Abstract
Population demographic history may be learned from contemporary genetic variation data. Methods based on aggregating the statistics of many single loci into an allele frequency spectrum (AFS) have proven powerful, but such methods ignore potentially informative patterns of linkage disequilibrium (LD) between neighboring loci. To leverage such patterns, we developed a composite-likelihood framework for inferring demographic history from aggregated statistics of pairs of loci. Using this framework, we show that two-locus statistics are more sensitive to demographic history than single-locus statistics such as the AFS. In particular, two-locus statistics escape the notorious confounding of depth and duration of a bottleneck, and they provide a means to estimate effective population size based on the recombination rather than mutation rate. We applied our approach to a Zambian population of Drosophila melanogaster. Notably, using both single- and two-locus statistics, we inferred a substantially lower ancestral effective population size than previous works and did not infer a bottleneck history. Together, our results demonstrate the broad potential for two-locus statistics to enable powerful population genetic inference.
Keywords: allele frequencies, demographic inference, diffusion approximation, linkage disequilibrium
PATTERNS of genetic variation within a population are shaped by the evolutionary and demographic history of that population, so observed variation encodes information about that history. Knowing population demographic history serves as an important control for learning about natural selection (Bustamante et al. 2001; Boyko et al. 2008) and understanding the relative efficacy of selection as populations change in size (Lohmueller et al. 2008; Henn et al. 2016). One particularly informative statistic used to summarize genetic polymorphism data is the allele frequency spectrum (AFS), which stores the distribution of observed single-locus allele frequencies from a sample of the population. The shape of the AFS is sensitive to demographic history, and fitting the expected AFS under parameterized demographic models to the observed AFS is a powerful approach for learning about demographic history (Marth et al. 2004; Williamson et al. 2005; Gutenkunst et al. 2009; Kamm et al. 2017).
For unlinked loci, the AFS is a sufficient statistic of the data and completely describes observed patterns of variation (Lohmueller et al. 2009). The expected AFS under arbitrary single- or multi-population histories can be efficiently calculated with either coalescent (Kingman 1982; Tajima 1983) or diffusion (Kimura 1964; Williamson et al. 2005; Gutenkunst et al. 2009) approaches. Poisson random field (PRF) theory (Sawyer and Hartl 1992) can then be used to calculate the likelihood of the data given model parameters. A key assumption of the PRF framework is that of independence between segregating loci, so that allele frequency trajectories are uncorrelated. However, neighboring loci are physically linked on the chromosome, and their allele frequencies are thus correlated. Recombination serves to reduce this correlation, with a higher rate of recombination between two loci more rapidly breaking down that association. For any two linked SNPs, their linkage disequilibrium (LD) is a measure of their nonindependence. Furthermore, as with allele frequencies, patterns of LD are shaped by historical demographic events such as bottlenecks, growth, and admixture, and therefore they are also informative about history (Pritchard and Przeworski 2001).
For linked sites, the distribution of LD carries additional information to the allele frequency spectrum about past demography (Myers et al. 2008), and the joint distribution of allele frequencies and LD between pairs of SNPs should afford greater power for demographic inferences than those based on allele frequencies alone. Characterizing two-locus allele frequency dynamics, and calculating their sampling probabilities, has attracted a large body of work. Kimura (1955) considered the case of genetic drift at multi-allelic loci using a diffusion approximation, and he calculated the time to fixation for one of the alleles when more than two alleles are present. This approach was expanded over the following decade to explicitly consider the two-locus setting with two alleles at each locus (Kimura 1963; Hill and Robertson 1966; Karlin and McGregor 1968; Ohta and Kimura 1969; Watterson 1970). These studies were generally interested in the probability and rates of fixation under arbitrary recombination between the two loci, and in characterizing the expectation and variance of LD.
More recently, sampling probabilities for two neutral linked loci were directly calculated under equilibrium demography (Golding 1984; Hudson 1985; Ethier and Griffiths 1990), often using the recursion approach due to Golding (1984). Hudson (2001) extended these results to generate those sampling probabilities with knowledge of the ancestral state and proposed a composite likelihood approach for fine-scale estimation of recombination rates across the genome, which has been implemented to infer recombination maps and identify hotspots in human and Drosophila populations (McVean et al. 2004; Auton and McVean 2007; Chan et al. 2012). Xie (2011) used a diffusion approach to calculate the allele frequency spectrum for two completely linked loci under neutrality or equal levels of selection, while Ferretti et al. (2016) recently used a coalescent approach to calculate the expected frequency spectrum for two completely linked neutral loci, and neutral sampling probabilities were developed under the coalescent with recombination for moderate to large recombination rates and constant population size (Jenkins and Song 2009, 2010, 2012; Bhaskar and Song 2012). Recently, Kamm et al. (2016) developed a coalescent approach to generate two-locus sampling probabilities under arbitrary demography and recombination, and found that accounting for demographic history improves accuracy in composite likelihood approaches for estimating fine-scale recombination rates.
Here, we characterize the increase in power of demographic inference from using two-locus allele frequency statistics vs. using the single-locus AFS. In particular, the depth and duration of a bottleneck are confounded when using the AFS, but we show they can be independently inferred using two-locus statistics. To enable our analyses, we developed a numerical solution to the diffusion approximation for two-locus allele frequencies with arbitrary recombination. We packaged this method in a two-locus composite likelihood framework that can be used to infer single-population demographic histories. Additionally, this framework allows for an estimate of the effective population size based on recombination that is independent from estimates based on levels of diversity. Using this approach, we inferred demographic history for a highly studied Zambian Drosophila melanogaster population, finding a smaller effective population size than previous analyses (), and a demographic history of recent modest growth with no severe bottlenecks.
By incorporating linkage between pairs of loci, our work extends previous demographic history inference approaches based on the AFS and Poisson Random Field theory. Recent approaches based on the sequentially Markovian coalescent (SMC) incorporate linkage information in an alternative manner. In particular, Li and Durbin (2011) used the SMC to model the distribution of expected times to the most recent common ancestor of segments of paired chromosomes, from which the history of effective population size can be inferred. This approach has since been extended to multiple chromosomes, increasing precision, and enabling inferences of population split times and gene flow (Harris and Nielsen 2013; Sheehan et al. 2013; Schiffels and Durbin 2014). Most recently, computational advances have scaled the SMC approach to hundreds of unphased whole genomes (Terhorst et al. 2017).
