Abstract
Development of resistance to chemotherapy treatments is a major challenge in the battle against cancer. Although a vast repertoire of chemotherapeutics is currently available for treating cancer, a technique for rapidly identifying the right drug based on the chemo-resistivity of the cancer cells is not available and it currently takes weeks to months to evaluate the response of cancer patients to a drug. A sensitive, low-cost diagnostic assay capable of rapidly evaluating the effect of a series of drugs on cancer cells can significantly change the paradigm in cancer treatment management. Integration of microfluidics and electrical sensing modality in a 3D tumour microenvironment may provide a powerful platform to tackle this issue. Here, we report a 3D microfluidic platform that could be potentially used for a real-time deterministic analysis of the success rate of a chemotherapeutic drug in less than 12 h. The platform (66 mm × 50 mm; L×W) is integrated with the microsensors (interdigitated gold electrodes with width and spacing 10 μm) that can measure the change in the electrical response of cancer cells seeded in a 3D extra cellular matrix when a chemotherapeutic drug is flown next to the matrix. B16-F10 mouse melanoma, 4T1 mouse breast cancer, and DU 145 human prostate cancer cells were used as clinical models. The change in impedance magnitude on flowing chemotherapeutics drugs measured at 12 h for drug-susceptible and drug tolerant breast cancer cells compared to control were 50552 ± 144 Ω and 28786 ± 233 Ω, respectively, while that of drug-susceptible melanoma cells were 40197 ± 222 Ω and 4069 ± 79 Ω, respectively. In case of prostate cancer the impedance change between susceptible and resistant cells were 8971 ± 1515 Ω and 3281 ± 429 Ω, respectively, which demonstrated that the microfluidic platform was capable of delineating drug susceptible cells, drug tolerant, and drug resistant cells in less than 12 h.
Keywords: Microfluidics, Drug Screening, Chemotherapy, Cancer Microenvironment, Electrical Sensing
1. Introduction
Cancer has become a universal health problem. It is presently the second major cause of death in the United States and is predicted to outpace heart diseases in the next few years. In 2016, 1,685,210 new cancer cases and 595,690 cancer deaths are projected to occur in the United States (Siegel et al., 2016). Selecting the correct chemotherapeutic agents for a particular patient is imperative in the effective treatment of cancer. First line therapy against cancer involves a standard set of treatments, such as surgery followed by chemotherapy and radiation. Imaging-based technologies using Computerized Tomography (CT) scan and Magnetic Resonance Imaging (MRI) to monitor tumour size are the current standard methods for evaluating treatment success. Sometimes, first line therapies take a long time to show progress followed by a stalling or continued growth of cancer (Hassan et al., 2010; Kuczynski et al. 2013). In addition, cancer cells may develop resistance to the chemotherapeutic agents. Currently, the therapy efficacy can be determined only after a few weeks to several months, which represents one of the major challenges in the timely management of cancer (Morabito et al., 2014; Gottesman et al., 2002). Therefore, there is a need to develop approaches for rapid screening techniques to evaluate the efficacy of anti-cancer drugs on tumours that would help in stratifying patient responders and non-responders early on.
In addition, currently available in-vitro models to study the efficacy of chemotherapy, which targets tumours in two-dimensional culture systems, do not replicate the tumour microenvironment in the human body. The failure of chemotherapy treatment can be attributed to drug resistant cells and involvement of pharmacological and biochemical mechanisms such as drug-degradation due to altered drug-metabolizing enzymes, decreased drug activation, and subcellular distribution. Therefore, instead of unification of cancer treatment, there is a need for personalization of cancer therapy, which requires new methods for efficient drug screening. Drug sensitivity data obtained from 2D-based cell culture systems are often ambiguous due to the variations observed in the morphology, growth pattern, and gene expression of tumour cells in a 3D matrix. In addition, compared to the planar cell culture, 3D cell culture has been shown to more accurately influence cell morphology, gene/protein expression, signal transduction, proliferation, migration, and drug tolerance (Asphahani et al., 2011; Arias et al., 2010; Gurski et al., 2010; Nyga et al., 2011; Hong et al., 2012; Ning et al., 2011; Ling et al., 2012; Forestier et al., 2012). Different drug sensitivities were observed for cells grown in 3D culture configurations compared with a 2D monolayer model (Serebriiskii et al., 2008; Doillon et al., 2004). Thus, a 3D platform for studying the response of chemotherapeutic agents is essential.
Electrical sensing is a simple, rapid, label-free, inexpensive, and sensitive modality which provides real-time kinetic information of the cell growth and necrosis pattern on the surface of the electrodes (Huang et al., 2013; Yamada et al., 2007; Picollet-D’hahan 2011). Electric cell-substrate impedance sensing (ECIS) has been used to measure cell proliferation, morphology, and motility (Tran et al., 2016). ECIS-based platforms have been extensively used for monitoring different phases of cancer cell growth in vitro due to its advantages over conventional assays such as fluorescence imaging, radioactive detection and antibody- or nucleic acid-based detection which are time-consuming, expensive, and laboratory-based (Tran et al., 2016). ECIS appears to be a fast and convenient strategy for evaluating different stages of cell adhesion i.e. spreading, attachment, migration, and death of cancer cells (Morabito et al., 2014; Yaofang et al., 2013; Tiruppathi et al., 1992). However, it does not recreate the in vivo environment in which tumours are found. Growing cells, which adhere onto the electrode surface provided with the regular supply of media, do not replicate the in vivo environment.
Integration of microfluidics and electrical sensing modality in a 3D tumour microenvironment may provide a powerful platform to accurately and rapidly monitor the response of cancer cells to a series of drugs (Pavesi et al., 2016). In the past decade, microfluidics has shown a great promise in developing tools for cancer research (Mukhopadhyay et al., 2007; Calleja et al., 2016; Wlodkowic et al., 2010). Microfluidic systems are strong candidates for the next generation of in vitro cancer models due to their capability of manipulating individual cells (Zare et al., 2010), and reduced number of cells required for each endpoint (~103 cells/endpoint rather than >106 cells/endpoint). This facilitates high content study and handling of scarce patient material such as breast tissue biopsies with <106 mammary fibroblasts/0.25cm (Hassan et al., 2010; Wlodkowic et al., 2010; Sung et al., 2014). The animal studies have the disadvantage over microfluidics as they contain non-human host cells while, in microfluidics, the studies can be done using human tissue biopsies (Calleja et al., 2016). Studies have been carried out to determine the optimum flow rate in microfluidic devices seeded with cancer cells (Couzon et al., 2009). An optimized flow rate should be selected in order to minimize the stress on the cancer cells seeded on a microfluidic bed (Ling et al., 2005; Tehranirokh et al., 2013). However, in the previous systems (King et al., 2007; Lee et al., 2006; Thompson et al., 2007; Ye et al., 2007; Hompland et al., 2012)), cancer cells were seeded directly inside microchannels allowing the cells to be in direct contact with the fluid flow which applies shear stresses on the cells. Such stress is not present in vivo except for duct epithelial cells and endothelial cells such as kidney epithelial cells and alveolar cells. Diffusion is the main transport mechanism for the drug in vivo between tissues and microvessels or capillaries.
