Abstract
The combination of experimental evolution with high-throughput sequencing of pooled individuals—i.e., evolve and resequence (E&R)—is a powerful approach to study adaptation from standing genetic variation under controlled, replicated conditions. Nevertheless, E&R studies in Drosophila melanogaster have frequently resulted in inordinate numbers of candidate SNPs, particularly for complex traits. Here, we contrast the genomic signature of adaptation following ∼60 generations in a novel hot environment for D. melanogaster and D. simulans. For D. simulans, the regions carrying putatively selected loci were far more distinct, and thus harbored fewer false positives, than those in D. melanogaster. We propose that species without segregating inversions and higher recombination rates, such as D. simulans, are better suited for E&R studies that aim to characterize the genetic variants underlying the adaptive response.
Keywords: experimental evolution, evolve and resequence, Drosophila simulans, Drosophila melanogaster, chromosomal inversions
Standing genetic variation in natural populations underlies their potential to adapt to novel environments. The evolve and resequence (E&R) approach (Turner et al. 2011), which combines experimental evolution with sequencing of pooled individuals (Pool-Seq) (Schlötterer et al. 2014), provides an excellent opportunity to understand how this standing genetic variation is being used to fuel adaptation of the evolving populations (Schlötterer et al. 2015; Long et al. 2015). Because experimental evolution permits the analysis of replicate populations, which have evolved from the same standing genetic variation under identical culture conditions, it is possible to distinguish selection from random, nondirectional changes (Kawecki et al. 2012; Schlötterer et al. 2015).
A short generation time and high levels of polymorphism, in combination with a small, well-annotated genome, has made Drosophila melanogaster the preferred sexual model organism to study the genomic response to truncating selection. Many traits such as aging (Remolina et al. 2012), courtship song (Turner et al. 2013), hypoxia (Zhou et al. 2011; Jha et al. 2016), body size (Turner et al. 2011), egg size (Jha et al. 2015), development time (Burke et al. 2010; Graves et al. 2017), and Drosophila C virus (DCV) resistance (Martins et al. 2014) have already been studied. The E&R approach has also been applied to laboratory natural selection experiments, in which differential reproductive success is the sole driver of adaptation to novel environments such as elevated temperature (Orozco-terWengel et al. 2012; Tobler et al. 2014; Franssen et al. 2015) and high cadmium and salt concentration (Huang et al. 2014). For traits with a simple genetic basis, such as DCV resistance, E&R has identified causal genes (Martins et al. 2014); on the other hand, identification of the genetic basis of polygenic traits has been considerably more challenging because of the large size of the genomic regions that have been identified (Burke et al. 2010; Turner et al. 2011; Zhou et al. 2011; Orozco-terWengel et al. 2012; Remolina et al. 2012; Tobler et al., 2014). These genomic regions often contain a substantial number of candidate SNPs that are mostly false positives (Nuzhdin and Turner 2013; Tobler et al. 2014; Franssen et al. 2015). The inflated numbers of false positives can be partly attributed to linkage disequilibrium (LD) and long-range hitchhiking caused by low frequency adaptive alleles (Tobler et al. 2014; Franssen et al. 2015). Other important factors contributing to the large number of false positives include (1) reduced recombination rates close to the centromeres, and (2) the presence of large chromosomal inversions that suppress recombination and occasionally also respond to selection (Kapun et al. 2014).
D. simulans, a sister species of D. melanogaster, lacks large segregating inversions (Aulard et al. 2004), has higher recombination rate, and the centromeric recombinational suppression is restricted to a much smaller part of the chromosomes (True et al. 1996). These characteristics make D. simulans potentially more suitable for E&R studies (Kofler and Schlötterer 2014; Tobler et al. 2014). While the availability of genomic and functional resources is not comparable to D. melanogaster, improved genome assemblies and annotations are available for D. simulans (Hu et al. 2013; Palmieri et al. 2015).
In this study we contrast the genomic response of a D. simulans E&R study spanning 60 generations to D. melanogaster populations that have been evolving for the same length of time in the same hot temperature environment. Consistent with the absence of segregating inversions and higher recombination rate, the selection signatures in D. simulans result in substantially smaller genomic regions carrying putatively selected variants.
Materials and Methods
D. simulans experimental populations and selection regimes
202 isofemale lines were established from a natural D. simulans population collected in Tallahassee, Florida in November 2010. The isofemale lines were maintained in the laboratory for nine generations prior to the establishment of the founder populations to rule out infections and determine the species. Five mated females from each isofemale line were used to establish 10 replicates of the founder population, three of which were used in our study. They were maintained as independent replicates with a census population size of 1000 and ∼50:50 sex ratio. Both temperature and light was cycled every 12 hr between 18 and 28°, corresponding to night and day.