Methods
A two-locus model with influx of new mutations
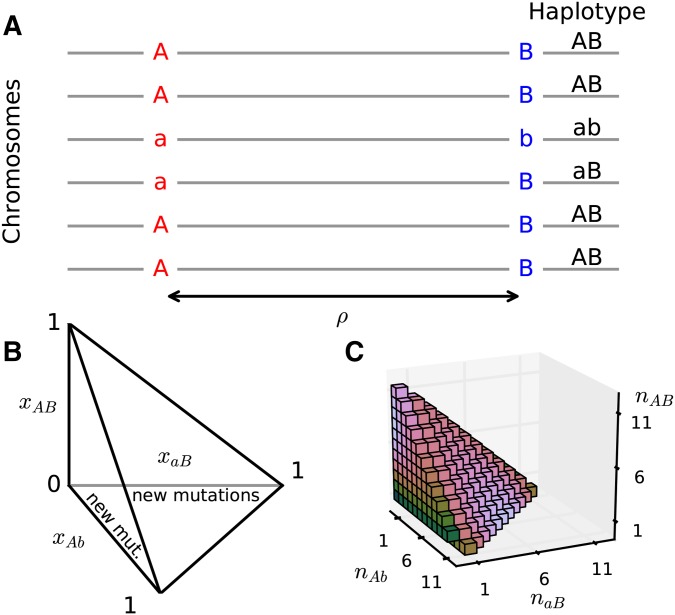
We used a diffusion approximation to a two-locus model that allows for two alleles at each locus, which are separated by recombination probability r (Karlin and McGregor 1968; Watterson 1970). We allow the left locus to carry alleles A and a, while the right locus permits alleles B and b. Then four haplotypes are possible, and with frequencies and that sum to (Figure 1A). Frequencies in the subsequent generation are found by considering the random pairing of haplotypes and the probability of a given pairing passing on each type to their offspring. These probabilities depend on current haplotype frequencies and the recombination rate, and are described in Table 1 of Watterson (1970). For example, a parent carrying haplotypes will pass on with probability and with probability even with recombination. On the other hand, a parent with will pass on or each with probability and or each with probability The numbers ( ) of each haplotype in the next generation are then pulled from the multinomial distribution for sampling haplotypes with probabilities found by considering random pairing of haplotypes and recombination.
Figure 1.
Two-locus model and frequency spectrum (A) Two loci with two alleles each are separated by recombination distance Four haplotypes are possible, and we track the frequencies of the three derived haplotypes. (B) Frequencies change within a tetrahedral domain, with corners of the domain corresponding to one of the four haplotypes fixed in the population. New two-locus pairs occur when a new mutation A occurs against the background, or when B occurs against the background, so we inject density along the or axes proportional to the background one-locus allele frequencies. (C) A sample two-locus haplotype frequency spectrum for a sample size of
Table 1. Point estimates from fits to Drosophila data. Reported log-likelihoods () are for two-locus data using the demographic history parameters from each fit. 95% confidence intervals are given in Table S1 in File S1.
| Data Statistics (Model) | |||||||
|---|---|---|---|---|---|---|---|
| One-locus (2-epoch) | 4.23 | 0.329 | 0.0476 | 302,900 | –1,368,000 | ||
| One-locus (3-epoch) | 2.35 | 10.7 | 0.388 | 0.0938 | 0.0496 | 291,500 | –1,629,500 |
| Two-locus (fix 2-epoch) | 4.05 | 0.371 | 0.0454 | 3 × 105 | –1.325,400 | ||
| Two-locus (fix 3-epoch) | 1.76 | 4.67 | 0.302 | 0.247 | 0.0469 | 3 × 105 | –1,289,400 |
| Two-locus (var. 2-epoch) | 4.05 | 0.371 | 0.0454 | 299,900 | –1,325,400 | ||
| Two-locus (var. 3-epoch) | 1.47 | 4.64 | 0.398 | 0.287 | 0.0474 | 286,600 | –1,283,740 |
New two-locus pairings, with two alleles segregating at both sites, arise when a new mutation occurs at one unmutated locus when the other locus is already polymorphic. Suppose, without loss of generality, that the right locus is already polymorphic, with derived allele B at frequency and ancestral allele b at frequency Then, a new A mutation at the left locus begins at frequency and occurs on the B haplotype with probability or on the b haplotype with probability Two-locus frequencies then evolve under the multinomial process described above until one or both loci are fixed for either the ancestral or derived allele, at which point we stop tracking that two-locus pair. The frequencies are drawn from the population distribution of one-locus frequencies which can be approximated using diffusion theory (Kimura 1964). Thus, new independent two-locus pairs enter the population with frequencies with rate proportional to and with rate proportional to
The density of two-locus haplotype frequencies, where and are the relative frequencies of haplotypes and respectively (Figure 1B), can be approximated using diffusion theory, as described in the next section. The two-locus haplotype frequency spectrum stores the counts of derived haplotypes in a sample, where one or both loci carry the derived allele. To obtain the two-locus spectrum F for n samples from the density function φ (Figure 1C), we sample against the multinomial sampling distribution:
| (1) |
Here, is the multinomial coefficient, defined as Because we assume that two-locus pairs are independent realizations of this process, PRF theory tells us that if we observe data each entry in the observed two-locus spectrum is a Poisson random variable with mean This allows the application of likelihood theory to compare observed data to model expectations.
Two-locus diffusion approximation
We solved the multiallelic diffusion equation for φ to obtain the expected sample two-locus spectrum. Measuring time τ in units of generations, where is the ancestral reference population size, the forward diffusion equation describes the evolution of the probability density of two-locus frequencies, and is written (Equation 3 in Hill and Robertson 1966) as
| (2) |
Here, is the LD, given haplotype frequencies and is a function for the relative population size to the ancestral population size at time τ. The population scaled recombination rate between the and loci is where r is probability of recombination between the two loci per generation. The action of recombination is readily interpretable in the diffusion equation: recombination acts directionally on the haplotype frequencies pushing them toward linkage equilibrium () at a rate directly proportional to the recombination rate ρ.
The domain of the two-locus diffusion equation is the tetrahedron with for and (Figure 1B). If the and there is no recurrent mutation, then all boundary surfaces of the domain are absorbing, so if one of the haplotypes is lost from the population it remains lost. However, with the boundary is not necessarily absorbing, as recombination may reintroduce a previously absent haplotype. For example, if only and haplotypes are found in the population, a recombination event may give rise to either an or haplotype in the next generation. Some of the edges of the domain are absorbing, since once one of either or fixes at the left or right locus, respectively, that two-locus pair remains fixed in the absence of recurrent mutation.