Here, we have engineered a microfluidic platform for rapid screening of chemotherapeutic drugs through a dynamic delivery of anti-cancer drugs to cancer cells seeded into a 3D gel matrix in a square chamber (2 mm × 2 mm) with interdigitated microelectrodes (Fig. 1). We evaluated the performance of the microchip using three cancer cell models including drug susceptible, drug tolerant, drug resistant breast cancer, melanoma and human prostate cancer cells as these are amongst the top causes of cancer related-deaths (American Chemical Society, 2016). We validated the microfluidic platform by comparing our chip-based electrical sensing results with the standard cell viability assay. The microchip better mimics the in vivo configuration as compared to the classical culture-based dishes and offers a relatively low-cost, rapid approach for real-time drug screening analysis (Pavesi et al., 2016). This study is a step towards an accurate, reliable, and effective screening of chemotherapeutics and is a significant development towards developing a microfluidic chip for personalized medicine in cancer treatment.
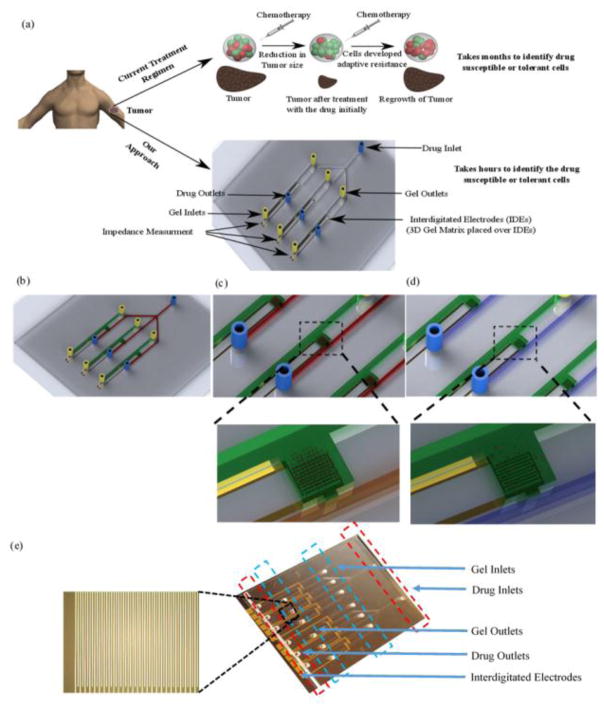
Figure 1. Lab-on-a-Chip for chemotherapy drug response screening.
(a) 3D schematic of the presented platform for multiplex drug susceptibility testing, (b) Cancer cells are placed in a 3D matrix in a separate microchannel (green) that is connected to a parallel microchannel for drug flow (red), (c) Cancer cells are intact and viable in the 3D gel structure and before the introduction of drugs. In the presence of effective drug flow, the cancer cells respond (lyse), (d) We observed that the number of intact cancer cells reduced after introducing effective drug flow in the chip, (e) Image of a fabricated device. The chip involves interdigitated electrodes with 10 μm width and spacing.
2. Materials and Methods
2.1 Microfluidic Device Fabrication
Microchips were fabricated using lithography and laser cutting. (i) A 4-inch glass wafer was used as a substrate, (ii) Layers of chrome/gold (Cr/Au) (0.02 μm/0.4 μm) were deposited on glass substrates using an e-beam evaporator. Using a spin-coater, a positive photoresist (1 μm thick) (AZ3312 Integrated Micromaterials™) was coated at 4000 rpm on the glass deposited with Cr/Au and prebaked at 90 °C for 1 min. Standard photolithography technique was used to pattern interdigitated electrodes (UV exposure (500 W lamp, 18 sec) using EVG®620), postbake (110 °C for 1 min), and photoresist development (AZ 726 MIF developer at 21 °C). The width and spacing of the electrodes were 10 μm. The area covered by electrodes was 2 mm2. The unwanted Cr/Au was etched through wet etching followed by photoresist stripping by dipping the glass wafers inside a beaker containing acetone placed in an ultrasonic bath for 3 min. (iii) SU-8 2075 was spin coated at 1000 rpm/min for 1 min on top of the glass wafer with metal electrodes to obtain a uniform layer of SU-8 with 150 μm thickness. The SU8 was prebaked at 65 °C for 7 min followed by 90 °C for 25 min. After prebaking, SU-8 film was exposed to 240 mJ/cm2 of UV for 15 minutes. Post exposure bake was performed at 95 °C for 12 min before immersing the wafer in MicroChem’s SU-8 developer for 15 minutes to obtain the channels with 600 μm width, chamber (2 mm × 2 mm) for gel loading, and slit (0.25 mm) for interaction between drug and cancer cells loaded in 3D gel. The width of the channel is close to the upper range (>500 μm) of pulmonary vessel’s diameter (Cavallotti et al. 2005). Holes for inlets and outlets were created on the thin sheet of PMMA (1.5 mm thick) using a laser cutter (Universal Laser System Inc., VLS 2.3). The power, scan speed, and pulse per inch rate were set to 25 W, 5 mm/s, and 500 pulses/inch, respectively. The PMMA sheet was then bonded to the glass wafer with patterned electrodes and SU-8 using a 180 KPa pressure at 100 °C for 30 min. (Fig. 1e). The length and width of the microfluidic device were 66 mm and 50 mm, respectively.
2.2 Generation of Drug-tolerant cancer cells
B16-F10 mouse melanoma, 4T1 mouse breast cancer and DU 145 human prostate cancer cell lines were purchased from American Type Culture Collection and were grown in Dulbecco’s Modified Eagle Media supplemented with 10% Fetal Bovine Serum and 0.1% antibiotic-antimycotic all procured from Thermo Fisher Scientific. Carboplatin is a platinum chemotherapeutic agent - a cisplatin analogue, currently being used in the treatment of various tumours (Solly et al., 2004). It is one of the most common chemotherapeutic drugs being used for melanoma (Ehret et al., 1998). Carboplatin is also being used in combination with other drugs for enhanced efficacy in triple-negative breast cancer (TNBC) in Murine models (Solly et al., 2004). Thus, we have used Carboplatin for B16-F10 and 4T1 cancer cell lines. Taxane drugs which include Paclitaxel and Docetaxel are the standard drugs being used for chemotherapy for prostate cancer treatment (Yang et al., 2011; Hua et al., 2010). Both in vivo and in vitro enhanced cancer cell death and suppression of human prostate cancer cell lines were observed when the cells were treated with Paclitaxel prior to radiation (Hua et al. 2010; Shafiee et al., 2013). We have used Paclitaxel for prostate cancer cell lines.
