Abstract
Here, we develop and test a method to address whether DNA samples sequenced from a group of fossil hominin bone or tooth fragments originate from the same individual or from closely related individuals. Our method assumes low amounts of retrievable DNA, significant levels of sequencing error, and contamination from one or more present-day humans. We develop and implement a maximum likelihood method that estimates levels of contamination, sequencing error rates, and pairwise relatedness coefficients in a set of individuals. We assume that there is no reference panel for the ancient population to provide allele and haplotype frequencies. Our approach makes use of single nucleotide polymorphisms (SNPs) and does not make assumptions about the underlying demographic model. By artificially mating genomes from the 1000 Genomes Project, we determine the numbers of individuals at a given genomic coverage that are required to detect different levels of genetic relatedness with confidence.
Keywords: ancient DNA, population genetics, relatedness
OVER the past few years, the amount of ancient DNA (aDNA) recovered from fossilized bones, teeth, and hair has grown rapidly (Prüfer et al. 2014; Mathieson et al. 2015; Sawyer et al. 2015). Despite significant advances in sequencing technology, laboratory practices, and computational methods, problems still arise because of low amounts of endogenous nuclear DNA, short degraded fragments, contamination from present-day humans, and sequencing errors. Nevertheless, data from ancient remains are a precious source of information, providing insights about the history of humans and their closest relatives that are unavailable from any other source. DNA from several ancient individuals found in the same location is especially important because it can provide clues about relatedness within groups. This information is valuable for downstream analyses that make assumptions about relatedness among individuals.
In sexually reproducing species, the coefficient of relatedness (r) is twice the probability that two sites sampled at random from autosomes (one from each individual) are identical by descent (IBD). With that definition, for two samples from the same individual or from monozygotic twins, for first-degree relatives (parents and offspring or full siblings), and for second-degree relatives (aunt or uncle and nephew or niece, half siblings, grandparent and grandchild, or double first cousins), etc.
Information about the genetic relatedness between individuals is of significance in the fields of forensic sciences, agriculture, human genetics, and ecological sciences. A variety of approaches have been developed to infer relatedness, each suited to specific types of data. For comprehensive reviews on statistical methods and available approaches see Weir et al. (2006) and Speed and Balding (2015). The general concept underlying relatedness analyses is that of IBD, but this quantity cannot be observed directly in data. Instead, allelic states at a particular locus are used to make inferences about IBD and relatedness. When good quality, high-coverage genomes from individuals are available, inferring relatedness is relatively easy and many methods have been developed (Purcell et al. 2007; Browning and Browning 2010; Pemberton et al. 2010; Huff et al. 2011; Wang 2011; Li et al. 2014)). However, for aDNA, the quality and amount of data are often sufficiently limited that existing methods cannot be applied.
There have been some attempts to deal with the problems posed by aDNA. For example, Vohr et al. (2015) developed an approach to identify whether two DNA samples with extremely low coverage originate from the same or different individuals. The authors introduced a likelihood method that uses information from SNPs and patterns of linkage disequilibrium. However, this method relies on a reference panel of phased haplotypes from the same population to infer allele and haplotype frequencies. This method can be used for human fossils that are sufficiently recent that a present-day population can be used as a reference panel. However, for older human fossils and for Neanderthals and Denisovans, no reference panels are yet available.
In another recent study, M. D. Martin, F. Jay, S. Castellano, and M. Slatkin (unpublished results) presented a method to infer close genetic relatedness using low-coverage, next-generation sequencing samples from ancient individuals. They did not assume that a reference panel was available and they accounted for contamination from modern humans and sequencing errors. Their method investigates the overlap of pairwise genetic distance distributions calculated under certain realistic scenarios to identify the relatedness between pairs of individuals.
In this study, we extend the work of M. D. Martin, F. Jay, S. Castellano, and M. Slatkin (unpublished results) by using a maximum likelihood framework applied to each polymorphic site and determine whether this approach provides improved accuracy. In addition to inferring relatedness, our method provides estimates of allele frequencies and contamination levels for each sample. By artificially mating individual sequences from the publicly available 1000 Genomes Project, we determine the number of individuals at a given genomic coverage that are required to distinguish different levels of genetic relatedness.
Methods
Model notation
The relatedness r of two individuals is twice the probability of identity by descent of two chromosomes chosen at random. Individuals are denoted by and sites are denoted by We further assume the sequencing error rate e is the same for every site k in every sequence. e is the probability that a site is misread during the sequencing; if it is misread at a site that is actually monomorphic then it creates a false SNP, but if it is misread at a site that is actually polymorphic then it is misread as the alternative allele. The contamination rate for an ancient individual i is the probability that a randomly chosen read from an individual i is derived from a present-day human. The average contamination rate over all sequenced ancient individuals is denoted We use only sites that are polymorphic in the contaminant panel and we will assume that we observe only ancestral or derived (non-chimpanzee) alleles at every site, thereby ignoring triallelic sites.