We numerically solved Equation 2 using finite differencing in a framework similar to Ragsdale et al. (2016). We split the diffusion operator into mixed and nonmixed terms, using an implicit alternating direction scheme for the nonmixed spatial derivatives (Chang and Cooper 1970), and a standard explicit scheme for the mixed spatial derivatives. We used equal numbers of uniformly spaced grid points for each spatial dimension, so that grid points coincided directly on the off-axes surface of the domain. This allowed for density to be accurately integrated along the surface and interior of the domain. As discussed in Ragsdale et al. (2016), and detailed in Supplemental Material, File S1, naively applying finite differencing along the off-axes surface led to numerical error in the solution to φ. Thus, we instead accounted for density moving between the interior of the domain and that surface by directly moving density between the two each timestep. To solve for the two-locus spectrum under a nonequilibrium demographic model we first solved for φ at equilibrium, and then integrated forward according to ν. We then sampled φ against the multinomial sampling distribution with sample size n (Equation 1) to obtain the two-locus spectrum.
Because it is three-dimensional, numerically integrating the two-locus diffusion equation requires computation proportional to where is the number of grid points used in discretization. In practice, must be larger than the data sample size n for accurate solution, and extrapolating from several settings dramatically improves accuracy (File S1). By contrast, solving the single-population one-locus diffusion equation requires computation proportional to In most cases, analysis of two-locus statistics will thus be much more computationally intensive than analysis of one-locus statistics.
Extension of the PRF to two loci
Because the diffusion equation is linear, it can be used to solve for the density of all two-locus frequencies in the population by allowing for the continuous influx of new pairs of loci. In the single locus case, we assume an infinite sites model and that mutations evolve independently. New mutations arise at rate proportional to where μ is the per-base mutation rate, and N is the population size. Mutations begin at frequency which suggests that as in the diffusion limit, any new mutation would immediately vanish. Sawyer and Hartl (1992) describe how to compensate for this by taking the scaled mutation rate so that the expected allele frequency spectrum remains proportional to where is the ancestral effective population size. The AFS for a sample of size n is then found by integrating against the binomial sampling function
| (3) |
Because is proportional to θ, is also proportional to θ. Moreover, the likelihood of an observed AFS is the product of Poisson likelihoods across bins, with means given by (Sawyer and Hartl 1992). In the numerical solution to the single-locus diffusion equation, we approximate the limit by adding density to the smallest interior grid point at rate as in Gutenkunst et al. (2009).
In the two-locus model, a new linked pair of polymorphic sites arises when a mutation occurs at one locus (suppose the left locus) when the other is already polymorphic (). The frequency of B at the right locus is determined by and the probability that it is polymorphic is proportional to the population-scaled mutation rate θ, as in single-locus PRF theory. For the left locus, in the diffusion limit () we again allow so that new A mutations enter the population proportional to Observed pairs of loci, with both sites segregating in the population, thus occur at rate proportional to Numerically, we handled this influx of density by injecting mass into the two-locus diffusion equation. We simultaneously tracked the single-locus allele frequency density function f, and set the influx of density into φ proportional to along the and axes (Figure 1B and File S1).
Composite likelihood estimation and demographic inference
We follow the composite likelihood approach outlined by Hudson (2001), in which we consider pairs of loci and their sampling distribution. Reducing the full likelihood for more than two linked loci to the composite likelihood over all possible pairs of polymorphisms leads to the loss of information. However, computing two-locus sampling statistics retains a considerable amount of information regarding both allele frequencies and patterns of LD between them. For recombination distances we consider all pairs of loci separated by each value of ρ within this range and then store the sampled frequencies in the appropriate two-locus frequency spectrum. In practice, recombination distances vary continuously over any interval, so we are required to bin our data within subintervals of ρ by defining intervals For fine enough subintervals, we approximated the expected two-locus spectrum for an interval using our diffusion approach with the mean recombination rate over that interval
For a given ρ-interval, we made the assumption that all pairs of loci contributing to the two-locus spectrum are independent, approximating the full likelihood by the composite likelihood across all pairs of loci. The two-locus frequency spectrum then forms a Poisson random field, so for sample data D and expected model M calculated under model parameters the likelihood of the data can be calculated by assuming each data entry is a Poisson random variable with mean Thus, the likelihood function for a single ρ-bin is
| (4) |
We allowed the population mutation rate θ to be an implicit parameter for each bin, which scales the total size of the frequency spectrum, while retaining its shape. The maximum likelihood value for θ is then where is the model spectrum with θ set to one. The square root arises because mutations that are paired to existing variant sites arise at rate proportional to θ, but those existing mutations also arise at rate proportional to θ, so that the total rate of influx of new two-locus pairs occurs at a rate proportional to as described in the previous section.
We simultaneously considered all bin intervals of and so for bin centers the likelihood function is
| (5) |
where j indexes the ρ-bins, and i indexes the frequency spectrum entries for a given In reality, pairs of loci are not independent, so we used the Godambe Information Matrix (GIM) to estimate parameter uncertainties (Coffman et al. 2016), which adjusts the composite likelihood statistics to account for linkage between data. This required bootstrapping the data, and we did so by dividing the autosomal genome into 1000 bins of equal length and resampling these regions with replacement.
We fit single-population demographic models to the data, which are defined by parameterized population size history functions (Equation 2). We considered simplified demographic models that may be described by a handful of parameters, unlike the many-parameter functions used in Liu and Fu (2015). For example, in an instantaneous expansion model, the parameters are the relative change in size ν and the time T in the past that the population changed size. In principle, our approach may be used to simulate any size function including piece-wise constant and exponential functions. In practice, however, complex many-parameter functions may be unidentifiable from the available data (Bhaskar and Song 2014; Lapierre et al. 2017).
Phased and unphased data
For data with phased chromosomes, determining haplotype frequencies is a straightforward exercise of counting observed types. Using an aligned outgroup, the ancestral state for each SNP may be determined, so that the two-locus spectrum stores derived two-locus allele frequencies. The ancestral state for each locus may be misidentified, potentially due to sequencing error or recurrent mutation along the lineage leading to the outgroup, and this can distort the two-locus spectrum (Hernandez et al. 2007). To account for ancestral misidentification, we included the probability that a given SNP had a misidentified state in our model fitting. Thus, with probability the A allele was misidentified but the B allele was correctly identified, and with the same probability the B allele was misidentified and the A allele was correctly identified. Both alleles A and B were misidentified with probability In fitting a demographic model to data, we fit along with the parameters from the demographic model.
When data are unphased, as is the case for many genomic datasets, observed haplotypes can not be tallied. Rather, we are left with counts of genotypes in individuals, The composite linkage disequilibrium statistic is an unbiased estimator for D (Weir 1979; Zaykin 2004),
| (6) |
where n is the number of sampled individuals. One possible approach to summarize observed data might be to work with the joint statistics and Instead, we directly used genotype counts in the “genotype frequency spectrum” G. In genotype data, individuals may carry or at the left locus, and or at the right locus. Thus, there are nine possible two-locus genotypes ( …) that could be observed to be carried by an individual. G is an eight-dimensional object whose state space has size but it is sparse so it can be stored efficiently. Each genotype can only be formed by the pairing of two specific haplotypes (e.g., can only be from one haplotype of each and ), except for which could be formed by or Thus, we expected G to still carry information about demography through the joint patterns of allele frequencies and LD. Expected genotype frequencies can be calculated from expected haplotype frequencies, and we detail our approach in File S1.
