Abstract
Cells fine-tune their metabolic programs according to nutrient availability in order to maintain homeostasis. This is achieved largely through integrating signaling pathways and the gene expression program, allowing cells to adapt to nutritional change. Dbp2, a member of the DEAD-box RNA helicase family in Saccharomyces cerevisiae, has been proposed to integrate gene expression with cellular metabolism. Prior work from our laboratory has reported the necessity of DBP2 in proper gene expression, particularly for genes involved in glucose-dependent regulation. Here, by comparing differentially expressed genes in dbp2∆ to those of 700 other deletion strains from other studies, we find that CYC8 and TUP1, which form a complex and inhibit transcription of numerous genes, corepress a common set of genes with DBP2. Gene ontology (GO) annotations reveal that these corepressed genes are related to cellular metabolism, including respiration, gluconeogenesis, and alternative carbon-source utilization genes. Consistent with a direct role in metabolic gene regulation, loss of either DBP2 or CYC8 results in increased cellular respiration rates. Furthermore, we find that corepressed genes have a propensity to be associated with overlapping long noncoding RNAs and that upregulation of these genes in the absence of DBP2 correlates with decreased binding of Cyc8 to these gene promoters. Taken together, this suggests that Dbp2 integrates nutrient availability with energy homeostasis by maintaining repression of glucose-repressed, Cyc8-targeted genes across the genome.
Keywords: helicase, DEAD-box, metabolism, Cyc8, Dbp2
Rapidly proliferating cells, such as exponentially growing budding yeast Saccharomyces cerevisiae, must meet high demands on both energy production and biosynthesis of anabolic precursors. Budding yeast thrive in a wide range of environments with different nutrient availability. In order to optimize cell growth, yeast can adjust their metabolism in response to nutrient availability to balance the energy production and biomass synthesis. Signaling pathways such as protein kinase A (PKA)/Ras, AMP-activated protein kinase (AMPK)/Snf1, and target of rapamycin (TOR) pathways are involved in sensing nutrient availability and regulating metabolic decisions at transcriptional, post-transcriptional, translational, and post-translational levels (Broach 2012; Kayikci and Nielsen 2015). How cells integrate information about environmental nutrients with the gene expression program has long been an active area of research as it not only has huge implications on the chemical industry, but also sheds light on metabolism in human diseases (Diaz-Ruiz et al. 2011; Borodina and Nielsen 2014).
The budding yeast S. cerevisiae preferentially uses glucose as a carbon source. In the presence of high levels of glucose, yeast cells prefer to ferment glucose into ethanol instead of respiration despite the presence of oxygen (Broach 2012). In the presence of glucose, genes involved in gluconeogenesis, respiration, and utilization of alternative carbon sources are repressed; a process referred to as carbon catabolite or glucose repression (Gancedo 1998; Kayikci and Nielsen 2015). Glucose repression depends on transcription factor Mig1 and the Cyc8-Tup1 corepressor complex as well as the inactivation of the kinase Snf1 (Gancedo 1998; Kayikci and Nielsen 2015). However, budding yeast has the capability to use alternative fermentable carbon sources, such as galactose and sucrose; or nonfermentable carbon sources, such as ethanol and acetate (Schüller 2003; Turcotte et al. 2010). A shift from glucose to a nonfermentable carbon source leads to reprogramming of gene expression such that gluconeogenic, respiratory, and alternative carbon-source utilization genes are induced (DeRisi et al. 1997; Soontorngun et al. 2007). This switch is achieved through transcriptional activators such as Cat8 and the Hap2/3/4/5 complex as well as the activation of the kinase Snf1 (Schüller 2003; Broach 2012). Activated Snf1 influences transcription in many aspects, including phosphorylating the repressor Mig1 (Treitel et al. 1998; Broach 2012). The phosphorylation of Mig1 leads to its relocalization from the nucleus to the cytoplasm, thus derepressing its target, glucose-repressed genes (DeVit et al. 1997; DeVit and Johnston 1999).
Dbp2, a member of the DEAD-box RNA helicase family, functions as a key player in gene expression programs related to cellular metabolism in S. cerevisiae (Cloutier et al. 2012; Beck et al. 2014). We have previously reported that the nuclear localization of Dbp2 is dependent on glucose availability, suggestive of a broad role in regulating gene expression in response to nutrient availability (Beck et al. 2014). However, the mechanism of Dbp2 in regulating a specific cellular pathway was unknown. Herein, we show that Dbp2 corepresses Cyc8-targeted genes, representing a major branch of the metabolic pathway, and that loss of DBP2 correlates with derepression of respiration and gluconeogenesis. This suggests that Dbp2 integrates nutrient availability with metabolic adaptation through regulation of Cyc8-targeted genes.
Materials and Methods
Bioinformatics analyses
RNA sequencing data processing and differential gene expression analyses:
RNA sequencing (RNA-seq) data were previously deposited in the Gene Expression Omnibus (GSE58097; Beck et al. 2014). Color space reads were mapped to the yeast reference genome (R64-2-1) using the SHort Read Mapping Program 2 (SHRiMP2, version 2.2.3) with the option “–single-best-mapping” (David et al. 2011). The aligned reads were processed as previously described (Cloutier et al. 2016) with the following modifications. Briefly, the aligned reads were imported into R using Rsamtools and GenomicAlignments (Morgan et al. 2010; Lawrence et al. 2013; R Development Core Team 2016). The first mapped read in a proper pair (flags = 83, 99) with a mapping quality (mapq) ≥ 40 was retained. Reads overlapping rDNA regions were excluded from the analysis. Remaining reads were counted against the genomic features using GenomicAlignments from Bioconductor to determine the number of reads for sense RNA transcripts (Gentleman et al. 2004; Lawrence et al. 2013). The mode of “IntersectionNotEmpty” was used for the summarizeOverlaps function. EdgeR was used to determine differential gene expression (Robinson et al. 2010; McCarthy et al. 2012; GSE99097). Genes with very low counts (<10) across all samples were filtered from the analysis. Transcripts with gene expression change >2.2-fold and FDR ≤ 0.01 were determined as differentially expressed.
Comparison of differentially expressed genes in dbp2∆ and other strains with genetic perturbations:
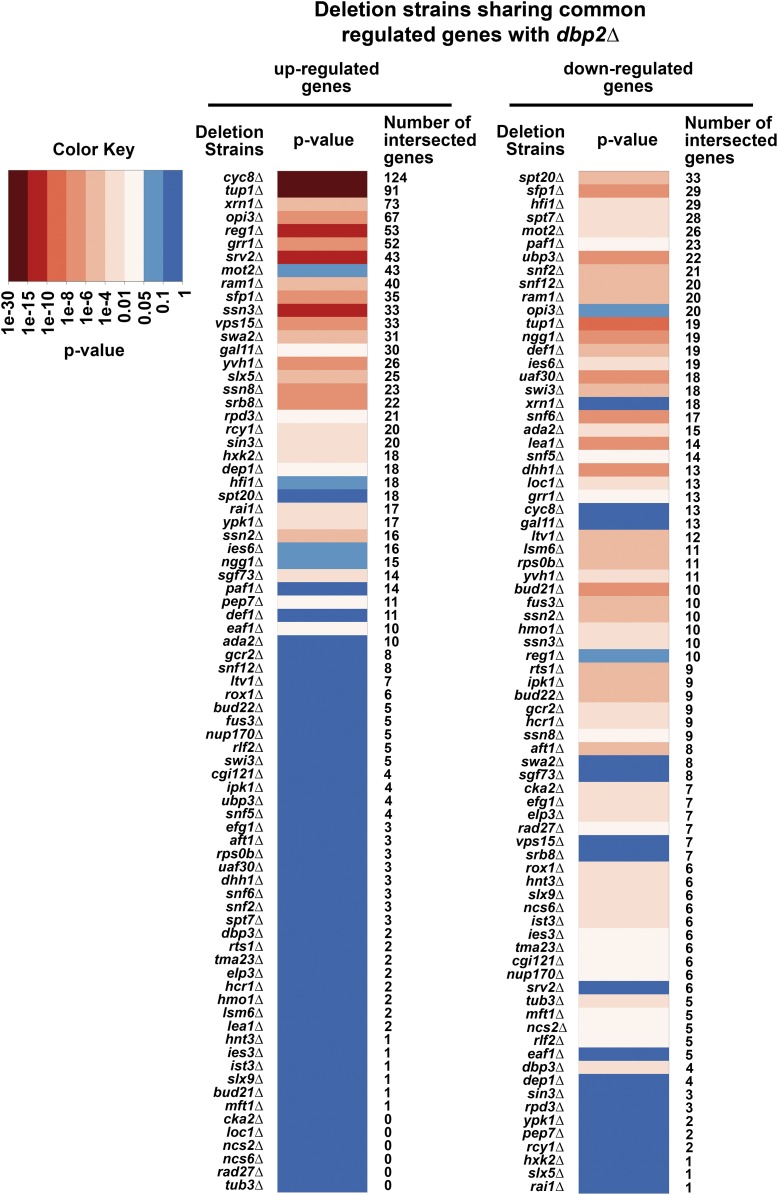
The gene expression profiles of 700 responsive deletion mutants identified in Kemmeren et al. (2014) that have at least four differentially expressed transcripts compared to the wild-type strain were used for transcription profile analysis. As previously described, a cutoff of fold change (FC) > 1.7 and P < 0.05 was applied to determine the list of significantly changed genes of each of the 700 deletion mutants. To look for the genes that may act in concert with DBP2 in gene expression, the list of up- and downregulated genes in each of the 700 deletion mutants was compared to the up- and downregulated sense (protein-coding) transcripts in dbp2∆. In addition, the dbp2∆ strain has a slow growth phenotype. This phenotype was also observed for many other deletion mutants, and a slow growth signature was identified (O’Duibhir et al. 2014). The slow growth-related genes (SGGs) (n = 633) identified from O’Duibhir et al. (2014) were filtered from the lists of differentially expressed genes (DEGs) to avoid identifying genes that are affected by the slow growth phenotype. One-sided Fisher’s exact tests were conducted for each comparison using R. Bonferroni correction was applied to correct for multiple comparisons. Lists of genes that have an effect similar to DBP2 (P ≤ 0.05 after correction) in repressing (share common upregulated genes between dbp2∆ and the deletion mutant) or activating (share common downregulated genes between dbp2∆ and the deletion mutant) gene expression were extracted and P-values from the Fisher’s exact tests are presented as a heatmap (Figure 1).