Furthermore, let be the derived allele frequency (daf) at site k in the putative contaminating population (e.g., modern humans). The observed daf at site k in the ancient samples is and is a weighted average of the endogenous and contaminating allele frequencies (note that the model assumes exactly one allele per genomic position per individual):
| (1) |
where is the endogenous daf at site k in the ancient samples (unobservable because the alleles sequenced at a site might either be endogenous or from the contaminating population). We use because it is, in principle, impossible to determine which of the reads at a given site comes from the contaminating population. Therefore, our best estimate of is:
| (2) |
To summarize, the model input parameters are the allelic (ancestral/derived) states at each site from each of the ancient reads. The observed parameter is is the only parameter used from a contaminating reference data set. The more individuals that are available in the contaminating reference data set the closer these values approach true population frequencies resulting in more accurate parameter estimates. A parameter that cannot be directly observed from the ancient data is but it is calculated at each step based on , and The parameters that we will aim to estimate are the relatedness coefficient for each pair of individuals the contamination rate for each individual , and the overall sequencing error rate e.
For a pair of individuals i and j with relatedness there are three sets of parameters that need to be modeled.
- Endogenous frequencies - the probabilities of allelic configurations 11,10,01,00 in the aDNA (1 being derived, 0 being ancestral):
(3) - Contaminated frequencies - the probabilities of allelic configurations in the contaminated sample:
(4) - Sequenced frequencies - the probabilities of allelic configurations in the sequences themselves, allowing for sequencing error:
(5)
Parameter estimation
Assume a data set of N ancient individuals and L aligned sites. For each pair of individuals i and j (out of total pairs) the log likelihood is calculated as:
| . | (6) |
The log likelihood for the entire data set is then the sum over all log likelihoods for all pairs of individuals. We refer to this approach as the “complete method” (note that this is a composite likelihood because a single individual contributes N−1 times to the calculation and the pairs of individuals are not independent).
Overall, the number of values that need to be estimated are parameters for the relatedness coefficients N parameters for contamination rates , and one parameter for the sequencing error rate e. A method to maximize the log-likelihood of these input parameters is implemented in C++ using the nonlinear optimization routine L-BFGS from the dlib C++ library (King 2009). The software we generated is available online at https://github.com/christoph-theunert/. Lower and upper bounds for the parameters r, C, and e are set to [0.001, 0.9999], [0.0, 0.25], and [0.001, 0.25] respectively.
As mentioned in the results, a slightly different procedure of using only a subset of all available individuals to calculate the likelihood for the entire data set was tested. In this case individuals are used to calculate the overall likelihood as the sum over likelihoods For example, if one is only interested in a certain pair of individuals i and j, then and only one needs to be estimated. However, at site k is still estimated using all N individuals. Depending on the actual value of n this approach may result in faster computation times. We refer to this approach as the “subset method.”
We simulated 50 independent data sets for each combination of N, L, and r, and separately performed the parameter estimation for each of them. Therefore, the final estimates of r are given as an average, and the accuracy of our method is evaluated by the root-mean-square error (rmse) and the mean absolute error (mae). When used together, rmse and mae can characterize the errors in a set of forecasts. The magnitude of the difference between them is informative about the amount of variance in the individual errors in the sample.
We checked the convergence of the L-BFGS routine by running the optimization for the same data set 15 times. Each time, the parameter vector was initialized with a random set of parameter values. We did that for multiple different data sets and for every case we found the same final optimal value.
Simulations
For the initial evaluation of the model we generated sets of 2N sequences of length L sites. For each sequence, alleles at each position k were either derived (1) with probability or ancestral (0) with probability where was randomly drawn from To generate contaminated reads from our simulated genotypes, we adopted a method used in Racimo et al. (2016). For each simulated individual i, the number of derived and ancestral fragments at a particular site follows a binomial distribution that depends on the true ancient genotype, the sequencing error rate, and the contamination rate [see Equations 3–6 in Racimo et al. (2016)]. Contamination rate for individual i was randomly drawn from a uniform distribution between and separately for each simulation (i.e., in each simulation individuals have different rates of contamination ). Sequencing error rate e was set to 0.001 throughout all simulations. To systematically study the behavior of our method we assume one read per individual at each simulated genomic position. We further assume for each site from a putative reference panel to be randomly drawn from
Furthermore, we simulated a scenario where reads are only available from a random subset of individuals (out of a total of N) at each genomic site. Supplemental Material, Tables S3 and S4 in File S1 summarize the results.
