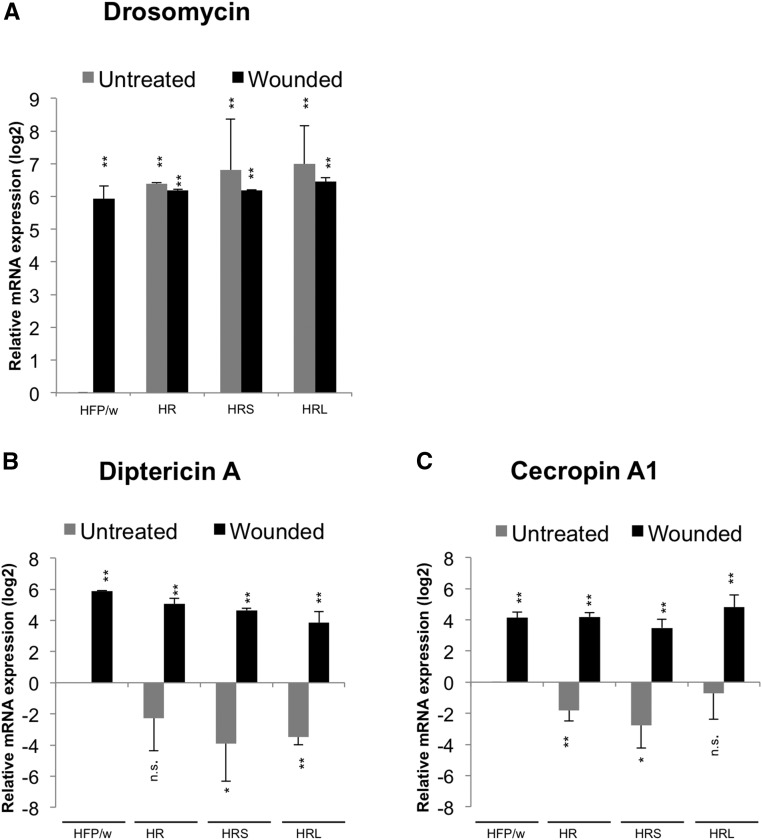
Figure 4.
Toll and Imd signaling are regulated in opposite ways in leukemic larvae. Compared to control (HFP/w), expression (A) is found upregulated while Dpt A and Cec A1 (B and C) mRNA levels are downregulated in leukemic larvae compared to untreated larvae (normalized to Rpl32 after RT-qPCR, all levels are shown relative to nontreated controls, which are set to 1). Upon wounding, Drs is also upregulated in control larvae to levels similar to nonwounded leukemic lines [HFP/w in (A)]. However, no further Drs induction is observed after wounding leukemic larvae. In contrast, Dpt A and Cec A1 levels are induced after wounding in all lines. Data represent means of SD; Student’s t-test: ** P < 0.01, * P < 0.05. n.s., not significant.
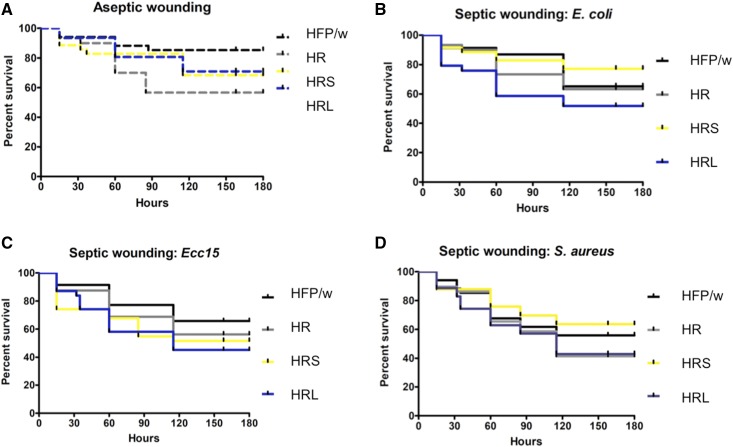
Figure 5.