Carboplatin and Paclitaxel were purchased from Sigma-Aldrich and LC laboratories, respectively. 4T1 and B16-F10 cells were seeded with the density of 250,000 cells/mL in a 10 mL tissue culture treated dish and were allowed to reach 60% confluency. The cells were then treated with Carboplatin to a final concentration of 100 nM for 48 h. After the treatment, the media was aspirated and the cells were washed with cold PBS (4 °C) to remove any dead cells that were still attached to the plate. The remaining Carboplatin-tolerant, live cells were collected by trypsinization and were used for further experiments. Similar procedure was performed to prepare Paclitaxel-resistant DU 145 cells for experiments.
2.3 Live/Dead cell counting
DMEM cell culture media containing drug with a final concentration of 30 μM was allowed to flow at a rate of 4 μL/min in the microfluidic device containing tumour cells for 3 h, 5 h, 7 h, 9 h, and 12 h. After each time point, the cells were collected from the gel using Collagenase Type I and their viability was tested using Trypan Blue exclusion. Equal volumes of Trypan blue and cell suspension were mixed, loaded on a hemaocytometer and examined under a light microscope to observe the number of viable cells (cells which did not take up the dye).
2.4 Cell viability assay
4T1, B16F10, DU145 and their respective tolerant cells were seeded at a density of 10,000 cells/well in a 96 well plate. Tolerant cells were prepared as mentioned earlier. 4T1 and B16 cells were treated with different concentrations (101 – 105 nM) of Carboplatin and DU145 were treated with different concentrations of Paclitaxel (101 – 105 nM). After 24 h of treatment, the media was aspirated and the cells were washed with cold PBS (4 °C). They were then exposed to the MTS reagent acquired from Promega and incubated for 2 h in 5% CO2 atmosphere at 37 °C.
The absorbance was measured at 490 nm using an Epoch (BioTek) plate reader. Results were quantified by subtracting the blank value from each value then normalizing against the control values and results were analyzed by using Prism software (GraphPad).
2.5 Impedance measurement
We performed impedance spectroscopy at 1 V for frequencies between 100 Hz and 1 MHz using a LCR impedance meter (LCR8110G, GW Instek, CA). For statistical analysis, we chose 1000 Hz as we observed maximum impedance change at this frequency, which is in agreement with our previous findings on electrical sensing of target pathogens on-chip (Shafiee et al., 2013). The electronic circuit modeling for a microchip with electrodes for impedance sensing has been presented in our previously published paper (Shafiee et al., 2013).
3. Results
3.1 Simulation
The in vivo interstitial flow for non-metastatic cancer is between 0.1–1 μm/s (Tarbell et al., 2012; Hompland et al., 2012). The drug flow in the microfluidic device should match this interstitial flow (Hompland et al., 2012). Our numerical modelling showed that 4 μl/min flow rate in our microchip would satisfy the interstitial flow criterion (flow velocity of 0.86 μm/s < 1 μm/s). We used COMSOL Multiphysics to simulate the fluid and particle flow for velocity of 4 μl/min in the designed chip (Fig. 2). The flow was treated as incompressible stokes flow, neglecting inertial components of flow and the particles were set to have a diameter of 312 nm, in accordance with the optimum size of Carboplatin nanoparticles (Wlodkowic et al., 2010). The flow characteristics were solved using the simplified Navier-Stoke equation and continuity equation ((1) and (2)).
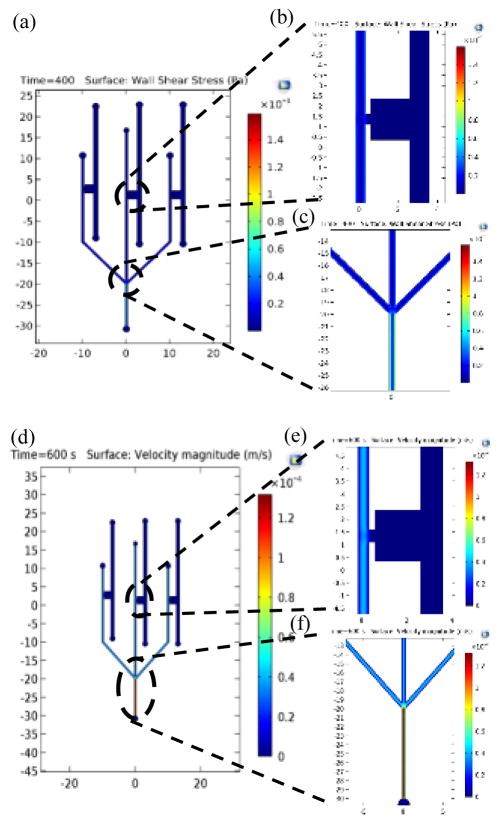
Figure 2. Simulation for drug flow in the microfluidic device.
(a) Shear stress plot in the microchannels, (b) Shear stress plot at the interaction area of the device. The region filled with collagen is separated by a boundary wall and is under no shear stress due to fluid flow, hence is shown to be at 0 Pa. The shear stress at the interaction area is found to be 0.04 Pa, (c) Shear stress plot at the three junctions, (d) Velocity plot in the microchannels, (e) Velocity plot at the interaction area of the device. The velocity near the interaction area is found to be 0.86 μm/s allowing proper time for interaction between drug and cancer cells seeded in collagen gel, (f) Velocity plot of the junction area.
| (1) |
and
| (2) |
where, ∇ is the gradient, p is pressure, μ is the dynamic viscosity of the fluid, u is the velocity vector matrix, F is the external force applied on the fluid, and ρ is the density of the fluid. I is the identity matrix and T is the matrix transpose function. The wall conditions were set to no slip and inlet velocity of fluid at 4 μl/min. These equations were utilized through COMSOL’s inbuilt pre-sets as the model is an accurate simplification of the complex creep flow through the channel. The wall shear stress was solved using the formula:
| (3) |
where τ is the shear stress, μ is the dynamic viscosity of the fluid, Vx is the x-component of velocity and Vy is the y-component of velocity. The shear stress and velocity plots for the microfluidic device, interaction area and inlets are shown in Fig. 2(a) – 2(f). The shear stress near the interaction area was 0.04 Pa which is lower than the critical stress which causes the change in the cell morphology (Couzon et al., 2009). The velocity drops at the interaction area as the pressure drops, slowing the particles down as they approach the interaction area. Diffusion velocity of the particles tends to be lower than flow velocities (Zlatoš et al., 2010), and thus for an effective penetration of drug particles, the velocity of flow near the interaction area should allow for sufficient diffusion into the gel. For the present design of the microfluidic device, the flow rate of 4 μL/min yielded the flow velocity of 0.86 μm/s which falls within the range of interstitial flow for cancer (Hompland et al., 2012).
