Abstract
In the framework of a gene flow assessment, we investigated the natural hybridization rate between Gossypium hirsutum (AADD genome) and G. herbaceum (AA genome). The latter species, a diploid progenitor of G. hirsutum, is spontaneously present in South Africa. Reciprocal crosses were performed without emasculation between G. herbaceum and G. hirsutum. Neither examination of the morphological characteristics nor flow cytometry analysis of the 335 plants resulting from the G. hirsutum × G. herbaceum cross showed any hybrid features. Of the 148 plants produced from the G. herbaceum × G. hirsutum cross, three showed a hybrid phenotype, and their hybrid status was confirmed by SSR markers. Analysis of DNA content by flow cytometry and morphological traits clearly showed that two of these plants were triploid (AAD). The third plant had a flow cytometry DNA content slightly higher than G. hirsutum. In addition, its morphological characteristics (plant architecture, presence and size of petal spots, leaf shape) led us to conclude that this plant was AAAD thus resulting from fertilization with an unreduced AA gamete of the female G. herbaceum parent. Fluorescent In Situ Hybridization (FISH) and meiotic behavior confirmed this hypothesis. To the best of our knowledge, this is the first description of such gametes in G. herbaceum, and it opens new avenues in breeding programs. Furthermore, this plant material could provide a useful tool for studying the expression of genes duplicated in the A and D cotton genome.
Keywords: Gene flow, natural hybridization, unreduced gamete, Gossypium hirsutum, Gossypium herbaceum
Assessment of gene flow from a crop to a wild relative is particularly relevant in the case of allopolyploid species, which are formed by hybridization between related species, when one of its progenitors is present in cultivated areas. In certain geographical areas, this may be the case for the cultivated tetraploid cotton (Gossypium hirsutum, AADD, 2n = 4x = 52) and a wild diploid species (Gossypium herbaceum, AA, 2n = 2x = 26). Cultivated cotton, the most important source of natural fiber, has been the long-standing subject of a number of taxonomical studies (Watt 1907; Hutchinson et al. 1947; Saunders 1961; cited in Wendel et al. 2010; Fryxell 1979; Fryxell et al. 1992). The cotton genus (Gossypium L.) includes 50 diploid species (2n = 2x = 26) that can be differentiated cytogenetically into eight genome groups (A–G and K), and seven allotetraploid species (2n = 4x = 52) (Fryxell 1979; Fryxell et al. 1992; Stewart 1995; Wendel and Cronn 2003; Grover et al. 2015; Gallagher et al. 2017). Remarkably, among the four domesticated species, there are two allotetraploids (the New World tetraploids G. hirsutum and G. barbadense) and two diploids (the Old World diploids G. arboreum and G. herbaceum). The tetraploid species originated from a single polyploidization event between an A-genome diploid species and a D-genome diploid species (Wendel and Cronn 2003). The two African-Asian A-genome cottons, G. arboreum and G. herbaceum, are the equivalent of the maternal genome donor to polyploid cotton (Wendel 1989), although some evidence suggests that G. herbaceum may more closely resemble the A-genome donor than G. arboreum (Wendel et al. 2010). Recent results have suggested an independent domestication of these two Old World cotton species (Renny-Byfield et al. 2016). Grover et al. (2015) demonstrated that this combination of a D-genome progenitor, resembling modern G. raimondii, and an A-genome progenitor, much like modern G. arboreum and G. herbaceum, occurred as a single hybridization event (genomic merger and doubling) that gave rise to all polyploid cottons. Interspecific hybridizations have been used extensively to improve yield, fiber quality, or other agronomic traits, and numerous studies have focused on the introgression of traits of interest from wild diploid cotton into cultivated cotton G. hirsutum (Meyer 1974; Stewart 1995; Mergeai 2003; Chee et al. 2016). Colchicine doubling is an essential step in producing an improved cultivar through interspecific crossing, as the recovered triploid hybrids are sterile. Unreduced gametes have proven useful in enabling crosses between plants of different ploidy levels, which otherwise often fail due to unbalanced parental contributions in the developing seed (Barcaccia et al. 2003; Carputo et al. 2003). If the plant of the lower ploidy level can be induced to produce unreduced gametes, such limitations can be overcome. This strategy has been successfully used in various species such as potato, alfalfa, and manioc (reviewed in Brownfield and Kohler 2011; Dewitte et al. 2012).
The large-scale development of transgenic insect-resistant and/or herbicide-tolerant cotton has promoted studies about the ability of native diploid A or D species to produce fertile hybrids with tetraploid cotton (Stewart 1995; Brubaker et al. 1999). In South Africa, and particularly in the KwaZulu Natal Province, where the commercialization of transgenic Bt cotton began in 1998, a wild species (G. herbaceum), one of the progenitors of G. hirsutum, is found in areas neighboring the cultivated cotton fields. Consequently, we have undertaken a study to evaluate the likelihood of gene flow between the allotetraploid cultivated cotton (G. hirsutum) and its wild diploid relative (G. herbaceum). Transgene escape depends on the generation of fertile hybrids, and the possible selective advantage that would result in the development of weedy derivatives. In this paper, we present our analysis of natural cotton gene flow in South Africa, which furthermore provides evidence for unreduced gametes in the species G. herbaceum.
Materials and Methods
Plant material
Reciprocal crosses were performed without emasculation between G. herbaceum Indian accession Boumi Aria (A1A1, 2n = 26) and G. hirsutum (AtAtDtDt, 2n = 52; lower case letter “t” referring to the “tetraploid” genome) Delta Opal RR from Deltapine, South Africa. Forty plants from each accession were maintained in individual pots in a greenhouse at Pretoria University and divided into two batches: donor plants for pollen harvest and recipient plants for hybrid production. To simulate both pollination occurring in the field and regular visits by pollinators to plants, pollen harvested from newly opened flowers was spread on the recipient flowers without emasculation two or three times between 9 and 11 am. All the subsequently produced seeds were sown (one seed per pot) in greenhouses at the Versailles Centre of the French National Institute of Agricultural Research (INRA) to screen for hybrids.