Figure 1.
Dbp2 and other gene regulatory factors share coregulated genes. A heatmap of P-values of the transcriptional profile comparisons between dbp2∆ and other deletion strains is shown and sorted by the number of intersected genes. Lists of DEGs in dbp2∆ were compared to that of 700 deletion strains (Beck et al. 2014; Kemmeren et al. 2014). One-sided Fisher’s exact test was used to estimate the significance of the observed intersection (Bonferroni correction). Common genes between the two studies (Beck et al. 2014; Kemmeren et al. 2014) were used as background. SGGs (O’Duibhir et al. 2014) were filtered from the DEG lists to help identify more direct effects of the deleted genes. A heatmap of the P-values indicating significant similarities (P ≤ 0.05, light to dark red; color key) or no similarities (P > 0.05, light to dark blue; color key) of the coregulated genes between deletion strains and dbp2∆ was generated. Deletion strains sharing significant up- and/or downregulated genes with dbp2∆ are included in the heatmap. The heatmap was sorted by the number of intersected up- (left) or downregulated (right) genes between dbp2∆ and the number of intersected genes in each comparison is listed on the right-hand side of each heatmap.
Comparison of DEGs in dbp2∆ and upon glucose deprivation:
RNA-seq data of wild-type cells cultivated in the presence or absence of glucose were retrieved from the NCBI’s Gene Expression Omnibus (GSE43747; Freeberg et al. 2013). Data were processed as previously described (Freeberg et al. 2013) with the following modifications. First, nucleotides with low quality (<30) were trimmed from the end of the reads using FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Next, sequences of the barcodes and adaptors were trimmed from the reads using Cutadapt (Martin 2011). Reads <18 nt were removed at the same time. The processed reads were mapped to the reference yeast genome (R64-2-1) using Tophat and Bowtie2 (Trapnell et al. 2009; Langmead and Salzberg 2012). The aligned reads were read in R using Rsamtools (Morgan et al.2010). Only reads with mapq ≥40 were kept. Read counts per feature were determined using GenomicAlignments from Bioconductor (Gentleman et al. 2004; Lawrence et al. 2013). EdgeR was used to determine differential gene expression between strains grown in the glucose-replete or -deprived media (Robinson et al. 2010; McCarthy et al. 2012). Genes with very low counts (<10) across all samples were filtered. A cutoff of FC > 2.2 and FDR ≤ 0.01 was used to determine the significant change in gene expression. In addition, as the cells used for the RNA-seq above were treated with 4sU, the effects of 4sU were removed by removing genes that are significantly changed upon 4sU treatment (FC > 2.2 and FDR ≤ 0.01, n = 41) from the DEG lists upon glucose deprivation (Freeberg et al. 2013). SGGs were then filtered from the DEG lists. One-sided Fisher’s exact tests were used to estimate the significance of the intersection between the lists of DEGs in dbp2∆ and that upon glucose deprivation. Venn diagrams were generated by the online program BioVenn (Hulsen et al. 2008).
Functional annotation clustering analysis:
Functional annotation clustering of biological processes was conducted using DAVID (version 6.7) (Huang et al. 2009a,b). Functional clusters with enrichment score ≥1.3 (equivalent to P ≤ 0.05) are shown.
Experimental procedures
Plasmids and strains:
Yeast strains were constructed using classical yeast genetics and are listed in Table 1. To construct the CYC8/CEN plasmid, a DNA fragment with 888 bp upstream and 460 bp downstream of the CYC8 open reading frame (ORF) was produced with 5′ SmaI and a 3′ BamHI restriction sites flanking the fragment using genomic DNA as templates. The fragment was then cloned into pRS315 using the standard molecular cloning method. The TUP1/CEN plasmid was constructed in the same way except that the inserted DNA fragment was flanked with 1000 bp upstream and 499 bp downstream of the TUP1 ORF with 5′ SmaI and 3′ SpeI restriction sites. Oligos used for cloning are listed in Table 2.
Table 1. Yeast strains.
| Strains | Genotype | Source |
|---|---|---|
| Wild type (BY4741) | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 | Open Biosystems |
| dbp2∆ | MATa dbp2::KanR ura3∆0 leu2∆0 his3∆0 TRP1 met- lys? | Cloutier et al. (2012) |
| CYC8-3xFLAG | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 CYC8-3xFLAG | Cloutier et al. (2013) |
| dbp2∆ CYC8-3xFLAG | MATa dbp2::HygB his3∆1 leu2∆0 met15∆0 ura3∆0 CYC8-3xFLAG | Cloutier et al. (2013) |
| cyc8∆ | MATa cyc8::KanR his3∆1 leu2∆0 met15∆0 ura3∆0 | Open Biosystems |
| tup1∆ | MATa tup1::KanR his3∆1 leu2∆0 met15∆0 ura3∆0 | Open Biosystems |
| dbp3∆ | MATa dbp3::KanR his3∆1 leu2∆0 met15∆0 ura3∆0 | Open Biosystems |
Table 2. Oligos used for plasmid construction.
| Sequence | |
|---|---|
| SmaI-CYC8 F | TCCCCCGGGCTAAAGCACATCCGATCTGAG |
| BamHI-CYC8 R | CGCGGATCCCGTAGAACCCAAAGCATTAGG |
| SmaI-TUP1 F | TCCCCCGGGGTAAGTCTGCGGAATCGATCTG |
| SpeI-TUP1 R | GGACTAGTAAGTGCGACGTGGACGAATC |
Respiration profile measurements:
The oxygen consumption assays were conducted as described (Yao et al. 2016). Briefly, yeast strains were grown to log phase in YPD and shifted to assay medium (1.67% yeast nitrogen base, 0.5% ammonium sulfate, and 2% ethanol or glucose). Cells were seeded in the 24-well microplate (Seahorse Bioscience) at 5 × 105 cells per well and incubated at 30° for 1 hr. The oxygen consumption rates were measured using the Seahorse XFe24 analyzer. The cyc8∆ and tup1∆ strains were confirmed with plasmid-based complementation. Briefly, yeast cells were grown in SC-LEU media to midlog phase and spotted on the SC-LEU agar plates in fivefold serial dilutions. Plates were incubated at 37° until wild-type cells were fully grown.
Gene induction analyses:
Yeast cells were first grown at 30° to an OD600 of 0.4–0.6 in SC with 2% glucose medium. The cells were then spun at 3000 rpm, washed twice with SC and transferred to SC medium containing 1% potassium acetate and grown for another 6 hr. 15 OD units were collected prior to (0 min) and at 30, 60, 90, 120, 240, and 360 min after the nutritional shift. Cells were flash frozen and stored at −80° before use.