To ensure that the simulation method mentioned above does not introduce any biases, we carried out simulations where we artificially mated unrelated European (EUR) sequences from the 1000 Genomes Project Phase 3 (1000 Genomes Project Consortium et al. 2015). A similar approach was introduced by M. D. Martin, F. Jay, S. Castellano, and M. Slatkin (unpublished results). This population was chosen only to demonstrate the workflow and performance of our method on real data and not with the intention of drawing any conclusions about European populations. We focused on phased genomes and extracted all biallelic polymorphic sites from single chromosomes from EUR individuals. Contamination from a putative contaminant panel was implemented in the same way as described before. We restricted our analyses to SNPs that passed the basic 1000 Genomes Project filtering criteria and for which ancestral allele information was available. The ancestral states were determined by using information from the inferred human–chimpanzee ancestor at each site. We filtered sites with a Map20 < 1 (Duke uniqueness tracks of 20 bp) and we removed deletions and insertions. The method behaves exactly the same for both data sets (simulated sequences and sequences from the 1000 Genomes Project).
In both cases, we performed artificial meioses of pairs of individuals for single chromosomes. The recombination rate was assumed to be uniform along the genome and set to be /bp per generation (Kong et al. 2002; Prüfer et al. 2014). We implemented a minimal number of one recombination event per chromosome per generation. Relatedness among individuals was then simulated by artificially mating them with other individuals to produce offspring.
To investigate the effect of different types of genomic sites, we analyzed each data set (not the present-day reference panel) filtered for: (1) fixed and polymorphic sites, (2) only polymorphic sites, and (3) polymorphic sites that were either changed to being fixed or remained polymorphic after allowing for contamination and sequencing error. This way, we could study the effect of different classes of sites on the accuracy of our estimates.
Simulated pedigrees
Three different pedigrees (denoted f1, f2, and f3) were generated with the mating method described above:
f1: father + mother = child
f2: father + mother = child1; child1 + X0 = child2
f3: father + mother = child1; father + mother = child2; child1 + X1 = child3; child2 + X2 = child4
where X0, X1, and X2 represent unrelated partner individuals. Throughout the manuscript each data set is further represented by an additional number which denotes the absence (0.0) or presence (0.1) of contamination and sequencing errors (e.g., f2.1 is the second pedigree f2 with contamination and error). In each data set all remaining individuals were kept unrelated. The very last individual in each data set is a direct copy of another individual before contamination and error. This allows us to test the method for different degrees of relatedness: in f1; and in f2; and in f3; and in all three.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Accuracy when and
First, we studied the accuracy of our method to identify genetic relatedness simulated in pedigree f1.0 in the absence of contamination and sequencing errors by using the subset approach with
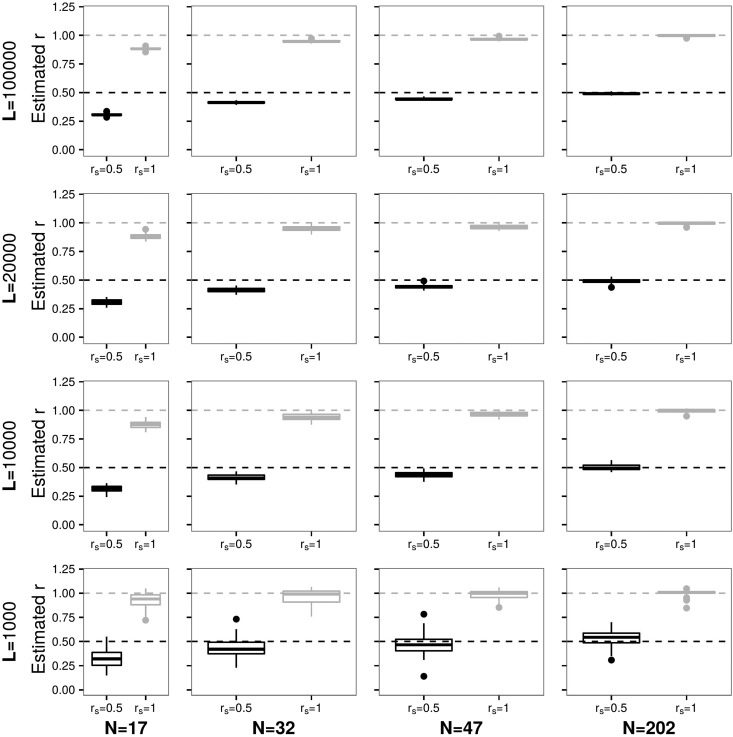
In Figure 1, each subfigure of boxplots represents a different combination of N individuals (rows) and L sites (columns) and shows the distribution of r for a pair of related individuals over 50 independent data sets (see Figure S1 in File S1 for more details and error values). Note that we refer to the true simulated relatedness coefficient as the point estimates of it as r, and the estimated average over 50 independent data sets as
Figure 1.