Seeds with a thick seed coat harvested from G. herbaceum were scraped with sandpaper to facilitate germination, and then placed on wet paper at 24°. Germinating seeds were planted in soil in a greenhouse following radicle emergence.
Morphological examinations
Morphological examinations were made both at the vegetative stage (six leaves) and at the reproductive stage. Details of flower morphology were recorded. In plants that exhibited hybrid characteristics, particular attention was paid to flower size and color, and especially to the presence and intensity of red petal spots, as G. hirsutum has no spot, and G. herbaceum shows large, bright red spots.
Pollen viability
Pollen viability was assessed after Alexander dye staining (Alexander 1969) with a 3-hr incubation period at 50°. Several samples were taken in the greenhouse between 10 and 11 am on several distinct days.
Estimation of nuclear DNA amount by flow cytometry
Flow cytometry analyses (Partec ploidy analyzer, Munster, Germany) were performed for all progeny on leaf tissue to assess their nuclear DNA amount, as described by Eber et al. (1997). Barley leaves served as the internal reference with set value of 100. Preliminary experiments confirmed that the measure was not affected neither by leaf age or conservation delay (5–20 min) of shredded leaves in the buffer (Kit Partec, Munster, Germany). Three measurements with independent leaf samples were performed for each control and potentially hybrid plant. Twenty plants were analyzed for the G. hirsutum and G. herbaceum parents. The twofold size difference between the two genomes A and D was taken into account to determine chromosome numbers.
Meiotic behavior and chromosome counting
Young squares (flower buds) were sampled in the greenhouse from the two parental lines and for deemed hybrid plants according to flow cytometric analysis and morphological observations. They were fixed in Carnoy’s solution (ethanol:chloroform:acetic acid, 6:3:1) for 24 hr, and preserved in 70% ethanol until characterized. For the observation of pollen mother cells (PMCs), anthers were squashed in 1% acetocarmin, and cells inspected with a light microscope. PMCs were observed at metaphase I to establish the meiotic behavior of the hybrids.
SSR marker analysis
Microsatellite (SSR) markers were used to verify the hybrid status of progenies obtained from the different hybridizations. Six SSRs (BNL 3264, BNL 3563, BNL 409, BNL 4049, NAU 0915, and NAU 1246) that demonstrated clear polymorphisms between G. hirsutum and G. herbaceum were selected, based on prior screens of diploid and tetraploid accessions (J.M. Lacape, unpublished data). Their primer sequences are available from CottonGen (https://www.cottongen.org/). Sixty-six DNAs were analyzed, including 64 progenies from the two reciprocal crosses (46 for G hirsutum × G herbaceum, and 18 for G. herbaceum × G. hirsutum), and from control DNAs of each parental species. Polymorphic markers between the two species were kept for hybrid characterization. DNA extraction, PCR amplification and electrophoresis-based allele separation were performed according to Lacape et al. (2007). The progenies were assigned an allelic “hybrid” coding when alleles of both G. hirsutum (could be one or two amplicons) and G. herbaceum (an additional unique amplicon) were detected.
Fluorescence in situ hybridization (FISH)
Mitotic chromosomal squashes were prepared with standard techniques. FISH experiments were carried out with two DNA probes: the 45S rDNA probe (pTa 71) is a cloned 9-kb EcoRI fragment of a rDNA repeat unit (18S-5.8S-26S genes and spacers) isolated from Triticum aestivum (Gerlach and Bedbrook 1979) and the pTa 794 probe is a 410 bp BamHI fragment of the 5S rDNA from wheat (Gerlach and Dyer 1980). The two probes were labeled by random priming with Alexa-488 dUTP (Invitrogen, Life Technologies) and Alexa 594 (Invitrogen, Life Technologies) respectively. Genomic in situ hybridization (GISH) was performed with probe DNAs from the progenitors of the allotetraploids. Total genomic DNA from G. herbaceum (A-genome) was labeled by nick translation with biotin-16-dUTP (Roche Diagnostics, Indianapolis, IN). Total genomic DNAs from G. raimondi (D-genome) were autoclaved to yield fragments of 100–300 bp and used as a blocking DNA. A series of test assessments determined the amount of blocking DNA that was required to discriminate between the two genomes. Optimum results were obtained with a ratio of 1:50 for A genome probe and D genome blocking DNA in the hybridization mixture. Biotinylated probes were immunodetected by Texas-red-conjugated with avidin antibodies (Vector Laboratories, Burlingame, CA). The chromosomes were mounted and counterstained in Vectashield (Vector Laboratories) containing 2.5 µg/ml 4,6-diamidino-2-phenylindole (DAPI). Fluorescence images were captured using a CoolSnap HQ camera (Photometrics, Tucson, AZ) on an Axioplan 2 microscope (Zeiss, Oberkochen, Germany), and analyzed with the MetaVueTM software (Universal Imaging Corporation, Downington, PA).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Interspecific F1 hybrid production
G. hirsutum (AtAtDtDt) × G. herbaceum (A1A1):
After pollination, moderate boll shedding was observed, as 60 mature bolls were obtained from 68 pollinated flowers. A total of 335 plants was obtained from 504 sown seeds. Flow cytometry analysis revealed that all of them were identical to the female allotetraploid parent, G. hirsutum. This observation was confirmed by their morphological characteristics including plant architecture, leaf shape as well as flower size, shape, and color. No interspecific hybrid was obtained.
G. herbaceum (A1A1) × G. hirsutum (AtAtDtDt):
Significant boll abscission was observed from the hybridizations involving G. herbaceum as the female (29 mature bolls out of 146 pollinated flowers). Similarly, a large number of aborted seeds were harvested (163 aborted seeds out of a total 498 harvested). Out of 335 seeds sown, 148 developed into plants: among these, three interspecific hybrids were detected. Flow cytometric analysis showed that two of them (namely H1 and H2) had an intermediate value possibly corresponding to A1AtDt hybrids. The third hybrid (H3) exhibited a slightly higher DNA content than G. hirsutum.