3.2 Bonding Test
Weak bonding between SU-8 and PMMA may result in leakage of the sample flowing in the microchannels. To determine the adhesion strength between SU8 and PMMA, a shear test was carried out on three microfabricated chips using Instron®(Jubery et al., 2012). The test speed was 500 μm/s and the force was applied by the tester tip on one side of the chip. We observed that the average debonding pressure for the chips was 18 ± 1.3 MPa (n=3), and 10 MPa was needed for a shear movement of 100 μm.
3.3 Evaluating Electrical Response of Cancer Cells
To evaluate the electrical response of cancer cells in the presence of drugs, we conducted experiments using drug-susceptible and tolerant B16-F10 mouse melanoma, 4T1 mouse breast cancer, and DU 145 human prostate cancer cells and compared the microchip results with hemocytometer-based cell counting and the MTS assay analysis. The microchip setup was kept in an incubator to maintain the level of humidity (>80 %), temperature (37 °C), and carbon dioxide (5 %) constant during the period of 12 h experiments. In this study, cells were encapsulated in hydrogel scaffold in microchannels and assumed to be uniformly distributed. Here, we used four sets of controls: (i) collagen gel on electrodes, (ii) collagen gel mixed with drug on electrodes, (iii) collagen gel seeded with cancer cells, and (iv) collagen gel seeded with cancer cells along with media flow. The impedance spectra were measured for the control and cancer cells (susceptible, tolerant, and resistant cells) for frequencies ranging from 100 Hz to 1 MHz (Fig. 3). We did not observe any significant change in the bulk impedance magnitude of the control samples over a period of 12 hrs testing (Figs. 4, 5, n=3, p>0.1). We observed that the impedance magnitude of both drug-susceptible and tolerant breast cancer and melanoma cell samples decreased over a period of 12 hrs when Carboplatin was introduced into the chips, however, the signal change for drug-susceptible cells was higher compared to drug-tolerant cancer cells. The impedance magnitude change of drug-susceptible breast cancer cells in static and dynamic modes were 48756 ± 516 Ω and 50552 ± 144 Ω respectively at t=12 h, which was significantly higher than the impedance magnitude change of drug-tolerant breast cancer cells in static 8687±88 Ω and dynamic modes 28786 ± 233 Ω (Fig. 5a,b, n=3, P<0.0001). Similarly, the impedance magnitude change of drug-susceptible melanoma cancer cells in static and dynamic modes were 46639 ± 252 Ω and 40197 ± 222 Ω, respectively at t=12 h, which was significantly higher than the impedance magnitude change of drug-tolerant melanoma cancer cells in static 5998 ± 324 Ω and dynamic modes 4069 ± 79 Ω (Fig. 4c,d, n=3, P<0.0026).
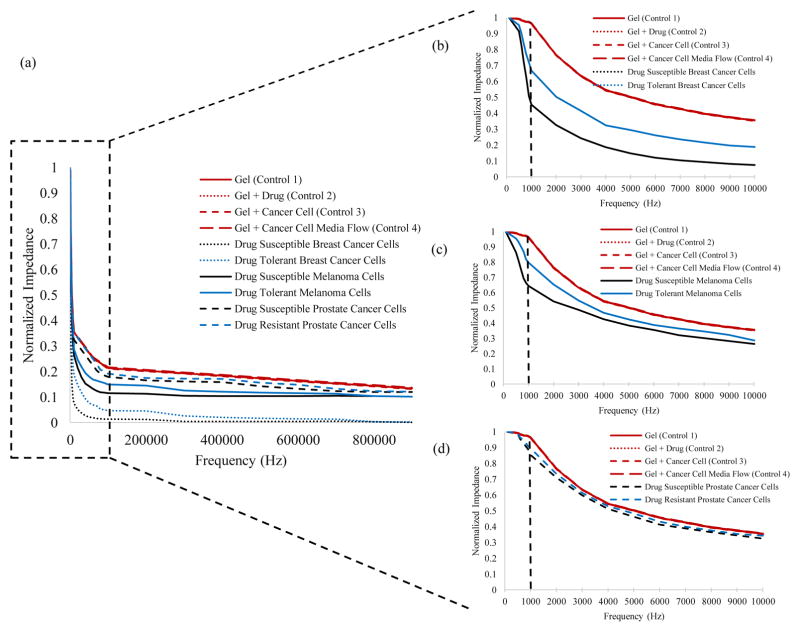
Figure 3. Impedance Spectra.
(a) The impedance spectra for the control and cancer cells (Breast cancer, Melanoma, and Prostate cancer), (b–d) shows that the maximum impedance magnitude shift occurs at 1000 Hz.
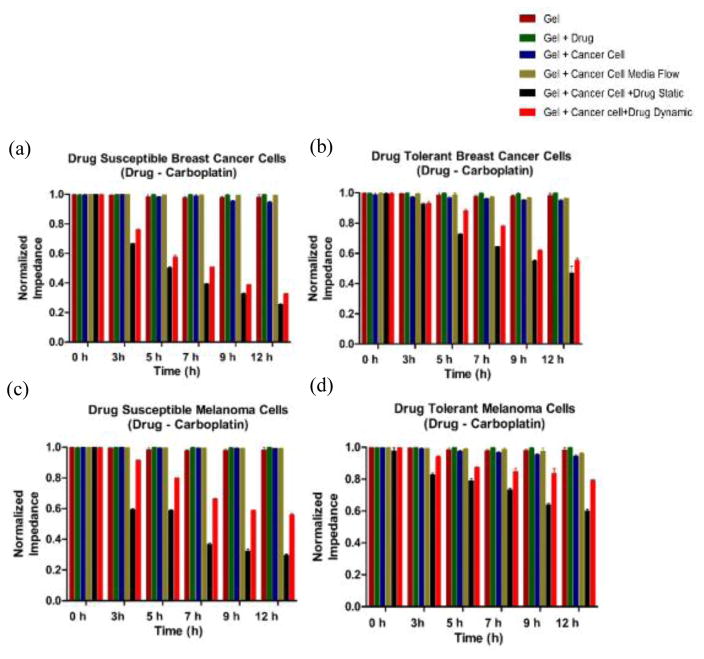
Figure 4. Electrical response of drug susceptible and tolerant murine cancer cells.