Genome sequencing, mapping, and SNP calling of D. simulans data
Genomic DNA was prepared for three founder replicates (females only) and three replicates of the evolved populations at generation 60 (mixed sexes). Details of DNA extraction and library preparation are summarized in Supplemental Material, Table S1. The average genome-wide sequence coverage across the founder and evolved population replicates was ∼259× and ∼100×, respectively.
Reads were trimmed using ReadTools version 0.2.1 (https://github.com/magicDGS/ReadTools) to remove low quality bases (Phred score <18) at 3′ end of reads (parameters: --minimum-length 50 --no-5p-trim --quality-threshold 18 --no-trim-quality). The trimmed reads were mapped using bwa (version 0.5.8c; aln algorithm; parameters: -o 1 -n 0.01 -l 200 -e 12 -d 12) (Li and Durbin 2009) to the D. simulans reference genome (Palmieri et al. 2015) on a Hadoop cluster with Distmap version 1.0 (Pandey and Schlötterer 2013). Reads in the bam files were sorted and duplicates were removed with Picard version 1.140 (http://broadinstitute.github.io/picard). Reads with low mapping quality and improper pairing were removed (parameters: -q 20 -f 0x0002 -F 0x0004 -F 0x0008) and the bam files were converted to mpileup files using SAMtools version 1.2 (Li et al. 2009). The mpileup files were converted to a synchronized pileup file using PoPoolation2 (parameter: --min-qual 20) (Kofler et al. 2011). Furthermore, repeats (identified by RepeatMasker, http://www.repeatmasker.org) and 5-bp regions flanking indels (identified by PoPoolation2: identify-genomic-indel-regions.pl --indel-window 5 --min-count 5) were masked using PoPoolation2 (identify-indel-regions.pl --min-count 2% of the average coverage across all founder libraries).
SNPs were called from the founder populations; in brief, initially the SNPs with minimum base quality of 40 present in at least one replicate of the three founder populations were selected for further analyses. To improve the reliability of the pipeline, the polymorphic sites lying in the upper and lower 1% tails of the coverage distribution (i.e., ≥423× and ≤11×, respectively; upper tail based on the library with the highest sequencing depth, lower tail estimated from total coverage of all replicates and time points) were removed. Further, we masked 200-bp flanking SNPs specific to autosomal genes translocated to the Y chromosome (R. Tobler, V. Nolte, and C. Schlötterer, unpublished data). In total, 4,391,296 SNPs on chromosomes 2 and 3 were used for subsequent analysis [644,423 SNPs on X chromosome were used for effective population size (Ne) estimation but were excluded from other analyses, see below]. For all SNP sites remaining after the filtering steps, we determined the allele frequencies using only reads with a quality score of at least 20 at the SNP position.
Genome sequencing and mapping of D. melanogaster data
The D. melanogaster data used in this study are part of an ongoing experiment (founder population from Orozco-terWengel et al. 2012; F59 populations from Franssen et al. 2015). Similar to D. simulans, temperature and light was cycled every 12 hr between 18 and 28°, corresponding to night and day. To increase the coverage of libraries for the founder populations, additional sequencing was performed (see Table S1 for details of DNA extraction and library preparation). The final average genome-wide coverage of the founder and evolved populations was ∼190× and ∼83×, respectively. Read processing and mapping are described in Tobler et al. (2014). Similar to the D. simulans data set, SNPs were called from the base population with base quality of 40. Then, SNPs lying in the upper and lower 1% tails of the coverage distribution (i.e., ≥328× and ≤9×, respectively; upper tail based on the library with the highest sequencing depth, lower tail estimated separately from total coverage of all replicates and time points) were removed. 2,934,945 SNPs on chromosomes 2 and 3 were used for further analyses. SNPs on the X chromosome (408,982) were only used for Ne estimation. Allele frequencies were determined based on reads with base quality of at least 20.