Our results indicate a hybridization rate of 0.02 hybrid seed per pollinated flower, compared to ∼30 seeds per boll observed after selfing of G. hirsutum. Among the 148 plants observed, the interspecific hybridization rate detected was 0.02.
Characterization of interspecific F1 hybrids
Morphological traits:
Among the 148 plants obtained with G. herbaceum as the female parent, observations 20 d after sowing (at the three-leaf stage) led to the morphological identification of the three hybrids. Already at a young stage (three-leaf, 20 d after sowing), three plants exhibited general morphological traits similar to G. hirsutum. Several weeks later, two types of plants could be distinguished. Among the three hybrids, H1 and H2 displayed identical morphological characteristics, and exhibited the phenotype typical of a triploid hybrid (A1AtDt), with traits intermediate between the two parents (Figure 1). For the third hybrid (H3), the overall plant phenotype was intermediate between G. hirsutum and the triploid (Figure 1), with individual plant parts varying between the triploid and G. herbaceum. Hybrid H3 leaves had less marked serrations, but more lobes, and were intermediate between G. herbaceum and the triploid (Figure 2). In its first stages of development, the hybrid H3 displayed density of stem hairs and gossypol glands similar to those of G. herbaceum. The two types of hybrid phenotypes were also differentiated by flower characteristics: corolla size, petal red spot intensity and size, and stamen color and size (Figure 3). The corollas of the H1 and H2 hybrid flowers had the same size as G. hirsutum, but they exhibited light red spots (Figure 3). H3 had larger, brighter red spots, intermediate between A1AtDt and G. herbaceum (Figure 3), and its corolla size was slightly smaller than those of H1 and H2. These observations can support the hypothesis that the H3 hybrid carries more genome A than the G. herbaceum parent. Moreover, it has been reported that spot presence is controlled by a single gene carried by the A genome (Stephens 1974).
Figure 1.
Plants at 70 d of culture. (A) Left to right G. hirsutum, G herbaceum, (B) left to right, hybrid H1, hybrid H2, and hybrid H3.
Figure 2.
Leaf shapes. Left to right G. herbaceum, hybrid H1, hybrid H3, and G. hirsutum. Bar, 1 cm.
Figure 3.
Flower characteristics: Left to right: G. herbaceum, hybrid H1, hybrid H3, and G. hirsutum. Bar, 1 cm.
Microsatellite-based allelic status of the interspecific hybrids:
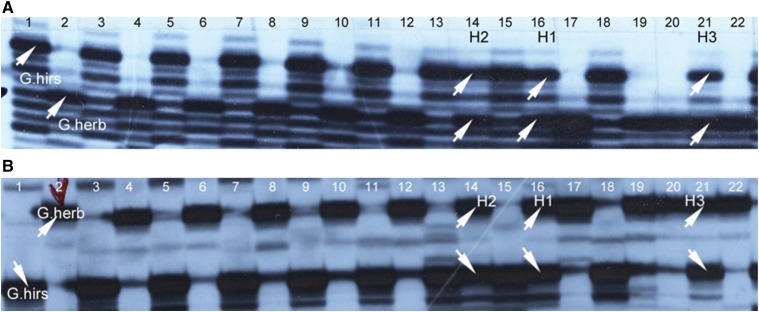
One of the six SSRs tested was not exploited because of ambiguous allele assignment. For the five remaining SSRs (BNL3563, BNL409, BNL4049, NAU0915, and NAU1246), the hybrid status of hybrids H1, H2, and H3 was confirmed (Figure 4). The three hybrids exhibited the alleles from G. hirsutum as well as from G. herbaceum. However, the amplification profiles did not reveal any quantitative difference between these hybrids.
Figure 4.
Molecular profiles of parents G. hirsutum, G. herbaceum, and 20 progenies showing the hybrid profile of H1, H2, and H3. (A) Marker BNL409, (B): marker NAU1246.
Identification of the genomic structure of hybrids:
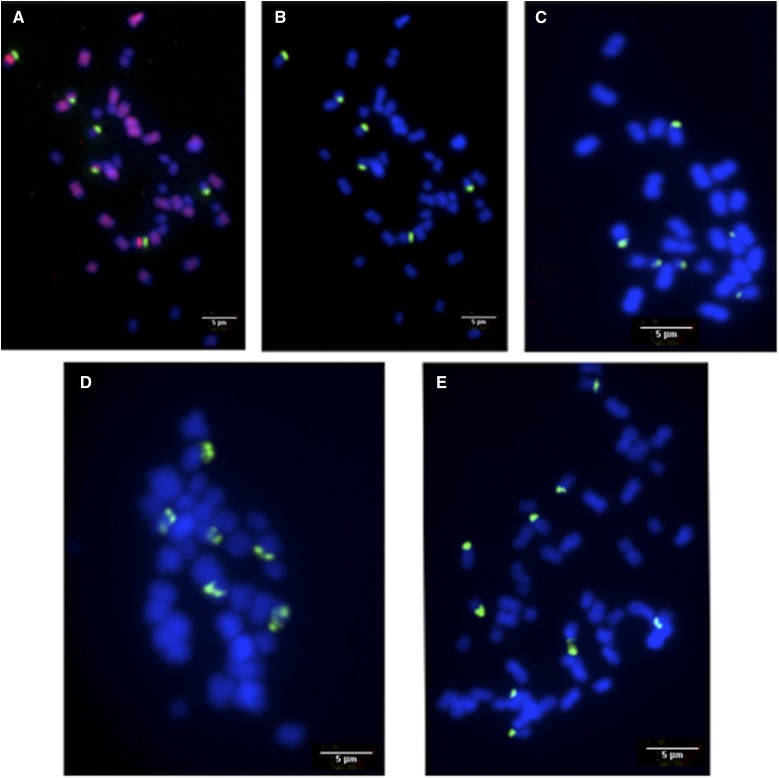
Results of the FISH analysis at mitosis showed that G. hirsutum (AtAtDtDt) carried six 45S loci with two loci on At genome (Figure 5, A and B), in accordance with previous reports (Hanson et al. 1996; Gan et al. 2013). However, six 45S loci were also observed on the A genome of G. herbaceum (A1A1) (Figure 5C). The structure of H1 and H2 hybrids was confirmed with 39 chromosomes, and we found the expected number of 45S loci for an A1AtDt genomic structure, i.e., six in total with three from G. herbaceum and three from G. hirsutum (Figure 5D). Analysis of H3 resulted in a count of 52 chromosomes carrying nine 45S loci six from G. herbaceum, plus three from G. hirsutum thus compatible with the genome structure A1A1AtDt (Figure 5E).