Each panel represents a different combination of N (columns) and L (rows) and shows a boxplot for the estimates of simulated relatedness of (same individual) and (parent–offspring or full siblings) over 50 independent data sets for pedigree f1.0. Dashed horizontal lines denote the simulated values of
For example, in the upper right corner we simulated reads for 202 diploid individuals and 100,000 overlapping polymorphic sites. For the two samples that result from the same individual (), estimates are with rmse = 0.01 and mae = 0.01. In the same data set for a pair of parent–offspring individuals (), is 0.49 with rmse = 0.01 and mae = 0.01.
The variation of the parameter estimates, is given in more detail in Figure S2 in File S1 showing that the range in estimates for this data set is rather small (for r = [0.98, 1.01]; for r = [0.47, 0.49]). We note here that values of are possible as the final step of the parameter inference is In the majority of cases, the method underestimates the value of As expected, the fewer overlapping sites and individuals that are available, the more the estimates deviate from the true value of and the higher the error estimates become. For example, for N = 17 and L = 1000, is 0.94 with rmse = 0.11 and mae = 0.09 (for ); and with rmse = 0.2 and mae = 0.18 (for ). It is worth mentioning that the distribution of estimates for different do not overlap with each other in any of the data sets.
Figure S3 in File S1 shows the comparison of estimated and simulated contamination rates for each related individual with the rmse shown in the legend of each graph (note that the true ). The method overestimates the contamination rates, but the majority of is estimated to be when the number of individuals and sites increase.
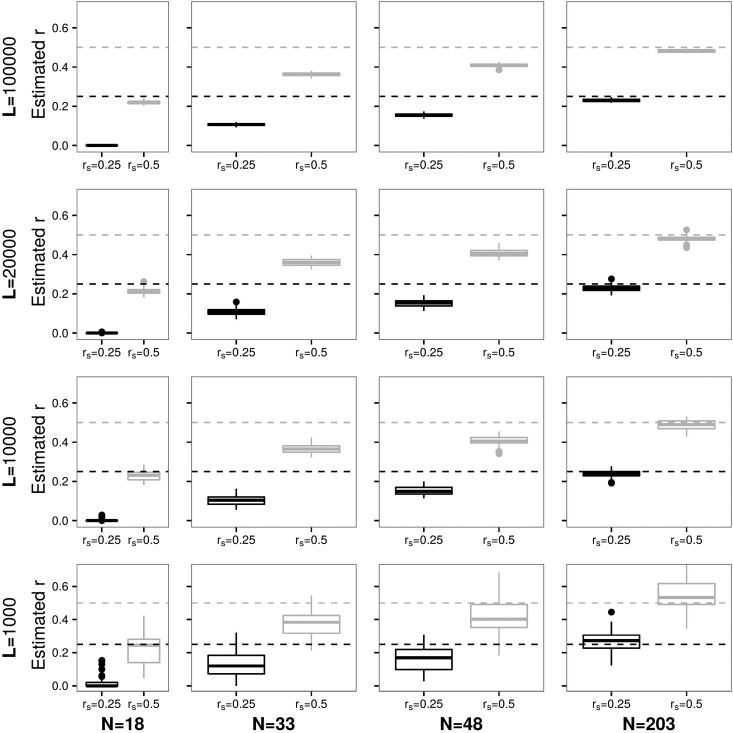
The accuracy of the method to identify relatedness coefficients from pedigree f2.0 is presented in Figure 2. Again, with reads from 202 individuals and the results are highly accurate, and simulated as well as second-degree relatedness are estimated to be and respectively (see Figure S4 in File S1 for more details and error values).
Figure 2.
Each panel represents a different combination of N (columns) and L (rows) and shows a boxplot for the estimates of simulated relatedness of (parent–offspring or full siblings) and (e.g., grandparent–grandchild or half siblings) over 50 independent data sets for pedigree f2.0. Dashed horizontal lines denote the simulated values of
As expected, the smaller N and L, the less accurate results become, e.g., and error estimates ∼0.10 for when N = 48 and L = 10,000. Furthermore, with N = 18 the method does not pick up the signal of anymore. Although for more distantly related individuals parameter inference may be less accurate, distributions of estimates do not overlap and so provide valuable information about differences in relatedness (see Figures S5 and S6 in File S1).