Candidate SNP inference in D. simulans and D. melanogaster
To compare the selected genomic regions between D. simulans and D. melanogaster, the sequencing reads were downsampled using Picard (DownsampleSam, http://broadinstitute.github.io/picard) to obtain similar mean genome-wide coverage of the libraries in both species (Table S2). To identify SNPs with pronounced allele frequency changes (AFC), we contrasted the founder and evolved populations (at generation 60 for D. simulans and 59 for D. melanogaster) using the Cochran–Mantel–Haenszel (CMH) test (Agresti 2002). For each species, we estimated Ne in windows of 1000 SNPs across all chromosomes and replicates using Nest (function estimateWndNe, method Np.planI; Jónás et al. 2016). Averaging the medians of the Ne values across replicates, we obtained the Ne estimate for autosomes and the X chromosome of each species. The estimated Ne for the X chromosome (Ne = 224) was approximately three-quarters of autosomes (Ne = 285) in D. simulans, whereas in D. melanogaster, the Ne of the X chromosome (Ne = 301) was 1.5 times higher than that of the autosomes (Ne = 201). This discrepancy in D. melanogaster has been noted before (Orozco-terWengel et al. 2012; Jónás et al. 2016) and has been attributed to an unbalanced sex ratio, background selection, and a larger number of SNPs being affected by selection on the autosomes. Because it is not clear whether these pronounced differences reflect differences in selection or mating patterns, we excluded the X chromosome from the analysis. Since the CMH test does not account for drift, we inferred candidate SNPs by simulating drift based on the inferred autosomal Ne estimates and determined an empirical CMH cutoff using a 2% false positive rate. Forward Wright–Fisher simulations were performed with independent loci using Nest (function wf.traj; Jónás et al. 2016). The simulation parameters (i.e., number of SNPs, allele frequencies in the founder populations, coverage of libraries, Ne, and number of replicates and generations) matched the experimental data. To infer the genomic regions under selection, we computed the average p-value of all candidate SNPs (above the empirical CMH cutoff: 31 for D. simulans and 27.32 for D. melanogaster) in 200-kb sliding windows with 100-kb overlap. Adjacent windows with the average p-value above the CMH cutoff were merged.
Data availability
The raw reads for all populations are available from the European Sequence Read Archive under the accession numbers mentioned in Table S1. SNP data sets in sync format (Kofler et al. 2011) are available from the Dryad Digital Repository under http://dx.doi.org/10.5061/dryad.p7c77.
Results
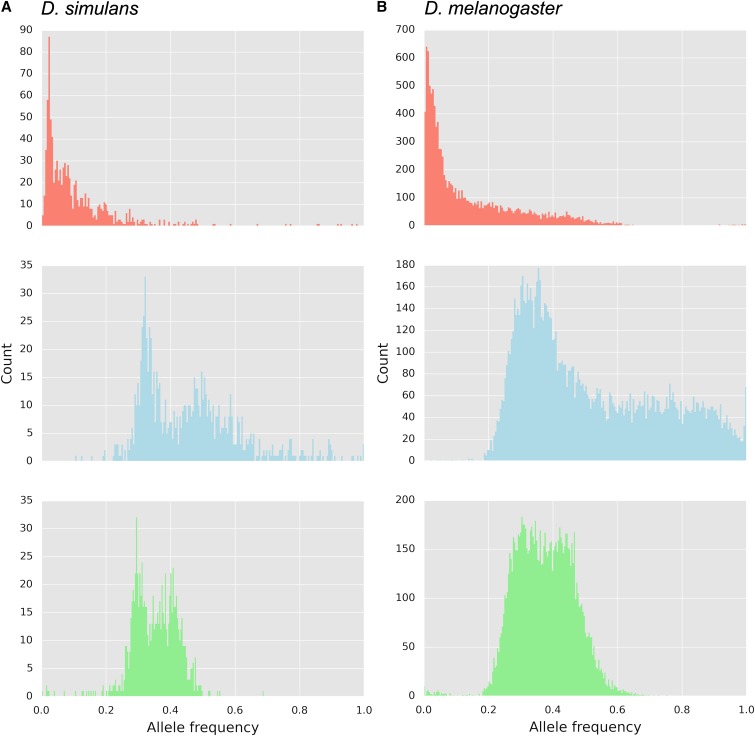
Three replicates of a D. simulans founder population were maintained in a hot temperature environment for 60 nonoverlapping generations. We sequenced pooled individuals of the three founder populations and three evolved populations, and compared these data to D. melanogaster, which evolved for 59 generations under the identical selection regime (Orozco-terWengel et al. 2012; Franssen et al. 2015). SNPs with pronounced AFC across the three replicates were identified with the CMH test by contrasting the founder and evolved populations of each species. While the CMH test is a powerful tool for identifying putative targets of selection (Kofler and Schlötterer 2014), it is not sufficient for determining which outlier loci are deviating from neutral expectations. Consequently, we estimated the Ne for each species based on genome-wide AFC between founder and evolved populations (Jónás et al. 2016). We then performed neutral simulations with the predicted Ne for autosomes to derive an empirical CMH cutoff based on a 2% false positive rate. We identified 918 candidate SNPs in D. simulans; whereas in D. melanogaster, 11,115 SNPs were identified as outliers (Figure 1 and Figure 2). In both species, the majority of candidate SNPs start from a low frequency. D. melanogaster has more SNPs starting at intermediate frequencies that reach higher frequencies after 59 generations (Figure 1). Nonetheless, despite the rapid frequency change in response to the hot environment, only a small fraction of candidate SNPs (1.2% in D. simulans and 6.3% in D. melanogaster) approached fixation (major allele frequency ≥0.9) after 60 generations.