On-chip impedance magnitude of: (a) Drug susceptible 4T1 mouse breast cancer cells, (b) Drug tolerant 4T1 mouse breast cancer cells, (c) Drug susceptible B16-F10 mouse melanoma cells, and (d) Drug tolerant B16-F10 mouse melanoma cells. The impedance was measured at 0 h, 3 h, 5 h, 7 h, 9 h, and 12 h time points. Gel, Gel + Drug, Gel + Cancer Cell, and Gel + Cancer Cell + Media were used as controls. Error bars represent standard error of the mean.
Figure 5. Electrical response of drug susceptible and drug-resistant human prostate cancer cells.
On-chip impedance magnitude of (a) Drug susceptible and (b) Drug Resistant human prostate (DU 145) cancer cells at 0 h, 3 h, 5 h, 7 h, 9 h, and 12 h time points. Gel, Gel + Drug, Gel + Cancer Cell, and Gel + Cancer Cell + Media were used as controls. Error bars represent standard error of the mean (n=3).
These results also show that the impedance magnitude change in static mode was significantly higher compared to dynamic mode for all cells, which is due to constant direct contact between the drug and cancer cells in the static mode resulting in more cell lysate. We also observed a similar trend in electrical response for prostate cancer cells, however, the impedance magnitude change for drug-resistant prostate cancer cells were lesser compared to the change for drug-tolerant breast cancer and melanoma cells (Figs. 4, 5). This is due to inherent property of the resistant cancer cell-lines in which the response to drug is minimal even at higher concentrations. The impedance magnitude change of drug-susceptible prostate cancer cells in static and dynamic modes were 8693 ± 379 Ω and 8971 ± 1515 Ω, respectively at t=12 h, while the impedance magnitude for drug-resistant prostate cancer cells for t=12 h in static mode and dynamic modes were 4450 ± 491 Ω and 3281 ± 429 Ω, respectively. (Fig. 4a, b, n=3, p<0.0001). The statistical analysis was performed using t-test.
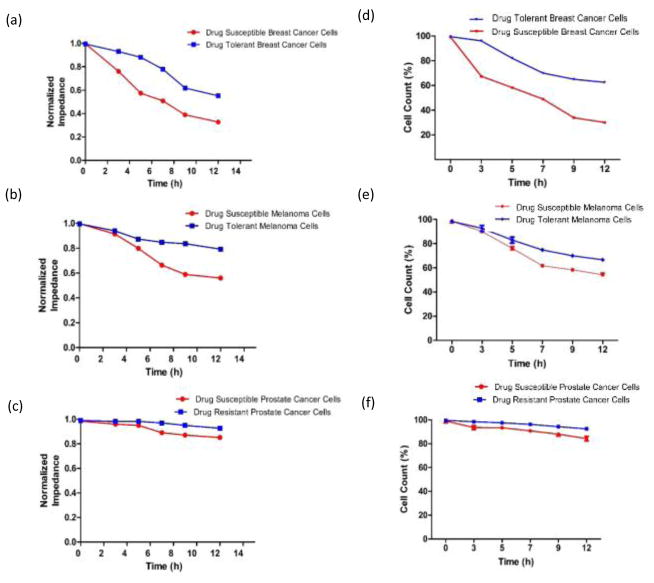
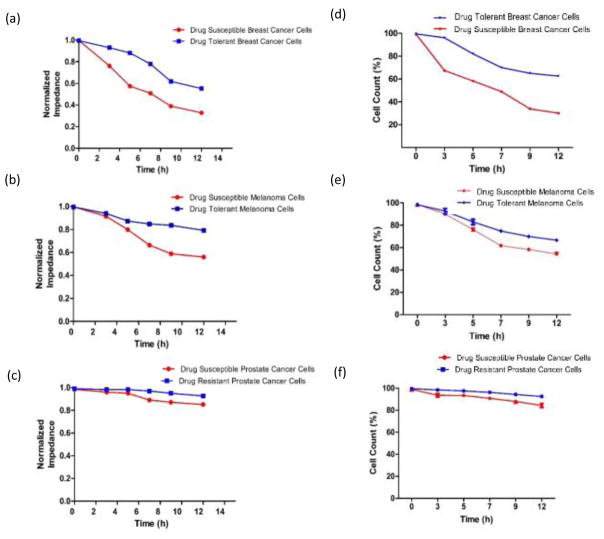
Figure 6 shows the normalized impedance magnitude and cell count results for drug susceptible and resistant cancer cells over a period of 12 h when the microchip was used. The cancer cells seeded in the gel were collected after performing the on-chip experiments using collagenase and were manually counted with an inverted microscope using a hemocytometer. The cell count results are in agreement with electrical sensing results obtained from microchips, which shows that target cells were lysed during the process of drug introduction to the chips. The cell count of drug susceptible cancer cells were significantly lower than drug tolerant/resistant cancer cells at different time points between 5 and 12 h (Figs. 6d, e, f, n=3, p<0.0001).
Figure 6. Normalized impedance and cell count results.
(a, b, c) Normalized impedance magnitude for drug-susceptible and tolerant/resistant breast cancer, melanoma, and prostate cancer cells at the drug concentration of 30 μM at 0 h, 3 h, 5 h, 7 h, 9 h, and 12 h time intervals, (d, e, f) Cell viability of drug-susceptible and tolerant/resistant breast cancer, melanoma, and prostate cancer cells after flowing the drug for 0 h, 3 h, 5 h, 7 h, 9 h, and 12 h. The viability was measured by Trypan Blue dye exclusion. Error bars represent standard error of the mean (n=3).
3.4 Cell Viability Testing
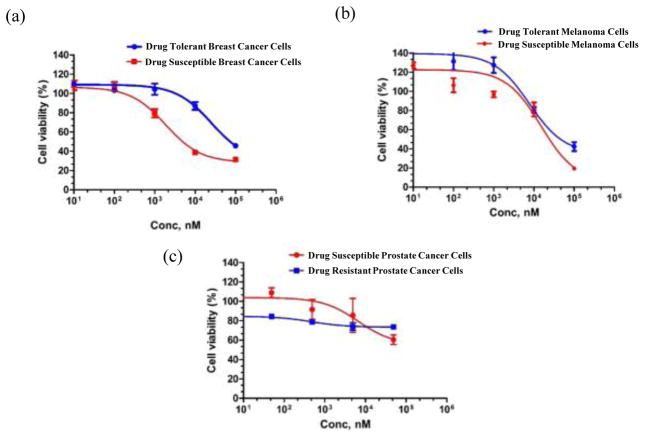
The cell viabilities of breast cancer, melanoma, and prostate cancer cells were also measured when five different drug concentrations were used using MTS assay and IC50 values were measured (Fig. 7). The absorbance was obtained at 490 nm using a 96 well plate reader. We observed that for the same concentrations of Carboplatin, cell viability was greater in Carboplatin-tolerant 4T1 compared to susceptible 4T1 which is similar to previous published results on similar platinum-based drug (cisplatin) with breast cancer cells (Prabhakaran et al. 2013). Cell viability was greater in drug-tolerant B16/F10 than in drug-susceptible B16/F10 for all concentrations of Carboplatin drug which follows the published results on drug-tolerant melanoma cancer cell lines (Figure 7a, b) (Quirt et al. 2007; McDermott et al., 2014). In case of prostate cancer it was observed that for lower concentrations of drug the DU145 resistant cells showed negligible change in the cell viability while at higher concentration a slight decrease in the cell viability was observed (Fig. 7c). These results are in agreement with literature in which the drug-resistant prostate cancer cells have higher cell viability compared to drug-susceptible cells when treated with different concentrations of docetaxel drug (O’Neill et al., 2011). Additionally, the impedance measurements (Fig. 6a, b, c) are in agreement with the cell viability results obtained from MTS (Fig. 7a, b, c).