Survival of septic wounded leukemic larvae is similar to wild type. (A) Aseptic wounding: control wild-type larvae show a slight increased survival compared to leukemic larvae; however, this is not significant. (B–D) Septic wounding with both Gram-negative (E. coli and Erwinia carotovora carotovora) and Gram-positive bacteria (S. aureus). None of the septic wounds caused significant increases in mortality for the leukemic lines compared with controls. Survival was monitored until eclosion. Log-rank test on GraphPad 6.0 was performed to determine statistical significance for the survival curves. (A) P = 0.1182, (B) P = 0.2074, (C) P = 0.3682, and (D) P = 0.3491.
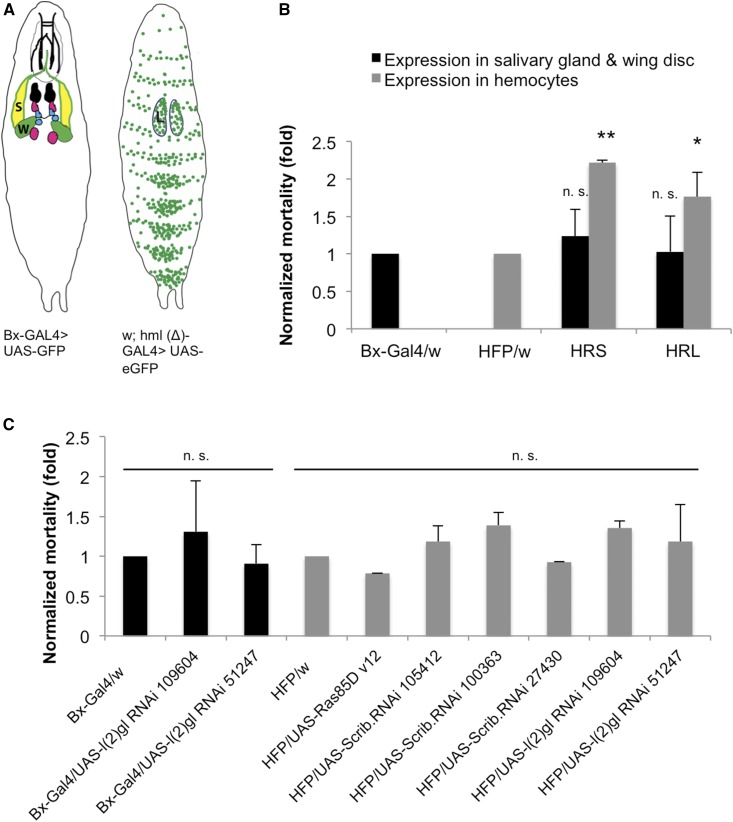
Figure 6.
Leukemic lines combined with tumor suppressor knockdowns are more susceptible to nematode infection. (A) Schematic diagrams of the expression pattern of the two different GAL4 drivers used. Bx-GAL4 expression is in the salivary glands and wing discs, whereas hml (Δ)-GAL4 expression is in the larval lymph glands and in both circulating and sessile hemocytes. (B) Upon infection with entomopathogenic Heterorhabditis bacteriophora, the combination of Ras85DV12 expression and knockdown of tumor suppressors in hemocytes showed increased mortality compared to larvae expressing either construct alone (UAS- Ras85DV12 and UAS-RNAi of tumor suppressors) or in salivary glands and wing discs. Comparisons were made with respective controls (Bx-GAL4/w and HFP/w). Mortality was normalized to controls which were set to 1. Data represent means of SD; Student’s t-test: ** P < 0.01, * P < 0.05. (C) Knocking down of different RNAi lines using two GAL4 driver lines (Bx-GAL4 and HFP) and expression of Ras85DV12 alone with HFP only. When compared to respective controls, none of them showed significant mortality upon nematode infection. L, lymph gland; n.s., not significantm, S, salivary gland; W, wing disc.