Figure 1.
Allele frequency distribution of candidate SNPs averaged across replicates in (A) D. simulans and (B) D. melanogaster. Founder population (top panels), generation 60/59 (middle panels), and frequency change (bottom panels) of candidate SNPs. Candidate SNPs were determined from an empirical 2% false positive rate determined by neutral simulations assuming no linkage.
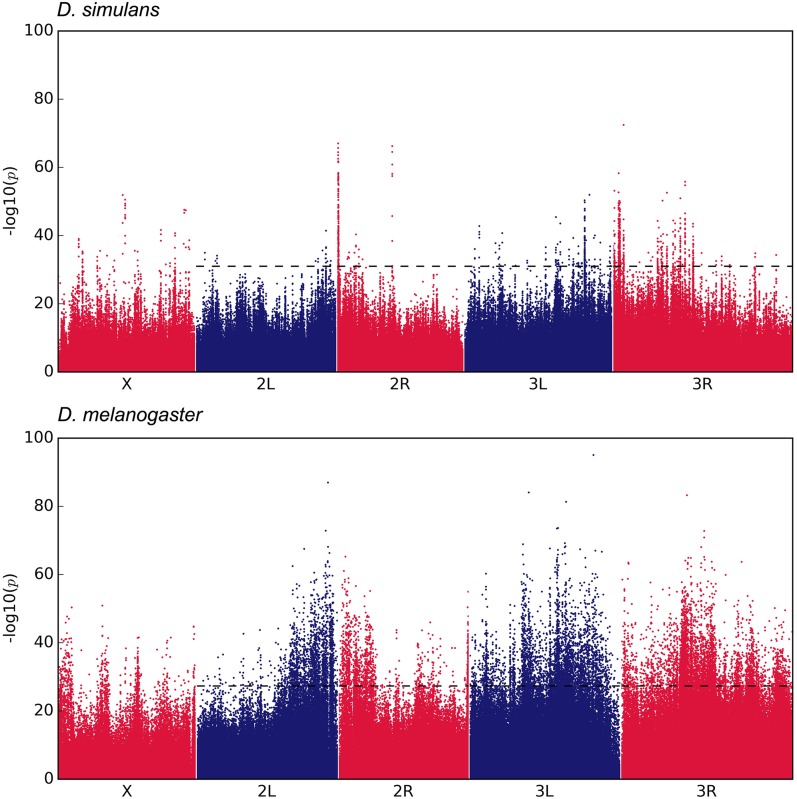
Figure 2.
The genomic distribution of candidate SNPs in D. simulans (top panel) and D. melanogaster (bottom panel): The Manhattan plots show the negative log10-transformed p-values of SNPs corresponding to the genomic positions. The p-values were determined using CMH test by comparing the founder and evolved populations using the same sequencing coverage for both species. The dotted lines show the CMH cutoff based on empirical 2% false positive rate determined by neutral simulations assuming no linkage. Because the relative Ne estimates of X chromosomes and autosomes were nonconcordant between both species, we did not determine outlier SNPs for the X chromosome.