Figure 5.
In situ hybridization analysis. (A) The GISH probe hybridized specifically to all A genome chromosomes (pink) in G. hirsutum (AtAtDtDt); 5SrDNA in red, (B–E) FISH with 45S rDNA in green. FISH analyses of somatic metaphase chromosomes from G. hirsutum (B), G. herbaceum (C), hybrid H2 (D), and hybrid H3 (E). Chromosomes were counterstained with DAPI (blue).
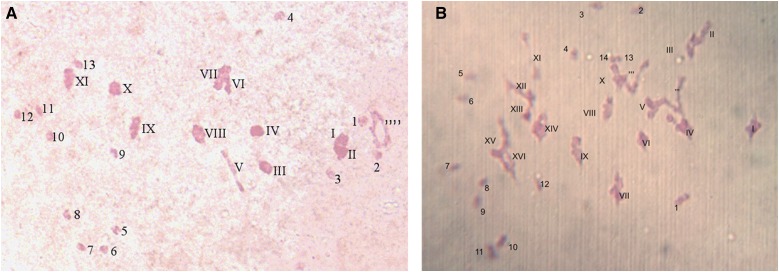
Therefore, this hybrid was formed from a female unreduced gamete of G. herbaceum and a reduced gamete of G. hirsutum (Table 1). According to the genomic composition (A1AtDt) of H1 and H2 plants, the expected meiotic behavior is 13 univalents corresponding to the D genome and 13 bivalents between the homologous chromosomes of the A genomes. Of the 26 analyzed PMCs, 35% matched this configuration, but 57.7% revealed the presence of multivalents, trivalents, or quadrivalents (Figure 6A and Table 2). In the third H3 hybrid, all the 15 PMCs observed indicated the presence of multivalents, likely due to the presence of three homologous A genomes (Figure 6B and Table 2). Tetrads harboring 3–7 microspores have been observed from young flowers of the triploid hybrids H1 and H2.
Table 1. 45S rDNA signal numbers and genome structures.
| Chromosomes Numbers | 45srDNA Signal Numbers | Genome Structure | |
|---|---|---|---|
| G. hirsutum | 52 | 6: 2 (At At), 4 (Dt Dt) | At At Dt Dt |
| Hybrid H1 | 39 | 6: 3 A1, 1 At, 2 Dt | A1 At D |
| Hybrid H2 | 39 | 6: 3 A1, 1 At, 2 Dt | A1 At D |
| Hybrid H3 | 52 | 9: 6 (A1A1), 1At, 2Dt | A1 A1 At Dt |
Figure 6.
PMC in metaphase I. (A) from A1AtDt hybrid H1, 13 univalents (1–13), 11 bivalents (I–XI), and one quadrivalent; (B) from A1A1AtDt hybrid H3: 14 univalents (1–14), 16 bivalents (I–XVI), and two trivalents.
Table 2. Meiotic behavior of hybrids H1, H2, and H3.
| 2n | PMC | Univ. | Bival. | Trival. | Quadrival. | |
|---|---|---|---|---|---|---|
| Hybrid H1–H2 | 39 | 26 | 12.61a (9–14)b | 11.81 (7–14) | 0.31 (0–2) | 0.17 (0–2) |
| Hybrid H3 | 52 | 15 | 14.67 (12–17) | 16.8 (14–20 | 1.07 (0–4) | 0.13 (0–1) |
Average per cell.
Range.
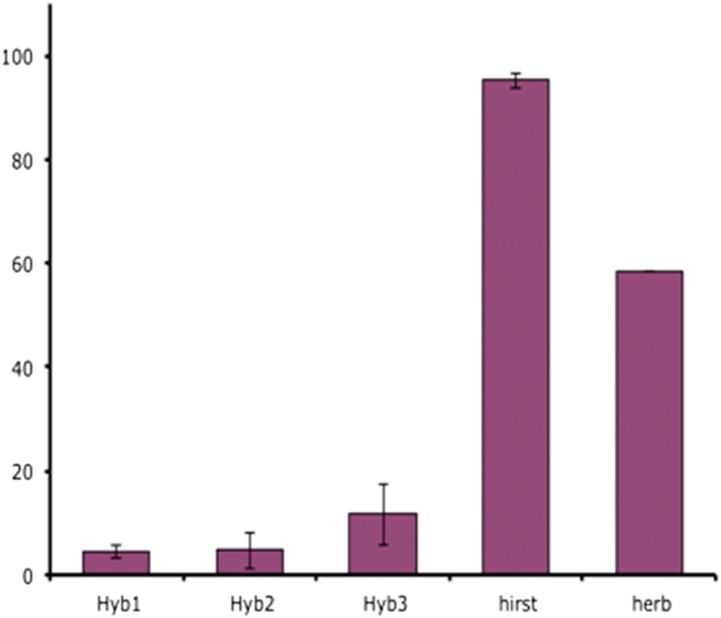
Fertility of interspecific hybrids:
As expected, pollen grains from the parental lines had a high viability (Figure 7). In contrast, very few pollen grains (<5%) from hybrids H1 and H2 were stained by Alexander dye (Figure 7). Pollen grains from the hybrid H3 exhibited a range of viability, up to 20%. Sampling was performed on all the plants at the same time, and the fertility of G. herbaceum was observed to be lower than that of G. hirsutum. This difference in the viability of the pollen grains from G. hirsutum and G. herbaceum could be explained by the time of the sampling. Flowers were collected at the same time in the morning, which might not have been optimal for the sampling of G. herbaceum pollen.