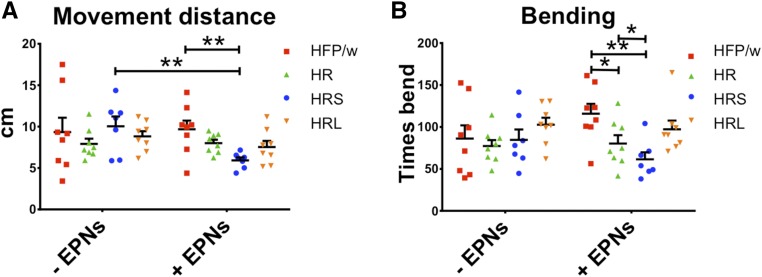
Figure 7.
Larval locomotion is impaired in HRS compared to control (HFP/w) in the presence of nematodes. (A) In the absence of nematodes no differences were observed in the distance covered by leukemic lines and control crosses whereas in the presence of nematodes HRS lines showed reduced mobility. (B) Bending frequencies do not differ in the absence of nematodes whereas in the presence of nematodes HRS larvae show reduced bending compared to controls. Each dot represents the mean value for a replicate and the middle line represents the mean of the replicates. Error bar represents SEM; sample size was at least 115 larvae. Fisher’s LSD test: ** P < 0.01, * P < 0.05.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Three Drosophila leukemia models
To induce leukemic states in Drosophila hemocytes, we expressed a dominant-active form of Ras (RasV12) in hemocytes. To increase hemocyte-specific expression, we relied on the hemolectin driver (HmlΔ-GAL4) instead of the previously used collagen IV driver (Cg-GAL4) (Asha et al. 2003). To further increase the tumorigenic potential, we combined RasV12 with knockdowns of l(2)gl and scrib, which had previously been used in combination with RasV12 to successfully induce tumors in tissues other than hemocytes (Brumby and Richardson 2003; Pagliarini and Xu 2003). We observed a substantial increase in hemocyte counts both on expression of RasV12 alone and when coexpressed with one of the RNAi constructs for tumor suppressors (Figure 1, A–E). The increase was ∼30-fold for RasV12-overexpression (HR in Figure 1) compared with the control crosses and even more pronounced upon knockdown of tumor suppressors. L(2)gl RNAi; Ras85DV12 –larvae (HRL) showed the strongest effect with a ∼170-fold increase in hemocyte numbers while scribRNAi; Ras85DV12 (HRS) larvae contained ∼90 times more hemocytes when compared to control (HFP/w). At 25°, all combinations were viable up to the pupal stage and migrated along the wall of culture vials to prepare for pupation (Figure 1F), but only RasV12-expressing pupae show eclosion rates comparable to wild type. In contrast, only a fraction of the combinations with tumor suppressors managed to eclose (Figure 1G). When the temperature was raised to 29° to allow for a higher Gal4 activity, all three tumor lines died either at the larval or pupal stage (Figure 2A). While upon bleeding, control pupae were shown to contain precursors for adult structures (Figure 2B), most likely partially unfolded imaginal discs, larvae from all tumor lines were desiccated (Figure 2C) and the remaining hemolymph contained aggregates of GFP-expressing hemocytes (Figure 2, C–E). Moreover, adult tissue formation was not observed in leukemic pupae (Figure 2, C–E).
The immune status of leukemic larvae
To address the physiology and immune status of leukemic larvae we first assessed their phagocytic capacity by counting phagocytosed Texas red–conjugated bacteria and found that none of the leukemic lines displayed a reduction in phagocytosis (Figure 3, A–C). Similarly, upon wounding, hemocytes were attracted to wound sites in all lines and larvae survived wounding equally well, although there was a trend for HRS larvae to survive slightly worse than the other lines (Figure 3, D–G). To check whether the proliferation of hemocytes had any proinflammatory effects, we analyzed the expression of antimicrobial peptide gene reporters for two major Drosophila immune pathways, including Drs (a proxy for the Toll pathway, Figure 4A), Diptericin A (Dpt A, an Imd reporter, Figure 4B), and Cecropin A1 (Cec A1, which receives input from both pathways, Figure 4C). Compared to untreated control larvae, Drs expression was activated in leukemic larvae and both Dpt A and Cec A1 reduced, although to a lesser extent. Wounding nonleukemic larvae induced Drs to the same degree as the leukemic stage (Figure 4A). Dpt A and Cec A1, which are more dependent on Imd signaling, were induced to a similar extent upon wounding all leukemic and control lines (Figure 4B). Taken together, this indicates that several aspects of cellular immunity appear normal in leukemic lines but there is a shift in the immune status, which mimics some aspects of wounding.