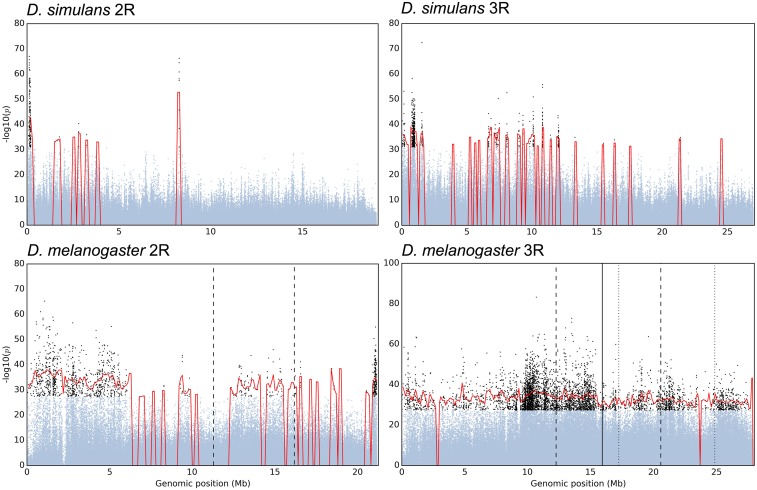
Neutral SNPs linked to a target of selection change their frequencies more than expected by chance, which results in a characteristic peak structure observed in Manhattan plots. Such selection peaks can be recognized above the dotted line, which separates candidate SNPs in D. simulans based on the empirical 2% false positive rate from nonselected SNPs (Figure 2, upper panel). Visual inspection of the Manhattan plots for both species revealed that D. simulans had narrower and more distinct peak structures than D. melanogaster (Figure 2). While a pronounced peak structure narrows the genomic region affected by selection, a peak also indicates that SNP-based analyses are not informative and can be misleading: many nonselected SNPs show a selection signature due to linkage with the target of selection. Thus, the identification of peak structures is a prerequisite for determining selection targets. To this end, we explored several peak finding procedures (e.g., Futschik et al. 2014; Beissinger et al. 2015), but complex selection signatures in our data sets makes this a challenging task, even for D. simulans which has much clearer peak structures. One of the challenges we encountered in our efforts to separate distinct peaks was that very narrow peaks were not recognized due to too few candidate SNPs. We therefore employed an approximate method to determine the fraction of the genome affected by selection. Averaging the p-value of all candidate SNPs in 200-kb sliding windows, we distinguished between regions influenced by directional selection from those evolving neutrally (Figure 3 and Figures S1 and S2 in File S1). Using this method, we detected 46 peaks covering ∼22.6 Mb (25.3% of chromosomes 2 and 3 of the reference genome) in D. simulans, and 31 peaks covering 84.4 Mb (87.4%) in D. melanogaster. Particularly striking is the difference between the two species on chromosome 3R, which contains three segregating, overlapping inversions [In(3R)Payne, In(3R)Mo, and In(3R)C] in the D. melanogaster population (Kapun et al. 2014). Almost the entire D. melanogaster 3R chromosome was characterized as a genomic region affected by selection, while in D. simulans several distinct peaks could be recognized (Figure 3). Moreover, in chromosome 2 of D. melanogaster, the regions near the centromere spanning to both chromosome arms contained numerous candidate SNPs forming broad peaks, probably due to a reduced recombination in this region (Figure 2, Figure 3, and Figure S1 in File S1).
Figure 3.
Identification of selected regions on two chromosome arms: Manhattan plots of chromosome arms 2R (left panels) and 3R (right panels) are shown for D. simulans (top panels) and D. melanogaster (bottom panels). The CMH p-values of candidate SNPs (black dots) were averaged across 200-kb windows, over sliding intervals every 100 kb. Adjacent windows with average p-values above CMH cutoffs (see Materials and Methods) were merged (red lines). Boundaries of the inversion in 2R [In(2R)Ns] are shown in dashed lines. Three overlapping inversions in 3R, i.e., In(3R)Payne, In(3R)Mo, and In(3R)C are indicated with dashed, dotted, and solid lines, respectively.
Discussion
The different genomic signatures of D. simulans and D. melanogaster induced by adaptation to high temperature can be attributed to species-specific characteristics and the design of the experimental study. Factors such as chromosomal inversions and low recombination regions can be associated with broad genomic regions determined to be under selection in D. melanogaster. Large chromosomal inversions are common in natural D. melanogaster populations, suppressing recombination over extensive genomic regions (Kirkpatrick 2010). In the D. melanogaster experimental populations, these inversions have contributed in two ways to the large number of observed candidate SNPs: first, the suppression of recombination has resulted in the association of SNPs effectively across entire chromosomes—even under the influence of drift alone. Second, inversion In(3R)C showed a consistent increase in frequency across multiple replicates, suggesting that it harbors, or is linked to, some selection targets (figure 4 in Kapun et al. 2014), exacerbating the impact of the inversions on chromosome 3R. On top of this, in D. melanogaster large parts of the chromosomes are affected by the reduced recombination rate toward the centromeres (True et al. 1996 and Comeron et al. 2012). Consistent with low recombination affecting the selection signature, we observed a broad peak near the centromere in chromosome 2 spanning both chromosome arms (Figure 2, bottom panel; Figure 3; and Figure S1 in File S1). The impact of recombination was also noted previously (Franssen et al. 2015), where low recombination regions were associated with high LD (1–10 Mb) in a D. melanogaster experimental evolution data set.