Drosophila sequence data and recombination map
As an application, we considered a single Zambian population of fruit flies, using data from phase 3 of the Drosophila Population Genomics Project (DPGP3), available from the Drosophila Genome Nexus (Lack et al. 2015). The data consisted of 197 sequenced haploid embryos, so genomes were necessarily phased. We used Annovar (Wang et al. 2010) to annotate all biallelic SNPs across the genome, and we used intronic and intergenic regions in our two-locus analysis. We determined the ancestral allele for each SNP using the alignment to D. simulans (April 2006, dm3 aligned to droSim1, downloaded from the UCSC genome browser), by assuming the D. simulans allele was ancestral. If the D. melanogaster site had no alignment, or if the D. simulans allele was different than the two melanogaster alleles, we discarded that site.
For each chromosome, we considered all pairs of biallelic SNPs in intergenic and intronic regions for which an ancestral state could be determined, within recombination distance We determined recombination distances using the recombination map inferred by Comeron et al. (2012), which reports cumulative recombination rates in units of centimorgan over 100,000 bp intervals along each chromosome per female meiosis. Because recombination only occurs in females in D. melanogaster, we halved the reported rates to find the effective recombination rate per generation. We converted to by taking the map distance d (in centimorgan) separating the two SNPs and multiplying by This required an estimate for so we used neutral demographic fits to intronic and intergenic single-locus data, which provided an estimate for Here, we assumed a mutation rate of (Schrider et al. 2013). The total length of sequences that were included in our analysis was Then, For each two-locus pair, we counted the number of and haplotypes across all 197 samples, and then subsampled to a sample size of These data could be projected down to a sample size (File S1), but we chose to subsample, because caching in memory the many projection matrices necessary to account for different numbers of successful calls was infeasible. Subsampling allowed for more pairs to be included in the data, as any pair of loci without missing haplotype data for at least 20 samples was included. Additionally, the smaller sample size allowed for more rapid evaluation of the expected frequency spectrum for optimization, because coarser grids could be used to obtain an accurate numerical solution to φ.
Independent inference of
The effective population size is scaled out of the diffusion equation (Equation 2), but the likelihood of the data does depend on because two-locus statistics are binned by the population-scaled recombination rate where r is the per-generation recombination rate. Thus can be inferred from two-locus statistics if the per-site rate of recombination r is known, similar to how can be inferred from one-locus statistics and if the per-site mutation rate μ is known. Given a recombination map, we then require an accurate estimate for to appropriately bin the data. In the case that the effective population size is unknown, may be left as a parameter to be fit during optimization of the model to the data. In this approach, we guess an initial effective population size to first bin the data by (for example, for human populations, or for Drosophila) and then allow the ρ-value for each bin to be rescaled by as If the best fit then turned out to be the best fit effective population size, while if is larger or smaller than one, then the best fit is inferred to be larger or smaller than by that factor. We rescaled the ρ value for each bin of data instead of reassigning data to fixed bins for fair comparison of likelihoods across varying values of and because reassigning two-locus data each iteration of optimization would be computationally burdensome. Because of this rebinning, likelihood calculations for different values of require integrating Equation 2 for different values of ρ. For computational efficiency, we cached equilibrium φ densities over a fine grid of ρ values, and used the cached φ from the closest ρ as the initial condition for each integration.
Inaccuracies in the assumed recombination map may bias our inferences. In the simplest case, recombination rates may be systematically underestimated or overestimated relative to the true rates. In the fixed analysis, pairs of loci will thus be assigned to incorrect ρ bins, biasing inferences. In the free analysis, systematic errors in the map will be absorbed into the estimate of so demographic parameters will not be biased when expressed in genetic units, but will be biased when expressed in physical units. In a more complex case, the assumed recombination map may be too coarse to capture hotspots of recombination. The assumed recombination rate between a pair of loci may thus be higher or lower than the true rate, depending on whether the pair spans a hotspot (Figure S1 in File S1). This would add noise to the binning of pairs of loci by recombination rate, which may bias inference. The magnitude of this effect will depend on the density and strengths of hotspots, the density of polymorphisms, and potentially the underlying demographic history, all which may be particular to the species or population being studied.
Data availability
The Drosophila data we analyzed (Lack et al. 2015) are available from the Drosophila Genome Nexus at http://www.johnpool.net/genomes.html. Our methodology for solving the two-locus diffusion equation and fitting data are integrated into dadi, available at https://bitbucket.org/gutenkunstlab/dadi. Supplemental text, figures, and a table that further detail the methodology and Drosophila application are available in File S1.
Results and Discussion
Numerical accuracy of solution to two-locus allele frequency spectrum
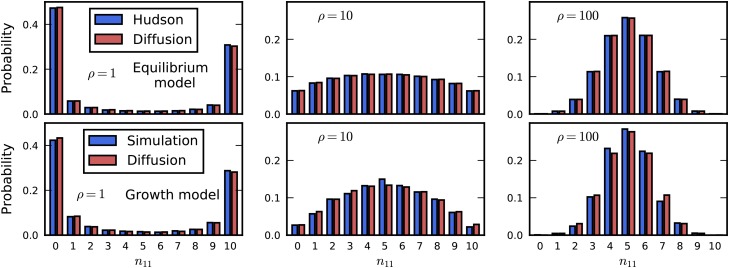
We first compared our numerical solution for two-locus statistics for a population in demographic equilibrium to those calculated by Hudson (2001). Our solution matched those using Hudson’s algorithm across all values of ρ, from completely linked () to loose linkage () (Figure 2, top row). To verify our numerical solution for nonequilibrium demography, we compared it to simulations of the discrete two-locus process with an influx of mutations. We simulated a population of diploid, randomly mating individuals for independent pairs of loci separated by a given recombination rate. New two-locus pairs entered the population at a rate proportional to Eq. S3 and S4 in File S1. We allowed the simulation to proceed for generations, and then applied specified population size changes, sampling two-locus haplotype frequencies from the population after each simulation completed. Our nonequilibrium solution matched the simulated two-locus statistics (Figure 2, bottom row). See File S1 for further details regarding simulation and numerical accuracy.
Figure 2.
Verification of numerical solution. For sample size the distribution of is shown, when the frequencies of A and B are and and ρ is varied. Top row: Comparison to equilibrium statistics from Hudson (2001). Bottom row: Comparison to discrete simulation under growth model.
