Abstract
How sex is determined in insects is diverse and dynamic, and includes male heterogamety, female heterogamety, and haplodiploidy. In many insect lineages, sex determination is either completely unknown or poorly studied. We studied sex determination in Psocodea—a species-rich order of insects that includes parasitic lice, barklice, and booklice. We focus on a recently discovered species of Liposcelis booklice (Psocodea: Troctomorpha), which are among the closest free-living relatives of parasitic lice. Using genetic, genomic, and immunohistochemical approaches, we show that this group exhibits paternal genome elimination (PGE), an unusual mode of sex determination that involves genomic imprinting. Controlled crosses, following a genetic marker over multiple generations, demonstrated that males only transmit to offspring genes they inherited from their mother. Immunofluorescence microscopy revealed densely packed chromocenters associated with H3K9me3—a conserved marker for heterochromatin—in males, but not in females, suggesting silencing of chromosomes in males. Genome assembly and comparison of read coverage in male and female libraries showed no evidence for differentiated sex chromosomes. We also found that females produce more sons early in life, consistent with facultative sex allocation. It is likely that PGE is widespread in Psocodea, including human lice. This order represents a promising model for studying this enigmatic mode of sex determination.
Keywords: genome exclusion, segregation distortion, sex determination, sex ratio, genomic imprinting
FEMALES and males are ubiquitous across the animal kingdom, yet how the sexes are determined is incredibly dynamic (Bachtrog et al. 2014; Beukeboom and Perrin 2014). Insects are an excellent demonstration of this diversity. For example, while the ancestral sex determination in insects is thought to be male heterogamety (i.e., XY or XO males), there have been several transitions to other modes, such as female heterogamety (i.e., ZW or ZO females, for example in butterflies and moths), and haplodiploidy (i.e., diploid females and haploid males, for example, in wasps, bees, and thrips) (Blackmon et al. 2017). Even in lineages where the mode of sex determination is conserved, sex chromosomes and sex determining genes can change rapidly. For example, Vicoso and Bachtrog (2015) have recently found that although dipterans (i.e., flies) typically exhibit male heterogamety, there have been numerous gains and losses of sex chromosomes. Perhaps the most striking example of rapid evolution and diversity of sex determination systems in insects is that of the housefly, Musca domestica, which is polymorphic for male heterogamety, female heterogamety, and even temperature-dependent sex determination, driven largely by a highly mobile and variable master sex determining locus (Dübendorfer et al. 2002).
While there has been exciting progress on the genetics and evolution of sex determination in insects, there are enormous gaps in our knowledge. The factors that drive the rapid turnover of sex determination systems are not well understood, although it is likely that conflicts over transmission and sexually antagonistic genes both play important roles (Normark 2003; Kozielska et al. 2010; Bachtrog et al. 2014). The master sex determining gene has been identified in only a handful of insects (Bell et al. 1988; Beye et al. 2003; Kiuchi et al. 2014; Hall et al. 2015; Krzywinska et al. 2016). Furthermore, there remain entire lineages of insects for which the mode of sex determination is not known (Beukeboom and Perrin 2014).
In this paper, we fill this gap by studying sex determination in Psocodea—a species-rich (∼10,000 extant described species) order of insects that includes parasitic lice, barklice, and booklice, and that is related to true bugs and thrips (insects with incomplete metamorphosis and piercing, sucking mouthparts) (Li et al. 2015). Until recently, Psocodea consisted of two separate orders: Psocoptera (barklice and booklice) and Phthiraptera (parasitic lice). However, molecular and morphological phylogenetic analyses clearly demonstrate that Phthiraptera emerged from within Psocoptera (Yoshizawa and Johnson 2003; Li et al. 2015), and are most closely related to Liposcelididae—wingless, flattened booklice that include a number of cosmopolitan stored grain pests.
