Abstract
SNP arrays are enabling tools for high-resolution studies of the genetic basis of complex traits in farmed and wild animals. Oysters are of critical importance in many regions from both an ecological and economic perspective, and oyster aquaculture forms a key component of global food security. The aim of our study was to design a combined-species, medium density SNP array for Pacific oyster (Crassostrea gigas) and European flat oyster (Ostrea edulis), and to test the performance of this array on farmed and wild populations from multiple locations, with a focus on European populations. SNP discovery was carried out by whole-genome sequencing (WGS) of pooled genomic DNA samples from eight C. gigas populations, and restriction site-associated DNA sequencing (RAD-Seq) of 11 geographically diverse O. edulis populations. Nearly 12 million candidate SNPs were discovered and filtered based on several criteria, including preference for SNPs segregating in multiple populations and SNPs with monomorphic flanking regions. An Affymetrix Axiom Custom Array was created and tested on a diverse set of samples (n = 219) showing ∼27 K high quality SNPs for C. gigas and ∼11 K high quality SNPs for O. edulis segregating in these populations. A high proportion of SNPs were segregating in each of the populations, and the array was used to detect population structure and levels of linkage disequilibrium (LD). Further testing of the array on three C. gigas nuclear families (n = 165) revealed that the array can be used to clearly distinguish between both families based on identity-by-state (IBS) clustering parental assignment software. This medium density, combined-species array will be publicly available through Affymetrix, and will be applied for genome-wide association and evolutionary genetic studies, and for genomic selection in oyster breeding programs.
Keywords: Pacific oyster, flat oyster, single nucleotide polymorphism (SNP), array, aquaculture
Oyster farming is one of the most important aquaculture activities worldwide, providing a socioeconomic contribution to many coastal communities. Among the numerous farmed oyster species, the Pacific oyster (Crassostrea gigas) is one of the most widely cultivated, with a global annual production estimated at 583 K tons in 2015 (FAO 2017). Starting in the 1960s, C. gigas was successfully introduced from Japan to all continents for cultivation (Troost 2010) due to its high acclimation ability, rapid growth, and high production, and as an alternative to replace the flat oyster farms affected by persistent disease outbreaks (Pernet et al. 2016). Accordingly, the European flat oyster (Ostrea edulis), an endemic species to Europe, has suffered a decrease in global production from 30 K tons in 1960 to 3 K tons produced in 2014. O. edulis is now a target for conservation efforts to help restore native populations (Lallias et al. 2010), and is also a niche aquaculture product, particularly in Europe and the USA.
In the past decade, there has been increasing interest from researchers and industry in the development of genomic resources for oysters, mainly because of the economic and ecological importance of both C. gigas and O. edulis. The genomic toolbox for C. gigas includes a moderate number of genetic markers, such as microsatellites (Li et al. 2003; Sekino et al. 2003) and SNPs (Fleury et al. 2009; Sauvage et al. 2007; Wang et al. 2015). Low density linkage maps have been developed, containing both microsatellites and SNPs (Hedgecock et al. 2015; Hubert and Hedgecock 2004). In addition, quantitative trait loci (QTL) analyses have been carried out to identify genomic regions associated with desirable traits for aquaculture (Sauvage et al. 2010; Guo et al. 2012; Zhong et al. 2014). In addition, a reference genome sequence assembly is available for C. gigas (Zhang et al. 2012), although a number of putative assembly errors have been identified (Hedgecock et al. 2015). In contrast, genomic tools and resources are scarce for O. edulis, and only a limited number of markers, mostly microsatellites and amplified fragment length polymorphisms (AFLPs), have been utilized for the development of a linkage map (Lallias et al. 2007, 2009). Recently, the generation of genomic resources led to the development of a database containing genomic and transcriptome resources for O. edulis (Pardo et al. 2016; Vera et al. 2016).
SNPs have become the marker of choice in genetics research due to their high abundance, codominant mode of inheritance, ease of high-throughput discovery, and low cost of genotyping per locus. Next-generation sequencing technologies enable efficient identification of many thousands of SNPs in a single experiment using either WGS or reduced representation approaches such as RAD-Seq (Baird et al. 2008; Davey et al. 2011). While the medium density SNP arrays typically generated by direct genotyping-by-sequencing approaches have been widely applied in aquaculture species (Robledo et al. 2017), SNP arrays can offer a higher density genotyping platform that is simpler to use. SNP arrays have been developed for most terrestrial livestock species such as cattle, pig, and chicken (Matukumalli et al. 2009; Ramos et al. 2009; Kranis et al. 2013), and also for farmed finfish species such as Atlantic salmon, rainbow trout, catfish, and carp among others (Houston et al. 2014; Yáñez et al. 2016; Palti et al. 2015; Liu et al. 2014; Xu et al. 2014). These arrays have formed the basis of genome-wide association studies for traits of economic importance such as resistance to pathogens (Geng et al. 2015; Correa et al. 2015; Tsai et al. 2016) and the application of genomic selection in aquaculture breeding (Ødegård and Meuwissen 2014; Tsai et al. 2015, 2016; Vallejo et al. 2016).
For oyster species, low density SNP arrays for C. gigas and O. edulis have been developed, with 384 markers per species (Lapègue et al. 2014), and these have been applied for parentage assignment. In addition, a C. gigas-specific high density array was recently developed, which contains ∼134 K SNP markers shown to be polymorphic across populations sampled from China, Japan, Korea, and Canada (Qi et al. 2017). However, a medium density, combined-species platform is a worthy addition to the genomic toolbox for oysters because: (i) the performance of the higher density (133 K) array in farmed C. gigas populations from other global regions (e.g., Europe) is not known, (ii) medium density arrays are adequate for many genetics and breeding studies at substantially lower cost than high density arrays, and (iii) there is not yet a medium or high density genotyping platform for O. edulis. The major aim of the current study was to design and test a medium density, combined-species SNP array for two key oyster species, C. gigas and O. edulis, and to test the performance of the array on hatchery and wild populations from multiple locations, as well as nuclear families from pair-crosses.
Materials and Methods
Sample collection and sequencing
The DNA sequencing protocols for SNP discovery were tailored to the status of genomic tools available for the two species. Since C. gigas has a reference genome sequence (Zhang et al. 2012), a whole-genome resequencing approach was taken with reads subsequently aligned to the reference assembly as described below. There was no reference sequence available for O. edulis, so a RAD-Seq approach was taken since this is suitable for de novo assembly and discovery of SNPs within RAD loci (Baird et al. 2008).
Samples from eight C. gigas populations from different geographical locations (primarily from hatcheries in the UK and France) were obtained, each comprising 13–47 individuals (Table 1). These included a population of 16 samples from lines of oysters that had been selected for resistance to Oyster Herpes Virus by Ifremer (France). Genomic DNA from all individuals was extracted via the CTAB (cetyl trimethylammonium bromide) protocol described by Richards et al. (2013). Briefly, oyster tissue was incubated at 56° in lysis solution (3% CTAB, 100 mM Tris-HCl, pH 7.5, 25 mM EDTA, and 2 mM NaCl) with 0.2 mg/ml proteinase K and 5 μl of RNase (10 mg/ml). After lysis, a chloroform extraction was performed twice and three volumes of CTAB dilution solution were added (1% CTAB, 50 mM Tris-HCl, pH 7.5, and 10 mM EDTA, pH 8). The pellet was then washed in 0.4 M NaCl in TE, resuspended in 1.42 M NaCl in TE, and finally precipitated overnight in 1 ml ethanol (99%) at −4 C. Within each population, DNA samples were then pooled in equimolar concentrations, and these pools were prepared for WGS using the TruSeq Nano DNA Library Prep kit (Illumina, San Diego). Libraries were sequenced across five lanes of Illumina Hisequation 2500 to produce 125 bp paired end reads.