Identifying a relatedness of from data set f3.0 is even more difficult. Shown in Figures S7, S8, and S9 in File S1 are estimates for It can be seen that, only with reads from 205 individuals, results are rather accurate at and errors of ∼0.03.
Accuracy when C > 0 and e > 0
Under a more realistic scenario, contamination from modern humans and sequencing error may create bias. Therefore, we tested the method on simulated data sets that are affected by these factors. As described before, each is drawn from and e is set to 0.001 for all data sets.
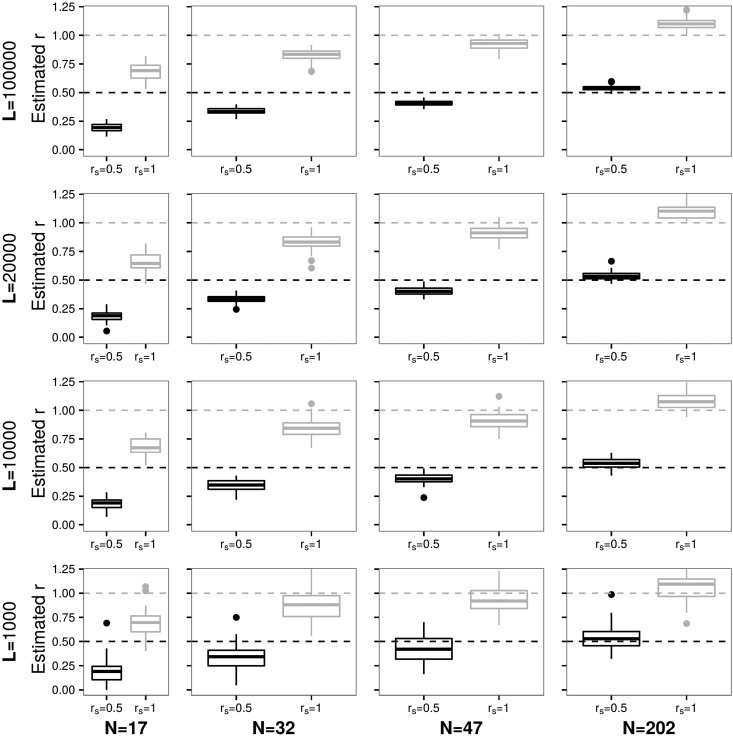
Summarizing the information for pedigree f1.1 from Figure 3, Figure 4, and Figures S10 and S11 in File S1, the observations are similar to what we reported before but and e affect the accuracy of the results. The method is still able to identify the same or related individuals while the amount of available data has a more pronounced effect on the accuracy. Estimates are less accurate than in the absence of and e. However, when comparing the results for and from the same data set, the distributions of estimates for do not overlap. This does provide valuable information (see Figures S10 and S11 in File S1). For example, for 32 individuals and 10,000 sites, estimates of the relatedness coefficient range between when and when With and , estimates of r and are highly accurate with small error values. See Supplemental Material text and Table S1 in File S1 for a comparison between the subset method and the complete method for this data set.
Figure 3.
Results are presented for estimates of (same individual) and (parent–offspring or full siblings) as in Figure 1, but with being between and sequencing error set to 0.001 as in pedigree f1.1.
Figure 4.
Each panel represents a different combination of N (columns) and L (rows) and shows an x–y plot for estimated (y-axis) and simulated (x-axis) contamination rates for pedigree f1.1. Dashed lines indicate rmse of 0. was simulated to be between and
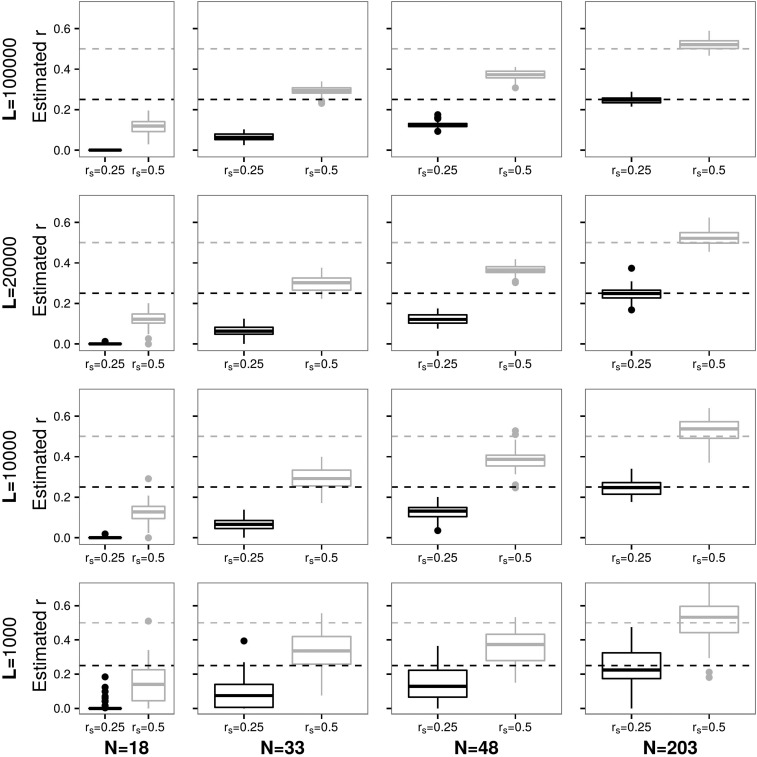
The more distant the genetic relatedness between two individuals the more data are needed to identify it. Figure 5 shows results for pedigree f2.1. Again, note that with 203 individuals and sites, of 0.25 and are accurately inferred (see Figures S12, S13 and S14 in File S1 for more details and error values). The same is true for pedigree f3.1 as seen in Figures S15, S16, and S17 in File S1. The likelihood landscape under the presence and absence of and e is shown in Figure S30 in File S1.