RNA isolation and Northern blotting:
RNA was isolated from cells using standard acid phenol purification. 30 μg total RNA was resolved on a 1.2% formaldehyde-agarose gel and transferred to a nylon membrane (Brightstar Hybond N+, Invitrogen). Northern blotting was performed as previously described (Cloutier et al. 2012). Radiolabeled double-stranded DNA (dsDNA) probes were generated using PCR products using wild-type genomic DNA as a template. Oligos used for Northern blotting probes are listed in Table 3. Target transcripts were visualized using a PhosphorImager (GE Healthcare) and quantified by densitometry using ImageQuant (GE Healthcare).
Table 3. Oligos used for Northern blotting (dsDNA probes).
| Sequence | |
|---|---|
| PCK1 F | ATGGTCTATCAACCGTGAAAGAGC |
| PCK1 R | TCAACAATCTATGTGGGTCTGCG |
| SFC1 F | CATCCAGCCATCAATCTCATGG |
| SFC1 R | TAGTATCCAATGGAGCGTTGGA |
| JEN1 F | CTGGTGAACAGCAACAACCTG |
| JEN1 R | GGTGCATCTTCAATCGCTGTT |
| YAT1 F | ATACATCGAGCAGTTCTGGTATGAC |
| YAT1 R | GGATCGAGTCTGTGTAGATGTCTG |
| SCR1 F | GGATACGTTGAGAATTCTGGCCGAGG |
| SCR1 R | AATGTGCGAGTAAATCCTGATGGCACC |
Chromatin immunoprecipitation:
Chromatin immunoprecipitation (ChIP) was performed as described previously (Cloutier et al. 2013) with the following modifications. Chromatin from ∼5 × 106 cells was immunoprecipitated with 2 μl of FLAG M2 monoclonal antibody (F3165; Sigma) and 24 μl of Protein G Dynabeads (30 mg/ml; Invitrogen) at 4° for 2 hr. Immunoprecipitated DNA was isolated and quantitative PCR (qPCR) was performed using PrimeTime Assay primers listed in Table 4 and TaqMan qPCR mix (Invitrogen). All ChIP experiments were performed with three biological replicates. Error bars represent the SD from the mean.
Table 4. PrimeTime assays for ChIP.
| Name | Forward | Reverse | Probe |
|---|---|---|---|
| PCK1 | ATTATGGATAGGCGGATAAAGGG | ACGTACCATTGTCCAACCAG | CCCAAACAGGATTGTAAAGCTTAGACGC |
| SFC1 | CTGCTCGAGGTGCTATCTTTT | TGTGACATTACGGTTTGTAAAAGG | CTTTGAGATTCTTGTCGCCACGGAGT |
| YAT1 | GCGGAATGACAAAACCATCAG | TCCTTGCCCCTTTACTTTGG | CACGATCTCTCCAGTCACAATGGCAA |
| JEN1 | CTTCTGCACACCATCCGG | ACCAGGTAACGTTCTAATGCAT | TCTTCACTTAACGGTCTTTTGCCCCC |
Data availability
Data reference numbers deposited in the Gene Expression Omnibus are as follows: GSE58097 (Beck et al. 2014), GSE42527 (Kemmeren et al. 2014), and GSE43747 (Freeberg et al. 2013), GSE99097.
Results
Analysis of transcription profiles reveals similarities between DBP2 and other gene regulatory factors
Nutritional status and gene expression programs are highly integrated in yeast (Broach 2012). Gene expression profiling has revealed that genes necessary for nutrient utilization and metabolism require DBP2 for normal expression (Beck et al. 2014). This specificity suggests that Dbp2 may function similar to or in concert with known transcription factors involved in nutrient utilization. A recent study of 1484 yeast deletion strains used gene expression profiling by microarray to identify 700 genes that affect gene expression in S. cerevisiae (Kemmeren et al. 2014). To test our hypothesis, we compared lists of DEGs in dbp2∆ to those for each of the 700 strains from this study. Importantly, as the dbp2∆ strain has a slow growth phenotype that is also observed for other deletion strains (O’Duibhir et al. 2014), we removed SGGs from the DEG lists to avoid analysis of indirect effects from the growth defects (O’Duibhir et al. 2014). The remaining DEGs were then compared and one-sided Fisher’s exact tests were used to determine the significance of the observed intersection of the lists between dbp2∆ and other deletion strains. This analysis identified 77 deletion strains that have similar up- or downregulated genes to dbp2∆ (Figure 1, P value ≤ 0.05 after a Bonferroni correction). Interestingly, strains harboring the deletion of CYC8 or TUP1 share the most upregulated genes with dbp2∆, with cyc8∆ and tup1∆ sharing 124 and 91 upregulated ones with dbp2∆, respectively (Figure 1, left). Cyc8 and Tup1 form a heteropentameric complex required for transcriptional repression of various genes, including glucose-repressed genes, cell-type-specific genes and hypoxic genes (Williams and Trumbly 1990; Keleher et al. 1992; Balasubramanian et al. 1993; Varanasi et al. 1996; Smith and Johnson 2000). Whereas tup1∆ significantly shares both up- and downregulated genes with dbp2∆, cyc8∆ only significantly shares upregulated genes. This suggests that TUP1 and CYC8 may not always act in concert.
Besides tup1∆, only a small subset of deletion strains that share similar upregulated genes with dbp2∆ also share downregulated genes. These include strains harboring deletions of the RNA polymerase II (RNAPII) mediator complex (SSN2, SSN3, SSN8), a component of SCF ubiquitin-ligase complex (GRR1), ribosome biosynthesis genes (SFP1, YVH1), and a component of the CAAX farnesyltransferase complex (RAM1) (Figure 1, left and right) (He et al. 1991; Hengartner et al. 1995; Skowyra et al. 1997; Marion et al. 2004; Kemmler et al. 2009; Lo et al. 2009). We also observed common upregulated genes between dbp2∆ and strains with deletions in components of the Rpd3 histone deacetylase complex, such as RPD3, SIN3, and DEP1, as well as RNA decay factor XRN1 (Figure 1, left) (Larimer and Stevens 1990; Hengartner et al. 1995; Carrozza et al. 2005). This suggests that DBP2 represses gene expression through regulating transcription and/or RNA stability. Interestingly, SSN3, SSN8, and SRB8 are involved in glucose repression (Balciunas and Ronne 1995). Several other genes required for glucose repression, such as REG1, GRR1, and HXK2, were also identified as similar to DBP2 (Figure 1, left) (Flick and Johnston 1991; Gancedo 1998). The overlap between genes repressed by DBP2 and those repressed by factors involved in glucose repression is consistent with a link between DBP2 and glucose metabolism (Beck et al. 2014).
CYC8 and TUP1 share corepressed genes with DBP2
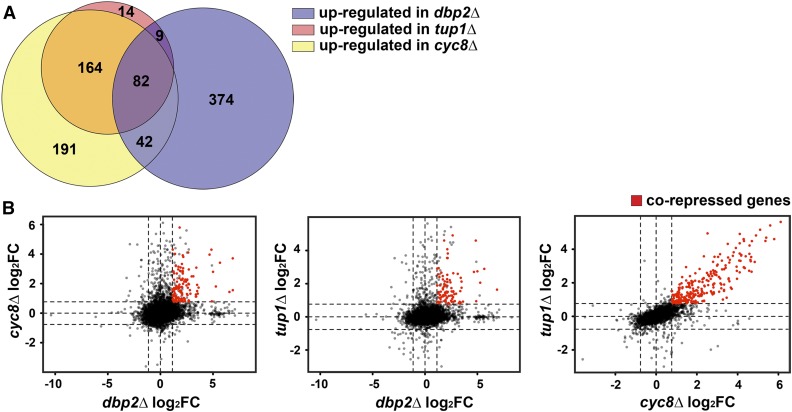
The GAL protein-coding genes are part of the galactose metabolic switch that allows yeast cells to use galactose when glucose is absent (Sellick et al. 2008). Our prior studies demonstrated that Dbp2 maintains Cyc8-dependent repression of the GAL cluster genes (Cloutier et al. 2013). The fact that deletion of CYC8 or TUP1 also results in a similar set of derepressed genes as loss of DBP2, however, suggests that this role may be more widespread than just at this locus. To see how many genes are commonly upregulated between dbp2∆, cyc8∆, and tup1∆, a Venn diagram was plotted using the lists of upregulated genes in each deletion strain. This revealed that 82 genes are corepressed by CYC8, TUP1, and DBP2 (Figure 2A). To determine if loss of CYC8 or TUP1 affects gene expression to a similar extent as loss of DBP2, we plotted the log2FC of all genes in dbp2∆ against those of cyc8∆ and tup1∆ (Figure 2B). Consistent with a function in gene repression, the majority of the common DEGs in dbp2∆ and cyc8∆ (Figure 2B, left) or dbp2∆ and tup1∆ (Figure 2B, middle) are upregulated in the mutants. However, the common upregulated genes between dbp2∆ and tup1∆ or dbp2∆ and cyc8∆ do not show a linear relationship. This is in contrast to TUP1 and CYC8, whose corepressed genes show a linear relationship in the deletion strains (Figure 2B, right). This suggests that even though TUP1 and CYC8 share corepressed genes with DBP2, Dbp2 and Cyc8 or Tup1 act in a biochemically distinct manner. Alternatively, the lack of correlation could be due to the different methods of gene expression profiling, i.e., microarray vs. RNA-seq (Zhao et al. 2014). We also observed that many genes are specifically repressed by CYC8 but not TUP1 [Figure 2B, right; cyc8∆ log2FC > log2(1.7) and tup1∆ log2FC < log2(1.7)], suggesting that these two factors may not always function in concert. This is consistent with a prior study showing additive effects from simultaneous deletion of CYC8 and TUP1 (Williams and Trumbly 1990).