Very little is known about sex determination in Psocodea. Cytological studies concluded that male barklice have an XO (or rarely XY) karyotype (Wong and Thornton 1966; Golub and Nokkala 2001, 2009). Nothing is known about sex determination in booklice (Liposcelididae), and sex determination in parasitic lice is mysterious and as yet unresolved, but they do not appear to have heteromorphic sex chromosomes (Tombesi and Papeschi 1993; Golub and Nokkala 2004). Recently, the first genetic study of reproduction in parasitic lice (or in Psocodea for that matter) found a puzzling result. McMeniman and Barker (2006) followed the inheritance of microsatellite markers in human lice, Pediculus humanus, and found that some heterozygous males transmit their genes in Mendelian fashion, while other males only transmit genes inherited from their mother.
We investigated the reproductive mode of a recently discovered species of Liposcelis (Liposcelididae), collected from the Chiricahua Mountains in Arizona (Perlman et al. 2015). Liposcelis occupies an interesting place in the psocodean evolutionary tree, as it is a member of the family that is the closest free-living relative (and sister group) of parasitic lice (Yoshizawa and Johnson 2003; Li et al. 2015). We used controlled crosses, immunohistochemistry, and genomic analysis to demonstrate that this lineage exhibits paternal genome elimination (PGE), an unusual mode of reproduction that has evolved independently in at least six clades of arthropods, including scale insects, phytoseiid mites, and fungus gnats and their relatives (Blackmon et al. 2017). We also show that females produce more sons early in life, consistent with the facultative sex allocation found in other species that exhibit paternal genome elimination. In organisms with paternal genome elimination, males arise from fertilized eggs (in contrast to arrhenotokous haplodiploidy), but only transmit the genes they inherited from their mother. Much is still unknown about the mechanism of paternal genome elimination; however, genomic imprinting seems to be at the heart of this unusual form of reproduction (Herrick and Seger 1999). Altogether, this study fills a large gap in the insect tree of life in terms of how sex is determined, and documents a new case of paternal genome elimination—an interesting and unusual mode of sex determination.
Materials and Methods
Culture information
Individuals of Liposcelis sp. were initially collected from the Chiricahua Mountains, Arizona, in 2010 (Perlman et al. 2015), and laboratory cultures were established. Individuals from our laboratory culture have been deposited in the insect collection at the Royal British Columbia Museum, Victoria, BC, while this species awaits formal description. [A maternally transmitted sex ratio distortion was previously reported in this species (Perlman et al. 2015), but note that this polymorphism is not present in the cultures used in this study.]
Colonies are maintained at ∼27° and 75% relative humidity. We keep Liposcelis sp. in small glass canning jars (125 ml) with the lid replaced with 70 mm Whatman filter paper (Sigma-Aldrich). We rear them on a diet of 1:10 (w:w) mixture of Rice Krispies (Kellogg’s) to cracked red wheat (Planet Organic). We check the colonies every second week, and replace food with new food as needed to avoid crowding in the colonies. It takes ∼40 days for individuals to be reproductively mature. To obtain virgin females, we isolate them at their final nymphal stage (they are larger and have a rounder abdomen than males at this point). Males develop faster than females so we collect virgin males by isolating them before females of the same age develop into adults.
Inheritance experiment
We used controlled crosses over two successive generations to test for paternal genome elimination and departures from Mendelian inheritance. Our crossing scheme took advantage of a two allele polymorphism in the cAMP-specific IBMX-insensitive 3′, 5′-cyclic phosphodiesterase gene (Phos1 for short) in our laboratory culture of Liposcelis sp. (Perlman et al. 2015). By following the inheritance of Phos1 alleles, we were able to test two specific predictions: (a) heterozygous females will transmit both alleles, and (b) heterozygous males will only transmit the allele they inherited from their mother. We extracted DNA from single booklice using 30 μl Prepman Ultra (Thermo Fisher Scientific) according to manufacturer instructions (to yield 15 μl of product). Individuals were genotyped after PCR amplification with the primers Phos1F (5′-TCCCTTCCGTCAATAAATGC-3′) and Phos1R (5′-AATGTTCGAAATGCCGAGTC-3′) using the following thermocycling conditions: 95° × 3 min, (94° × 1 min, 56° × 1 min, 72° × 2 min) × 35, 72° × 10 min. Sequencing was performed by Sequetech (CA). We scored individuals as either homozygous or heterozygous by examining chromatograms for double peaks, using Geneious 6.1.8. See Supplemental Material, Figure S1 and Figure S2 in File S1 for visualization of Phos1 alleles and an example of our crossing setup.