Because the selection signature in D. melanogaster extends to linked neutral SNPs over large genomic regions, almost no specific selected targets could be distinguished. However, several regions with presumably distinct selection targets could be identified for D. simulans (Figure 2 and Figure 3), a species that lacks large segregating inversions (Aulard et al. 2004) and has a 1.3× higher genome-wide recombination rate, with a much less pronounced recombination depression close to centromeres and telomeres (True et al. 1996). Contrasting the patterns of nucleotide polymorphism in natural populations of both species (Nolte et al. 2013), the difference in recombination landscape between D. melanogaster and D. simulans is evident (Figures S3–S6 in File S1), suggesting the smaller genomic region with suppressed recombination toward the centromeres in D. simulans contributes to a clearer selection signature.
In D. melanogaster, it has been proposed that LD and long-range hitchhiking caused by low frequency adaptive alleles result in a large number of false positive candidate SNPs (Tobler et al. 2014; Franssen et al. 2015). The D. simulans founder population had more LD than the D. melanogaster population (D. Gómez-Sánchez, R. Poupardin, V. Nolte, and C. Schlötterer, unpublished data; Figure S7 in File S1), which is most likely a consequence of their different demographic histories (Hamblin and Veuille 1999 and the references therein). This increased LD is expected to have the opposite effect, resulting in a higher mapping accuracy in D. melanogaster. However, our results indicate that despite higher LD in D. simulans, the genomic regions under selection in this species are still narrower than in D. melanogaster. One alternative explanation for the difference in mapping resolution of D. melanogaster and D. simulans may be that the genetic architecture of adaptation differs between the two species. Nevertheless, we consider this unlikely. First, most of the candidates in both D. melanogaster and D. simulans (Figure 1) start from low frequency, indicating that selection is acting on rare variants in both species. Hence, it is not likely that the selected alleles occurring at lower frequency in D. melanogaster would result in more hitchhiking of linked variants than in D. simulans. Second, in natural populations the genetic architecture seems to be similar between the two species. In North America and Australia, parallel clines have been described for D. simulans and D. melanogaster (Reinhardt et al. 2014; Zhao et al. 2015; Machado et al. 2016; Sedghifar et al. 2016). Remarkably, more genes share the pattern of clinal variation in both species than expected by chance (Reinhardt et al. 2014; Machado et al. 2016; Sedghifar et al. 2016). Furthermore, several clinal genes were also differentially expressed at high and low temperatures in both D. melanogaster and D. simulans (Zhao et al. 2015). While the strength and stability of the clines differ between D. melanogaster and D. simulans, the observation that both species share genes with clinal variation and differential expression in response to temperature treatment, strongly suggests that there are similarities in the genomic architecture of temperature adaptation between both species.
Computer simulations indicated that the mapping accuracy increases with the number of founder chromosomes (Baldwin-Brown et al. 2014; Kofler and Schlötterer 2014; Kessner and Novembre 2015). The founder population of D. melanogaster encompassed only 113 isofemale lines, while the D. simulans experiment was started from 202 isofemale lines. Notably, while it is difficult to determine to what extent the various factors have contributed to the higher resolution of the D. simulans E&R study, the presence of distinct peaks in D. simulans suggests that the large segregating chromosomal inversions, low recombination rate, and, likely, fewer founder chromosomes, among other factors, contributed most to the low resolution of the D. melanogaster E&R experiment.
While the higher recombination rate in D. simulans and the absence of segregating inversions support our observation that D. simulans may be better suited for E&R studies than D. melanogaster, it is important to keep in mind that we tested only a single selection regime, and for other traits D. melanogaster may have a cleaner selection response. While possible, we do not consider this very likely because other studies selecting for different traits in D. melanogaster also identified a large number of loci deviating from neutral expectations (Burke et al. 2010; Turner et al. 2011; Zhou et al. 2011; Orozco-terWengel et al. 2012; Remolina et al. 2012; Tobler et al. 2014).