Table 1. Detail of populations sampled for sequencing and SNP discovery.
| C. gigas | O. edulis | ||||
|---|---|---|---|---|---|
| Population | Location (Lat, Long) | N | Population | Location (Lat, Long) | N |
| Guernsey, England | 49.497, −2.502 | 47 | Croatia | 42.855, 17.688 | 14 |
| Maldon, England | 51.724, 0.710 | 15 | Lough Foyle, Ireland | 55.130, −7.087 | 15 |
| Sea Salter, England | 51.378, 1.212 | 13 | Lake Grevelingen, The Netherlands | 51.709, 4.017 | 15 |
| Ifremer, France | n/a | 16 | Larne, Northern Ireland | 54.817, −5.751 | 14 |
| Hatchery 1 (Marinove), France | 46.987, −2.238 | 29 | Mersea, England | 51.776, 0.9646 | 15 |
| Hatchery 2 (SATMAR), France | 46.948, −2.052 | 26 | Baie de Quiberon, France | 47.548, −2.996 | 15 |
| Hatchery 3 (France Naissain), France | 47.514, −2.666 | 29 | Rossmore (Cork), Ireland | 51.883, −8. 247 | 15 |
| Hatchery 4 (Novostrea), France | 46.954, −2.044 | 28 | Sveio, Norway | 59.519, 5.227 | 15 |
| Swansea Bay, England | 51.604, −3.981 | 15 | |||
| Tralee, Ireland | 52.316, −10. 028 | 13 | |||
| Damariscotta, Maine | 44.028, −69.534 | 14 | |||
Lat, latitude; Long, longitude.
Samples from 11 O. edulis wild populations from diverse geographical locations were obtained (Table 1). Each population sample comprised 13–15 individuals, and genomic DNA had previously been extracted from these samples using a phenol-chloroform method. Equimolar pools of genomic DNA were generated for each population and the pooled genomic DNA was digested using the endonuclease PstI. Standard RAD libraries were constructed in three replicates following the standard protocol described by Baird et al. (2008). Equimolar amounts of all libraries were combined and sequenced on a single Illumina Hisequation 2500 lane to produce 125 bp paired end reads.
SNP identification and filtering
C. gigas WGS reads were aligned to the C. gigas genome (GCA_000297895.1) using BWA-mem (v0.7.10) (Li and Durbin 2009) with the -M flag. Potential duplicated reads originating from PCR were then removed using Picard Tools (v1.69) MarkDuplicates and SAMtools (v1.2) (Li et al. 2009). Local realignment around indels was performed using the GATK (v3.4.0) (McKenna et al. 2010) and alignments with a quality phred score > 20 were retained. SNP calling was performed using PoPoolation2 (Kofler et al. 2011), filtering to discard bases with a call quality phred score < 30.
O. edulis RAD-Seq reads were trimmed with Cutadapt (v1.7.1) (Martin 2011). Data from each of the three replicates described above were combined. Read 1 reads were clustered using ustacks (v1.30) with the parameters “-m 2 -M 5 -H,” followed by cstacks (Catchen et al. 2013) with the parameter “-n 2,” to create consensus sequences for each locus. RAD loci absent from ≥8 of the 11 pooled samples were discarded. Read 1 trimmed reads from each of the samples were then aligned to the set of RAD consensus sequences using BWA (v.0.7.9a) (Li and Durbin 2009) (step 1). Reads mapping to each separate consensus sequence were then identified, and the corresponding read 2 sequences extracted from the trimmed data. These read 2 sequences for each locus were then assembled using IBDA-UD (Peng et al. 2012) (step 2). The read 1 consensus sequences and the associated assembled read 2 sequences for each locus were merged using flash (v1.2.2) (Magoč and Salzberg 2011). For SNP discovery, the trimmed sequences corresponding to each locus were then mapped to the merged consensus sequence using smalt (v0.7.6). Duplicate reads were marked using Picard tools (v1.115) and realignments around indels performed using GATK indel realigner (v3.4.0) (McKenna et al. 2010).
SNPs were identified and genotyped using PoPoolation2 and SAMtools (v1.3) pileup. Reads with a mapping quality phred score < 20 and bases with a call quality phred score < 20 were discarded.
SNP selection for Axiom array design
A list of candidate SNPs from both species (containing 1,691,005 and 117,235 priority SNPs from C. gigas and O. edulis, respectively) was provided to Affymetrix as 71-mer nucleotide sequences from the forward strand, with the alleles at the target SNP highlighted at position 36. A “p-convert” value (representing the probability of a given SNP converting to a reliable SNP assay on the Axiom array system) was computed by Affymetrix for each submitted SNP sequence. Probes are assessed for each SNP in both the forward and reverse direction, in return each strand is designated as “recommended,” “neutral,” or “not recommended” based on p-convert values.
The list of recommended markers (1,316,870 SNPs for C. gigas and O. edulis combined) was much greater than the total capacity of the Axiom MyDesign custom array. Therefore, additional filtering steps were carried out. For C. gigas, starting from the 1,216,467 Affymetrix-recommended SNPs, those with evidence for a 20 bp flanking monomorphic region covered by at least 36 reads from each pooled sample were retained (n = 186,948). For O. edulis, the Affymetrix-recommended SNPs (n = 100,403) were filtered so that each RAD locus contained a maximum of one SNP. When a RAD locus had multiple recommended SNPs, only the best SNP (based on the p-convert scores) was included (resulting in 59,976 candidate SNPs). Subsequently, to filter the SNPs to the required number for the array, SNPs for both species were selected according to the following additional filtering criteria: (i) highest p-convert values, (ii) even distribution across the reference genome (with at least 1000 bp distance between pairs of SNPs for C. gigas), and (iii) preference for those with a positive hit (minimum e-value 10E−4) against the BLASTx NCBI NR database or against the C. gigas genome (for O. edulis). In addition, most A/T and C/G SNP transversions were discarded since these require double the space on the Affymetrix Axiom array platform. Additionally, 463 SNPs identified and validated by Hedgecock et al. (2015) passed the SNP filtering and scoring process and were included in the final array design.
SNP array validation
A plate of 384 individual genomic DNA samples (274 C. gigas and 110 O. edulis) was sent to Edinburgh Genomics (Edinburgh, UK) for genotyping using the array. Of these 384 samples, 219 were used for testing and validating the array’s performance and quantifying the number of segregating SNPs in the various sampled populations. These included 109 C. gigas samples of individuals of unknown relatedness from eight populations (the same eight populations used for SNP discovery, plus an additional set of 28 broodstock oysters from Guernsey Sea Farms (Guernsey, UK). The validation samples also included 110 O. edulis samples corresponding to the 11 population samples used for SNP discovery (Table 1), with n = 10 from each population. The remaining 165 samples were offspring of three nuclear families derived from parents from Guernsey Sea Farms, reared at the Centre for Environment, Fisheries and Aquaculture Science (Cefas, UK). These were analyzed separately to test parentage assignment, genetic structure, and within-family linkage LD levels (see below).