For the first generation of the experiment, we set up 15 small petri dishes, each containing one virgin male and three virgin females, along with 0.5 g of food. Females were left with the male for 2 weeks, after which the male was removed and his DNA extracted. We transferred the females into individual dishes with the same amount of food and left them for 2 weeks to lay eggs, when we transferred them into new dishes and left them for another 2 weeks before extracting their DNA. We sequenced the Phos1 region of each male and female, and noted the possible offspring genotypes each cross could produce.
We sequenced the F1 offspring from several types of parental crosses to determine whether all expected offspring genotypes were present in the F1 generation. The three types were: (1) heterozygous male mated to homozygous female, (2) homozygous male mated to heterozygous female, and (3) heterozygous male mated to heterozygous female. Offspring from pairings in which the male parent was heterozygous (type 1 and 3) should be missing an expected genotype if the male is only transmitting one allele, as expected if PGE is present in the system. Pairings in which the male parent was homozygous but the female parent was heterozygous (type 2) were screened to assess whether the female is transmitting both alleles.
Finally, we set up crosses between F1 individuals, ensuring that they were isolated before they mated. Here, we only used males that were potentially informative, i.e., we did not use males whose parents had the same homozygous genotype. We checked dishes weekly, removing the F1 father once F2 nymphs were observed, and preserving him in 95% EtOH. We left the F1 female in the dish for another 2 weeks, then removed and preserved her in 95% EtOH. We allowed the offspring to develop for another 2 weeks. We then sequenced the Phos1 region of all of the F2 offspring whose F1 fathers were heterozygous. We only sequenced individuals from crosses that had produced >8 offspring. We analyzed whether the F1 males transmitted both the alleles they inherited to F2 offspring. To do this, we determined which allele each F1 male inherited from his mother or father, and used Fisher’s exact tests to determine whether F2 offspring exhibited deviations in the expected allele frequencies inherited from their paternal grandmother and grandfather. Crosses were pooled based on the expected genotypes in the F2 offspring (i.e., whether three or two genotypes were possible in the F2 generation).
Screening for differentiated sex chromosomes in Liposcelis sp.
We compared read coverage from high-throughput sequencing of males and females to test for the presence of differentiated sex chromosomes in Liposcelis sp., and to ensure the Phos1 marker used for the inheritance study is not associated with a sex chromosome. To do this, we assembled a draft genome of Liposcelis sp., and mapped reads to the assembled contig set (GenBank accession: BioProject ID PRJNA355858). Briefly, DNA was extracted from separate pools of male and female Liposcelis sp. (∼80 individuals, DNeasy kit; Qiagen), and sequenced using 100 bp PE Illumina HiSeq following library construction at Genome Quebec; these reads were combined with previously generated sequence (Perlman et al. 2015) for assembly. Assembly was done using Ray v 2.2.0 (k = 31; Boisvert et al. 2012), with ∼123 M 100 bp PE reads to generate an assembly of ∼264 Mb and a contig N50 of 4617 bp. Raw reads from female-specific (∼44 M) and male-specific (∼53 M) libraries were mapped to the assembly using bwa mem (Li 2013), and high quality read mappings (mapq > 10) retained and quantified using samtools (Li et al. 2009). Raw read mappings were normalized as counts per million mapped reads (CPM), with contigs >1000 bp retained in the analysis (Vicoso and Bachtrog 2015).
Immunofluorescence microscopy
Paternal genome elimination often results in condensation of paternal chromosomes in male somatic and/or germ tissue (Brun et al. 1995; Bongiorni et al. 2004, 2007). To test for the presence of condensed chromosomes in male booklice, we conducted immunofluorescence microscopy with an antibody for H3K9me3, a conserved marker for heterochromatin (Cowell et al. 2002). We conducted immunofluorescence staining on female and male Liposcelis abdominal tissue. The abdomen was used for staining since we wanted to include both reproductive tissue and somatic tissue in the preparations to explore the specificity of heterochromatinization.