Figure 7. Cell viability results using MTS assay.
Cell viability curves comparing the drug response of (a) 4T1 breast cancer, (b) B16 melanoma cells with their Carboplatin-tolerant variants, and the drug response of (c) DU145 prostate cancer cells and Paclitaxel–resistant DU145 to Paclitaxel after 24 h of treatment. Error bars represent standard error of the mean (n=3).
3.5 Discussion
In the present study, we have shown a microfluidic platform capable of delineating the drug-susceptible and tolerant/resistant cancer cell lines in a rapid and label-free manner. This method can potentially be used for studying patient samples and could be a significant step toward the development of a biomedical device for personalized medicine. The impedance magnitude represents the total impedance combining live cells, dead cells, and hydrogel. Our main hypothesis in this study was that the cancer cells show a significant electrical response in 3D microenvironment in the presence of the hydrodynamic flow of effective drugs due to electrical conductivity change after cell lysis. For our study, we seeded around one million cells in the 3D gel matrix. We found that the impedance of the 3D gel matrix for each cell line was different and reflected overall cellular activities including cell growth, cell morphology, and cell density. The difference in the initial impedance can be further attributed to cell shape, cell-cell was greater in Carboplatin-tolerant 4T1 compared to susceptible 4T1 which is similar to previous published results on similar platinum-based drug (cisplatin) with breast cancer cells (Prabhakaran et al., 2013). Cell viability was greater in drug-tolerant B16/F10 than in drug-susceptible B16/F10 for all concentrations of Carboplatin drug which follows the published results on drug-tolerant melanoma cancer cell lines (Figure 7a, b) (Quirt et al., 2007; McDermott et al.,; 2014). In case of prostate cancer it was observed that for lower concentrations of drug the DU145 resistant cells showed negligible change in the cell viability while at higher concentration a slight decrease in the cell viability was observed (Fig. 7c). These results are in agreement with literature in which the drug-resistant prostate cancer cells have higher cell viability compared to drug-susceptible cells when treated with different concentrations of docetaxel drug (O’Neill et al., 2011). Additionally, the impedance measurements (Fig. 6a, b, c) are in agreement with the cell viability results obtained from MTS (Fig. 7a, b, c).
In this study, we seeded around one million cells in the 3D gel matrix. We found that the impedance of the 3D gel matrix for each cell line was different and reflected overall cellular activities including cell growth, cell morphology, and cell density. The difference in the initial impedance can be further attributed to cell shape, cell-cell interactions, cell size, and cellular activities due to external stimulations (Carvalho et al., 2015; Liu et al., 2009; Zhu et al., 2015; Asphahani et al., 2011). It is known that the drug molecules, less than approximately 500 Da, are absorbed into PDMS (Gomez et al., 2007), which complicates the interpretation of some studies. Liu et al., 2010 reported that attachment and cell spreading were substantially impaired on PDMS, compared to polystyrene. Additionally, the wetting of the polymer to the mould or the post removal flowing of the polymer does not hold the shape of the mold due to polymer relaxation (Mukhopadhyay et al., 2007). In this study we have used Carboplatin drug which has molecules approximately 371 Da. To avoid the complexity arising with PDMS we have used PMMA-SU8-glass based microfluidic platform instead of PMMA-PDMS or PDMS-glass.
4. Conclusions
We report an engineered microfluidic platform that could be used for the pre-screening of chemotherapeutics by measuring an electrical response of the cancer cells in a 3D microenvironment. Using this platform we were able to delineate the drug susceptible and tolerant/resistant cells in less than 12 h. The microfluidic platform is incorporated with multiple chambers and perfusion channels that enables screening different chemotherapeutics in different cancer samples to determine the right drug that can overcome chemo-resistance in a patient-specific manner.
Highlights.
A microfluidic device was developed for drug screening in a 3D cancer microenvironment.
Device can be used for a real-time deterministic analysis of the success rate of a chemotherapeutic drug in less than 12 h.
The microfluidic device was able to delineate between drug-susceptible and drug-tolerant cancer cells.
Acknowledgments
Hardik J. Pandya acknowledges the Department of Electronic Systems Engineering, Indian Institute of Science for providing facilities to complete the manuscript. Research reported in this publication was partially supported by the National Institute of Allergy and Infectious Disease of the National Institute of Health under award number R01AI118502 (H.S.); Brigham and Women’s Hospital and Harvard Medical School through Bright Futures Prize and Innovation Evergreen Award (H.S.); and Center for Nanotechnology at the King Abdul-Aziz University, Saudi Arabia (H.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We acknowledge the support of Maryland Nanocenter for sensor fabrication.