Figure 7.
Pollen viability (percentage). Seven repetitions (40 to >1000 pollen grains per sampling) for G. hirsutum. and H1, H2, and H3. One observation on 120 pollen grains for G. herbaceum.
None of the three hybrids produced any seed, either through open or self-pollination. In order to save the hybrid materials, the plants were reproduced through cutting when they became too large.
Discussion
The results showed a relatively high hybridization rate (three hybrids among 148 progeny) between G. herbaceum and G. hirsutum without emasculation of the female parent, and when the plant with the lower ploidy level was the female. This result is similar to those observed with Brassica napus, which is also an allotetraploid species; when one of its progenitors, B. rapa was used as female, the spontaneous interspecific hybridization ranged from 3 to 8% (Warwick et al. 2003). When the diploid parent G. herbaceum was used as male, no hybrids were obtained. These results are not in accordance with those of Brubaker et al. (1999), working on the improvement of cultivated cotton by crosses with diploid Australian species (genomes C, G, and K). They found that it was more difficult to obtain triploid hybrids when the female parent was diploid. Similarly, Beasley (1940) concluded that the likelihood of successful hybridization is higher if the female parent has the higher ploidy level, reflecting the traditional recommendations concerning the critical role of ploidy ratio of endosperm and zygote for successful embryo development (Beasley 1942; Stephens 1942 cited in Ahoton et al. 2003). Similarly, it has been observed among Brassicaceae species that the production of interspecific hybrids using B. napus as female or male with different diploid species depends on the species combination (Chèvre et al. 2004). Our combined analyses based on cytometry, fluorescent in situ hybridization and chromosome counting indicated that the three hybrids obtained were of two types. Two, H1 and H2, were triploids (A1AtDt), and, remarkably, the third one (H3) was a tetraploid carrying three A genomes and a single D genome. The meiotic behavior of triploid hybrids is close to what can be expected with a majority of pollen mother cells with 13 bivalents corresponding to homologous pairing of the 13 A chromosomes, whereas the D chromosomes remained at univalent stage with 13 univalents. However, it is important to note that several cells showed the presence of multivalents, indicating the possibility of homeologous pairing between A and D genomes, even if these exchanges were difficult to detect after a long evolution in G. hirsutum (Page et al. 2016). In the third hybrid, the presence of >2 A genomes was shown by the morphological characteristics (leaf shape, flower size, and red spots on the corolla). Its genomic structure, A1A1AtDt, was confirmed by observation of metaphase in mitosis, revealing the presence of the nine 45S loci expected for this genomic structure. Additionally, the high frequency of multivalents was in agreement with the presence of three A genomes.
Our primary objective was to estimate the risk of cross-pollination between allotetraploid cultivated cotton (G. hirsutum) and wild diploid cotton (G. herbaceum), since, in the Makhathini flats area of South Africa, the diploid progenitor grows in proximity to the cultivated transgenic cotton fields. Most of published studies were concerned either with gene flow between transgenic and nontransgenic cultivated cotton in various areas of the world (Zhang et al. 2005; Van Deynze et al. 2005; Llewellyn et al. 2007; Heuberger et al. 2010; Loureiro et al. 2016), or between wild and cultivated tetraploid cotton (Wegier et al. 2011; Pereira et al. 2012). Our study on the hybridization rate between diploid and tetraploid cotton, showed that the three recovered hybrids produced no seeds, and that the viability of their pollen was very low. The single seed was produced after crossing a triploid hybrid, and G. hirsutum developed after hand-pollination on an emasculated flower of the triploid (data not shown). It is reasonable to conclude from the present study that the probability of obtaining offspring is very low. Moreover, only the species G. herbaceum var. africanum is endemic in South Africa (van Wyk and van Wyk 1997). Entomological investigations have shown that the beetle Astylus atromaculatus could be an unexpected but efficient pollinator on G. hirsutum in South Africa (Pierre and Hofs 2010). However, A. atromaculatus and domestic bees are not present on G. herbaceum (J. L. Hofs, personal communication). In contrast, Anthophoridae are common on both G. hirsutum and G. herbaceum and have a great capacity for carrying pollen, and may thus be a potential vector for pollen transfer between the two species (J. L. Hofs, personal communication). However, in the case of trials involving interspecific hybridization, reverting to a more fertile form required several generations of backcrosses with the cultivated species (Kammacher 1966; Mergeai 2003).