Raw data containing the results of the intensity calculations (CEL files) was imported into the Axiom Analysis Suite (v2.0.035, Affymetrix) for quality control analysis and genotype calling. Samples with a dish quality control (DQC) value > 0.82 and QC call rate > 0.97 threshold (following the “Best Practices Workflow” recommended by Affymetrix), were considered to have passed the quality control assessment. The quality control analysis classifies the SNPs into categories according to their clustering performance with respect to various Axiom-generated quality control criteria: (i) “polymorphic high resolution” where the SNP passes all QC, (ii) “monomorphic high resolution” where the SNP passes all QC except the presence of a minor allele in two or more samples, (iii) “call rate below threshold” where genotype call rate is < 97%, (iv) “no minor homozygote” where the SNP passes all QC but only two clusters are observed, (v) “off-target variant” where atypical cluster properties arise from variants in the SNP flanking region, and (vi) “other” where the SNP does not fall into any of the previous categories. For further analyses, only SNPs from categories (i) and (iv) were included and classified as “good quality,” as they are most likely to be reliable and informative SNPs.
Descriptive statistics and family assignment
Calculations of minor allele frequencies (MAF), levels of heterozygosity, discriminant analysis of principal components (DAPC), LD, and IBS followed by multi-dimensional scaling (MDS) were carried out using Plink (Purcell et al. 2007), adegenet 1.3-1 package in R (Jombart and Ahmed 2011), and Genepop (Rousset 2008). Family assignment for the C. gigas families was performed using Cervus 3.07 (Kalinowski et al. 2007). Cervus assigns offspring to their parent pairs based on the pair-wise likelihood comparison approach generating locus-by-locus likelihood scores for each candidate parent for each offspring, and assigns parentage to a candidate parent with the highest LOD score.
Data availability
The Illumina sequencing data for the pooled C. gigas and O. edulis samples have been deposited into the European nucleotide archive under accession number PRJEB20253 (http://www.ebi.ac.uk/ena/data/view/PRJEB20253). The details of the SNP markers on the array are given in (Supplemental Material, File S1). O. edulis markers with significant alignment to the C. gigas genome (e-value 1E−4) are given in File S2.
Results and Discussion
Sequencing and SNP selection
To discover and prioritize SNPs for inclusion on the combined-species oyster SNP array, species-specific DNA sequencing, SNP discovery, and filtering strategies were followed. For C. gigas, WGS data aligned to the oyster genome identified 12.4 million putative SNPs across all populations. The 1,216,467 putative SNPs that passed the Affymetrix evaluation were subsequently filtered using the criteria described above to 40,625 putative SNPs that were submitted for the final Axiom MyDesign array. For O. edulis, 588,266 putative SNPs were identified, of which 100,403 putative SNPs were recommended at least for one strand by Affymetrix. Further filtering based on the criteria described above reduced the set to 19,215 putative SNPs that were submitted for array design and production.
The final array contained 40,625 putative SNPs from C. gigas and 14,950 putative SNPs from O. edulis to give a total of 55,575 putative SNPs assayed by a total of 111,360 probes. There were a greater number of C. gigas SNPs placed on the array than O. edulis due to the anticipated greater future use of the array for genome-wide association studies and genomic prediction for economically important traits in breeding programs in this species. This includes an ongoing project to study host resistance to Oyster Herpes Virus based on genotyping samples collected from a large challenge experiment on oysters derived from Guernsey Sea Farm stocks. Nonetheless, it is anticipated that the ∼15 K putative O. edulis SNPs will be widely applied for population and conservation genetics in future studies of this species.
Evaluation of the SNP array in C. gigas and O. edulis
The oyster array was evaluated in C. gigas by analyzing the “validation populations” of 109 samples corresponding to eight distinct populations from France and the UK (Table 2). All but one sample passed the DQC and genotype call rate ≥97% threshold. The classification of SNPs according to their quality showed that 68.2% (n = 27,697) had probes classified as good quality (either “Poly High Resolution” or “No Minor Hom”), which is similar to the percentage of informative markers obtained by the recently published C. gigas 134 K array (Qi et al. 2017). The MAF of these good quality SNPs (MAF > 0) in the combined 108 samples varied between 0.005 and 0.5 with a median of 0.18 (Table 2). From the 110 O. edulis samples genotyped (Table 3), two samples failed the DQC and genotype call rate ≥97% threshold, resulting in genotypes for 108 samples. A total of 74.6% of SNPs (n = 11,151) were classified as good quality as described above. The MAF of these good quality SNPs (combining all the 108 samples and SNPs with a MAF > 0) also varied between 0.005 and 0.5 with a median of 0.21 (Table 3).
Table 2. Descriptive population genetic estimates for the sampled C. gigas populations included in the validation of the array.
| MAF > 0 | |||||
|---|---|---|---|---|---|
| Sample N | # SNPs | Average MAF | Ho | He | |
| UK (combined)a | 56 | 27,313 | 0.186 | 0.294 | 0.298 |
| GSF + parents | 38 | 26,549 | 0.19 | 0.308 | 0.304 |
| Maldon | 9 | 22,079 | 0.216 | 0.308 | 0.303 |
| Sea Salter | 9 | 22,821 | 0.214 | 0.317 | 0.302 |
| Average within UK populationsb | 23,816 | 0.207 | 0.311 | 0.303 | |
| France (combined)a | 52 | 26,891 | 0.182 | 0.240 | 0.254 |
| Ifremer | 13 | 23,010 | 0.203 | 0.312 | 0.328 |
| Hatchery 1 | 10 | 21,479 | 0.217 | 0.321 | 0.303 |
| Hatchery 2 | 10 | 20,141 | 0.221 | 0.322 | 0.307 |
| Hatchery 3 | 10 | 21,730 | 0.215 | 0.302 | 0.302 |
| Hatchery 4 | 9 | 22,052 | 0.214 | 0.317 | 0.301 |
| Average within French populationsb | 21,682 | 0.214 | 0.315 | 0.308 | |
| All populations (combined)a | 108 | 27,697 | 0.182 | 0.268 | 0.283 |
MAF, minor allele frequency; #, number; SNPs, single nucleotide polymorphisms; Ho, level of genetic variability in terms of observed heterozygosity; He, level of genetic variability in terms of expected heterozygosity; GSF, Guernsey Sea Farm.
Values were obtained by the analysis of the combined dataset, not the average of the individual populations.
Values represent the within-population average.