Leukemic lines survive selected bacterial infections
To assess the immune competence of leukemic lines, we infected larvae with either Gram-positive or Gram-negative bacteria (Figure 5). Leukemic lines survived equally well as controls upon infection with Staphylococcus aureus, which is primarily cleared via phagocytosis (Defaye et al. 2009). This is in line with our previous observation that phagocytic ability is not affected in leukemic hemocytes (Figure 3, A–C). Similarly, infection with E. coli and the natural pathogen Erwinia carotovora carotovora failed to induce higher mortality in leukemic larvae.
Upon knockdown of tumor suppressors, RasV12 expressing lines are more susceptible to nematodes
To analyze the immune status of leukemic lines in a more natural setting, we infected larvae with the EPN H. bacteriophora and its associated bacterium Photorhabdus luminescens. Heterorhabditis infects insect hosts by entering the hemocoel via the cuticle or the gut and subsequently releases Photorhabdus into the hemolymph. All three leukemic lines were infected using EPNs as well as control crosses and single knockdown lines for the tumor suppressors. We also included crosses where all combinations were expressed under control of the Beadex driver (Bx-GAL4) in the wing discs and the salivary glands [Figure 6A, see also Hauling et al. (2014)]. Although some lines with only one construct showed a trend toward increased susceptibility, none of these were significant (Figure 6C). Only the leukemic lines that expressed RasV12 in combination with tumor suppressor knockdowns were consistently more sensitive to nematode infections (Figure 6B). Of note, even crosses with the Beadex driver showed similar mortality when compared to their respective control, although immune activation had been observed upon expression of both RasV12 alone and in combination with tumor suppressor knockdowns (Hauling et al. 2014 and Krautz, R. Khalili, D. Hauling, T. and Theopold, U., unpublished data).
Larvae expressing RasV12 and reduced scrib show altered behavior in the presence of nematodes
Since we did not detect any gross developmental defects or deficiencies in the cellular response of leukemic larvae, we considered alternative explanations for the increased susceptibility of leukemic lines toward EPNs such as subtle differences in their behavior. To analyze larval locomotion, we used frustrated total internal reflection of infrared light (Risse et al. 2013), which we recently adapted for use in infection studies (Kunc et al. 2017). When exposed to nematodes, HRS larvae covered shorter distances than larvae from control crosses or the two other leukemic lines. However, no such difference was observed in the absence of nematodes (Figure 7A). The results from measuring the frequency of bending, which is part of larval avoidance behavior, were less clear but they showed that in the presence of nematodes, the bending frequency differed between HR and HRS larvae compared to controls (Figure 7B). This suggests that in the presence of nematodes, leukemic larvae are at least by some measures less mobile, which will decrease their chances of escaping infection.
Discussion
We show that by expressing dominant-active RasV12 in hemocytes, a substantial increase in hemocyte numbers is induced, confirming earlier results using a different driver (Asha et al. 2003). Hemocyte overproliferation is further exacerbated when tumor suppressors are knocked down (Figure 1). This establishes a set of three Drosophila leukemia models which display a graded increase in hemocyte numbers. All models show developmental defects at higher temperature but no such defects at the last larval instar at 25° (Figure 2). The severity of the developmental phenotype positively correlates with the increase in hemocyte numbers and may therefore be at least partially due to a redistribution of resources upon extensive hemocyte overproliferation. This is in line with the cachexia-like phenotype, which eventually leads to pupal death (Figure 2, C–E).