Our results have demonstrated the possibility of the formation of unreduced gametes. To our knowledge, this phenomenon as been poorly documented in cotton, while it is prevalent in diverse plant species, and considered to be an important mechanism for polyploidization (reviewed in Mason and Pires 2015). The formation of unreduced gametes in Gossypium interspecific hybrids has been reported in crosses between G. hirsutum and Australian K-genome diploids G. exiguum or G. nobile (Brubaker et al. 1999). Observations of meiosis in these hybrids suggested that some may have derived from unreduced gametes arising from dysfunctions during M2; hence, the duplication of the segregating chromosomes. Furthermore, Skovsted (1934), working on the production of hybrids between diploid Asiatic and tetraploid New World cotton, obtained a hybrid with 52 chromosomes, and hypothesized the production of 2n gametes in the diploid parent. Likewise, in cytogenetic studies of cotton cultivars grown in Iran, the occurrence of large pollen grains was assumed to be potential unreduced gametes (Sheidai 2008; Noormohammadi et al. 2012), although other methods are considered to be necessary to confirm the correlation between size and ploidy in pollen (Dewitte et al. 2012). For the same type of sterile hybrids (triploids 2n = 39, one tetraploid 2n = 52) obtained by crossing A-genome Asiatic cottons with tetraploid New World cottons, Amin (1940) reported that backcrossing to New World cottons gave occasional success in boll setting on the F1s. Such bolls contained very few seeds, and the plants developed from them had varying degrees of fertility. In this context, by crossing one of our triploid hybrid (H1) as female with G. hirsutum (25 flowers emasculated and hand pollinated with G. hirsutum pollen) we obtained a single large seed. Its germination gave rise to a plant with 2n = 65 chromosomes and a putative A1AtAtDt structure (data not shown). The structure may be the result of unreduced gametes from the triploid hybrid. This ability of Gossypium to produce such gametes from diploid parents and from F1 hybrids might point to an alternative explanation for the formation of the allotetraploid cultivated species, G. hirsutum. In fact, it has been hypothesized the formation of unreduced gametes offers the possibility to generate a triploid bridge, which can be at the origin of polyploid species (reviewed in Mason and Pires 2015). Interspecific hybridization has often been carried out to provide new sources of germplasm for incorporation into breeding programs (Chee et al. 2016). However, it is not possible to obtain fertile hybrids from crosses between cultivated tetraploid G. hirsutum and wild diploids species without treating the floral bud at the time of fertilization with growth substances in order to prevent boll abscission (Meyer 1974; Altman 1988). Brubaker et al. (1999) have shown that, in the best case, the natural crossing of diploid wild Australian species with tetraploid cotton cultivars resulted only in the production of sterile hybrids. However, fertility can be restored by doubling the number of chromosomes using colchicine to produce hexaploid plants (2n = 78 chromosomes). This approach has been widely used in interspecific hybridization to improve cotton plants, but such a doubling has never been observed occurring spontaneously in nature (Mergeai 2003, 2006). 2n gametes have already been exploited to create new cultivars at higher ploidy levels, but, more interestingly, they have also been used in various species to create a bridge for the transfer of desirable genes from wild diploid species into the cultivated polyploid gene pool in various species (reviewed in Dewitte et al. (2012)). This approach is particularly relevant for cultivated cotton, as the wild diploid species of Gossypium are rich in rare desirable attributes that are not available in the germplasm of cultivated species (Stewart 1995; Mergeai 2006). This methodology has been used to transfer genetic diversity from diploids through 2n gametes to polyploid crop varieties, as demonstrated for example in potato and alfalfa (Peloquin et al. 1999; Ramanna and Jacobsen 2003). The production of autopolyploids to overcome the difficulty of transferring desirable traits from wild or cultivated diploids to G. hirsutum was investigated by Mehetre et al. (2003). The use of unreduced gametes would facilitate the direct production of such AAAD hybrids either by direct crosses (H3 hybrid) or by crossing an AAD hybrid with G. hirsutum. Even though the production of unreduced gametes has rarely been described in cotton, various methods have been tested in other species to increase their occurrence (Brownfield and Kohler, 2011; Dewitte et al. 2012; Younis et al. 2014). New strategies (Ravi et al. 2008; Chan 2010; Ravi and Chan 2010) developed on the model plant Arabidopsis thaliana (reviewed in Crismani et al. 2013) are considered by the authors to be possibly applicable to crops.
The possibility of obtaining an interspecific hybrid between tetraploid cultivated and diploid wild cotton through fertilization with an unreduced gamete raises the question of its evolution in natural populations. We have shown that this hybrid produced no seeds and its pollen has very low viability. In conclusion, the likelihood of natural transfer of chromosomal material from cultivated cotton to the diploid G. herbaceum is very low; even though one hybrid with a particular genomic structure arose via an unreduced gamete of the diploid parent, it did not produce seeds after selfing. However, the assessment of the hybrids’ ability to produce a backcross progeny would require further experimentation, such as cultivating hybrids in the presence of G. herbaceum. Furthermore, the hybrid materials obtained in this study, with different genome dosages (triploids AAD and unreduced gamete-derived hybrid AAAD), could provide useful tools for the study of the expression of duplicated genes and the effects of allele dosage from A and D cotton genomes.
Acknowledgments
The authors offer sincere thanks to Jonathan Wendel for reading the manuscript and valuable exchanges about their work, and are thankful to Mark Tepfer and Pierre Hilson for critical reading of the manuscript. They acknowledge support for this project from the French Ministry for Education and Research/ ANR (Agence Nationale de la Recherche), grant MFD/AO OGM 0217. The Jean-Pierre Bourgin Institute (IJPB) benefits from the support of the LabEx Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS).