Two-locus statistics are sensitive to demographic history
To assess the increase in statistical power for demographic history inference using the two-locus spectrum vs. the single-locus spectrum, we used the information theoretical measure Kullback-Leibler (KL) divergence (Kullback and Leibler 1951). KL divergence measures the amount of information lost if an incorrect demographic model is used to approximate the true model and it can be interpreted as the expected likelihood ratio statistic for testing against For discrete distributions, such as frequency spectra, KL divergence is defined as
| (7) |
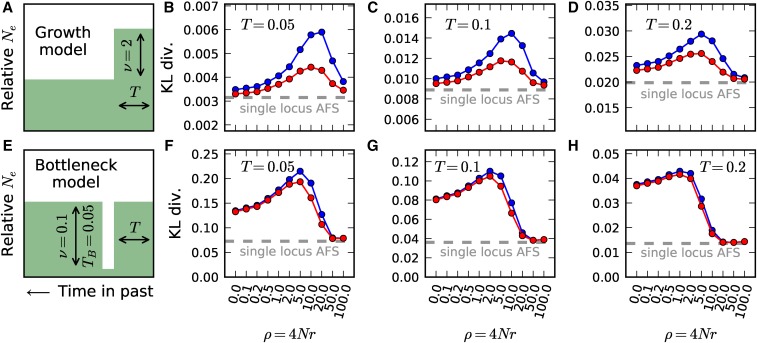
In our comparisons, we took to be a model of constant demography, and compared the KL divergence for two demographic models, an instantaneous growth model, and a bottleneck and recovery model, between two-locus and single-locus frequency spectra (Figure 3). A larger KL divergence indicated that more information is contained in the data to reject the constant size model. For the two model types, we considered varying recovery times T since the demographic event, so, in the growth model, T is the time since the instantaneous expansion (), and, in the bottleneck model, T is the time since recovery from the bottleneck (). In all cases, the two-locus spectrum is more informative about the demography per pair of linked loci than are two unlinked loci in the single-locus frequency spectrum.
Figure 3.
Sensitivity to demographic history. We compared KL divergence measures between two-locus statistics and the single-locus frequency spectrum for a simple growth model (A, top row) and a bottleneck model (E, bottom row). The blue curve shows the KL divergence for phased (haplotype) data, while the red curve is for unphased (genotype) data. In each comparison, we considered the KL divergence between the specified demographic model and a null model of constant population size. (A) In the instantaneous growth model, the population doubled in size some time T in the past, and we considered (B) (C) 0.1, and (D) 0.2. (E) In the bottleneck model, the population shrank to its original size for genetic time units, and then recovered to its original size T genetic units ago for (F) (G) 0.1, and (H) 0.2. In all cases, and across all values of ρ, KL divergence was greater for two-locus statistics than the corresponding single locus statistics of the same number of unlinked sites. The two-locus spectrum is thus more sensitive to demographic history than the single-locus spectrum.
We considered the KL divergence for varying values of recombination rate ρ from completely linked () to loose linkage (). For large ρ, KL divergence from two-locus statistics converged to the measure for unlinked single-locus data, which is to be expected since implies unlinked loci. The case of complete linkage () corresponds to the triallelic frequency spectrum (Jenkins et al. 2014; Ragsdale et al. 2016), because without recombination the fourth haplotype never occurs. Jenkins et al. (2014) have shown that triallelic loci are more informative about demographic history than biallelic loci, but our results show that pairs of sites separated by an intermediate recombination distance are often even more informative (Figure 3). Notably, the most informative recombination distance varied between demographic models and recovery times T since demographic events. As T increases, lower recombination rates are relatively more sensitive, because higher recombination rates will restore levels of LD faster than lower recombination rates. Therefore, loosely linked loci are more informative about recent demographic events, while tightly linked loci are more informative about deeper events.
We performed the KL divergence analysis on genotype data as well (Figure 3, red curves), and we found that two-locus statistics at the genotype level are also more sensitive than one-locus statistics. For the growth model, the KL divergence of genotype data were intermediate between the KL divergences of one-locus and haplotype data, but, for the bottleneck model, little sensitivity is lost when using genotype data instead of haplotype data.
Fits to simulated data
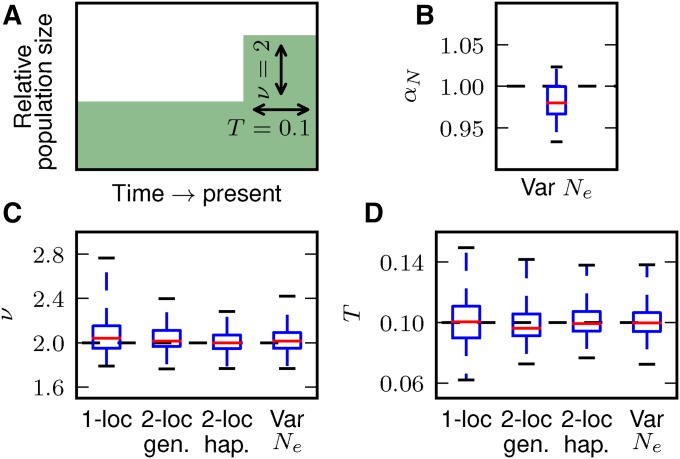
To further validate our model and to explore efficient and informative ways to collate two-locus statistics, we simulated single-population demographic history under neutrality with realistic human mutation and recombination rates using ms (Hudson 2002) (details in File S1). Each simulation consisted of 100, 1-Mb regions under a simple growth model (instantaneous expansion by a factor of 2, 0.1 time units before present), to which we then fit a two-epoch demographic model using both the single- and two-locus statistics of the simulation (File S1). We repeated this simulation and fitting process 50 times, and checked how well we recovered the simulated demographic parameters. We used the same simulations to check the accuracy of our fits to genotype data, by pairing chromosomes to create diploid individuals. Figure 4 shows our fits to simulated data, with two-locus genotype statistics more precisely recovering the true demographic model than single-locus statistics, and haplotype statistics more precisely than genotype statistics. When we allowed to vary, we also precisely recovered the simulated parameters, including (Figure 4B). The inferred parameter values were correlated between approaches (Figure S2 in File S1). For example, if the expansion factor was overestimated using single-locus statistics, it also tended to be overestimated using two-locus statistics.
Figure 4.