Our results show that using inversion-free D. simulans with low recombination depression toward the centromeres improves the resolution of E&R studies, resulting in identification of narrower and more precise genomic regions under selection than in D. melanogaster (Orozco-terWengel et al. 2012; Tobler et al. 2014; Franssen et al. 2015). Even though the selection signatures in D. simulans were substantially more distinct than those in D. melanogaster, we caution that subsequent characterization of the selection targets is still challenging. More refined methods need to be developed that separate the selection signatures from adjacent targets of selection by accounting for the differences in starting frequencies of the SNPs and selection intensities. Thus, the comparison of selection targets between both species is not informative unless at least for one species the target of selection can be further narrowed down using, for example, expression profiling in combination with Pool-Seq selection signatures. Furthermore, improvements in the experimental design, e.g., using more replicates and more founder chromosomes (Baldwin-Brown et al. 2014; Kofler and Schlötterer 2014; Kessner and Novembre 2015) can further increase the accuracy of mapping the selected targets.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.043349/-/DC1.
Acknowledgments
We acknowledge David Houle and Stephanie Schwinn (Florida State University) for providing resources and assistance in collecting the flies. We thank all members of the Institute of Population Genetics, in particular S. Franssen, P. Nouhaud, K. Otte, A. M. Langmüller, and T. Taus, for comments on an earlier version of manuscript. We also thank anonymous reviewers for highlighting the importance of the underlying genetic architecture. This work was supported by the European Research Council grant “ArchAdapt” and the Austrian Science Fund (FWF, W1225-B20).
Author contributions: C.S. designed the study and R.T. collected D. simulans flies and established the experimental evolution populations. V.N. performed DNA extractions and library preparations, and N.B. performed the analysis. N.B. and C.S. wrote the manuscript with contributions from R.T. and V.N. All authors approved the final manuscript.
Footnotes
Communicating editor: J. A. Birchler
Literature Cited
- Agresti A., 2002. Categorical Data Analysis. John Wiley & Sons, New York. [Google Scholar]
- Aulard S., Monti L., Chaminade N., Lemeunier F., 2004. Mitotic and polytene chromosomes: comparisons between Drosophila melanogaster and Drosophila simulans. Genetica 120: 137–150. [DOI] [PubMed] [Google Scholar]
- Baldwin-Brown J. G., Long A. D., Thornton K. R., 2014. The power to detect quantitative trait loci using resequenced, experimentally evolved populations of diploid, sexual organisms. Mol. Biol. Evol. 31: 1040–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissinger T. M., Rosa G. J. M., Kaeppler S. M., Gianola D., de Leon N., 2015. Defining window-boundaries for genomic analyses using smoothing spline techniques. Genet. Sel. Evol. 47: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke M. K., Dunham J. P., Shahrestani P., Thornton K. R., Rose M. R., et al. , 2010. Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature 467: 587–590. [DOI] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franssen S. U., Nolte V., Tobler R., Schlötterer C., 2015. Patterns of linkage disequilibrium and long range hitchhiking in evolving experimental Drosophila melanogaster populations. Mol. Biol. Evol. 32: 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futschik A., Hotz T., Munk A., Sieling H., 2014. Multiscale DNA partitioning: statistical evidence for segments. Bioinformatics 30: 2255–2262. [DOI] [PubMed] [Google Scholar]
- Graves J. L., Jr, Hertweck K. L., Phillips M. A., Han M. V., Cabral L. G., et al. , 2017. Genomics of parallel experimental evolution in Drosophila. Mol. Biol. Evol. 34: 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin M. T., Veuille M., 1999. Population structure among African and derived populations of Drosophila simulans: evidence for ancient subdivision and recent admixture. Genetics 153: 305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T. T., Eisen M. B., Thornton K. R., Andolfatto P., 2013. A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence. Genome Res. 23: 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Wright S. I., Agrawal A. F., 2014. Genome-wide patterns of genetic variation within and among alternative selective regimes. PLoS Genet. 10: e1004527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha A. R., Miles C. M., Lippert N. R., Brown C. D., White K. P., et al. , 2015. Whole-genome resequencing of experimental populations reveals polygenic basis of egg-size variation in Drosophila melanogaster. Mol. Biol. Evol. 32: 2616–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha A. R., Zhou D., Brown C. D., Kreitman M., Haddad G. G., et al. , 2016. Shared genetic signals of hypoxia adaptation in Drosophila and in high-altitude human populations. Mol. Biol. Evol. 33: 501–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jónás Á., Taus T., Kosiol C., Schlötterer C., Futschik A., 2016. Estimating the effective population size from temporal allele frequency changes in experimental evolution. Genetics 204: 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapun M., van Schalkwyk H., McAllister B., Flatt T., Schlötterer C., 2014. Inference of chromosomal inversion dynamics from Pool-Seq data in natural and laboratory populations of Drosophila melanogaster. Mol. Ecol. 23: 1813–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawecki T. J., Lenski R. E., Ebert D., Hollis B., Olivieri I., et al. , 2012. Experimental evolution. Trends Ecol. Evol. 27: 547–560. [DOI] [PubMed] [Google Scholar]