The immunofluorescence protocol we used was adapted from Bongiorni et al. (2007), who previously used this approach to study paternal genome elimination in the mealybug Planococcus citri. Briefly, virgin female and male Liposcelis were collected in Bradley-Carnoy fixative (4:3:1 chloroform:ethanol:acetic acid), followed by fixation and dissection in a drop of 45% glacial acetic acid on siliconized coverslips. After dissection to isolate abdominal tissues, siliconized coverslips were squashed on poly-l-lysine (P8920; Sigma) coated microscope slides (which transferred the tissue to the slide) followed by freezing in liquid nitrogen. Coverslips were removed with a razor blade, and tissues permeabilized by incubating the slide in 1× PBS containing 1% Triton X-100 and 0.5% acetic acid. Slides were washed three times in 1× PBS for 5 min, and blocked in 1% BSA in PBST (1× PBS + 0.1% Tween 20) for 30 min at room temperature, followed by incubation with a rabbit primary antibody targeting H3K9me3 (9754S-1:200; Cell Signaling Technology) in 1% BSA in PBST for 1 hr in a humid chamber. Slides were then washed three times in 1× PBS followed by incubation with anti-rabbit Alexa Fluor 488 secondary antibody (A-11008- 1:500; Invitrogen) for 1 hr in a humid chamber, and three washes in 1× PBS as above. DAPI-containing mounting media (F6057; Sigma) was used to counterstain for DNA, and slides were sealed with nail polish. Slides were imaged on a Leica DM IRE2 inverted fluorescent microscope.
Sex allocation in Liposcelis sp. females
A major prediction of systems with paternal genome elimination is maternal control over offspring sex ratio (Haig 1993; Varndell and Godfray 1996; Nagelkerke and Sabelis 1998; Sánchez 2010). We set up an experiment to test whether females exhibit facultative sex allocation by examining whether female age and rearing condition affect sex ratio. We placed ∼200 late instar female nymphs and 200 males into jars (125 ml, 70 mm diameter) containing a small amount of food. We left females for 7 days so they had an opportunity to mature and mate before transferring them into petri dishes (35 mm in diameter) containing 1.7 g of food. The experiment consisted of three treatments: a low-, medium-, and high-density treatment with two, 10, or 20 females in each dish, and five replicate dishes for each treatment. We also kept three males in each dish to ensure females were not sperm limited, replacing males when necessary. Adults were transferred into new dishes weekly for 4 weeks, upon which the experiment was terminated.
We measured the sex ratio (measured as the number of offspring of each sex reaching adulthood) produced by females in each replicate each week, which allowed us to measure both the total sex ratio for each treatment, and also how the sex ratio changed over time. If >20% of the females in a replicate died, we stopped recording data from that replicate. This occurred for one replicate in the low-density treatment in week 3, and one replicate in the medium-density treatment in week 4. We analyzed data in RStudio v3.1.0 (R Core Team 2014) using a generalized linear mixed model with a binomial error distribution and logit link. We used a model selection process, choosing the model that minimized the AIC and including female density, and the week the data were collected, as explanatory variables, and replicate as a random variable.
Data availability
File S1 contains supplementary information including information on genotypes in the inheritance study, and additional results from the immunohistochemistry staining. Illumina sequence data are deposited in GenBank (NCBI) under BioProject ID PRJNA355858, and allele inheritance study sequence data under accessions KY454577 and KY454578.
Results
Transmission distortion of Phos1 allele in males
We sequenced 155 F1 offspring from 14 crosses with 10 males mated to up to three different females (Table 1). We found that heterozygous females mated with homozygous males (i.e., type 2 crosses 11-1, 9-1, and 6-3) produced offspring with both of the expected genotypes, indicating that females transmit both of their alleles. On the other hand, crosses involving heterozygous males (type 1 and 3) did not produce genotypes that would be expected under standard diploid Mendelian inheritance. These crosses were always missing one of the expected offspring genotypes. Heterozygous males mated to more than one female (for example in crosses 12-1 and 12-2) always transmitted the same allele to offspring.