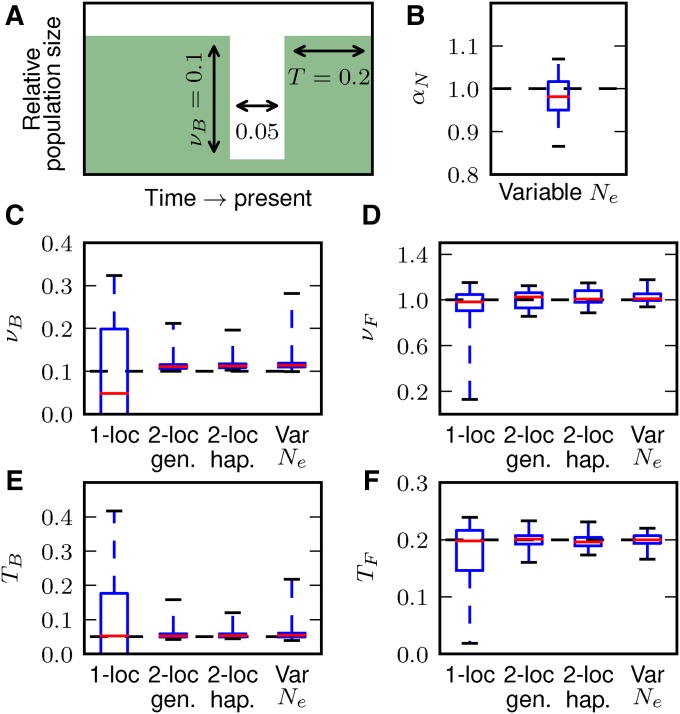
Fits to data from simulated growth model. (A) We simulated 50 replicate data sets with length 100 Mb under an instantaneous growth model using ms and checked how precisely we recovered the simulated parameters for both single- and two-locus data, including allowing to vary (B). (C, D) For both ν and T, fits to the two-locus frequency spectrum were more precise than single-locus fits. Here, the median values and top and bottom quartiles are indicated by the boxes, and the whiskers extend to the largest and smallest inferred values from the simulated datasets.
In an identical fashion, we also simulated a bottleneck model, in which the population size shrank by a factor of 0.1 for 0.05 genetic time units, and then recovered to its original size for 0.2 time units until sampling at present (Figure 5). For this demography, the fits to single-locus statistics were inconsistent, and many replicates did not converge to reasonable parameter values, with tending to 0. The two-locus haplotype fits more precisely recovered the modeled parameters, although the inferred values of were consistently slightly elevated. The fits to genotype data were also more precise than using single-locus data, consistent with our KL divergence results (Figure 3). Disentangling the depth and duration of a bottleneck from allele frequency data are notoriously challenging (Keinan et al. 2007; Bunnefeld et al. 2015), and jointly incorporating information about LD dramatically improves parameter identifiability.
Figure 5.
Fits to data from simulated bottleneck model. (A) We simulated 50 replicate data sets with length 100 Mb under a bottleneck and recovery demographic history, in which the population declined to 0.1 its original size for genetic time units and then recovered to its original size for 0.2 time units. (C–F) Demographic inferences using single-locus data alone could not consistently recover the true parameters. However, using genotype or haplotype two-locus data allowed for precise inference of model parameters, including when was allowed to vary (B).
Demographic inference of a Zambian Drosophila population
As an application of our approach, we considered the demographic history of a Zambian population of D. melanogaster, which is thought to be a close proxy to the ancestral population (Lack et al. 2015). We first fit two- and three-epoch single-population demographic models to intronic and intergenic single-locus data in order to estimate θ and (Table 1). We inferred the ancestral effective population size to be ∼ which is somewhat lower than previously suggested sizes for D. melanogaster (Keightley et al. 2014; Garud and Petrov 2016). Using the recombination map of Comeron et al. (2012), we determined distances in ρ between pairs of loci, assuming an effective population size of and we binned two-locus data as described above. We then fit the two- and three-epoch models to the two-locus data, with and without varying (Figure S3 in File S1 and Table 1) and calculated parameter uncertainties using the Godambe Information Matrix (Table S1 in File S1). For all fits, we subsampled the data to 20 samples for computational speed, and additional speed-up was afforded by calculating each ρ-bin’s expected frequency spectrum in parallel. Our models all included a parameter to account for potential misidentification of the ancestral state of an allele. As expected (Hernandez et al. 2007), for all fits, was inferred to be similar to the divergence along the D. simulans lineage from D. melanogaster (Begun et al. 2007).
For the two-epoch model, parameter values inferred using single- and two-locus data were similar (Table 1). For the three-epoch model, the two-locus data led to inferences of less dramatic growth deeper in the past than did the single-locus data. Notably, the three-epoch model fit the one-locus data better than the two-epoch model, but it produced a worse likelihood when the resulting demographic parameters were applied to two-locus data, perhaps indicating over-fitting (Figure 6 and Table 1). When we included the ancestral effective population size as a parameter in the two-locus fits, the best-fit value was similar to that inferred from single-locus data. In an earlier analysis, we mistakenly set fixed in the two-locus fits to be half the intended value, which led to dramatically different inferences of demographic parameters (Table S1 in File S1). Scaling the recombination map by a fixed estimate for may thus introduce significant bias into downstream parameter estimates when that estimate is incorrect.
Figure 6.
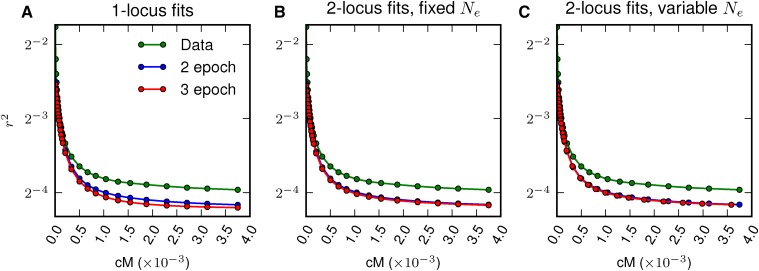
Fits to LD-decay from Drosophila data. LD-decay curves for two-locus models compared to observed decay curves from the data. (A) The two-locus model using the best fit parameters from single-locus data, (B) the two-locus model fit with set to and (C) the two-locus model with allowed to vary. Each of the models underestimates long-range LD decay, as also observed by Garud and Petrov (2016).
All the inferred models fit the single-locus frequency spectrum well (Figure S4 in File S1). All models, however, underestimated long-range LD, although the two-locus model fits performed better than the single-locus fits (Figure 6). Previous models of D. melanogaster demographic history also underestimated long-range LD (Garud and Petrov 2016). While a more complex demography might be able to better fit the LD curve, factors aside from single-population demography may be critical to generating the pattern of long-range elevated LD, including population substructure, recent admixture, or the effects of linked selection.