Figure 5.
Estimates of (parent–offspring or full siblings) and (e.g., grandparent–grandchild or half siblings) as in Figure 2, but with being between and , and sequencing error set to 0.001 as in pedigree f2.1.
In conclusion, the proposed method can accurately infer the degree of genetic relatedness even in the presence of contamination and sequencing error. However, first-, second-, and third-degree relatedness require more data to be identified than when identifying DNA sequences that originate from the same individual. For example, for a and N = 32, is still 0.84. In this case, the distributions of estimates in Figure S11 in File S1 show that the values do not drop below r = 0.7 in the majority of the cases (for ). Our method tends to underestimate the parameters without an overlap between the distributions of estimates for , and This fact supports the validity of results for even more, as it seems unlikely that an estimated value of r = 0.8 is seen when the DNA sequences do not originate from the same individual. Note that, overall, the individual contamination rates are more often under- than overestimated when
Application to real data
To illustrate the application of our program to a real data set, we analyzed a sample of seven ancient individuals from the Motala population in Sweden, previously analyzed by Haak et al. (2015). The individuals were all found in the same site in Sweden and are dated to around the same time (5898–5531 calBCE). Although the number of individuals is rather small and might limit the power of our method to detect relatedness, this data set is one of the biggest publicly available collections of ancient individuals from the same location and time and, therefore, allows us to demonstrate the necessary steps to apply our program:
Filter each individual BAM file according to a desired set of filters (e.g., mapping quality, base quality etc.)
Intersect the genomic positions of all seven filtered BAM files and create a file in BED format that contains positions that are covered in each of the seven individuals
For each genomic position, identify the ancestral and derived allele and their population frequencies in a modern human contaminating panel (we chose 96 Northern and Western European ancestry (CEU) individuals from the 1000 Genomes Phase 3 data set) and save the information in a separate file (note that we only use sites that are polymorphic in this contaminating panel)
Intersect the genomic positions from step 2 and the genomic positions from step 3 for which ancestral/derived allele frequencies are available and create a new file in BED format
Based on the genomic positions from step 4, transform each individual filtered BAM file into PILEUP format (see http://samtools.sourceforge.net/pileup.shtml)
Based on the individual pileup formats, for each genomic position pick the allele from each individual (here we pick one allele at random if multiple reads cover a position) and represent an ancestral allele by “0” and a derived allele by “1”
Create a file in ms format [see Hudson (2002)], i.e., allelic information as one row per individual and one column per genomic site
Create a meta information file with three tab separated columns for each position: site number (k); derived allele frequency in the sample of seven individuals (); and derived allele frequency in CEU ()
Run our program with the files from step 7 and 8 as input
Additionally, we generated a direct copy of one of the individuals and introduced it to the data set for a total of eight individuals (and ∼7000 polymorphic sites). Therefore, one pair of individuals (7 and 8) should show a higher relatedness coefficient (note that the copied individual is not completely identical to its “parent” since we picked one allele at random for each genomic position). We then analyzed the whole set of eight individuals together and did not expect to see any relatedness between pairs of individuals except for the artificially related pair. As shown in Table 1, we do not observe any related pairs of individuals except for the pair (7,8) that shows an estimated r of 0.65. Although this is a rather small data set and only serves as an example, the program is able to identify the related pair. The individual contamination rates are estimated to be 0.0–0.03. However, note that it is difficult to correctly interpret these numbers: first because the true contamination rates are not known; second because based on our simulation results this dataset is rather small and therefore these estimates are likely biased; and third because the ancient individuals are much younger and therefore less diverged from modern humans than, for example, Neanderthals. This means that contamination is more difficult to detect the less diverged the ancient individuals are from the contaminating population. We recommend applying our method to all individuals in the data set to get an idea of how many related and unrelated individuals are present.