Figure 2.
TUP1 and CYC8 share corepressed genes with DBP2. (A) Venn diagram showing the numbers of genes upregulated upon loss of CYC8, TUP1, and DBP2. The Venn diagram was generated by BioVenn (Hulsen et al. 2008). SGGs were removed from the gene lists. (B) CYC8 and TUP1 repress a common subset of genes with DBP2, but to a different extent. The log2FC of all genes except SGGs in cyc8∆ and tup1∆ was plotted against that of dbp2∆, as well as of each other. Genes that are significantly upregulated between dbp2∆ and cyc8∆ (left), dbp2∆ and tup1∆ (middle), or cyc8∆ and tup1∆ (right) are colored in red. Dashed lines mark the log2FC cutoff in dbp2∆ [±log2(2.2)], cyc8∆, and tup1∆ [±log2(1.7)].
CYC8, TUP1, and DBP2 corepressed genes are enriched in monosaccharide metabolism and transcriptional regulation processes
To determine if genes corepressed by CYC8, TUP1, and DBP2 fall into common biological processes, we conducted functional annotation clustering analyses using DAVID (Huang et al. 2009a,b). This revealed enrichment of transcripts whose products function in monosaccharide metabolic processes or regulation of transcription (Table 5 and Supplemental Material, Table S1). Interestingly, MIG1, THI2, ROX1, and SMP1, which have been shown to regulate genes involved in metabolism and stress response, were also identified (Nehlin et al. 1991; Zitomer and Lowry 1992; de Nadal et al. 2003; Nosaka et al. 2005). This suggests that CYC8, TUP1, and DBP2 may regulate cellular metabolism and response to stress, possibly through regulating gene expression.
Table 5. Gene corepressed by CYC8, TUP1, and DBP2 (82).
| Annotation Cluster | Enrichment Scorea | Description of GO Termsb | Unique Genesc | Gene Countd |
|---|---|---|---|---|
| 1 | 2.35 | Monosaccharide metabolic process | GAC1, ERR3, GAL3, YHR210C, NDE2, YNR071C, PCK1, SGA1 | 8 |
| 2 | 2.07 | Regulation of transcription | ROX1, GAC1, YOR338W, RPM2, CSR2, VHR1, YAP6, REG2, SMP1, MIG1, THI2, RGM1, SUT1, HAL1, GAL3, YPR196W, ACA1, CUP9, GIS1, POG1, ASK10 | 21 |
Table of GO term clusters of CYC8, TUP1, and DBP2 commonly repressed genes. Functional annotation clustering analysis was conducted with the 82 genes corepressed by CYC8, TUP1, and DBP2 in Figure 2A.
GO clusters with significant enrichment score of at least 1.3 (P ≤ 0.05).
GO term descriptions for the corresponding functional annotation cluster.
Unique genes included in each GO cluster.
Number of unique genes in each cluster.
DBP2 also shares unique corepressed genes with CYC8 and TUP1, individually (Table 6 and Table 7 and Table S2 and Table S3). Out of the 124 genes corepressed by CYC8 and DBP2, we observed enrichment of six functional annotation clusters including oxidative phosphorylation and ion transport, generation of precursor metabolites and energy and cell death, regulation of transcription, carbohydrate transport, and transmembrane transport (Table 6 and Table S2). The TUP1 and DBP2 corepressed genes (n = 91), on the other hand, showed enrichment of clusters including monosaccharide metabolic processes and regulation of RNAPII transcription (Table 7 and Table S3). This suggests that Cyc8 and Tup1, although thought to act exclusively as a complex, may have different functions individually in gene regulation. This is consistent with our prior observation that loss of CYC8 and TUP1 has different effects at some different gene loci (Figure 2B, right).
Table 6. Genes corepressed by CYC8 and DBP2 (124).
| Annotation Cluster | Enrichment Score | Description of GO Terms | Unique Genes | Gene Count |
|---|---|---|---|---|
| 1 | 2.31 | Phosphate metabolic process/oxidative phosphorylation/ion transport | ATP1, ATP6, COX9, CYC1, COX8, QCR6, ATP19, ATP18, GAL3, MRK1, VHS1, KIN82, FRK1, SKS1, SFC1, JEN1, FRE7 | 17 |
| 2 | 1.87 | Generation of precursor metabolites and energy/cell death | ATP1, ATP6, COX9, CYC1, COX8, QCR6, ATP19, ATP18, GAC1, YBR238C, ERR3, ISF1, NDE2, YJL045W, SGA1, FRE7 | 16 |
| 3 | 1.64 | Regulation of transcription | ROX1, GAC1, YOR338W, RPM2, CSR2, VHR1, YAP6, REG2, SMP1, MIG1, THI2, MIG2, RGM1, SUT1, HAL1, SKS1, GAL3, YPR196W, ACA1, SOK2, CUP9, MET32, GIS1, POG1, ASK10 | 25 |
| 4 | 1.63 | Sulfur metabolic process | THI2, MET17, THI22, SNZ1, VHR1, MET32, ADI1, PET18, YAT1 | 9 |
| 5 | 1.58 | Carbohydrate transport | MAL11, STL1, HXT4, HXT1, SKS1 | 5 |
| 6 | 1.55 | Monosaccharide metabolic process | GAC1, ERR3, GAL3, YHR210C, NDE2, YNR071C, PCK1, SGA1 | 8 |
Table of GO term clusters of CYC8 and DBP2 commonly repressed genes. Functional annotation clustering analysis was conducted with the 124 genes corepressed by CYC8 and DBP2 in Figure 2A.
Table 7. Genes corepressed by TUP1 and DBP2 (91).
| Annotation Cluster | Enrichment Score | Description of GO Terms | Unique Genes | Gene Count |
|---|---|---|---|---|
| 1 | 3.30 | Monosaccharide metabolic process | GAC1, ERR3, TOS3, GAL3, YHR210C, NDE2, PFK27, YNR071C, PCK1, SGA1 | 10 |
| 2 | 3.12 | Regulation of transcription | ROX1, GAC1, YOR338W, RPM2, CSR2, VHR1, YAP6, REG2, SMP1, MIG1, THI2, RGM1, SUT1, HAL1, GAL3, ZNF1, YPR196W, ACA1, CUP9, GIS1, POG1, ASK10 | 22 |
| 3 | 1.55 | Regulation of transcription from RNAPII promoter | ROX1, GAC1, YOR338W, RPM2, CSR2, VHR1, YAP6, REG2, SMP1, MIG1, THI2, RGM1, SUT1, HAL1, GAL3 | 15 |
Table of GO term clusters of TUP1 and DBP2 commonly repressed genes. Functional annotation clustering analysis was conducted with the 91 genes corepressed by TUP1 and DBP2 in Figure 2A.