Footnotes
Competing financial interests:
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- Hassan MSU, Ansari J, Spooner D, Hussain SA. Chemotherapy for breast cancer (Review) Oncol Rep. 2010;24:1121–1131. doi: 10.3892/or_00000963. [DOI] [PubMed] [Google Scholar]
- Kuczynski EA, Sargent DJ, Grothey A, Kerbel RS. Drug rechallenge and treatment beyond progression—implications for drug resistance. Nat Rev Clin Oncol. 2013;10(10):571–587. doi: 10.1038/nrclinonc.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morabito A, Carillio G, Daniele G, Piccirillo MC, Montanino A, Costanzo R, Sandomenico C, Giordano P, Normanno N, Perrone F, Rocco G, Maio MD. Treatment of small cell lung cancer. Crit Rev Oncol Hemat. 2014;91:257–270. doi: 10.1016/j.critrevonc.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Gottesman MM. Mechanisms of Cancer Drug Resistance. Ann Rev Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- Asphahani F, Wang K, Thein M, Veiseh O, Yung S, Xu J, Zhang M. Single-cell bioelectrical impedance platform for monitoring cellular response to drug treatment. Phys Biol. 2011;8(1):015006. doi: 10.1088/1478-3975/8/1/015006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias LR, Perry CA, Yang L. Real-time electrical impedance detection of cellular activities of oral cancer cells. Biosen Bioelectron. 2010;10:2225–2231. doi: 10.1016/j.bios.2010.02.029. [DOI] [PubMed] [Google Scholar]
- Gurski LA, Nicholas BS, Petrelli J, Jia X, Farach-Carson MC. 3D Matrices for Anti-Cancer Drug Testing and Development. Oncol Issues. 2010:20–25. [Google Scholar]
- Nyga A, Cheema U, Loizidou M. 3D tumour models: novel in vitro approaches to cancer studies. J Cell Commun Signal. 2011;5:239–248. doi: 10.1007/s12079-011-0132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JL, Lan KC, Jang LS. Electrical characteristics analysis of various cancer cells using a microfluidic device based on single-cell impedance measurement. Sens Actuat B. 2012;173:927–934. [Google Scholar]
- Ning Y, Manegold PC, Hong YK, et al. Interleukin-8 is associated with proliferation, migration, angiogenesis and chemosensitivity in vitro and in vivo in colon cancer cell line models. International Journal of Cancer. 2011;128(9):2038–2049. doi: 10.1002/ijc.25562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Z-Q, Qi C-J, Lu X-X, et al. Heterogeneity of chemosensitivity in esophageal cancer using ATP-tumor chemosensitivity assay. Acta Pharmacol Sin. 2012;33(3):401–406. doi: 10.1038/aps.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestier A, Sarrazy F, Caillat S, Vandenbrouck Y, Sauvaigo S. Functional DNA repair signature of cancer cell lines exposed to a set of cytotoxic anticancer drugs using a multiplexed enzymatic repair assay on biochip. PLoS ONE. 2012;7(12):e51754. doi: 10.1371/journal.pone.0051754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebriiskii I, Castello-Cros R, Lamb A, Golemis EA, Cukierman E. Fibroblast-derived 3D matrix differentially regulates the growth and drug-responsiveness of human cancer cells. Matrix Biol. 2008;27:573–585. doi: 10.1016/j.matbio.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doillon CJ, Gagnon E, Paradis R, Koutsilieris M. Three-dimensional culture system as a model for studying cancer cell invasion capacity and anticancer drug sensitivity. Anticancer Res. 2004;24:2169–2177. [PubMed] [Google Scholar]
- Huang S-B, Wang S-S, Hsieh C-H, Lin YC, Lai C-S, Wu M-H. An integrated microfluidic cell culture system for high-throughput perfusion three-dimensional cell culture based assays: effect of cell culture model on the results of chemosensitivity assays. Lab on a Chip. 2013;13(6):1133– 1143. doi: 10.1039/c2lc41264k. [DOI] [PubMed] [Google Scholar]
- Yamada KM, Cukierman E. Modeling Tissue Morphogenesis and Cancer in 3D. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Picollet-D’hahan N. Live Cell Analysis: When Electric Detection Interfaces Microfluidics. J Biochip Tissue chip. 2011:S1. [Google Scholar]
- Tran TB, Baek C, Min J. Electric Cell-Substrate Impedance Sensing (ECIS) with Microelectrode Arrays for Investigation of Cancer Cell – Fibroblasts Interaction. PLoS ONE. 2016;11(4):e0153813. doi: 10.1371/journal.pone.0153813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Frey O, Haandbaek N, Franke F, Rudolf F, Hierlemann A. Time-lapse electrical impedance spectroscopy for monitoring the cell cycle of single immobilized S pombe cells. Sci Reports. 2015;5:17180. doi: 10.1038/srep17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaofang H, Zuo P, Ye BC. Label-free electrochemical impedance spectroscopy biosensor for direct detection of cancer cells based on the interaction between carbohydrate and lectin. 2013;45:79–83. doi: 10.1016/j.bios.2012.11.028. [DOI] [PubMed] [Google Scholar]
- Tiruppathi C, Malik AB, Vecchio RJD, Keese CR, Giaver I. Electrical method for detection of endothelial cell shape change in real time: Assessment of endothelial barrier function. Proc Nati Acad Sci USA. 1992;89:7919–7923. doi: 10.1073/pnas.89.17.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A, Adriani G, Tay A, Warkiani ME, Yeap WH, Wong SC, Kamm RD. Engineering a 3D microfluidic culture platform for tumor-treating field application. Sci Rep. 2016;6:26584. doi: 10.1038/srep26584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R. When PDMS isn’t the best? What are its weaknesses, and which other polymers can researchers add to their toolboxes? Anal Chem. 2007:3249–3253. doi: 10.1021/ac071903e. [DOI] [PubMed] [Google Scholar]
- Calleja AB, Li R, Chen MB, Wong SC, Kamm RD. Microfluidics: A New Tool for Modeling Cancer–Immune Interactions Trends in Cancer. Trends Cancer. 2016;2(1):6–19. doi: 10.1016/j.trecan.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodkowic D, Cooper JM. Tumors on chips: oncology meets microfluidics. Curr Opin Chem Biol. 2010;14:556–567. doi: 10.1016/j.cbpa.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Zare RN, Kim S. Microfluidic Platforms for Single-Cell Analysis. Annu Rev Biomed Eng. 2010;12:187–201. doi: 10.1146/annurev-bioeng-070909-105238. [DOI] [PubMed] [Google Scholar]
- Sung KE, Beebe DJ. Microfluidic 3D models of cancer. Adv Drug Delivery Rev. 2014;79:68–78. doi: 10.1016/j.addr.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzon C, Duperray A, Verdier C. Critical stresses for cancer cell detachment in microchannels. Eur Biophys J. 2009;38:1035–1047. doi: 10.1007/s00249-009-0506-1. [DOI] [PubMed] [Google Scholar]