- Kessner D., Novembre J., 2015. Power analysis of artificial selection experiments using efficient whole genome simulation of quantitative traits. Genetics 199: 991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., 2010. How and why chromosome inversions evolve. PLoS Biol. 8: e1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R., Schlötterer C., 2014. A guide for the design of evolve and resequencing studies. Mol. Biol. Evol. 31: 474–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R., Pandey R. V., Schlötterer C., 2011. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 27: 3435–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long A., Liti G., Luptak A., Tenaillon O., 2015. Elucidating the molecular architecture of adaptation via evolve and resequence experiments. Nat. Rev. Genet. 16: 567–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado H. E., Bergland A. O., O’Brien K. R., Behrman E. L., Schmidt P. S., et al. , 2016. Comparative population genomics of latitudinal variation in Drosophila simulans and Drosophila melanogaster. Mol. Ecol. 25: 723–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins N. E., Faria V. G., Nolte V., Schlötterer C., Teixeira L., et al. , 2014. Host adaptation to viruses relies on few genes with different cross-resistance properties. Proc. Natl. Acad. Sci. USA 111: 5938–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte V., Pandey R. V., Kofler R., Schlötterer C., 2013. Genome-wide patterns of natural variation reveal strong selective sweeps and ongoing genomic conflict in Drosophila mauritiana. Genome Res. 23: 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin S. V., Turner T. L., 2013. Promises and limitations of hitchhiking mapping. Curr. Opin. Genet. Dev. 23: 694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-terWengel P., Kapun M., Nolte V., Kofler R., Flatt T., et al. , 2012. Adaptation of Drosophila to a novel laboratory environment reveals temporally heterogeneous trajectories of selected alleles. Mol. Ecol. 21: 4931–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri N., Nolte V., Chen J., Schlötterer C., 2015. Genome assembly and annotation of a Drosophila simulans strain from Madagascar. Mol. Ecol. Resour. 15: 372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R. V., Schlötterer C., 2013. DistMap: a toolkit for distributed short read mapping on a Hadoop cluster. PLoS One 8: e72614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt J. A., Kolaczkowski B., Jones C. D., Begun D. J., Kern A. D., 2014. Parallel geographic variation in Drosophila melanogaster. Genetics 197: 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remolina S. C., Chang P. L., Leips J., Nuzhdin S. V., Hughes K. A., 2012. Genomic basis of aging and life history evolution in Drosophila melanogaster. Evolution. 66: 3390–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer C., Tobler R., Kofler R., Nolte V., 2014. Sequencing pools of individuals — mining genome-wide polymorphism data without big funding. Nat. Rev. Genet. 15: 749–763. [DOI] [PubMed] [Google Scholar]
- Schlötterer C., Kofler R., Versace E., Tobler R., Franssen S. U., 2015. Combining experimental evolution with next-generation sequencing: a powerful tool to study adaptation from standing genetic variation. Heredity 114: 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedghifar A., Saelao P., Begun D. J., 2016. Genomic patterns of geographic differentiation in Drosophila simulans. Genetics 202: 1229–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler R., Franssen S. U., Kofler R., Orozco-terWengel P., Nolte V., et al. , 2014. Massive habitat-specific genomic response in D. melanogaster populations during experimental evolution in hot and cold environments. Mol. Biol. Evol. 31: 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True J. R., Mercer J. M., Laurie C. C., 1996. Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics 142: 507–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Stewart A. D., Fields A. T., Rice W. R., Tarone A. M., 2011. Population-based resequencing of experimentally evolved populations reveals the genetic basis of body size variation in Drosophila melanogaster. PLoS Genet. 7: e1001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Miller P. M., Cochrane V. A., 2013. Combining genome-wide methods to investigate the genetic complexity of courtship song variation in Drosophila melanogaster. Mol. Biol. Evol. 30: 2113–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Wit J., Svetec N., Begun D. J., 2015. Parallel gene expression differences between low and high latitude populations of Drosophila melanogaster and D. simulans. PLoS Genet. 11: e1005184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Udpa N., Gersten M., Visk D. W., Bashir A., et al. , 2011. Experimental selection of hypoxia-tolerant Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 108: 2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw reads for all populations are available from the European Sequence Read Archive under the accession numbers mentioned in Table S1. SNP data sets in sync format (Kofler et al. 2011) are available from the Dryad Digital Repository under http://dx.doi.org/10.5061/dryad.p7c77.