Table 3. Descriptive population genetic estimates for the sampled O. edulis populations included in the validation of the array.
| MAF > 0 | |||||
|---|---|---|---|---|---|
| Sample N | # SNPs | Average MAF | Ho | He | |
| Croatia | 9 | 8,474 | 0.234 | 0.323 | 0.320 |
| Foyle_IRL | 10 | 10,013 | 0.224 | 0.319 | 0.311 |
| Grevelingen_NLD | 10 | 9,946 | 0.224 | 0.319 | 0.310 |
| Larne_NIRL | 10 | 8,927 | 0.231 | 0.354 | 0.316 |
| Mersea_UK | 10 | 9,980 | 0.224 | 0.318 | 0.310 |
| Quiberon_FR | 10 | 9,973 | 0.226 | 0.315 | 0.312 |
| Rossmore_IRL | 10 | 9,846 | 0.228 | 0.327 | 0.314 |
| Sveio_NOR | 10 | 9,118 | 0.226 | 0.322 | 0.313 |
| Swansea_UK | 9 | 9,696 | 0.224 | 0.319 | 0.311 |
| Tralee_IRL | 10 | 9,980 | 0.219 | 0.317 | 0.306 |
| Maine_USA | 10 | 9,614 | 0.221 | 0.317 | 0.305 |
| Average within populationa | 9,597 | 0.225 | 0.323 | 0.312 | |
| All populations (combined)b | 108 | 11,151 | 0.210 | 0.292 | 0.311 |
MAF, minor allele frequency; #, number; SNPs, single nucleotide polymorphisms; Ho, level of genetic variability in terms of observed heterozygosity; He, level of genetic variability in terms of expected heterozygosity.
Values represent the within-population average.
Values were obtained by the analysis of the combined dataset, not the average of the individual populations.
Within-population segregation of SNPs:
The segregation of the SNPs was evaluated within each of the eight genotyped C. gigas population samples. From the 27,697 high quality SNPs defined across all population samples, the majority of SNPs (MAF > 0) were segregating within each of the populations (Figure S1), with an average of 22,486 SNPs segregating within each population, ranging from 20,141 (Hatchery 2) to 26,549 (Guernsey) (Table 2). Among the UK populations (sampled from Guernsey, Maldon, and Sea Salter), 19,613 SNPs were shared, while Guernsey had the highest number of exclusive SNPs (n = 2373) (Figure S2). This is likely to be due to the fact that the Guernsey population was the most highly represented within the sequenced populations used for SNP discovery (Table 1) and the validation samples (Table 2), giving a greater chance of detecting rare minor alleles. Among all the five French populations, 13,855 SNPs were shared, with few SNPs segregating exclusively in particular populations (Figure S3). Finally, 11,997 common SNPs were segregating in all the eight populations from both France and the UK (Figure S4). The average MAF (for markers showing a MAF > 0) was 0.207 across all UK populations, while it was 0.214 across all French populations. Analysis of the distribution of MAF values for polymorphic SNPs (MAF > 0) showed that the highest numbers of SNPs are located within a MAF value range between 0.01 and 0.2 in all populations, decreasing in frequency when the MAF approaches 0.5 (Figure S5). A similar situation was observed by Lapègue et al. (2014), who found a high proportion of low MAF SNPs within C. gigas populations. Based on an additional test of the array on a small number of Australian C. gigas samples (data not shown), the number of segregating SNPs was similar, indicating that the array is likely to perform comparably for geographically diverse populations.
From the 11,151 high quality SNPs segregating in the O. edulis populations, the average number of SNPs segregating (MAF > 0) in each population was 9597. The samples from Croatia showed the lowest number of segregating SNPs (n = 8474), while those from Foyle (Ireland) showed the highest (n = 10,013) (see Figure S6 and Table 3). A total of 4912 SNPs were shared between all (11) populations, with no particular population showing a high number of unique segregating SNPs. The average MAF value across the populations was 0.225, with Croatia showing the highest value of 0.234. Analysis of the distribution of MAF values for polymorphic SNPs (MAF > 0) showed that most populations have a large number of SNPs within a MAF value range between 0.05 and 0.2, with the exception of Croatia and Swansea, which show a greater number of SNPs with a MAF higher than 0.1 (Figure S7).
The levels of genetic variability in terms of observed (Ho) and expected (He) heterozygosity (according to HWE) showed that most populations (C. gigas and O. edulis) had higher observed levels of heterozygosity than expected. Overall, no strong evidence of heterozygous deficiency was detected, in contrast to some previous studies that have described heterozygous deficiency in oysters and bivalves in general, albeit typically using a much lower number of microsatellites, SNPs, and allozymes (Appleyard and Ward 2006; English et al. 2000; Li et al. 2003; Sekino et al. 2003; Lapègue et al. 2014; Yu and Li 2007; Sobolewska and Beaumont 2005; Vercaemer et al. 2006). This discrepancy may be due to the fact that genome-wide SNP markers were used in the current study at a density not previously tested. In a larger-scale SNP assay-based evaluation of the bivalve mollusc Chlamys farreri, no evidence for heterozygote deficiency was detected (Jiao et al. 2014). It is also possible that the strict filtering process led to SNPs on the array being enriched for stable genomic regions with lower levels of variation, while genomic regions with higher variability (and potentially more prone to null alleles) might have been discarded.
Assessing population structure using IBS:
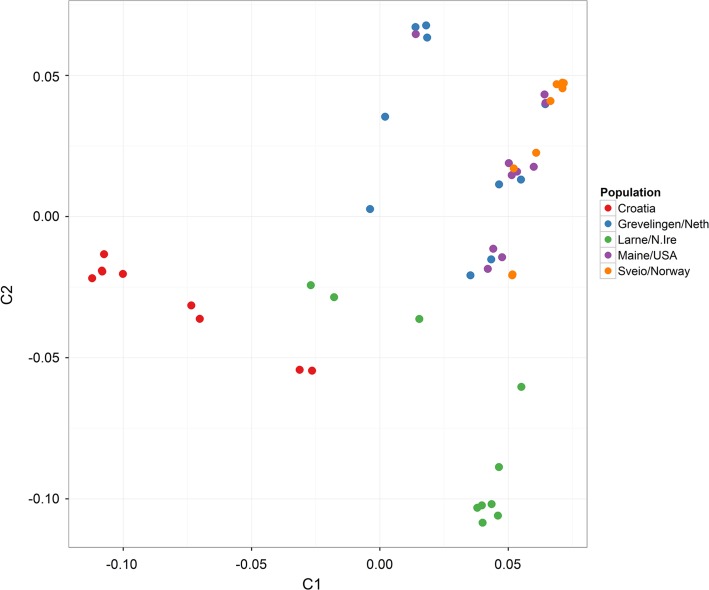
The overall genetic similarity of any two samples can be evaluated by calculating average measures of IBS of the marker loci, which was then summarized using MDS to give indications of population (sub)structure (IBS clustering was also confirmed by DAPC analysis, data not shown). There was some evidence of C. gigas samples clustering according to their hatchery origin, and French hatchery populations tended to cluster separately to UK hatchery populations (Figure S8). The O. edulis samples were typically from “wild” stocks from more diverse geographical locations than for the C. gigas samples (Figure S9 and Figure S10). Accordingly, certain populations did show evidence of genetic differentiation, notably Croatia, Larne (Northern Ireland), and Sveio (Norway), which are geographical outgroups (Figure 1 and Figure S10). Our results show evidence of a strong genetic similarity between Maine, Sveio (Norway), and Grevelingen (Netherlands) populations. Similarly, the origin of the Maine population has been linked to the Netherlands’ (Loosanoff 1955; Vercaemer et al. 2006), Netherlands populations have been linked to Denmark’s (Vera et al. 2016), and the genetic similarity between the Maine, Norway, Denmark, and Netherlands samples has also been observed using microsatellite markers (M. McCullough, personal communication). A lack of population structure according to geographical original was observed in the other O. edulis population samples tested, for example the majority of samples from the coast of the UK and Ireland (Figure S9). This is consistent with existing evidence that suggests that marine organisms with larval stages (such as bivalves) often show low genetic differentiation (Li et al. 2015; Shabtay et al. 2014; Rohfritsch et al. 2013; Giantsis et al. 2014), with temporal factors rather than geographical factors often playing the major role in population structure. It is also possible that historical stock translocations might have also played an important role in the lack of genetic structure and admixture of the O. edulis populations (Bromley et al. 2016).