Upon infection with Gram-positive or Gram-negative bacteria, none of the leukemic lines displayed an increase in mortality and thus appeared immune-competent. Supporting this, phagocytosis and recruitment of hemocytes to wounds were similar in leukemic lines and controls. On the contrary, when induction of the two major immune pathways was checked using pathway-specific reporters, we found that Toll-specific reporters were constitutively activated, while Imd-dependent signaling was reduced. Activation of Toll is in line with several recent findings where activation of this pathway was observed upon damage to cells or tissues. Such settings include the release of intracellular components (Kanoh et al. 2015), failure to induce apoptosis (Ming et al. 2014), epidermal wounding (Carvalho et al. 2014), induction of a protumorous state (Hauling et al. 2014; Parisi et al. 2014), induction of fibrotic lesions (Zang et al. 2015), chromosomal instability (Liu et al. 2015), and the deregulation of hemocyte development (Arefin et al. 2015).
When larvae were infected with EPNs, those expressing RasV12 in combination with either knockdown for tumor suppressors were more susceptible. This was most significant for the scrib knockdown. As expected for a complex model such as EPN infections, our data do not fully explain why leukemic lines are more susceptible but the dysregulation of the two major immune pathways provides a partial explanation: precocious activation of Toll-dependent genes may induce a proinflammatory state which hampers the response toward EPNs. Alternatively, or in addition, the repression of Imd targets may delay the response against the symbiotic bacteria that are released by EPNs after their entry into the hemocoel (Hallem et al. 2007; Kucerova et al. 2015; Arefin et al. 2014; Castillo et al. 2015). We also observed behavioral changes in the leukemic lines, some of which can be linked to the presence of nematodes (Figure 7) and a trend for decreased survival after injury (Figure 5A), which together may exacerbate the susceptibility toward EPNs. Taken together our data indicate that Drosophila leukemia models, which combine overexpression of RasV12 with knockdown of tumor suppressors, are more susceptible toward a naturally relevant infection despite a lack of developmental, behavioral, and immune defects in standard assays. This mimics the specific defects observed in some leukemias (Forconi and Moss 2015) and provides useful tools to study subtle interactions between tumor progression, inflammation, and immunity. Further screens for modifiers will permit a detailed genetic dissection of the genes and pathways involved. These will include genetic screens as well as screens for potential chemical modifiers (McCubrey et al. 2008) and will address candidate pathways involved in cachexia, stress, and sensors for cellular integrity (Sonoshita and Cagan 2017).
Acknowledgments
We thank Traimate Sangsuwan, Stockholm University, for helping with the extraction of mRNA of uninfected samples. Our work is supported by the Swedish Research Council (VR-2010-5988 and VR 2016-04077), the Swedish Foundation for International Cooperation in Research and Higher Education (IG2011-2042), the Knut and Alice Wallenberg Foundation (KAW2012.0058), and the Swedish Cancer Foundation (CAN 2010/553).
Footnotes
Communicating editor: B. H. Reed
Literature Cited
- Anderl I., Vesala L., Ihalainen T. O., Vanha-Aho L. M., Ando I., et al. , 2016. Transdifferentiation and proliferation in two distinct hemocyte lineages in Drosophila melanogaster larvae after wasp infection. PLoS Pathog. 12: e1005746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arefin B., Kucerova L., Dobes P., Markus R., Strnad H., et al. , 2014. Genome-wide transcriptional analysis of Drosophila larvae infected by entomopathogenic nematodes shows involvement of complement, recognition and extracellular matrix proteins. J. Innate Immun. 6: 192–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arefin B., Kucerova L., Krautz R., Kranenburg H., Parvin F., et al. , 2015. Apoptosis in hemocytes induces a shift in effector mechanisms in the Drosophila immune system and leads to a pro-inflammatory state. PLoS One 10: e0136593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asha H., Nagy I., Kovacs G., Stetson D., Ando I., et al. , 2003. Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics 163: 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangi E., 2013. Drosophila at the intersection of infection, inflammation, and cancer. Front. Cell. Infect. Microbiol. 3: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumby A. M., Richardson H. E., 2003. Scribble mutants cooperate with oncogenic Ras or notch to cause neoplastic overgrowth in Drosophila. EMBO J. 22: 5769–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumby A. M., Richardson H. E., 2005. Using Drosophila melanogaster to map human cancer pathways. Nat. Rev. Cancer 5: 626–639. [DOI] [PubMed] [Google Scholar]
- Carvalho L., Jacinto A., Matova N., 2014. The Toll/NF-kappaB signaling pathway is required for epidermal wound repair in Drosophila. Proc. Natl. Acad. Sci. USA 111: E5373–E5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo J. C., Creasy T., Kumari P., Shetty A., Shokal U., et al. , 2015. Drosophila anti-nematode and antibacterial immune regulators revealed by RNA-Seq. BMC Genomics 16: 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofi T., Apidianakis Y., 2013. Ras-oncogenic Drosophila hindgut but not midgut cells use an inflammation-like program to disseminate to distant sites. Gut Microbes 4: 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozatier M., Vincent A., 2011. Drosophila: a model for studying genetic and molecular aspects of haematopoiesis and associated leukaemias. Dis. Model. Mech. 4: 439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantoft W., Davis M. M., Lindvall J. M., Tang X., Uvell H., et al. , 2013. The Oct1 homolog nubbin is a repressor of NF-kappaB-dependent immune gene expression that increases the tolerance to gut microbiota. BMC Biol. 11: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defaye A., Evans I., Crozatier M., Wood W., Lemaitre B., et al. , 2009. Genetic ablation of Drosophila phagocytes reveals their contribution to both development and resistance to bacterial infection. J. Innate Immun. 1: 322–334. [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. [DOI] [PubMed] [Google Scholar]
- Forconi F., Moss P., 2015. Perturbation of the normal immune system in patients with CLL. Blood 126: 573–581. [DOI] [PubMed] [Google Scholar]
- Galko M. J., Krasnow M. A., 2004. Cellular and genetic analysis of wound healing in Drosophila larvae. PLoS Biol. 2: E239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganan-Gomez I., Wei Y., Starczynowski D. T., Colla S., Yang H., et al. , 2015. Deregulation of innate immune and inflammatory signaling in myelodysplastic syndromes. Leukemia 29: 1458–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gateff E., 1978. Malignant neoplasms of genetic origin in Drosophila melanogaster. Science 200: 1448–1459. [DOI] [PubMed] [Google Scholar]
- Gateff E., Gissman L., Shrestha R., Plus N., Pfister H., et al. , 1980. Characterization of two tumorous blood cell lines of Drosophila melanogaster and the viruses they contain, in Invertebrate Systems in Vitro, edited by Kurstak E., Maramorosch K., Duebendorfer A. Elsevier, New York. [Google Scholar]
- Gold K. S., Bruckner K., 2014. Drosophila as a model for the two myeloid blood cell systems in vertebrates. Exp. Hematol. 42: 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C., 2013. Drosophila melanogaster: a model and a tool to investigate malignancy and identify new therapeutics. Nat. Rev. Cancer 13: 172–183. [DOI] [PubMed] [Google Scholar]
- Hallem E. A., Rengarajan M., Ciche T. A., Sternberg P. W., 2007. Nematodes, bacteria, and flies: a tripartite model for nematode parasitism. Curr. Biol. 17: 898–904. [DOI] [PubMed] [Google Scholar]
- Hauling T., Krautz R., Markus R., Volkenhoff A., Kucerova L., et al. , 2014. A Drosophila immune response against Ras-induced overgrowth. Biol. Open 3: 250–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honti V., Csordas G., Kurucz E., Markus R., Ando I., 2014. The cell-mediated immunity of Drosophila melanogaster: hemocyte lineages, immune compartments, microanatomy and regulation. Dev. Comp. Immunol. 42: 47–56. [DOI] [PubMed] [Google Scholar]
- Kanoh H., Kuraishi T., Tong L. L., Watanabe R., Nagata S., et al. , 2015. Ex vivo genome-wide RNAi screening of the Drosophila Toll signaling pathway elicited by a larva-derived tissue extract. Biochem. Biophys. Res. Commun. 467: 400–406. [DOI] [PubMed] [Google Scholar]