Footnotes
Communicating editor: K. M. Devos
Literature Cited
- Ahoton L., Lacape J. M., Baudoin J. P., Mergeai G., 2003. Introduction of Australian diploid cotton genetic variation into upland cotton. Crop Sci. 43: 1999–2005. [Google Scholar]
- Alexander M. P., 1969. Differential staining of aborted and nonaborded pollen. Stain Technol. 44: 117–122. [DOI] [PubMed] [Google Scholar]
- Altman D. W., 1988. Exogenous hormone applications at pollination for in vitro and in vivo production of cotton interspecific hybrids. Plant Cell Rep. 7: 257–261. [DOI] [PubMed] [Google Scholar]
- Amin K. C., 1940. Interspecific hybridization between Asiatic and new world cottons. Indian J. Agric. Sci. 10: 404–413. [Google Scholar]
- Barcaccia G., Tavoletti S., Mariani A., Veronesi F., 2003. Occurrence, inheritance and use of reproductive mutants in alfalfa improvement. Euphytica 133: 37–56. [Google Scholar]
- Beasley J. O., 1940. The production of polyploids in Gossypium. J. Hered. 31: 39–48. [Google Scholar]
- Beasley J. O., 1942. Meiotic chromosome behavior in species, species hybrids, haploids and induced polyploids of Gossypium. Genetics 27: 25–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownfield L., Kohler C., 2011. Unreduced gamete formation in plants: mechanisms and prospects. J. Exp. Bot. 62: 1659–1668. [DOI] [PubMed] [Google Scholar]
- Brubaker C. L., Brown A. H. D., Stewart J. M., Kilby M. J., Grace J. P., 1999. Production of fertile hybrid germplasm with diploid Australian Gossypium species for cotton improvement. Euphytica 108: 199–213. [Google Scholar]
- Carputo D., Frusciante L., Peloquin S. J., 2003. The role of 2n gametes and endosperm balance number in the origin and evolution of polyploids in the tuber-bearing solanums. Genetics 163: 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. W. L., 2010. Chromosome engineering: power tools for plant genetics. Trends Biotechnol. 28: 605–610. [DOI] [PubMed] [Google Scholar]
- Chee P. W., Paterson A. H., Udall J. A., Wendel J. F., 2016. Interspecific hybridization for upland cotton improvement, pp. 1–20 in Polyploidy and Interspecific Hybridization for Crop Improvement, edited by Mason A. S. CRC Press, Boca Raton, FL. [Google Scholar]
- Chèvre A.-M., Ammitzbøll H., Breckling B., Dietz-Pfeilstetter A., Eber F., et al. , 2004. A review on interspecific gene flow from oilseed rape to wild relatives, pp. 235–251 in Introgression from Genetically Modified Plants into wild relatives, edited by Den Nijs H. C. M., Bartsch D., Sweet J. CABI Publishing, Cambridge. [Google Scholar]
- Crismani W., Girard C., Mercier R., 2013. Tinkering with meiosis. J. Exp. Bot. 64: 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte A., Van Laere K., Van Huylenbroeck J., 2012. Use of 2n gametes in plant breeding. Plant Breed. 69: 59–86. [Google Scholar]
- Eber F., Letanneur J. C., Chèvre A.-M., 1997. Chromosome number of oilseed rape (Brassica napus L.) - wild radish (Raphanus raphanistrum) spontaneous hybrids and of their progeny estimated by flow cytometry. Cruciferae Newsletter 19: 17–18. [Google Scholar]
- Fryxell P. A., 1979. The Natural History of Cotton Tribe, Texas A&M University Press, College Station, TX. [Google Scholar]
- Fryxell P. A., Craven L. A., Stewart J. M., 1992. A revision of gossypium sect. Grandicalyx (Malvaceae), including the description of 6 new species. Syst. Bot. 17: 91–114. [Google Scholar]
- Gallagher J. P., Grover C. E., Rex K., Moran M., Wendel J. F., 2017. A new species of cotton from Wake Atoll, Gossypium stephensii (Malvaceae). Syst. Bot. 42: 115–123. [Google Scholar]
- Gan Y. M., Liu F., Peng R. H., Wang C. Y., Li S. H., et al. , 2012. Individual chromosome identification, chromosomal collinearity and genetic-physical integrated map in Gossypium darwinii and four D genome cotton species revealed by BAC-FISH. Genes Genet. Syst. 87: 233–241. [DOI] [PubMed] [Google Scholar]
- Gerlach W. L., Bedbrook J. R., 1979. Cloning and characterization of ribosomal RNA genes in wheat and barley. Nucleic Acids Res. 7: 1869–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach W. L., Dyer T. A., 1980. Sequence organisation of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res. 8: 4851–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover C. E., Gallagher J. P., Jareczek J. J., Page J. T., Udall J. A., et al. , 2015. Re-evaluating the phylogeny of allopolyploid Gossypium L. Mol. Phylogenet. Evol. 92: 45–52. [DOI] [PubMed] [Google Scholar]
- Hanson R. E., IslamFaridi M. N., Percival E. A., Crane C. F., Ji Y., et al. , 1996. Distribution of 5S and 18S–28S rDNA loci in a tetraploid cotton (Gossypium hirsutum L) and its putative diploid ancestors. Chromosoma 105: 55–61. [DOI] [PubMed] [Google Scholar]
- Heuberger S., Ellers-Kirk C., Tabashnik B. E., Carriere Y., 2010. Pollen- and seed-mediated transgene flow in commercial cotton seed production fields. PLoS One 5: e14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammacher P., 1966. Etudes des relations genetiques et caryologiques entre genomes voisins du genre Gossypium. Coton et Fibres Tropicales 21: 263–290. [Google Scholar]
- Lacape J. M., Dessauw D., Rajab M., Noyer J. L., Hau B., 2007. Microsatellite diversity in tetraploid Gossypium germplasm: assembling a highly informative genotyping set of cotton SSRs. Mol. Breed. 19: 45–58. [Google Scholar]
- Llewellyn D., Tyson C., Constable G., Duggan B., Beale S., et al. , 2007. Containment of regulated genetically modified cotton in the field. Agric. Ecosyst. Environ. 121: 419–429. [Google Scholar]
- Mason A. S., Pires J. C., 2015. Unreduced gametes: meiotic mishap or evolutionary mechanism? Trends Genet. 31: 5–10. [DOI] [PubMed] [Google Scholar]