Previous studies have also fit demographic models with a potential bottleneck to data from African D. melanogaster populations. Duchen et al. (2013) used a Zimbabwean population to infer demographic history, and reported and an extremely severe bottleneck over 200,000 years ago. Using the same set of Zambian individuals as our study, Sheehan and Song (2016) inferred a bottleneck between 10,000 and 100,000 years ago, with on either side of the bottleneck, and during the bottleneck. In contrast, we infer modest stepwise increases in population size over the last 50,000 years, with no bottleneck. Our approach does have power to detect bottlenecks (Figure 5), even at modest sample sizes, so the origin of these differences is unclear.
Both our one-locus and two-locus estimates of the ancestral effective population size of D. melanogaster are notably smaller than previous estimates. Keightley et al. (2014) estimated the spontaneous mutation rate by sequencing a family of two parents and 12 full-sibling offspring, and used this estimate to infer The effective population size may also be estimated from observed levels of diversity, and Charlesworth (2015) estimated using observed synonymous site diversity. is often assumed, or estimated, to be at least and sometimes much larger, in many population genetic studies of D. melanogaster (Thornton and Andolfatto 2006; Sella et al. 2009; Garud et al. 2015; Garud and Petrov 2016). Our estimates for were substantially lower. Using levels of diversity for intronic and intergenic loci, we estimated through our demographic fits to the single-locus AFS (Table 1). In an alternative approach, we allowed to vary in the two-locus inference, and we again estimated a value of This approach is based on the scaling of the recombination map without assuming a fixed mutation rate, so it provides an independent inference of the effective population size. Together, our results suggest that ancestral for D. melanogaster may be lower than previously estimated, and studies that require an assumed effective population size should consider a range of possible values that include small sizes. Notably, it has been suggested that linked selection is common throughout the genome of D. melanogaster (Garud and Petrov 2016), and linked selection is known to increase the variance in offspring distribution, which, in turn, decreases the effective population size (Leffler et al. 2012).
Conclusions
Based on the continuous approximation to a two-allele two-locus discrete Wright-Fisher model with recombination, we developed a numerical solution to the two-locus diffusion equation that handles arbitrary recombination rates and demographic history. We used this method to develop a composite likelihood framework to infer demographic history from observed two-locus data, which can handle data sampled as either haplotypes or genotypes. While two-locus statistics have been used successfully and extensively to infer fine-scale recombination maps for many organisms, we focused on quantifying the additional power afforded by two-locus over single-locus statistics for demographic history inference. We found that two-locus statistics do provide substantial additional power. For example, while inferring the parameters of a bottleneck model from single-locus data are notoriously difficult (Keinan et al. 2007), we were able to precisely and consistently recover the correct demographic parameters using two-locus statistics. For at least some scenarios, little power is lost when data are unphased and genotype frequencies are fit. Finally, we turned to data from a Zambian fruit fly population, and we inferred recent modest population size growth. The demographic history that we inferred still underestimates the observed long-range levels of LD, which has been previously observed in this population (Garud and Petrov 2016). Moreover, using two independent approaches, one based on levels of diversity, and the other based on scaling the recombination map, we inferred the ancestral effective population size to be substantially lower than previous inferences. It is likely that additional factors to single population demography are at play, including potentially complicated demographic features such as substructure and admixture, and the effects of linked selection. The methods described here are integrated into dadi, available at https://bitbucket.org/gutenkunstlab/dadi.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.201251/-/DC1.
Acknowledgments
A.P.R. thanks Gleb Zhelezov for helpful discussions regarding various numerical approaches for this class of partial differential equations. We thank Nandita Garud for sharing recombination map data and useful discussion regarding the demographic history of the fly population. This work was supported by the National Science Foundation (DEB-1146074 to R.N.G.).
Footnotes
Communicating editor: Y. S. Song
Literature Cited
- Auton A., McVean G., 2007. Recombination rate estimation in the presence of hotspots. Genome Res. 17: 1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun D. J., Holloway A. K., Stevens K., Hillier L. D. W., Poh Y. P., et al. , 2007. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 5: 2534–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar A., Song Y. S., 2012. Closed-form asymptotic sampling distributions under the coalescent with recombination for an arbitrary number of loci. Adv. Appl. Probab. 44: 391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar A., Song Y. S., 2014. Descartes’ rule of signs and the identifiability of population demographic models from genomic variation data. Ann. Stat. 42: 2469–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A. R., Williamson S. H., Indap A. R., Degenhardt J. D., Hernandez R. D., et al. , 2008. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 4: e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnefeld L., Frantz L. A. F., Lohse K., 2015. Inferring bottlenecks from genome-wide samples of short sequence blocks. Genetics 201: 1157–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C. D., Wakeley J., Sawyer S., Hartl D. L., 2001. Directional selection and the site-frequency spectrum. Genetics 159: 1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A. H., Jenkins P. A., Song Y. S., 2012. Genome-wide fine-scale recombination rate variation in Drosophila melanogaster. PLoS Genet. 8: e1003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. S., Cooper G., 1970. A practical difference scheme for Fokker-Planck equations. J. Comput. Phys. 6: 1–16. [Google Scholar]
- Charlesworth B., 2015. Causes of natural variation in fitness: evidence from studies of Drosophila populations. Proc. Natl. Acad. Sci. USA 112: 1662–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman A. J., Hsieh P. H., Gravel S., Gutenkunst R. N., 2016. Computationally efficient composite likelihood statistics for demographic inference. Mol. Biol. Evol. 33: 591–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchen P., Živković D., Hutter S., Stephan W., Laurent S., 2013. Demographic inference reveals African and European admixture in the North American Drosophila melanogaster population. Genetics 193: 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethier S. N., Griffiths R. C., 1990. On the two-locus sampling distribution. J. Math. Biol. 29: 131–159. [Google Scholar]
- Ferretti, L., A. Klassmann, E. Raineri, T. Wiehe, S. E. Ramos-Onsins et al., 2016 The expected neutral frequency spectrum of linked sites. arXiv: 1604.06713 [q-bio.PE]. [DOI] [PubMed]
- Garud N. R., Petrov D. A., 2016. Elevated linkage disequilibrium and signatures of soft sweeps are common in Drosophila melanogaster. Genetics 203: 863–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garud N. R., Messer P. W., Buzbas E. O., Petrov D. A., 2015. Recent selective sweeps in North American Drosophila melanogaster show signatures of soft sweeps. PLoS Genet. 11: 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding G. B., 1984. The sampling distribution of linkage disequilibrium. Genetics 108: 257–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst R. N., Hernandez R. D., Williamson S. H., Bustamante C. D., 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5: e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K., Nielsen R., 2013. Inferring demographic history from a spectrum of shared haplotype lengths. PLoS Genet. 9: e1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn B. M., Botigué L. R., Peischl S., Dupanloup I., Lipatov M., et al. , 2016. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc. Natl. Acad. Sci. USA 113: E440–E449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez R. D., Williamson S. H., Bustamante C. D., 2007. Context dependence, ancestral misidentification, and spurious signatures of natural selection. Mol. Biol. Evol. 24: 1792–1800. [DOI] [PubMed] [Google Scholar]