Table 1. F1 offspring genotypes.
| Parents | F1 Offspring | |||||||
|---|---|---|---|---|---|---|---|---|
| Male | Male Genotype | Female | Female Genotype | Cross Type | AA | Aa | Aa | Total |
| 1 | Aa | 1-2 | Aa | 3 | 0 | 3 | 6 | 9 |
| 4 | Aa | 4-1 | AA | 1 | 0 | 6 | − | 6 |
| 4-2 | Aa | 3 | 0 | 4 | 4 | 8 | ||
| 4-3 | Aa | 3 | 0 | 6 | 9 | 15 | ||
| 5 | Aa | 5-2 | AA | 1 | 0 | 6 | − | 6 |
| 6 | AA | 6-3 | Aa | 2 | 6 | 2 | − | 8 |
| 8 | Aa | 8-1 | Aa | 3 | 0 | 5 | 5 | 10 |
| 9 | aa | 9-1 | Aa | 2 | − | 2 | 7 | 9 |
| 10 | Aa | 10-3 | Aa | 3 | 0 | 8 | 6 | 14 |
| 11 | AA | 11-1 | Aa | 2 | 7 | 9 | − | 16 |
| 12 | Aa | 12-1 | AA | 1 | 10 | 0 | − | 10 |
| 12-2 | aa | 1 | − | 15 | 0 | 15 | ||
| 14 | Aa | 14-1 | AA | 1 | 14 | 0 | − | 14 |
| 14-3 | Aa | 3 | 6 | 9 | 0 | 15 | ||
Only crosses that produced more than six offspring were included in the table. Cross type indicates whether only the male (type 1), the female (type 2), or both parents (type 3) were heterozygous. Dashes indicate genotypes that are not expected to be present in the offspring. Every cross in which the male parent is heterozygous is missing an expected offspring genotype.
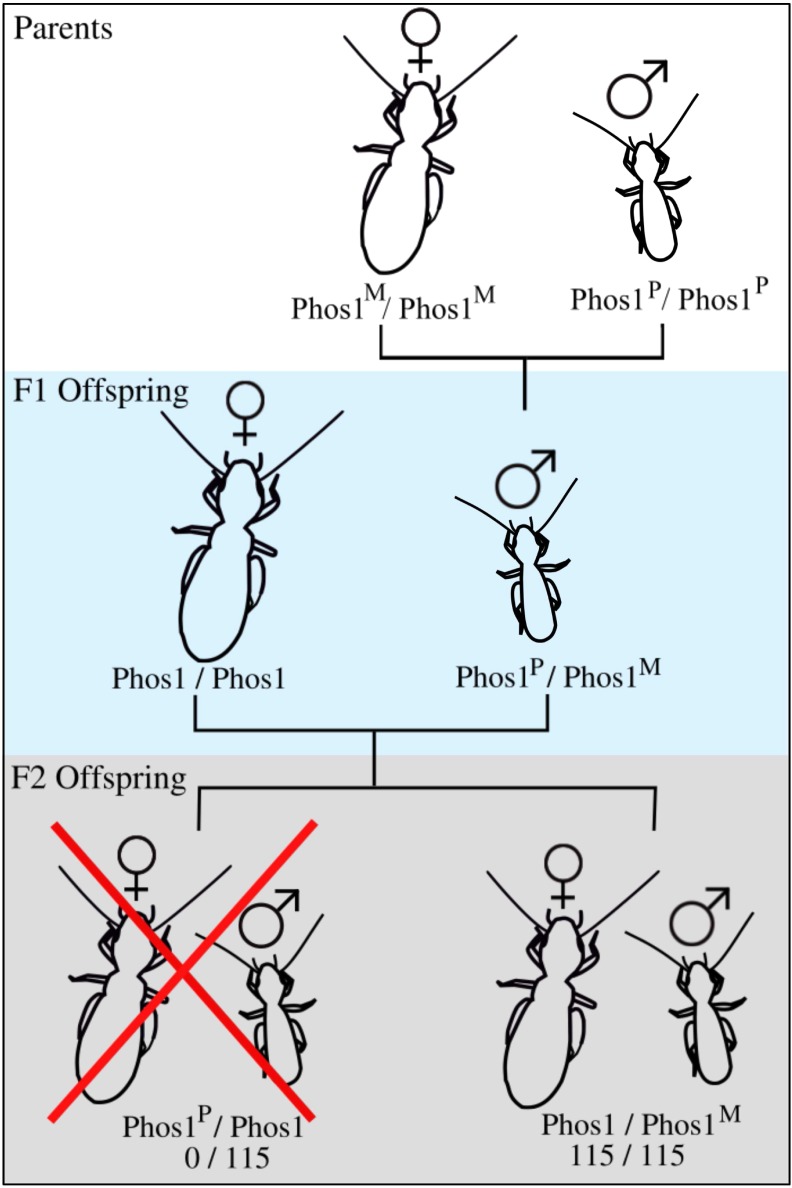
Our F2 crosses, using heterozygous F1 males, confirmed that males only transmit one allele to offspring, and allowed us to determine that allele’s parent-of-origin. We sequenced 115 F2 offspring from 11 crosses, and found that, in all cases, males transmitted exclusively the allele that they inherited from their mother to offspring (Figure 1 and Table 2) (P < 0.0001 for all comparisons).