- Kocks C., Cho J. H., Nehme N., Ulvila J., Pearson A. M., et al. , 2005. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell 123: 335–346. [DOI] [PubMed] [Google Scholar]
- Kucerova L., Broz V., Arefin B., Maaroufi H. O., Hurychova J., et al. , 2015. The Drosophila chitinase-like protein IDGF3 is involved in protection against nematodes and in wound healing. J. Innate Immun. 8: 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunc M., Arefin B., Hyrsl P., Theopold U., 2017. Monitoring the effect of pathogenic nematodes on locomotion of Drosophila larvae. Fly 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurucz E., Markus R., Zsamboki J., Folkl-Medzihradszky K., Darula Z., et al. , 2007. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr. Biol. 17: 649–654. [DOI] [PubMed] [Google Scholar]
- Liu D., Shaukat Z., Saint R. B., Gregory S. L., 2015. Chromosomal instability triggers cell death via local signalling through the innate immune receptor Toll. Oncotarget 6: 38552–38565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhijani K., Alexander B., Tanaka T., Rulifson E., Bruckner K., 2011. The peripheral nervous system supports blood cell homing and survival in the Drosophila larva. Development 138: 5379–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubrey J. A., Steelman L. S., Abrams S. L., Bertrand F. E., Ludwig D. E., et al. , 2008. Targeting survival cascades induced by activation of Ras/Raf/MEK/ERK, PI3K/PTEN/Akt/mTOR and Jak/STAT pathways for effective leukemia therapy. Leukemia 22: 708–722. [DOI] [PubMed] [Google Scholar]
- Mills C. D., Ley K., 2014. M1 and M2 macrophages: the chicken and the egg of immunity. J. Innate Immun. 6: 716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming M., Obata F., Kuranaga E., Miura M., 2014. Persephone/Spatzle pathogen sensors mediate the activation of Toll receptor signaling in response to endogenous danger signals in apoptosis-deficient Drosophila. J. Biol. Chem. 289: 7558–7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muallem G., Hunter C. A., 2014. ParadYm shift: Ym1 and Ym2 as innate immunological regulators of IL-17. Nat. Immunol. 15: 1099–1100. [DOI] [PubMed] [Google Scholar]
- Neyen C., Bretscher A. J., Binggeli O., Lemaitre B., 2014. Methods to study Drosophila immunity. Methods 68: 116–128. [DOI] [PubMed] [Google Scholar]
- Niemeyer C. M., 2014. RAS diseases in children. Haematologica 99(11): 1653–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini R. A., Xu T., 2003. A genetic screen in Drosophila for metastatic behavior. Science 302: 1227–1231. [DOI] [PubMed] [Google Scholar]
- Parisi F., Stefanatos R. K., Strathdee K., Yu Y., Vidal M., 2014. Transformed epithelia trigger non-tissue-autonomous tumor suppressor response by adipocytes via activation of Toll and Eiger/TNF signaling. Cell Reports 6: 855–867. [DOI] [PubMed] [Google Scholar]
- Pastor-Pareja J. C., Xu T., 2013. Dissecting social cell biology and tumors using Drosophila genetics. Annu. Rev. Genet. 47: 51–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan J. C., Brandao A. S., Leitao A. B., Mantas Dias A. R., Sucena E., et al. , 2013. Steroid hormone signaling is essential to regulate innate immune cells and fight bacterial infection in Drosophila. PLoS Pathog. 9: e1003720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risse B., Thomas S., Otto N., Lopmeier T., Valkov D., et al. , 2013. FIM, a novel FTIR-based imaging method for high throughput locomotion analysis. PLoS One 8: e53963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoshita M., Cagan R. L., 2017. Modeling human cancers in Drosophila. Curr. Top. Dev. Biol. 121: 287–309. [DOI] [PubMed] [Google Scholar]
- Theopold U., Krautz R., Dushay M. S., 2014. The Drosophila clotting system and its messages for mammals. Dev. Comp. Immunol. 42: 42–46. [DOI] [PubMed] [Google Scholar]
- Tipping M., Perrimon N., 2014. Drosophila as a model for context-dependent tumorigenesis. J. Cell. Physiol. 229: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Y., Wan M., Liu M., Ke H., Ma S., et al. , 2015. Plasma membrane overgrowth causes fibrotic collagen accumulation and immune activation in Drosophila adipocytes. Elife 4: e07187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.