- Hill W. G., Robertson A., 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269. [PubMed] [Google Scholar]
- Hudson R. R., 1985. The sampling distribution of linkage disequilibrium under an infinite allele model without selection. Genetics 109: 611–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., 2001. Two-locus sampling distributions and their application. Genetics 159: 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338. [DOI] [PubMed] [Google Scholar]
- Jenkins P. A., Song Y. S., 2009. Closed-form two-locus sampling distributions: accuracy and universality. Genetics 183: 1087–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins P. A., Song Y. S., 2010. An asymptotic sampling formula for the coalescent with recombination. Ann. Appl. Probab. 20: 1005–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins P. A., Song Y. S., 2012. Padé approximants and exact two-locus sampling distributions. Ann. Appl. Probab. 22: 576–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins P. A., Mueller J. W., Song Y. S., 2014. General triallelic frequency spectrum under demographic models with variable population size. Genetics 196: 295–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamm J. A., Spence J. P., Chan J., Song Y. S., 2016. Two-locus likelihoods under variable population size and fine-scale recombination rate estimation. Genetics 203: 1381–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamm J. A., Terhorst J., Song Y. S., 2017. Efficient computation of the joint sample frequency spectra for multiple populations. J. Comput. Graph. Stat. 26: 182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., McGregor J., 1968. Rates and probabilities of fixation for two locus random mating finite populations without selection. Genetics 58: 141–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Ness R. W., Halligan D. L., Haddrill P. R., 2014. Estimation of the spontaneous mutation rate per nucleotide site in a Drosophila melanogaster full-sib family. Genetics 196: 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A., Mullikin J. C., Patterson N., Reich D., 2007. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat. Genet. 39: 1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1955. Random genetic drift in multi-allelic locus. Evolution 9: 419–435. [Google Scholar]
- Kimura M., 1963. A probability method for treating inbreeding systems, especially with linked genes. Biometrics 19: 1–17. [Google Scholar]
- Kimura M., 1964. Diffusion models in population genetics. J. Appl. Probab. 1: 177–232. [Google Scholar]
- Kingman J., 1982. The coalescent. Stochastic Process. Appl. 13: 235–248. [Google Scholar]
- Kullback S., Leibler R. A., 1951. On information and sufficiency. Ann. Math. Stat. 22: 79–86. [Google Scholar]
- Lack J. B., Cardeno C. M., Crepeau M. W., Taylor W., Corbett-Detig R. B., et al. , 2015. The Drosophila genome nexus: a population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population. Genetics 199: 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre M., Lambert A., Achaz G., 2017. Accuracy of demographic inferences from the site frequency spectrum: the case of the Yoruba population. Genetics DOI: 10.1534/genetics.116.192708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler E. M., Bullaughey K., Matute D. R., Meyer W. K., Ségurel L., et al. , 2012. Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biol. 10: e1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2011. Inference of human population history from individual whole-genome sequences. Nature 475: 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Fu Y.-X., 2015. Exploring population size changes using SNP frequency spectra. Nat. Genet. 47: 555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller K. E., Indap A. R., Schmidt S., Boyko A. R., Hernandez R. D., et al. , 2008. Proportionally more deleterious genetic variation in European than in African populations. Nature 451: 994–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller K. E., Bustamante C. D., Clark A. G., 2009. Methods for human demographic inference using haplotype patterns from genomewide single-nucleotide polymorphism data. Genetics 182: 217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marth G. T., Czabarka E., Murvai J., Sherry S. T., 2004. The allele frequency spectrum in genome-side human variation data reveals signals of differential demographic history in three large world populations. Genetics 166: 351–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G., Myers S., Hunt S., 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304: 581–584. [DOI] [PubMed] [Google Scholar]
- Myers S., Fefferman C., Patterson N., 2008. Can one learn history from the allelic spectrum? Theor. Popul. Biol. 73: 342–348. [DOI] [PubMed] [Google Scholar]
- Ohta T., Kimura M., 1969. Linkage disequilibrium due to random genetic drift. Genet. Res. 13: 47–55. [Google Scholar]
- Pritchard J. K., Przeworski M., 2001. Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet. 69: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale A. P., Coffman A. J., Hsieh P., Struck T. J., Gutenkunst R. N., 2016. Triallelic population genomics for inferring correlated fitness effects of same site nonsynonymous mutations. Genetics 203: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S. A., Hartl D. L., 1992. Population genetics of polymorphism and divergence. Genetics 132: 1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S., Durbin R., 2014. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 46: 919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D. R., Houle D., Lynch M., Hahn M. W., 2013. Rates and genomic consequences of spontaneous mutational events in Drosophila melanogaster. Genetics 194: 937–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G., Petrov D. A., Przeworski M., Andolfatto P., 2009. Pervasive natural selection in the Drosophila genome? PLoS Genet. 5: e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan S., Song Y. S., 2016. Deep learning for population genetic inference. PLoS Comput. Biol. 12: e1004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan S., Harris K., Song Y. S., 2013. Estimating variable effective population sizes from multiple genomes: a sequentially Markov conditional sampling distribution approach. Genetics 194: 647–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhorst J., Kamm J. A., Song Y. S., 2017. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 49: 303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K., Andolfatto P., 2006. Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics 172: 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M., Hakonarson H., 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G., 1970. The effect of linkage in a finite population. Theor. Popul. Biol. 1: 72–87. [DOI] [PubMed] [Google Scholar]
- Weir B. S., 1979. Inferences about linkage disequilibrium. Biometrics 35: 235–254. [PubMed] [Google Scholar]
- Williamson S. H., Hernandez R., Fledel-Alon A., Zhu L., Nielsen R., et al. , 2005. Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc. Natl. Acad. Sci. USA 102: 7882–7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., 2011. The site-frequency spectrum of linked sites. Bull. Math. Biol. 73: 459–494. [DOI] [PubMed] [Google Scholar]
- Zaykin D. V., 2004. Bounds and normalization of the composite linkage disequilibrium coefficient. Genet. Epidemiol. 27: 252–257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Drosophila data we analyzed (Lack et al. 2015) are available from the Drosophila Genome Nexus at http://www.johnpool.net/genomes.html. Our methodology for solving the two-locus diffusion equation and fitting data are integrated into dadi, available at https://bitbucket.org/gutenkunstlab/dadi. Supplemental text, figures, and a table that further detail the methodology and Drosophila application are available in File S1.