Table 1. Results for pairwise relatedness coefficients for all pairs of 7 + 1 Motala individuals.
| i/j | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| 2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ||
| 3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |||
| 4 | 0.0 | 0.0 | 0.0 | 0.0 | ||||
| 5 | 0.0 | 0.0 | 0.0 | |||||
| 6 | 0.0 | 0.0 | ||||||
| 7 | 0.65 |
Discussion
In this study, we present a method to infer the relatedness coefficients from aDNA samples sequenced from a group of fossil hominin bone or tooth fragments. Our method accounts for sequencing error and for contamination from present-day humans. By artificially mating simulated sequences as well as sequences from the 1000 Genomes Project, we determine how many overlapping reads and how many individuals are required to obtain estimates of relatedness coefficients with confidence. The likelihood model we developed for this purpose differs from existing methods in that we directly model the (hidden) ancient derived allele frequencies and do not require a reference panel for the ancient population.
In our simulations, we assumed that each polymorphic site is sequenced in every individual. With that assumption, the number of overlapping sites is a parameter under our control. The actual number of overlapping sites when there is low-coverage sequence data is a random variable whose distribution depends on the sequencing method used. For shotgun sequencing, the simplest assumption is that the number of times a polymorphic site is sequenced is a Poisson distributed random variable with the mean equal to the coverage level, for individual i. The probability that the site is sequenced at least once in individuals i and j is . For example, if (i.e., 0.1 × coverage in both individuals), the probability that a site is covered by at least one read is roughly 0.009. Therefore, if there are polymorphic sites, there would be roughly 27,000 overlapping sites in two individuals. Different sets of sites would overlap in different pairs of individuals. Hence the expected number of samples that contribute to estimates of allele frequencies at each site in a sample of N individuals is where is the average coverage level. In the Figures S18–S29 and Tables S3 and S4 in File S1, we allowed for this possibility in simulations by assuming that fully overlapping sites in all individuals are not available. As expected, the accuracy of the method decreases when compared to using all individuals. However, the more individuals in total available in a data set, the higher the accuracy even when only using read information from a random subset (e.g., 5 or 10 individuals) of them at each genomic site. An alternative to shotgun sequencing is genomic capture (Mamanova et al. 2010; Rohland and Reich 2012). With a capture method that targets sites known to be polymorphic in the same or a closely related population, the probability that two sites are sequenced in two individuals depends on a number of factors, including the closeness of the population or populations used for ascertainment and the complexity of the genomic library. However, the success of targeted capture methods can be quite high. For example, Castellano et al. (2014) used exome capture on two Neanderthal samples. In the El Sidron sample that had endogenous DNA, of targeted sites were covered at least once. In the Vindija 33.15 sample that had endogenous DNA, of the targeted sites were covered. Therefore, if exome or SNP capture methods are used there is a good chance of high levels of overlap in different individuals.
As can be seen from the results, the accuracy of the method strongly depends on the number of genomes available. It is not particularly well suited to analyzing a small number of genomes. With only a few individuals, r is underestimated in all cases we have studied, regardless of how many genomic sites L we added (see Figure 3 for N = 17). The underlying problem is that we do not have information from a reference panel from the same population as the individuals under study to obtain accurate estimates of allele frequencies. Therefore, our estimates of the allele frequency strongly depend on the number of genomes sampled. Using a reference panel from another population might help, but the accuracy would depend on the divergence between the reference panel and the population under study. For example, using a reference panel from modern humans when analyzing Neanderthals would introduce an uncertainty in the allele frequency estimates, since Neanderthals and modern humans are quite diverged. This uncertainty would probably be as great as when using allele frequencies estimated from only a small number of Neanderthal individuals.
We analyzed the runtime of our program for different data sets (see Table S5 in File S1). From the results, it is obvious that the computational time of the method depends heavily on the number of individuals tested and, to a smaller extent, on the number of genomic sites. For example, for a data set of N = 17 and L = 10,000, it takes 5 and 8 sec to analyze n = 2 and n = 4 individuals, respectively. Analyzing n = 17 individuals already takes 25 min. The overall runtime increases because of the n(n−1)/2 individual pairs that need to be calculated but also because of the increase in the number of underlying parameters that the optimization method has to take into account. For the example data set, increasing L to 100,000 increases the runtime to 14 and 51 sec when analyzing n = 2 and n = 4 individuals, respectively. So an increase in sites by a factor of 10 has an effect on the runtime that is orders of magnitudes smaller than increasing the number of individuals tested. Therefore, using the subset method with n < N is computationally much more efficient than using n = N.