Figure 1.
Schematic of cross experiment design, as well as the results from the F2 generation. Phos1 indicates the cAMP-specific IBMX-insensitive 3′, 5′-cyclic phosphodiesterase gene region used for sequencing and the superscripts M and P indicate that the allele is maternal or paternal in the parental generation, respectively. All offspring in the F2 generation carry the allele transmitted to them from their paternal grandmother.
Table 2. F2 offspring genotypes produced by heterozygous F1 males mated to F1 females.
| F1 Parents | F2 Offspring | |||||
|---|---|---|---|---|---|---|
| Male | Male Genotype | Female Genotype | AA | Aa | Aa | Total |
| 4-1M2 | Aa | aa | − | 9 | 0 | 9 |
| 4-1M4 | Aa | aa | − | 13 | 0 | 13 |
| 4-3M5 | Aa | AA | 10 | 0 | − | 10 |
| 5-2M1 | Aa | AA | 6 | 0 | − | 6 |
| 5-2M4 | Aa | Aa | 5 | 6 | 0 | 11 |
| 6-2M1 | Aa | aa | − | 0 | 10 | 10 |
| 8-1M2 | Aa | AA | 14 | 0 | − | 14 |
| 9-1M1 | Aa | AA | 10 | 0 | − | 10 |
| 12-2M5 | Aa | Aa | 0 | 3 | 7 | 10 |
| 12-2M9 | Aa | Aa | 0 | 7 | 8 | 15 |
| 14-3M6 | Aa | Aa | 0 | 1 | 6 | 7 |
Each male parent received the allele in boldface type from his mother and the underlined one from his father. Dashes indicate offspring genotypes not expected to be present under standard diploid Mendelian inheritance. In every case, the male only transmitted the allele he inherited from his mother to offspring. (Note that cross 6-2 is not included in Table 1 as all offspring from this cross were expected to be heterozygous—parents were AA*aa).
No evidence for a differentiated sex chromosome in Liposcelis sp.