Figure 1.
Identity-by-state clustering of selected O. edulis populations. Neth, The Netherlands; N.Ire, Northern Ireland.
Evaluation of the SNP array in pair crosses of C. gigas:
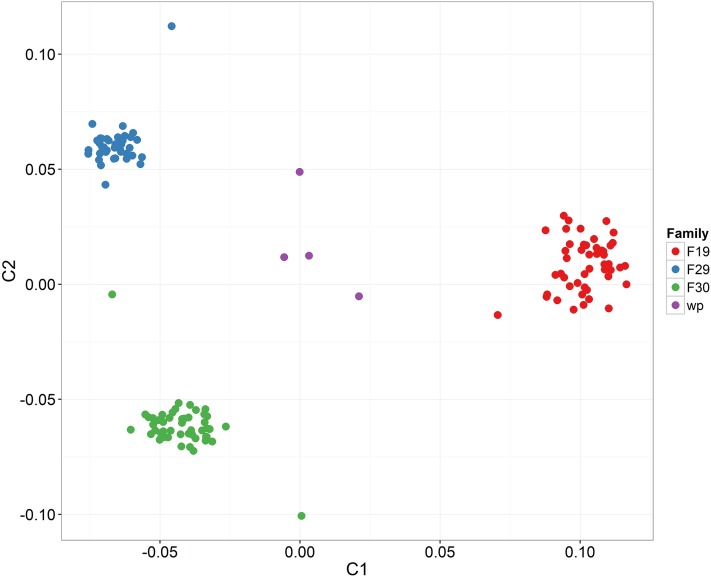
Three pair crosses between Guernsey Sea Farm parents were created, reared separately, and genotyped using the SNP array. Two of these nuclear families were half-siblings sharing a dam (F29 and F30). A total of 165 samples (161 offspring and their five parents) were genotyped. These families were analyzed separately from the population samples used to validate the array described above. In part, this was due to the difficulty in obtaining high quality genomic DNA from the juvenile oysters. From the 165 samples, 139 passed the DQC and genotype call rate ≥97% threshold, resulting in a total of 25,629 SNPs that were classified as good quality in these families. The vast majority of SNPs showed stable Mendelian inheritance in all samples, although there was an average of 395 SNPs (∼2% of total informative SNPs) with evidence of a Mendelian error per individual.
Since the offspring from each nuclear family were physically tracked throughout the experiment, such that their family structure was known a priori, the utility of the SNP array to differentiate between families was assessed using IBS clustering with MDS scaling. The MDS scaling plot based on IBS clustering clearly shows a clear separate cluster for each of the families, as shown in Figure 2. Interestingly, the clustering and separation of the three nuclear families was more obvious than for the population samples, even for populations from very distant geographical locations. Four individuals were distant to any of the family clusters, which may suggest incorrect pedigree assignment according to the physical animal tracking. Family assignment successfully assigned all the individuals to their correspondent parents using 3000 randomly chosen SNPs, and confirmed that the four aforementioned individuals were not members of any of these three families. Microsatellites and SNP panels for parentage assignment have been described previously for oysters (Wang et al. 2010; Li et al. 2010; Lapègue et al. 2014; Jin et al. 2014). However, the successful parentage assignment in these physically tracked nuclear families, and the clear IBS-based differentiation of these families, bodes well for the utility of this SNP array for high resolution genetic mapping studies and selective breeding programs for oysters.
Figure 2.
Identity-by-state-based clustering of the three nuclear C. gigas families. Samples in purple (wrong pedigree “wp”) were not assigned to any of the three families.
Distribution of SNPs in the pacific oyster genome:
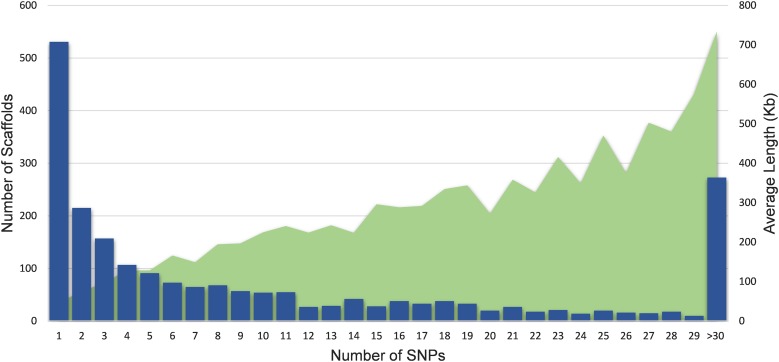
To assess the distribution of SNPs in the C. gigas genome (Zhang et al. 2012), SNPs were annotated according to the publicly available Ensembl oyster genome assembly (NCBI accession number: GCA_000297895.1). The oyster genome contains 7658 scaffolds (N50 = 401,585), 30,459 contigs (N50 = 31,239), and a total of ∼558 Mb of assembled sequence. All 27,697 SNPs are mapped to the oyster genome according to BLAST alignment using their flanking region(s), with at least one SNP on 2007 of the scaffolds, which in total covered 501 Mb (89.6% of the total assembled genome sequence). The number of SNPs per scaffold was positively associated with scaffold length (Figure 3), with approximately one-fifth of the scaffolds containing only one SNP. Additionally, harnessing the publicly-available oyster genome annotation (GCA_000297895.133), the SNPs on the array were grouped into putative positional and functional categories using SNPeff (Cingolani et al. 2012). A total of 14.6, 13.1, 18.7, 17.6, and 2.8% of the SNPs were located in intergenic, intron, downstream, upstream, and exon regions, respectively. The remaining SNPs (33%) were identified as transcript, splice site donor, splice site acceptor, and splice site region.
Figure 3.
Distribution of SNPs on the C. gigas genome. Number of scaffolds containing SNPs (primary axis) and the average length of the scaffolds holding an increasing number of SNPs (secondary axis). SNP, single nucleotide polymorphism.