In our analysis, we assume that the sampled (ancient) population is in Hardy–Weinberg equilibrium. That assumption allows us to derive the genotype frequencies from allele frequencies. If a population is made up of inbred individuals, then our method would not yield accurate results.
We do not make any inferences about the time of separation of the contaminating (present-day) population from the sampled (ancient) population. If the contaminating population is closely related to the sampled population, then allele frequencies in the two populations will be similar and the estimated allele frequencies in the ancient sample will depend only weakly on the contamination rate. The estimate of the contamination rate would not be accurate but the error in estimating that rate would not strongly affect estimates of relatedness coefficients. If the contaminating population has quite different allele frequencies, the estimates of contamination rate will be more accurate.
Finally, admixture from the ancient (e.g., Neanderthal) population into the contaminating (e.g., modern human) population will not affect our method. Admixture will make some of the contaminating allele frequencies slightly more similar to Neanderthals than they would be in the absence of admixture, which should not affect the estimates of contamination rates or relatedness coefficients.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.200600/-/DC1.
Acknowledgments
The authors gratefully acknowledge the help of Mike Martin. This work was supported in part by the Max Planck Society (as a salary for C.T.) and in part by a US National Institutes of Health grant (R01-GM40282 to M.S. and F.R.).
Footnotes
Communicating editor: N. A. Rosenberg
Literature Cited
- 1000 Genomes Project Consortium. Auton A., Brooks L. D., Durbin R. M., Garrison E. P., et al. , 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S. R., Browning B. L., 2010. High-resolution detection of identity by descent in unrelated individuals. Am. J. Hum. Genet. 86: 526–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano S., Parra G., Sánchez-Quinto F. A., Racimo F., Kuhlwilm M., et al. , 2014. Patterns of coding variation in the complete exomes of three Neandertals. Proc. Natl. Acad. Sci. USA 111: 6666–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., et al. , 2015. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522: 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics 18: 337–338. [DOI] [PubMed] [Google Scholar]
- Huff C. D., Witherspoon D. J., Simonson T. S., Xing J., Watkins W. S., et al. , 2011. Maximum-likelihood estimation of recent shared ancestry (ERSA). Genome Res. 21: 768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D. E., 2009. Dlib-ml: a machine learning toolkit. J. Mach. Learn. Res. 10: 1755–1758. [Google Scholar]
- Kong A., Gudbjartsson D. F., Sainz J., Jonsdottir G. M., Gudjonsson S. A., et al. , 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31: 241–247. [DOI] [PubMed] [Google Scholar]
- Li H., Glusman G., Hu H., Shankaracharya J., Caballero, et al. , 2014. Relationship estimation from whole-genome sequence data. PLoS Genet. 10: e1004144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamanova L., Coffey A. J., Scott C. E., Kozarewa I., Turner E. H., et al. , 2010. Target-enrichment strategies for next-generation sequencing. Nat. Methods 7: 111–118. [DOI] [PubMed] [Google Scholar]
- Mathieson I., Lazaridis I., Rohland N., Mallick S., Patterson N., et al. , 2015. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528: 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton T. J., Wang C., Li J. Z., Rosenberg N. A., 2010. Inference of unexpected genetic relatedness among individuals in HapMap phase III. Am. J. Hum. Genet. 87: 457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., et al. , 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racimo F., Renaud G., Slatkin M., 2016. Joint estimation of contamination, error and demography for nuclear DNA from ancient humans. PLoS Genet. 12: e1005972 (erratum: PLoS Genet. 12: e1006444). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N., Reich D., 2012. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22: 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S., Renaud G., Viola B., Hublin J. J., Gansauge M.-T., et al. , 2015. Nuclear and mitochondrial DNA sequences from two Denisovan individuals. Proc. Natl. Acad. Sci. USA 112: 2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed D., Balding D. J., 2015. Relatedness in the post-genomic era: is it still useful? Nat. Rev. Genet. 16: 33–44. [DOI] [PubMed] [Google Scholar]
- Vohr S., Buen Abad Najar C., Shapiro B., Green R., 2015. A method for positive forensic identification of samples from extremely low-coverage sequence data. BMC Genomics 16: 1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., 2011. Coancestry: a program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol. Ecol. Resour. 11: 141–145. [DOI] [PubMed] [Google Scholar]
- Weir B. S., Anderson A. D., Hepler A. B., 2006. Genetic relatedness analysis: modern data and new challenges. Nat. Rev. Genet. 7: 771–780. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.