DBP2 and CYC8, but not TUP1, repress cellular respiration
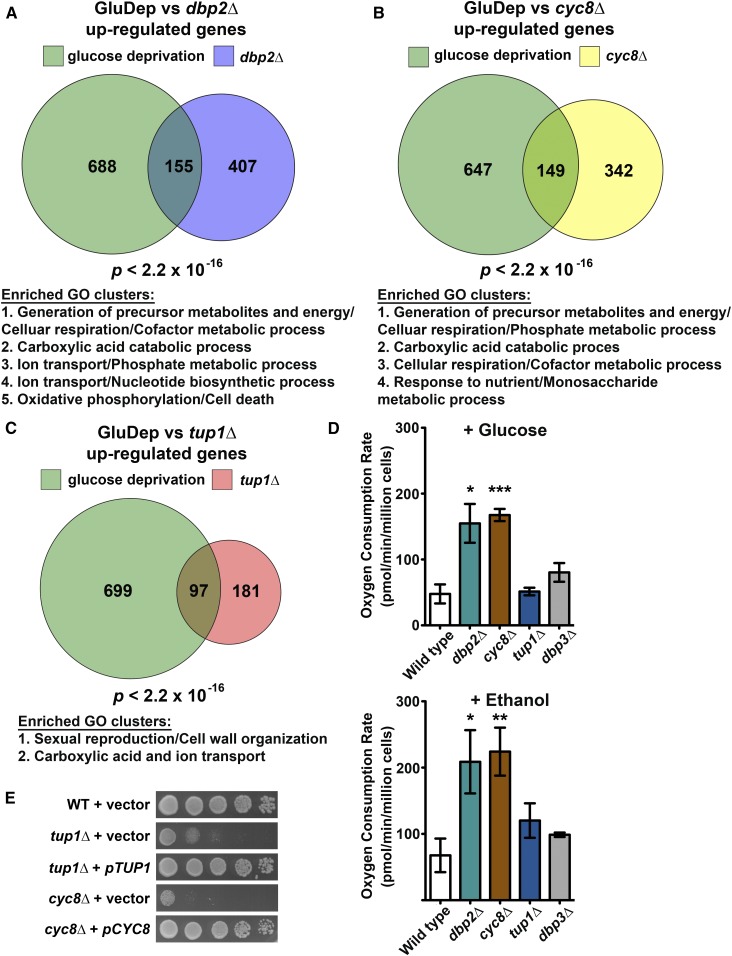
Yeast cells preferentially use glucose as a carbon source for energy production through aerobic fermentation (Broach 2012). The presence of glucose suppresses respiration, gluconeogenesis, and usage of nonfermentable carbon sources, referred to as carbon catabolite or glucose repression (Gancedo 1998; Broach 2012). It is known that CYC8 and TUP1 function as key components in glucose repression (Gancedo 1998). To determine if DBP2 is also required for glucose repression, we estimated the degree of overlap between genes upregulated in dbp2∆ and upon glucose deprivation by comparing the list of genes from our dbp2∆ RNA-seq to a previously published study of glucose-deprived wild-type cells (Freeberg et al. 2013). This revealed an overrepresentation of glucose-repressed genes in dbp2∆ cells (Figure 3A, one-sided Fisher’s exact test, P < 2.2 × 10−16). We also compared upregulated genes in cyc8∆ or tup1∆ to those in glucose-deprived cells (Figure 3, B and C). In agreement with roles for CYC8 and TUP1 in glucose repression, carbon catabolite-repressed genes are also overrepresented in both cyc8∆ and tup1∆ cells (Figure 3, B and C). Interestingly, functional annotation clustering revealed several commonly repressed gene categories between glucose-deprived cells and dbp2∆ (Figure 3A and Table S4) or cyc8∆ (Figure 3B and Table S5), including generation of precursor metabolites and cellular respiration. In contrast, genes upregulated in tup1∆ and glucose-deprived cells are enriched in sexual reproduction, cell wall organization, and carboxylic acid and ion transport categories (Figure 3C and Table S6). This suggests that Cyc8 and Dbp2 may modulate glucose repression of different cellular pathways than Tup1.
Figure 3.
DBP2 and CYC8, but not TUP1, are linked to cellular respiration. (A–C) Glucose repressed genes are enriched in DBP2 (A), CYC8 (B), and TUP1 (C) targets. A Venn diagram showing the number of genes upregulated in dbp2∆, cyc8∆, tup1∆, and upon glucose deprivation for 2 hr is shown (Freeberg et al. 2013). The P-value originates from one-sided Fisher’s exact tests conducted to estimate the significance of the intersection. Descriptions of enriched GO clusters for the intersected genes in each Venn diagram are listed below each of the Venn diagrams. See Table S4, Table S5, and Table S6 for complete results of the functional annotation clustering analysis. (D) Loss of DBP2 or CYC8 leads to increased respiration, even in the presence of glucose. Oxygen consumption in wild-type cells and indicated mutant cells was measured using the Seahorse XFe24 analyzer in the presence of 2% glucose or 2% ethanol. The dbp3∆ strain harbors a deletion of the DEAD-box protein Dbp3 involved in rRNA processing, and serves as a negative control. Data show the mean ± SD of three independent biological replicates. *P < 0.05, **P < 0.01, ***P < 0.001. (E) Ectopic expression of CYC8 or TUP1 from a CEN plasmid rescues the heat sensitivity of cyc8∆ and tup1∆, respectively. Serial dilutions of wild-type (WT), cyc8∆, and tup1∆ strains harboring an empty vector (pRS315), cyc8∆ strain harboring pCYC8, and tup1∆ strain harboring pTUP1 were spotted on SC-LEU plates and incubated at 37°.
To test whether DBP2, CYC8, and/or TUP1 repress respiration in the presence of glucose, we measured the oxygen consumption rate in wild-type, dbp2∆, cyc8∆, and tup1∆ cells as a readout for cellular respiration (Figure 3D). Strains harboring the CYC8 or TUP1 deletion were obtained from the null collection and were confirmed by PCR (data not shown) and plasmid-based complementation (Figure 3E). We also assayed the oxygen consumption in the absence of DBP3, an unrelated DEAD-box protein involved in rRNA processing (Weaver et al. 1997). Briefly, yeast cells were cultivated in media with glucose to log phase and shifted to assay medium with 2% ethanol or 2% glucose to promote respiration or fermentation, respectively. Cells were then seeded in a microplate, incubated at 30° for 1 hr, and the oxygen consumption rates were measured using a Seahorse XFe24 analyzer to measure metabolic flux. Interestingly, the oxygen consumption rate in both dbp2∆ and cyc8∆ was higher than in wild-type cells in the presence of glucose, indicating active respiration (Figure 3D, top). We then measured the oxygen consumption rate in the presence of ethanol to stimulate cellular respiration (Figure 3D, bottom). Similar to glucose, both dbp2∆ and cyc8∆ showed increased oxygen consumption as compared to wild type. In contrast, neither tup1∆ nor dbp3∆ showed differences as compared to wild type in either carbon source (Figure 3D), suggesting that Dbp2 and Cyc8 play specific roles in glucose repression of respiration, independent of Tup1.