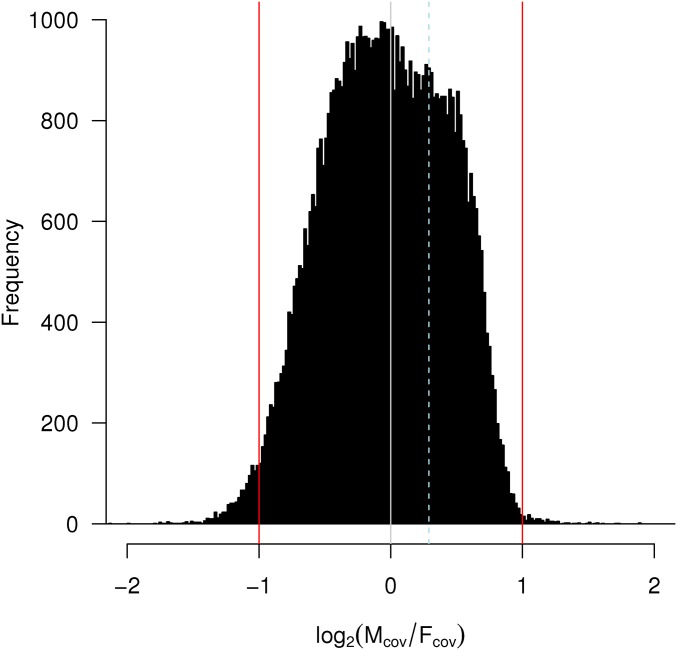
Following the logic of recent studies using next-generation sequencing approaches to characterize sex-determination systems (Vicoso and Bachtrog 2015), we assembled a genome combining female- and male-derived reads. We mapped raw reads to this assembly to identify contigs at 1/2 the coverage in males relative to females (and vice versa) that may represent portions of sex chromosomes. A histogram of the log2 male/female read coverage for contigs in this assembly (as read counts per million reads mapped) had a single discernible peak with a median near 0 (Figure 2; median = −0.03), representing equal read coverage in male and female libraries, lending little support to the existence of a differentiated sex chromosome. Importantly, our Phos1 marker does not show differential read coverage between males and females, suggesting that it does not lie in an atypical (or sex-linked) part of the genome.
Figure 2.
Histogram comparing the coverage of male to female reads mapping back to the Liposcelis sp. genome contigs. Reads at zero have the same coverage in males and females. Reads mapping to −1 are found at double the frequency in females than males (as would be expected for sex-restricted contigs under male heterogamety). The dashed line represents the position of the Phos1 marker used in inheritance experiments.
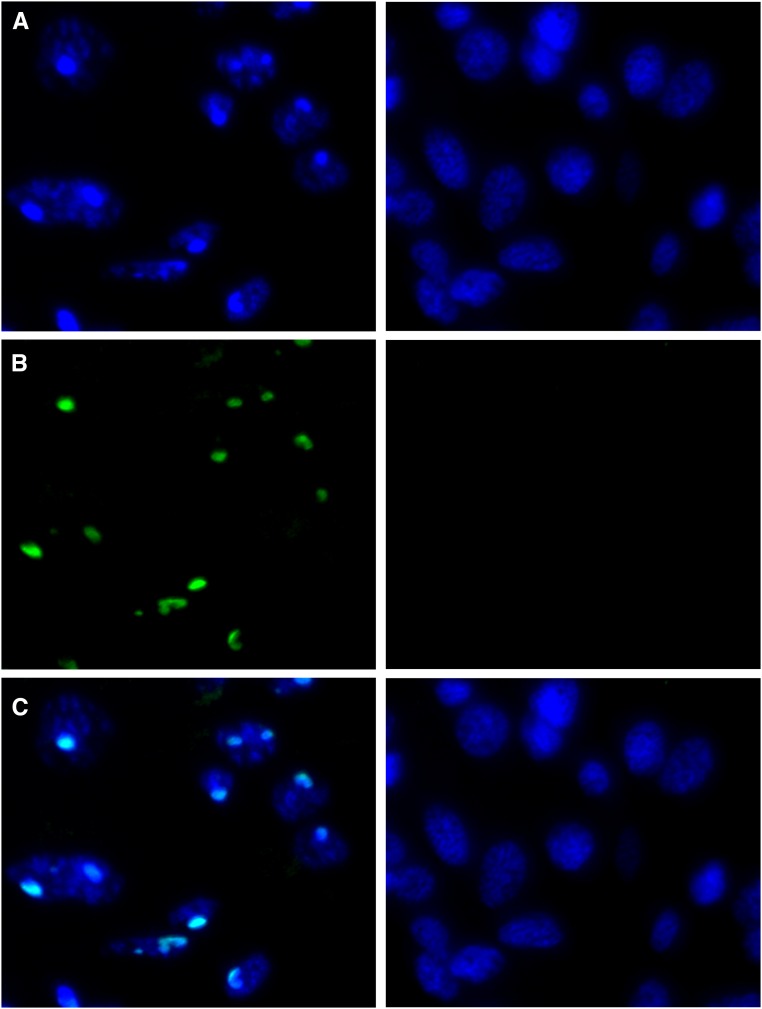
Heterochromatic chromocenters are present in males
DAPI staining revealed condensed regions of intense fluorescence (i.e., chromocenters) present throughout male abdominal tissue but not female abdominal tissue (Figure 3 and Figure S3 in File S1). Additionally, H3K9me3 fluorescence that colocalized with DAPI staining was present in male but not female cells (Figure 3). This indicates that these regions are likely heterochromatinized in males. Condensed heterochromatic regions were also present in head and thoracic tissue in Liposcelis sp. (Figure S4 in File S1).