- Mehetre S. S., Aher A. R., Gawande V. L., Patil V. R., Mokate A. S., 2003. Induced polyploidy in Gossypium: a tool to overcome interspecific incompatibility of cultivated tetraploid and diploid cottons. Curr. Sci. 84: 1510–1512. [Google Scholar]
- Mergeai, G., 2003 Forty years of genetic improvement of cotton through interspecific hybridisation at Gembloux Agricultural University: Achievement and prospects. in Proc. World Cotton Research Conference - 3. Cotton Production for the New Millennium, edited by A. Swanepoel. (Cape Town, South Africa: Agricultural Research Council, Institute for Industrial Crops, Pretoria), pp. 120–133. [Google Scholar]
- Mergeai G., 2006. Cotton improvement through interspecific hybridization. Cah. Agric. 15: 135–143. [Google Scholar]
- Meyer V. G., 1974. Interspecific cotton breeding. Econ. Bot. 28: 56–60. [Google Scholar]
- Noormohammadi Z., Sheidai M., Shojaei F., Jeshvaghani F. S., Alishah O., 2012. Cytogenetic analysis of mehr cotton cultivar and its crossing progenies: a search for unreduced pollen grains. Cytologia (Tokyo) 77: 107–112. [Google Scholar]
- Page J. T., Liechty Z. S., Alexander R. H., Clemons K., Hulse-Kemp A. M., et al. , 2016. DNA sequence evolution and rare homoeologous conversion in tetraploid cotton. PLoS Genet. 12: e1006206 (erratum: PLoS Genet. 12: e1006012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peloquin S. J., Boiteux L. S., Carputo D., 1999. Meiotic mutants in potato: valuable variants. Genetics 153: 1493–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G. S., Sousa R. L., Araujo R. L., Hoffmann L. V., Silva E. F., et al. , 2012. Selective fertilization in interspecific crosses of allotetraploid species of Gossypium. Botany-Botanique 90: 159–166. [Google Scholar]
- Pierre J., Hofs J. L., 2010. Astylus atromaculatus (Coleoptera: Melyridae): abundance and role in pollen dispersal in Bt and non-Bt cotton in South Africa. Environ. Entomol. 39: 1523–1531. [DOI] [PubMed] [Google Scholar]
- Ramanna M. S., Jacobsen E., 2003. Relevance of sexual polyploidization for crop improvement—a review. Euphytica 133: 3–18. [Google Scholar]
- Ravi M., Chan S. W., 2010. Haploid plants produced by centromere-mediated genome elimination. Nature 464: 615–618. [DOI] [PubMed] [Google Scholar]
- Ravi M., Marimuthu M. P. A., Siddiqi I., 2008. Gamete formation without meiosis in Arabidopsis. Nature 451: 1121–1124. [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S., Page J. T., Udall J. A., Sanders W. S., Peterson D. G., et al. , 2016. Independent domestication of two old world cotton species. Genome Biol. Evol. 8: 1940–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheidai M., 2008. Cytogenetic distinctiveness of sixty-six tetraploid cotton (Gossypium hirsutum L.) cultivars based on meiotic data. Acta Botanica Croatica 67: 209–220. [Google Scholar]
- Skovsted A., 1934. Cytological studies in Cotton. II. Two interspecific hybrids between Asiatic and new world cottons. J. Genet. 28: 407–424. [Google Scholar]
- Stephens S. G., 1942. Colchicine-produced polyploids in Gossypium I. And autotetraploid asiatic cotton and certain of its hybrids with wild diploid species. J. Genet. 44: 272–295. [Google Scholar]
- Stephens S. G., 1974. Geographic and taxonomic distribution of anthocyanin genes in new world cottons. J. Genet. 61: 128–141. [Google Scholar]
- Stewart, J., 1995 Potential for crop improvement with exotic germsplams and genetic engineering. In Challenging the Future - Proc. World. Cotton Research Conf. -1; Brisbane, Australia 14–17 February 1994. Constable GA and Forrester NW (Eds). CSIRO, Melbourne. pp 313–327 [Google Scholar]
- Van Deynze A. E., Sundstrom F. J., Bradford K. J., 2005. Pollen-mediated gene flow in California cotton depends on pollinator activity. Crop Sci. 45: 1565–1570. [Google Scholar]
- Van Deynze A. E., Hutmacher R. B., Bradford K. J., 2005. Gene flow between Gossypium hirsutum L. and Gossypium barbadense L. is asymmetric. Crop Sci. 51: 298–305. [Google Scholar]
- van Wyk B., van Wyk P., 1997. Field Guide to Trees of Southern Africa, Struik Publishers, Cape Town, South Africa. [Google Scholar]
- Warwick S. I., Simard M. J., Legere A., Beckie H. J., Braun L., et al. , 2003. Hybridization between transgenic Brassica napus L. and its wild relatives: Brassica rapa L., Raphanus raphanistrum L., Sinapis arvensis L., and Erucastrum gallicum (Willd.) OE Schulz. Theor. Appl. Genet. 107: 528–539. [DOI] [PubMed] [Google Scholar]
- Wegier A., Pineyro-Nelson A., Alarcon J., Galvez-Mariscal A., Alvarez-Buylla E. R., et al. 2011. Recent long-distance transgene flow into wild populations conforms to historical patterns of gene flow in cotton (Gossypium hirsutum) at its centre of origin Molecular Ecology 20: 4182–4194. [DOI] [PubMed] [Google Scholar]
- Wendel J. F., 1989. New world tetraploid cottons contain old-world cytoplasm. Proc. Natl. Acad. Sci. USA 86: 4132–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J. F., Cronn R. C., 2003. Polyploidy and the evolutionary history of cotton. Adv. Agron. 78(78): 139–186. [Google Scholar]
- Wendel J. F., Brubaker C. L., Seelanan T., 2010. The origin and evolution of Gossypium, pp. 1–18 in Physiology of Cotton, edited by Stewart J. M. D., Oosterhuis J. M., Heitholt J. J., Mauney J. R. Springer, Netherlands. [Google Scholar]
- Younis A., Hwang Y. J., Lim K. B., 2014. Exploitation of induced 2n-gametes for plant breeding. Plant Cell Rep. 33: 215–223. [DOI] [PubMed] [Google Scholar]
- Zhang B. H., Pan X. P., Guo T. L., Wang Q. L., Anderson T. A., 2005. Measuring gene flow in the cultivation of transgenic cotton (Gossypium hirsutum L.). Mol. Biotechnol. 31: 11–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.