- Ling S, Slattery MJ, Dong C. Shear stress and shear rate differentially affect the multi-step process of leukocyte-facilitated melanoma adhesion. Exp Cell Res. 2005;310(2):282–292. doi: 10.1016/j.yexcr.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehranirokh M, Kouzani AZ, Francis PS, Kanwar JR. Microfluidic devices for cell cultivation and proliferation. Biomicrofluidics. 2013;7:051502. doi: 10.1063/1.4826935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KR, Wang S, Irimia D, Jayaraman A, Toner M, Yarmush ML. A high-throughput microfluidic real-time gene expression living cell array. Lab Chip. 2007;7:77–85. doi: 10.1039/b612516f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PJ, Hung PJ, Rao VM, Lee LP. Nanoliter scale microbioreactor array for quantitative cell biology. Biotechnol Bioeng. 2006;94:5–14. doi: 10.1002/bit.20745. [DOI] [PubMed] [Google Scholar]
- Thompson DM, King KR, et al. Dynamic gene expression profiling using a microfabricated living cell array. Anal Chem. 2007;76:4098–4103. doi: 10.1021/ac0354241. [DOI] [PubMed] [Google Scholar]
- Ye N, Qin J, Shi W, Liu X, Lin B. Cell-based high content screening using an integrated microfluidic device. Lab Chip. 2007;7:1696–1704. doi: 10.1039/b711513j. [DOI] [PubMed] [Google Scholar]
- Tarbell JM, Shi ZD. Effect of the glycocalyx layer on transmission of interstitial flow shear stress to embedded cells. Biomech Model Mechanobiol. 2012;12:111–121. doi: 10.1007/s10237-012-0385-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hompland T, Ellingsen C, Øvrebø KM, Rofstad EK. Interstitial fluid pressure and associated lymph node metastasis revealed in tumors by dynamic contrast-enhanced MRI. Cancer Res. 2012;72(19):4899–4908. doi: 10.1158/0008-5472.CAN-12-0903. [DOI] [PubMed] [Google Scholar]
- American Cancer Society. Breast Cancer Facts & Figures 2015–2016. Atlanta: American Cancer Society, Inc; 2015. [Google Scholar]
- Cavallotti C, D’Andrea V, Cavallotti C, Cameroni M. Distribution of acetylcholinesterase and cholineacetyl-transferase activities in the human pulmonary vessels of younger and older adults. Geriatr Gerontol Int. 2005;5:286–292. [Google Scholar]
- Solly K, Wang X, Xu X, Strulovici B, Zheng W. Application of real-time cell electronic sensing (RT_CES) technology to cell-based assays. Assay Drug Dev Technol. 2004;2:363–372. doi: 10.1089/adt.2004.2.363. [DOI] [PubMed] [Google Scholar]
- Ehret R, Baumann W, Brischwein M, Schwinde A, Wolf B. On-line control of cellular adhesion with impedance measurements using interdigitated electrode structures. Med Biol Eng Comput. 1998;36:365–70. doi: 10.1007/BF02522485. [DOI] [PubMed] [Google Scholar]
- Yang L, Arias LR, Lane TS, Yancey MD, Mamouni J. Real-time electrical impedance-based measurement to distinguish oral cancer cells and non-cancer oral epithelial cells. Anal Bioanal Chem. 2011;399:1823–1833. doi: 10.1007/s00216-010-4584-9. [DOI] [PubMed] [Google Scholar]
- Hua MY, Yang HW, Chuang CK, Tsai RY, Chen WJ, Chuang KL, Chang YH, Chuang HC, Pang ST. Magnetic-nanoparticle-modified Paclitaxel for targeted therapy for prostate cancer. Biomaterials. 2010;31(28):7355–7363. doi: 10.1016/j.biomaterials.2010.05.061. [DOI] [PubMed] [Google Scholar]
- Zhang AL, Russell PJ, Knittel T, Milross C. Paclitaxel enhanced radiation sensitization for the suppression of human prostate cancer tumour growth via a p53 independent pathway. Prostate. 2007;67(15):1630–40. doi: 10.1002/pros.20638. [DOI] [PubMed] [Google Scholar]
- Shafiee H, et al. Acute On-Chip HIV Detection Through Label-Free Electrical Sensing of Viral Nano-Lysate. Small. 2013;9:2553–2563. doi: 10.1002/smll.201202195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzon C, Duperray A, Verdier C. Critical stresses for cancer cell detachment in microchannels. Eur Biophys J. 2009;38:1035–1047. doi: 10.1007/s00249-009-0506-1. [DOI] [PubMed] [Google Scholar]
- Zlatoš A. Diffusion in fluid flow: Dissipation enhancement by flows in 2D. Communications in Partial Differential Equations. 2010;(35)(3):496–534. [Google Scholar]
- Jubery TZ, Hossan MR, Bottenus DR, Ivory CF, Dong W, Dutta P. A new fabrication technique to form complex polymethylmethacrylate microchannel for bioseparation. Biomicrofluidics. 2012;6(1):016503-01–016503-13. doi: 10.1063/1.3683163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakaran P, Hassiotou F, Blancafort P, Filgueira L. Cisplatin Induces Differentiation of Breast Cancer Cells. Front Oncol. 2013;3(134):1–10. doi: 10.3389/fonc.2013.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirt I, Verma S, Petrella T, Bak K, Charette M. Temozolomide for the treatment of metastatic melanoma: a systematic review. Oncologist. 2007;12(9):1114–23. doi: 10.1634/theoncologist.12-9-1114. [DOI] [PubMed] [Google Scholar]
- McDermott M, Eustace1 AJ, Busschots S, Breen L, Crown J, Clynes M, O’Donovan N, Stordal B. In vitro development of chemotherapy and targeted therapy drug-resistant cancer cell lines: a practical guide with case studies. Front Oncol. 2014;4(14):1–15. doi: 10.3389/fonc.2014.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill AJ, Prencipe M, Dowling C, Fan Y, Mulrane L, Gallagher WG, O’Connor D, O’Connor R, Devery A, Corcoran C, Rani S, O’Driscoll L, Fitzpatrick JM, Watson RWG. Characterization and manipulation of docetaxel resistant prostate cancer cell lines. Molecular Cancer. 2011;10(126):1–13. doi: 10.1186/1476-4598-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho MR, Lima D, Reis RL, Corello VM, Oliveira JM. Evaluating Biomaterial- and Microfluidic-Based 3D Tumor Models Trends in Biotechnology. 2015;33(11):667–678. doi: 10.1016/j.tibtech.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Giaever I, Keese CR. A morphological biosensor for mammalian cells. Nature. 1993;366:591–592. doi: 10.1038/366591a0. [DOI] [PubMed] [Google Scholar]
- Liu QJ, Yu JJ, Xiao L, Tang JCO, Zhang Y, Wang P, Yang M. Impedance studies of bio-behavior and chemosensitivity of cancer cells by micro-electrode arrays. Biosens Bioelectron. 2009;24:1305–10. doi: 10.1016/j.bios.2008.07.044. [DOI] [PubMed] [Google Scholar]
- Gomez SR, Leyrat AA, Pirone DM, Chen CS, Quake SR. Versatile, Fully Automated, Microfluidic Cell Culture System. Anal Chem. 2007;79:8557–8563. doi: 10.1021/ac071311w. [DOI] [PubMed] [Google Scholar]
- Liu L, Luo C, Ni X, Wang L, Yamauchi K, Nomura SM, Nakatsuji N, Chen Y. A micro-channel-well system for culture and differentiation of embryonic stem cells on different types of substrate. Biomed Microdevice. 2010;12:505–511. doi: 10.1007/s10544-010-9407-4. [DOI] [PubMed] [Google Scholar]