DBP2, CYC8, and TUP1 corepressed genes are enriched in overlapping long noncoding RNAs
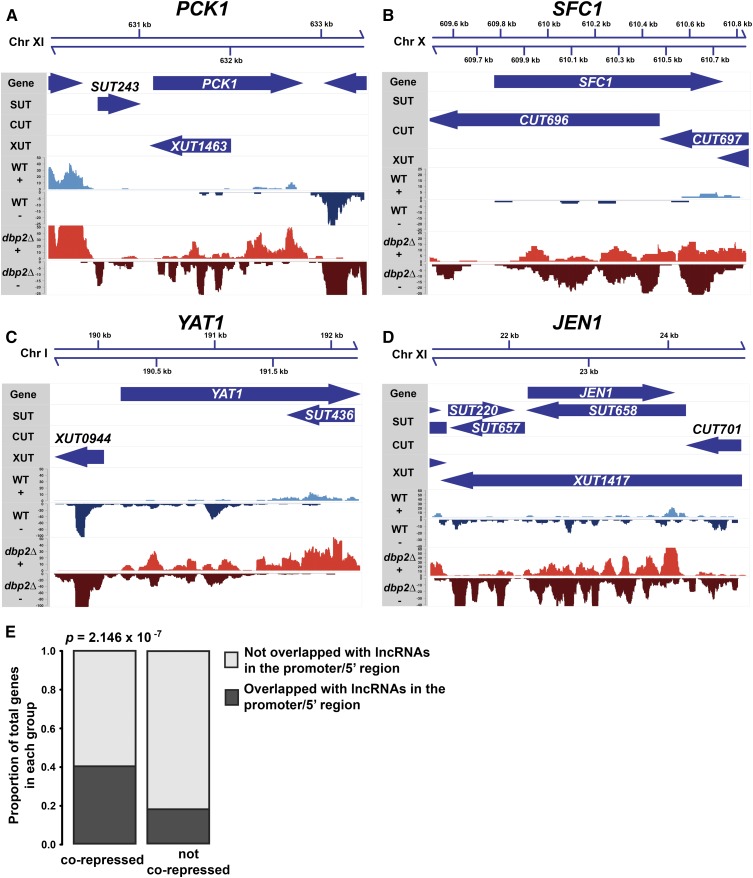
Besides respiration, genes involved in gluconeogenesis and utilization of nonfermentable carbon sources are also repressed in the presence of glucose and are corepressed by DBP2 and CYC8 or TUP1 (Table 5, Table 6, and Table 7) (Gancedo 1998; Broach 2012). These include PCK1, SFC1, YAT1, and JEN1 (Valdés-Hevia et al. 1989; Schmalix and Bandlow 1993; Palmieri et al. 1997; Casal et al. 1999; Akita et al. 2000; Turcotte et al. 2010). Work from our laboratory has shown that DBP2 promotes Cyc8-dependent repression at the GAL genes by antagonizing long noncoding RNAs (lncRNAs) that would otherwise promote induction (Cloutier et al. 2013, 2016). Interestingly, annotated lncRNAs are found at the promoter and/or 5′ regions of glucose-repressed genes in dbp2∆ cells including PCK1, SFC1, YAT1, and JEN1 (Figure 4, A–D). For example, the 5′ region of PCK1 is overlapped with XUT1463 in dbp2∆ (Figure 4A, dbp2∆ − strand) whereas this transcript is not readily detected in wild type (Figure 4A, WT − strand). Similarly, the promoter and/or 5′ regions of SFC1, YAT1, and JEN1 are overlapped with annotated lncRNAs including CUT696 (SFC1; Figure 4B, dbp2∆ − strand), XUT0944 (YAT1; Figure 4C, WT and dbp2∆ − strands) and SUT657 (JEN1; Figure 4D, WT and dbp2∆ − strands). To determine if there is a propensity for lncRNAs to cover the promoter and/or 5′ regions of genes corepressed by DBP2, CYC8, and TUP1, we analyzed the distribution of lncRNAs across the corepressed genes in comparison to unaffected genes. This revealed that ∼40% of the corepressed genes are overlapped with annotated lncRNAs within the promoter and/or 5′ end (Figure 4E). This ratio is significantly higher than that observed in genes not corepressed by CYC8, TUP1, and DBP2 (Figure 4E, proportion test, P = 2.146 × 10−7). This suggests that the expression of CYC8, TUP1, and DBP2 corepressed genes may be regulated by lncRNAs similarly to the GAL gene cluster (Cloutier et al. 2016).
Figure 4.
DBP2/CYC8/TUP1 corepressed genes are overlapped by lncRNAs. (A–D) Genomic tracks of PCK1, SFC1, YAT1, JEN1, and associated lncRNAs from dbp2∆ and wild type (WT). Normalized strand-specific genome tracks of PCK1 (A), SFC1 (B), YAT1 (C), and JEN1 (D) in wild type and dbp2∆ are presented at the bottom (GSE99097). Gene and annotated lncRNA (CUTs, SUTs, and XUTs) tracks (Xu et al. 2009; Wery et al. 2016) are shown at the top. The names of short transcripts are listed above the corresponding arrow. (E). Gene start region of corepressed genes are frequently overlapped by lncRNAs. Gene start region is defined as the region of 100 bp upstream plus the first 100 bp of ORF. The P-value indicates significant enrichment of genes overlapped with annotated lncRNAs among all of the corepressed genes (proportional test, one-sided). Chr, chromosome.
Loss of DBP2 results in increased expression of nonfermentable carbon utilization genes upon a nutrient shift
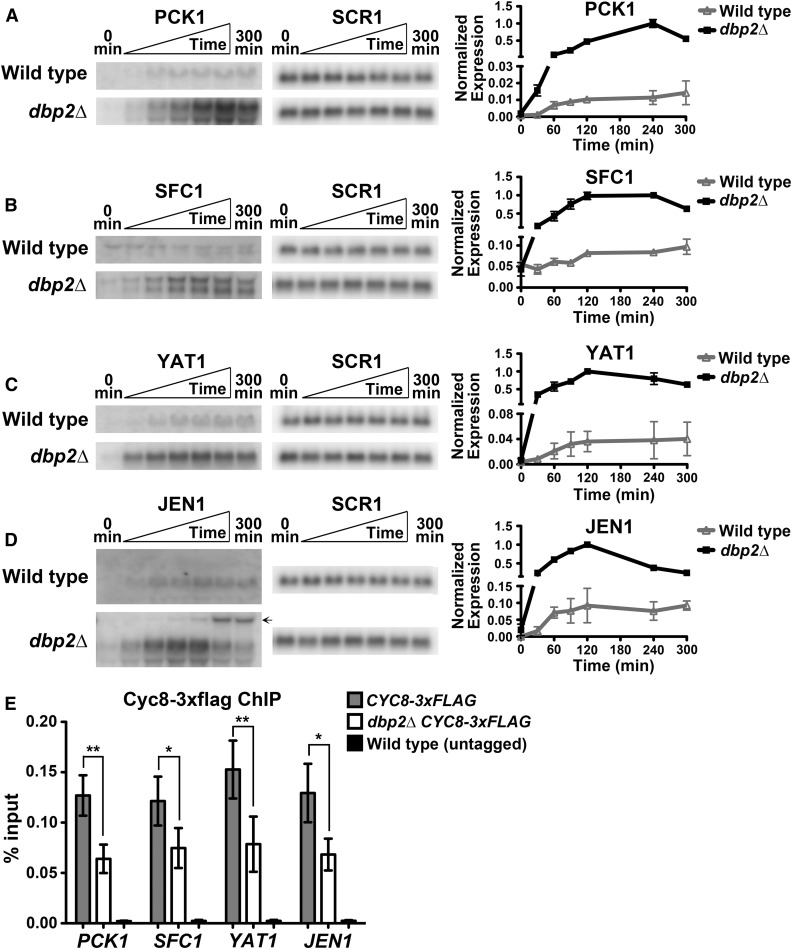
Upregulation of GAL lncRNAs in the absence of DBP2 causes reduced Cyc8 binding and derepression of the GAL cluster genes (Cloutier et al. 2013, 2016). To determine if these corepressed genes are also derepressed in the absence of DBP2, we conducted transcription induction assays prior to and at different time points following a nutritional shift from glucose to the nonfermentable carbon source acetate (Figure 5, A–D). This revealed increased expression of all four genes in dbp2∆ cells as compared to wild type, consistent with derepression. We also observed induction of a larger transcript overlapping JEN1 in dbp2∆ cells, presumably corresponding to XUT1417 (Figure 5D, arrow; see Figure 4D). Interestingly, levels of this transcript are inversely proportional to JEN1 mRNA, suggesting that transcription of these two RNAs is mutually exclusive.
Figure 5.
Loss of DBP2 results in reduced Cyc8 binding and rapid induction of genes involved in utilizing nonfermentable carbon source. (A–D) Transcriptional induction assays of PCK1 (A), SFC1 (B), YAT1 (C), and JEN1 (D) were conducted by shifting wild-type and dbp2∆ cells from SC+2% glucose to SC+1% potassium acetate media. RNA was collected from cells prior to (0 min) and at 30, 60, 90, 120, 240, and 300 min following nutritional shift. Transcripts were detected by Northern blotting using 32P-labeled, dsDNA probes. SCR1 serves as an internal control. Resulting transcript profiles from two biological replicates (error bars indicate ± SD) were normalized to SCR1 and plotted over time (right). The normalized expression of the time points with maximum transcript levels in each induction profile was set to one. The transcript, presumably XUT1417, is labelled ←. (E) ChIP of Cyc8-3xFLAG in wild-type and dbp2∆ cells. Wild-type and dbp2∆ strains harboring 3xFLAG-tagged CYC8 at the endogenous locus were cultivated in YP+2% glucose. ChIP was conducted using an anti-FLAG antibody. A wild-type strain with untagged CYC8 was used as negative control. Results were represented as percentage of immunoprecipitated DNA over input and shown as mean ± SD of three independent biological replicates. *P < 0.05, **P < 0.01.
To determine if loss of Cyc8 at gene promoters could account for derepression, ChIP of Cyc8-3xFLAG was conducted in wild type and dbp2∆ (Figure 5E). Consistent with our prior findings at the GAL gene cluster (Cloutier et al. 2016), we observed reduced Cyc8 binding to the promoters of PCK1, SFC1, YAT1, and JEN1 in dbp2∆ cells as compared to wild type (Figure 5E). This suggests that Dbp2 promotes efficient Cyc8 binding at target genes, providing a mechanism for coregulation of glucose repression. In conjunction with our prior studies showing that Dbp2 localization is responsive to nutrient availability (Beck et al. 2014), we propose that Dbp2 integrates nutritional status with metabolic gene regulation by maintaining Cyc8-dependent repression of lncRNA-targeted genes in the presence of glucose. This provides a putative mechanism for regulation of cellular metabolism by an RNA helicase, suggestive of a unique mechanism to regulate energy homeostasis in rapidly growing cells.