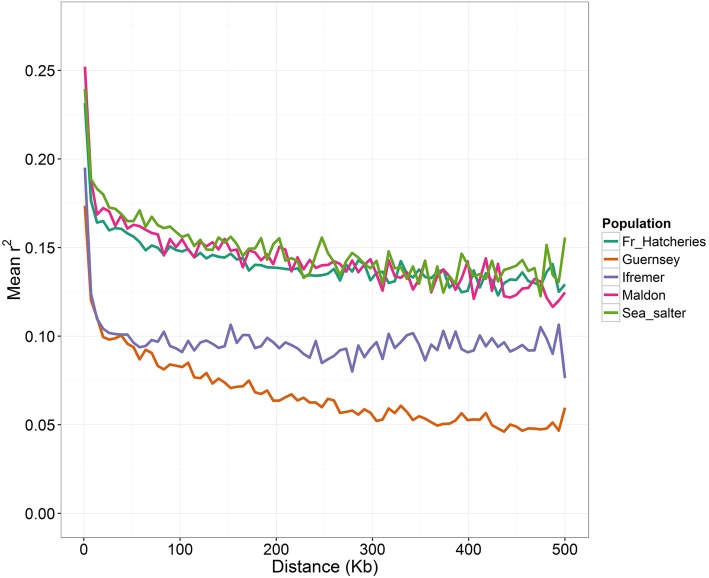
The extent of LD between SNP pairs was assessed relative to their physical distance for the C. gigas populations. Pairwise r2 was calculated using polymorphic SNPs with MAF ≥ 0.05 as shown in Table 2. The mean r2 was calculated for every kilobase and covering up to 500 kb, according to the physical distance on the oyster genome assembly, as shown in Figure 4. In general, low levels of LD with slow decay with increasing physical distance were observed. The Guernsey and Ifremer populations had lower levels of LD than the other populations. Although these LD levels are low compared to other aquaculture species, such as carp or tilapia (Hong Xia et al. 2015; Xu et al. 2014), they are in accordance with recent reports describing low levels and short extents of LD in wild C. gigas populations (Zhong et al. 2017). Moreover, differences in LD levels between populations can be related to the divergence of these populations and the number of generations for which they have been bred in isolation, as observed in cattle (de Roos et al. 2008).
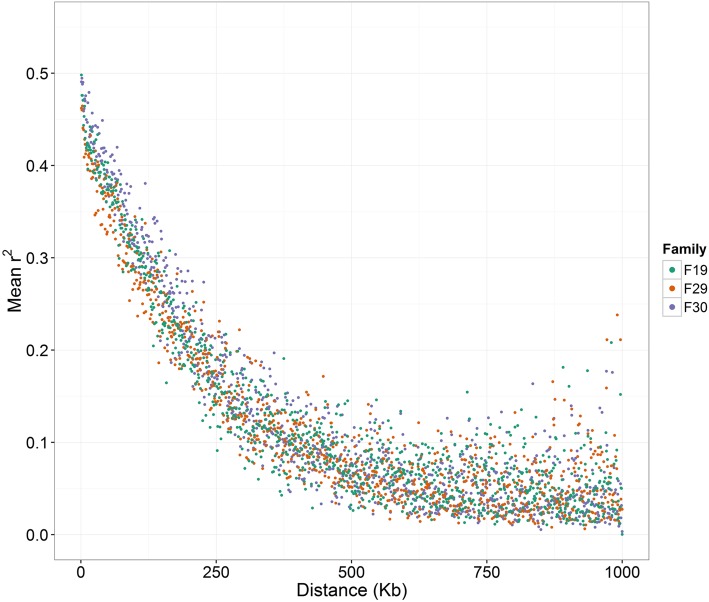
Figure 4.
Decay of linkage disequilibrium (LD) with physical distance between markers among all the sampled C. gigas populations. Fr, France.
There was a higher extent and slower decay of LD in the three nuclear families, and LD levels were substantially higher than those observed in the (presumably unrelated) validation populations, as would be expected (Figure 4 and Figure 5). A lower effective population size (Ne) brings higher levels of kinship between individuals and therefore a higher extent of LD (Sved 1971; Falconer and Mackay 1996).
Figure 5.
Decay of linkage disequilibrium (LD) among the three C. gigas families.
Conclusions
This article describes the development and analysis of a high density SNP array for two oyster species. A very large database of SNP markers was developed for both C. gigas and O. edulis, using WGA and RAD-Seq, respectively. Following extensive filtering, SNP assays for these two oyster species were combined on the array with 40,625 high quality SNPs for C. gigas and 14,950 for O. edulis. Testing of the array on genomic DNA samples from diverse locations revealed that the array contains a high number of SNPs that are shared between populations, and that the array can be applied to detect population and family structure. This oyster SNP array will be publicly available and will facilitate the study of important economic and ecological traits for these two oyster species, with possible applications for genomic selection, QTL mapping, evolutionary genetics, and conservation programs.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.041780/-/DC1.
Acknowledgments
The authors acknowledge Pierrick Haffray, Anastasia Bestin, Florian Enez, and Anne-Sophie Tyran from the Syndicat des Selectionneurs Avicoles and Aquacoles, France for coordinating the provision of the French hatchery samples used for single nucleotide polymorphism (SNP) discovery and genotyping; Lucie Buockellyoen from Marinove, Emilie Vetois from Satmar, Adeline Lange from France Naissain, and Fiz Dacosta from Novostrea for agreeing to provide samples; Lionel Degremont from Ifremer, France for coordinating the provision of the Ifremer selection line samples; and Dennis Hedgecock from the University of Southern California for providing details of validated SNPs to include on the array. P.A.P. and M.M.C. were supported by funding from the European Union (EU) INTERREG IVA Program (project 2859 “IBIS”) managed by the Special EU Programs Body. The authors acknowledge funding for this work under the Biotechnology and Biological Sciences Research Council (BBSRC)-Natural Environment Research Council UK Aquaculture Initiative (grant numbers: BB/M026140/1 and NE/P010695/1), via BBSRC Institute Strategic Funding grants to The Roslin Institute (BB/J004235/1, BB/J004324/1, and BB/J004243/1), and via the Centre for Environment, Fisheries and Aquaculture Science (Cefas) Seedcorn fund.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Appleyard S. A., Ward R. D., 2006. Genetic diversity and effective population size in mass selection lines of Pacific oyster (Crassostrea gigas). Aquaculture 254(1–4): 148–159. [Google Scholar]
- Baird N. A., Etter P. D., Atwood T. S., Currey M. C., Shiver A. L., et al. , 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3(10): e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromley C., McGonigle C., Ashton E. C., Roberts D., 2016. Bad moves: pros and cons of moving oysters – a case study of global translocations of Ostrea edulis Linnaeus, 1758 (Mollusca: Bivalvia). Ocean Coast. Manage. 122: 103–115. [Google Scholar]
- Catchen J., Hohenlohe P. A., Bassham S., Amores A., Cresko W. A., 2013. Stacks: an analysis tool set for population genomics. Mol. Ecol. 22(11): 3124–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 6(2): 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa K., Lhorente J. P., López M. E., Bassini L., Naswa S., et al. , 2015. Genome-wide association analysis reveals loci associated with resistance against Piscirickettsia salmonis in two Atlantic salmon (Salmo salar L.) chromosomes. BMC Genomics 16(1): 854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey J. W., Hohenlohe P. A., Etter P. D., Boone J. Q., Catchen J. M., et al. , 2011. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 12(7): 499–510. [DOI] [PubMed] [Google Scholar]
- de Roos A. P. W., Hayes B. J., Spelman R. J., Goddard M. E., 2008. Linkage disequilibrium and persistence of phase in Holstein–Friesian, Jersey and Angus cattle. Genetics 179: 1503–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English L. J., Maguire G. B., Ward R. D., 2000. Genetic variation of wild and hatchery populations of the Pacific oyster, Crassostrea gigas (Thunberg). Aust. Aquacult. 187(3–4): 283–298. [Google Scholar]
- Falconer D., Mackay T., 1996. Introduction to Quantitative Genetics, Pearson, Harlow, UK. [Google Scholar]
- FAO, 2017 Global aquaculture production: 1950–2015. Available at: http://www.fao.org/fishery/statistics/global-aquaculture-production/query/en. Accessed date: May 10, 2017