Discussion
Cells fine tune their metabolic programs according to nutrient availability in order to maintain homeostasis. Here, we demonstrate that the DEAD-box RNA helicase Dbp2 is a key component of this metabolic network, mediating glucose repression of metabolic genes through modulating Cyc8 association to the chromatin, and this role may involve lncRNAs. DEAD-box proteins are the largest group of RNA helicases in the helicase enzyme family and have been found in all domains of life (Linder and Jankowsky 2011). Dbp2, a DEAD-box protein in S. cerevisiae, functions as a key player of proper gene expression (Cloutier et al. 2012; Beck et al. 2014). It is proposed that Dbp2 promotes assembly of proteins onto nascent RNA during transcription (Ma et al. 2013), a role that may involve active remodeling of the RNA secondary structure and/or RNA-protein complexes. Interestingly, Dbp2 is actively shunted to the cytoplasm upon glucose deprivation or a switch to another carbon source (Beck et al. 2014), suggesting that Dbp2 may have a much more widespread role in metabolism, and its effects allow yeast cells to rapidly adapt to the environmental change.
The Cyc8-Tup1 complex in S. cerevisiae represses hundreds of genes including glucose-repressed genes, oxygen-regulated genes, and cell-type-specific genes (DeRisi et al. 1997; Green and Johnson 2004). Mutation or deletion of CYC8 or TUP1 leads to pleotropic effects including defects in carbon catabolite repression (Gancedo 1998; Smith and Johnson 2000). Interestingly, our study shows that CYC8 and DBP2, but not TUP1, repress respiration; one of several pathways that is repressed in the presence of sufficient extracellular glucose. This is surprising as Cyc8 and Tup1 are thought to function primarily as a complex. This suggests that Cyc8 may have functions outside of Tup1, a role supported by the unique distribution of transcript abundance between cyc8∆ and tup1∆ cells outside of those identified as coregulated (Figure 2B, right). A previous study has shown that simultaneous deletion of CYC8 and TUP1 results in higher invertase activity, as a result of SUC2 derepression, than either of the single deletions alone (Williams and Trumbly 1990). This suggests that these two factors have independent as well as shared activities.
lncRNAs are RNAPII products that lack an ORF and are >200 nt in length (Kornienko et al. 2013). Emerging evidence suggests many lncRNAs function in gene regulation of protein-coding gene transcripts by acting as decoy, guide, or scaffold of other regulatory factors or through transcription interference (Wang and Chang 2011; Kornienko et al. 2013). We have previously reported that Dbp2-dependent repression of the GAL cluster genes is dependent on expression of lncRNAs associated with the GAL cluster (Cloutier et al. 2016). By analyzing the presence of lncRNAs at corepressed genes by DBP2 and CYC8, we now show that a significant fraction of these genes are overlapped by lncRNAs encoded in cis; suggesting a genome-wide link between Dbp2, lncRNAs, and glucose repression. Interestingly, recent studies have connected several lncRNAs to glucose homeostasis in mammalian cells (Sun and Wong 2016). For example, H19 functions in glucose homeostasis through the bioavailability of microRNA (miRNA) let-7 that inhibits the expression of insulin receptor and lipoprotein lipase genes (Kallen et al. 2013; Gao et al. 2014). Another example is the lncRNA UCA1, which has been shown to promote glycolysis by activating mTOR-STAT3 and repressing miRNA143 in cancer cells (Li et al. 2014). A recent genome study has revealed >350 metabolically sensitive lncRNAs in mice (Yang et al. 2016). This suggests that lncRNAs can act as key regulators in metabolism.
Taken together, our study suggests that Dbp2 integrates nutritional availability with metabolic adaptation. This may also be the case for DDX5, the mammalian ortholog of Dbp2, as knockdown affects glucose metabolic genes, reduces glycolysis, and facilitates respiration in mammalian cells (Mazurek et al. 2014; Xing et al. 2017). Future studies are necessary to determine if the role of Dbp2 in cellular metabolism is conserved in mammalian cells. High fermentation rate suppresses the respiratory network, a phenomenon referred to as the Crabtree effect (De Deken 1966). Many tumor cells, on the other hand, exhibit enhanced glycolic activities and impaired respiratory activities, similar to the preference of S. cerevisiae in undergoing aerobic fermentation (Diaz-Ruiz et al. 2011). This drastic change in metabolic activities, which is referred to as the Warburg effect, is a hallmark of many cancer cells (Diaz-Ruiz et al. 2011). Thus, understanding how yeast cells control their metabolism has great implications on human diseases.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.041814/-/DC1.
Acknowledgments
We thank Scott Briggs for helpful comments on this manuscript. We also thank members of the Tran laboratory for scientific discussion. This work was supported by National Institutes of Health R01 GM-097332 to E.J.T. and P30 CA-023168 for core facilities at the Purdue University Center for Cancer Research.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Akita O., Nishimori C., Shimamoto T., Fujii T., Iefuji H., 2000. Transport of pyruvate in Saccharomyces cerevisiae and cloning of the gene encoded pyruvate permease. Biosci. Biotechnol. Biochem. 64: 980–984. [DOI] [PubMed] [Google Scholar]
- Balasubramanian B., Lowry C. V., Zitomer R. S., 1993. The Rox1 repressor of the Saccharomyces cerevisiae hypoxic genes is a specific DNA-binding protein with a high-mobility-group motif. Mol. Cell. Biol. 13: 6071–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas D., Ronne H., 1995. Three subunits of the RNA polymerase II mediator complex are involved in glucose repression. Nucleic Acids Res. 23: 4426–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck Z. T., Cloutier S. C., Schipma M. J., Petell C. J., Ma W. K., et al. , 2014. Regulation of glucose-dependent gene expression by the RNA helicase Dbp2 in Saccharomyces cerevisiae. Genetics 198: 1001–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodina I., Nielsen J., 2014. Advances in metabolic engineering of yeast Saccharomyces cerevisiae for production of chemicals. Biotechnol. J. 9: 609–620. [DOI] [PubMed] [Google Scholar]
- Broach J. R., 2012. Nutritional control of growth and development in yeast. Genetics 192: 73–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza M. J., Florens L., Swanson S. K., Shia W. J., Anderson S., et al. , 2005. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim. Biophys. Acta. 1731: 77–87. [DOI] [PubMed] [Google Scholar]
- Casal M., Paiva S., Andrade R. P., Gancedo C., Leão C., 1999. The lactate-proton symport of Saccharomyces cerevisiae is encoded by JEN1. J. Bacteriol. 181: 2620–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier S. C., Ma W. K., Nguyen L. T., Tran E. J., 2012. The DEAD-box RNA helicase Dbp2 connects RNA quality control with repression of aberrant transcription. J. Biol. Chem. 287: 26155–26166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier S. C., Wang S., Ma W. K., Petell C. J., Tran E. J., 2013. Long noncoding RNAs promote transcriptional poising of inducible genes. PLoS Biol. 11: e1001715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier S. C., Wang S., Ma W. K., Al Husini N., Dhoondia Z., et al. , 2016. Regulated formation of lncRNA-DNA hybrids enables faster transcriptional induction and environmental adaptation. Mol. Cell 61: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M., Dzamba M., Lister D., Ilie L., Brudno M., 2011. SHRiMP2: sensitive yet practical short read mapping. Bioinformatics 27: 1011–1012. [DOI] [PubMed] [Google Scholar]
- De Deken R. H., 1966. The Crabtree effect: a regulatory system in yeast. J. Gen. Microbiol. 44: 149–156. [DOI] [PubMed] [Google Scholar]
- de Nadal E., Casadomé L., Posas F., 2003. Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol. Cell. Biol. 23: 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi J. L., Iyer V. R., Brown P. O., 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680–686. [DOI] [PubMed] [Google Scholar]
- DeVit M. J., Johnston M., 1999. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr. Biol. 9: 1231–1241. [DOI] [PubMed] [Google Scholar]
- DeVit M. J., Waddle J. A., Johnston M., 1997. Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell 8: 1603–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Ruiz R., Rigoulet M., Devin A., 2011. The Warburg and Crabtree effects: on the origin of cancer cell energy metabolism and of yeast glucose repression. Biochim. Biophys. Acta 1807: 568–576. [DOI] [PubMed] [Google Scholar]
- Flick J. S., Johnston M., 1991. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol. Cell. Biol. 11: 5101–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeberg M. A., Han T., Moresco J. J., Kong A., Yang Y.-C., et al. , 2013. Pervasive and dynamic protein binding sites of the mRNA transcriptome in Saccharomyces cerevisiae. Genome Biol. 14: R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo J. M., 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62: 334–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Wu F., Zhou J., Yan L., Jurczak M. J., et al. , 2014. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res. 42: 13799–13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., et al. , 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green S. R., Johnson A. D., 2004. Promoter-dependent roles for the Srb10 cyclin-dependent kinase and the Hda1 deacetylase in Tup1-mediated repression in Saccharomyces cerevisiae. Mol. Biol. Cell 15: 4191–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Chen P., Chen S. Y., Vancura K. L., Michaelis S., et al. , 1991. RAM2, an essential gene of yeast, and RAM1 encode the two polypeptide components of the farnesyltransferase that prenylates a-factor and Ras proteins. Proc. Natl. Acad. Sci. USA 88: 11373–11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner C. J., Thompson C. M., Zhang J., Chao D. M., Liao S. M., et al. , 1995. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 9: 897–910. [DOI] [PubMed] [Google Scholar]
- Huang D. W., Lempicki R. A., Sherman B. T., 2009a Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A., 2009b Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen T., de Vlieg J., Alkema W., 2008. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen A. N., Zhou X. B., Xu J., Qiao C., Ma J., et al. , 2013. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 52: 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayikci Ö., Nielsen J., 2015. Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 15: fov068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher C. A., Redd M. J., Schultz J., Carlson M., Johnson A. D., 1992. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68: 709–719. [DOI] [PubMed] [Google Scholar]
- Kemmeren P., Sameith K., Van De Pasch L. A. L., Benschop J. J., Lenstra T. L., et al. , 2014. Large-scale genetic perturbations reveal regulatory networks and an abundance of gene-specific repressors. Cell 157: 740–752. [DOI] [PubMed] [Google Scholar]
- Kemmler S., Occhipinti L., Veisu M., Panse V. G., 2009. Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J. Cell Biol. 186: 863–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornienko A. E., Guenzl P. M., Barlow D. P., Pauler F. M., 2013. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 11: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimer F. W., Stevens A., 1990. Disruption of the gene XRN1, coding for a 5′ → 3′ exoribonuclease, restricts yeast cell growth. Gene 95: 85–90. [DOI] [PubMed] [Google Scholar]
- Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., et al. , 2013. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9: e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Li X., Wu S., Xue M., Chen W., 2014. Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci. 105: 951–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P., Jankowsky E., 2011. From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12: 505–516. [DOI] [PubMed] [Google Scholar]
- Lo K. Y., Li Z., Wang F., Marcotte E. M., Johnson A. W., 2009. Ribosome stalk assembly requires the dual-specificity phosphatase Yvh1 for the exchange of Mrt4 with P0. J. Cell Biol. 186: 849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W. K., Cloutier S. C., Tran E. J., 2013. The DEAD-box protein Dbp2 functions with the RNA-binding protein Yra1 to promote mRNP assembly. J. Mol. Biol. 425: 3824–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion R. M., Regev A., Segal E., Barash Y., Koller D., et al. , 2004. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. USA 101: 14315–14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. [Google Scholar]
- Mazurek A., Park Y., Miething C., Wilkinson J. E., Gillis J., et al. , 2014. Acquired dependence of acute myeloid leukemia on the DEAD-box RNA helicase DDX5. Cell Rep. 7: 1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D. J., Chen Y., Smyth G. K., 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40: 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, M., H. Pagès, V. Obenchain, and N. Hayden, 2010 Rsamtools: binary alignment (BAM), FASTA, variant call (BCF), and tabix file import. R Package version 1.22.0. Available at: http://bioconductor.org/packages/release/bioc/html.
- Nehlin J. O., Carlberg M., Ronne H., 1991. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 10: 3373–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosaka K., Onozuka M., Konno H., Kawasaki Y., Nishimura H., et al. , 2005. Genetic regulation mediated by thiamin pyrophosphate-binding motif in Saccharomyces cerevisiae. Mol. Microbiol. 58: 467–479. [DOI] [PubMed] [Google Scholar]
- O’Duibhir E., Lijnzaad P., Benschop J. J., Lenstra T. L., van Leenen D., et al. , 2014. Cell cycle population effects in perturbation studies. Mol. Syst. Biol. 10: 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L., Lasorsa F. M., De Palma A., Palmieri F., Runswick M. J., et al. , 1997. Identification of the yeast ACR1 gene product as a succinate-fumarate transporter essential for growth on ethanol or acetate. FEBS Lett. 417: 114–118. [DOI] [PubMed] [Google Scholar]
- R Development Core Team , 2016. R: A Language and Environment for Statistical Computing R Foundation Statistical Computuing, Vienna, Austria. [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalix W., Bandlow W., 1993. The ethanol-inducible YAT1 gene from yeast encodes a presumptive mitochondrial outer carnitine acetyltransferase. J. Biol. Chem. 268: 27428–27439. [PubMed] [Google Scholar]
- Schüller H.-J., 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 43: 139–160. [DOI] [PubMed] [Google Scholar]
- Sellick C. A., Campbell R. N., Reece R. J., 2008. Galactose metabolism in yeast-structure and regulation of the leloir pathway enzymes and the genes encoding them. Int. Rev. Cell Mol. Biol. 269: 111–150. [DOI] [PubMed] [Google Scholar]
- Skowyra D., Craig K. L., Tyers M., Elledge S. J., Harper J. W., 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91: 209–219. [DOI] [PubMed] [Google Scholar]
- Smith R. L., Johnson A. D., 2000. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25: 325–330. [DOI] [PubMed] [Google Scholar]
- Soontorngun N., Larochelle M., Drouin S., Robert F., Turcotte B., 2007. Regulation of gluconeogenesis in Saccharomyces cerevisiae is mediated by activator and repressor functions of Rds2. Mol. Cell. Biol. 27: 7895–7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Wong D., 2016. Long noncoding RNA-mediated regulation of glucose homeostasis and diabetes. Am. J. Cardiovasc. Dis. 6: 17–25. [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treitel M. A., Kuchin S., Carlson M., 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 6273–6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte B., Liang X. B., Robert F., Soontorngun N., 2010. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 10: 2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-Hevia M. D., de la Guerra R., Gancedo C., 1989. Isolation and characterization of the gene encoding phosphoenolpyruvate carboxykinase from Saccharomyces cerevisiae. FEBS Lett. 258: 313–316. [DOI] [PubMed] [Google Scholar]
- Varanasi U. S., Klis M., Mikesell P. B., Trumbly R. J., 1996. The Cyc8 (Ssn6)-Tup1 corepressor complex is composed of one Cyc8 and four Tup1 subunits. Mol. Cell. Biol. 16: 6707–6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. C., Chang H. Y., 2011. Molecular mechanisms of long noncoding RNAs. Mol. Cell 43: 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver P. L., Sun C., Chang T. H., 1997. Dbp3p, a putative RNA helicase in Saccharomyces cerevisiae, is required for efficient pre-rRNA processing predominantly at site A3. Mol. Cell. Biol. 17: 1354–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wery M., Descrimes M., Vogt N., Dallongeville A. S., Gautheret D., et al. , 2016. Nonsense-mediated decay restricts LncRNA levels in yeast unless blocked by double-stranded RNA structure. Mol. Cell 61: 379–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams F. E., Trumbly R. J., 1990. Characterization of TUP1, a mediator of glucose repression in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 6500–6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Z., Wang S., Tran E. J., 2017. Characterization of the mammalian DEAD-box protein DDX5 reveals functional conservation with S. cerevisiae ortholog Dbp2 in transcriptional control and glucose metabolism. RNA . 10.1261/rna.060335.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Münster S., et al. , 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Li P., Yang W., Ruan X., Kiesewetter K., et al. , 2016. Integrative transcriptome analyses of metabolic responses in mice define pivotal lncRNA metabolic regulators. Cell Metab. 24: 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W., King D. A., Beckwith S. L., Gowans G. J., Yen K., et al. , 2016. The INO80 complex requires the Arp5-Ies6 subcomplex for chromatin-remodeling and metabolic regulation. Mol. Cell. Biol. 36: 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Fung-Leung W. P., Bittner A., Ngo K., Liu X., 2014. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One 9: e78644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitomer R. S., Lowry C. V., 1992. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev. 56: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reference numbers deposited in the Gene Expression Omnibus are as follows: GSE58097 (Beck et al. 2014), GSE42527 (Kemmeren et al. 2014), and GSE43747 (Freeberg et al. 2013), GSE99097.