- Fleury E., Huvet A., Lelong C., de Lorgeril J., Boulo V., et al. , 2009. Generation and analysis of a 29,745 unique expressed sequence tags from the pacific oyster (Crassostrea gigas) assembled into a publicly accessible database: the GigasDatabase. BMC Genomics 10(1): 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X., Sha J., Liu S., Bao L., Zhang J., et al. , 2015. A genome-wide association study in catfish reveals the presence of functional hubs of related genes within QTLs for columnaris disease resistance. BMC Genomics 16(1): 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giantsis I. A., Mucci N., Randi E., Abatzopoulos T. J., Apostolidis A. P., 2014. Microsatellite variation of mussels (Mytilus galloprovincialis) in central and eastern Mediterranean: genetic panmixia in the Aegean and the Ionian seas. J. Mar. Biol. Assoc. U. K. 94(4): 797–809. [Google Scholar]
- Guo X., Li Q., Wang Q. Z., Kong L. F., 2012. Genetic mapping and QTL analysis of growth-related traits in the Pacific oyster. Mar. Biotechnol. (NY) 14(2): 218–226. [DOI] [PubMed] [Google Scholar]
- Hedgecock D., Shin G., Gracey A. Y., Den Berg D. V., Samanta M. P., 2015. Second-generation linkage maps for the Pacific oyster Crassostrea gigas reveal errors in assembly of genome scaffolds. G3 (Bethesda) 5(10): 2007–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Xia J., Bai Z., Meng Z., Zhang Y., Wang L., et al. , 2015. Signatures of selection in tilapia revealed by whole genome resequencing. Sci. Rep. 5: 14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston R. D., Taggart J. B., Cézard T., Bekaert M., Lowe N. R., et al. , 2014. Development and validation of a high density SNP genotyping array for Atlantic salmon (Salmo salar). BMC Genomics 15(1): 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert S., Hedgecock D., 2004. Linkage maps of microsatellite DNA markers for the Pacific oyster Crassostrea gigas. Genetics 168(1): 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao W., Fu X., Li J., Li L., Feng L., et al. , 2014. Large-scale development of gene-associated single-nucleotide polymorphism markers for molluscan population genomic, comparative genomic, and genome-wide association studies. DNA Res. 21(2): 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y.-L., Kong L.-F., Yu H., Li Q., 2014. Development, inheritance and evaluation of 55 novel single nucleotide polymorphism markers for parentage assignment in the Pacific oyster (Crassostrea gigas). Genes Genomics 36(2): 129–141. [Google Scholar]
- Jombart T., Ahmed I., 2011. adegenet 1.3–1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27(21): 3070–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski S. T., Taper M. L., Marshall T. C., 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16(5): 1099–1106. [DOI] [PubMed] [Google Scholar]
- Kofler R., Pandey R. V., Schlötterer C., 2011. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 27(24): 3435–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranis A., Gheyas A. A., Boschiero C., Turner F., Yu L., et al. , 2013. Development of a high density 600K SNP genotyping array for chicken. BMC Genomics 14(1): 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallias D., Beaumont A. R., Haley C. S., Boudry P., Heurtebise S., et al. , 2007. A first-generation genetic linkage map of the European flat oyster Ostrea edulis (L.) based on AFLP and microsatellite markers. Anim. Genet. 38(6): 560–568. [DOI] [PubMed] [Google Scholar]
- Lallias D., Stockdale R., Boudry P., Beaumont A. R., Lapègue S., 2009. Characterization of 27 microsatellite loci in the European flat oyster Ostrea edulis. Mol. Ecol. Resour. 9(3): 960–963. [DOI] [PubMed] [Google Scholar]
- Lallias D., Boudry P., Lapègue S., King J. W., Beaumont A. R., 2010. Strategies for the retention of high genetic variability in European flat oyster (Ostrea edulis) restoration programmes. Conserv. Genet. 11(5): 1899–1910. [Google Scholar]
- Lapègue S., Harrang E., Heurtebise S., Flahauw E., Donnadieu C., et al. , 2014. Development of SNP-genotyping arrays in two shellfish species. Mol. Ecol. Resour. 14(4): 820–830. [DOI] [PubMed] [Google Scholar]
- Li G., Hubert S., Bucklin K., Ribes V., Hedgecock D., 2003. Characterization of 79 microsatellite DNA markers in the Pacific oyster Crassostrea gigas. Mol. Ecol. Notes 3(2): 228–232. [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14): 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16): 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Li Q., Cornette F., Dégremont L., Lapègue S., 2010. Development of four EST-SSR multiplex PCRs in the Pacific oyster (Crassostrea gigas) and their validation in parentage assignment. Aquaculture 310(1–2): 234–239. [Google Scholar]
- Li S., Li Q., Yu H., Kong L., Liu S., 2015. Genetic variation and population structure of the Pacific oyster Crassostrea gigas in the northwestern Pacific inferred from mitochondrial COI sequences. Fish. Sci. 81(6): 1071–1082. [Google Scholar]
- Liu S., Sun L., Li Y., Sun F., Jiang Y., et al. , 2014. Development of the catfish 250K SNP array for genome-wide association studies. BMC Res. Notes 7(1): 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loosanoff V. L., 1955. The European oyster in American waters. Science 121(3135): 119–121. [DOI] [PubMed] [Google Scholar]
- Magoč T., Salzberg S. L., 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21): 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17(1): 10. [Google Scholar]
- Matukumalli L. K., Lawley C. T., Schnabel R. D., Taylor J. F., Allan M. F., et al. , 2009. Development and characterization of a high density SNP genotyping assay for cattle. PLoS One 4(4): e5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9): 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ødegård J., Meuwissen T. H., 2014. Identity-by-descent genomic selection using selective and sparse genotyping. Genet. Sel. Evol. 46(1): 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palti Y., Gao G., Liu S., Kent M. P., Lien S., et al. , 2015. The development and characterization of a 57K single nucleotide polymorphism array for rainbow trout. Mol. Ecol. Resour. 15(3): 662–672. [DOI] [PubMed] [Google Scholar]
- Pardo B. G., Álvarez-Dios J. A., Cao A., Ramilo A., Gómez-Tato A., et al. , 2016. Construction of an Ostrea edulis database from genomic and expressed sequence tags (ESTs) obtained from Bonamia ostreae infected haemocytes: development of an immune-enriched oligo-microarray. Fish Shellfish Immunol. 59: 331–344. [DOI] [PubMed] [Google Scholar]
- Peng Y., Leung H. C. M., Yiu S. M., Chin F. Y. L., 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28(11): 1420–1428. [DOI] [PubMed] [Google Scholar]
- Pernet F., Lupo C., Bacher C., Whittington R.J., 2016. Infectious diseases in oyster aquaculture require a new integrated approach. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 371: 20150213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81(3): 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Song K., Li C., Wang W., Li B., et al. , 2017. Construction and evaluation of a high-density SNP array for the Pacific oyster (Crassostrea gigas). PLoS One 12(3): e0174007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A. M., Crooijmans R. P. M. A., Affara N. A., Amaral A. J., Archibald A. L., et al. , 2009. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS One 4(8): e6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards P. M., Liu M. M., Lowe N., Davey J. W., Blaxter M. L., et al. , 2013. RAD-Seq derived markers flank the shell colour and banding loci of the Cepaea nemoralis supergene. Mol. Ecol. 22(11): 3077–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo D., Palaiokostas C., Bargelloni L., Martínez P., Houston R., 2017. Applications of genotyping by sequencing in aquaculture breeding and genetics. Rev. Aquacult. DOI: 10.1111/raq.12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohfritsch A., Bierne N., Boudry P., Heurtebise S., Cornette F., et al. , 2013. Population genomics shed light on the demographic and adaptive histories of European invasion in the Pacific oyster, Crassostrea gigas. Evol. Appl. 6(7): 1064–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F., 2008. GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour. 8(1): 103–106. [DOI] [PubMed] [Google Scholar]
- Sauvage C., Bierne N., Lapègue S., Boudry P., 2007. Single nucleotide polymorphisms and their relationship to codon usage bias in the Pacific oyster Crassostrea gigas. Gene 406(1–2): 13–22. [DOI] [PubMed] [Google Scholar]
- Sauvage C., Boudry P., De Koning D. J., Haley C. S., Heurtebise S., et al. , 2010. QTL for resistance to summer mortality and OsHV-1 load in the Pacific oyster (Crassostrea gigas). Anim. Genet. 41(4): 390–399. [DOI] [PubMed] [Google Scholar]
- Sekino M., Hamaguchi M., Aranishi F., Okoshi K., 2003. Development of novel microsatellite DNA markers from the Pacific oyster Crassostrea gigas. Mar. Biotechnol. (NY) 5(3): 227–233. [DOI] [PubMed] [Google Scholar]
- Shabtay A., Tikochinski Y., Benayahu Y., Rilov G., 2014. Preliminary data on the genetic structure of a highly successful invading population of oyster suggesting its establishment dynamics in the Levant. Mar. Biol. Res. 10(4): 407–415. [Google Scholar]
- Sobolewska H., Beaumont A. R., 2005. Genetic variation at microsatellite loci in northern populations of the European flat oyster (Ostrea edulis). J. Mar. Biol. Assoc. U. K. 85(04): 955–960. [Google Scholar]
- Sved J. A., 1971. Linkage disequilibrium and homozygosity of chromosome segments in finite populations. Theor. Popul. Biol. 2(2): 125–141. [DOI] [PubMed] [Google Scholar]
- Troost K., 2010. Causes and effects of a highly successful marine invasion: case-study of the introduced Pacific oyster Crassostrea gigas in continental NW European estuaries. J. Sea Res. 64(3): 145–165. [Google Scholar]
- Tsai H.-Y., Hamilton A., Tinch A. E., Guy D. R., Gharbi K., et al. , 2015. Genome wide association and genomic prediction for growth traits in juvenile farmed Atlantic salmon using a high density SNP array. BMC Genomics 16(1): 969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H.-Y., Hamilton A., Tinch A. E., Guy D. R., Bron J. E., et al. , 2016. Genomic prediction of host resistance to sea lice in farmed Atlantic salmon populations. Genet. Sel. Evol. 48(1): 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo R. L., Leeds T. D., Fragomeni B. O., Gao G., Hernandez A. G., et al. , 2016. Evaluation of genome-enabled selection for bacterial cold water disease resistance using progeny performance data in rainbow trout: insights on genotyping methods and genomic prediction models. Front. Genet. 7: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera M., Carlsson J., Carlsson J. E., Cross T., Lynch S., et al. , 2016. Current genetic status, temporal stability and structure of the remnant wild European flat oyster populations: conservation and restoring implications. Mar. Biol. 163(12): 239. [Google Scholar]
- Vercaemer B., Spence K. R., Herbinger C. M., Lapègue S., Kenchington E. L., 2006. Genetic diversity of the European oyster (Ostrea edulis L.) in Nova Scotia: comparison with other parts of Canada, Maine and Europe and implications for broodstock management. J. Shellfish Res. 25(2): 543–551. [Google Scholar]
- Wang J., Qi H., Li L., Que H., Wang D., et al. , 2015. Discovery and validation of genic single nucleotide polymorphisms in the Pacific oyster Crassostrea gigas. Mol. Ecol. Resour. 15(1): 123–135. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang X., Wang A., Guo X., 2010. A 16-microsatellite multiplex assay for parentage assignment in the eastern oyster (Crassostrea virginica Gmelin). Aquaculture 308(Suppl. 1): S28–S33. [Google Scholar]
- Xu J., Zhao Z., Zhang X., Zheng X., Li J., et al. , 2014. Development and evaluation of the first high-throughput SNP array for common carp (Cyprinus carpio). BMC Genomics 15(1): 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez J. M., Naswa S., López M. E., Bassini L., Correa K., et al. , 2016. Genomewide single nucleotide polymorphism discovery in Atlantic salmon (Salmo salar): validation in wild and farmed American and European populations. Mol. Ecol. Resour. 16(4): 1002–1011. [DOI] [PubMed] [Google Scholar]
- Yu H., Li Q., 2007. Genetic variation of wild and hatchery populations of the Pacific oyster Crassostrea gigas assessed by microsatellite markers. J. Genet. Genomics 34(12): 1114–1122. [DOI] [PubMed] [Google Scholar]
- Zhang G., Fang X., Guo X., Li L., Luo R., et al. , 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490(7418): 49–54. [DOI] [PubMed] [Google Scholar]
- Zhong X., Li Q., Guo X., Yu H., Kong L., 2014. QTL mapping for glycogen content and shell pigmentation in the Pacific oyster Crassostrea gigas using microsatellites and SNPs. Aquacult. Int. 22(6): 1877–1889. [Google Scholar]
- Zhong X., Li Q., Kong L., Yu H., 2017. Estimates of linkage disequilibrium and effective population size in wild and selected populations of the Pacific oyster using single-nucleotide polymorphism markers. J. World. Aquac. Soc. DOI: . 10.1111/jwas.12393 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Illumina sequencing data for the pooled C. gigas and O. edulis samples have been deposited into the European nucleotide archive under accession number PRJEB20253 (http://www.ebi.ac.uk/ena/data/view/PRJEB20253). The details of the SNP markers on the array are given in (Supplemental Material, File S1). O. edulis markers with significant alignment to the C. gigas genome (e-value 1E−4) are given in File S2.