Figure 3.
DAPI (A), H3K9me3 (B), and merged (C) images of male (left panels) and female (right panels) Liposcelis sp. abdominal tissue. Condensed regions of DAPI staining that colocalize with H3K9me3 staining are present in male tissue but absent from female tissue, indicating chromocenters are present in male cells. Bar, 5 μm.
Sex ratio varies with female age
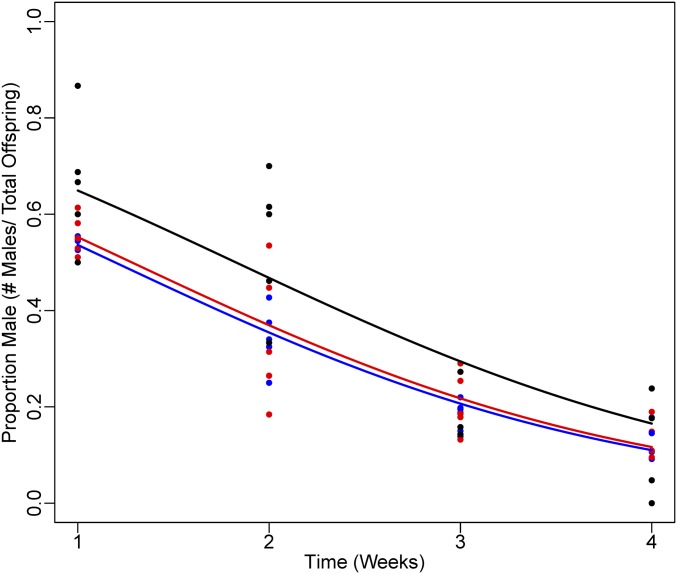
Females in all treatments produced offspring with a female biased sex ratio (Figure 4) [Sex ratio (# males/total offspring) = 0.40 ± 0.12, 0.32 ± 0.08, 0.30 ± 0.08 for low, medium, and high density treatments, respectively]. However, offspring sex ratio varied with maternal age. In all treatments, when females were young they produced more sons compared to when they aged (generalized linear model: P < 0.001). For example, in the first week of the experiment, when females had just become adults, the offspring sex ratio was 0.59 for all treatments, as opposed to the last week of the experiment when it averaged 0.13. These differences were unlikely to be due to differential offspring mortality, as females produced comparable numbers of offspring across treatments (mean offspring produced per female per week: 4.9, 5.4, and 7.1 for high, medium, and low density treatments, respectively) and over time (mean offspring produced per female per week: 4.5, 4.2, 5.4, and 5.9 for weeks 1–4 respectively). Finally, density had a small but significant effect on sex ratio, with females in the low density treatment producing a slightly more male biased sex ratio than females in the other density treatments (generalized linear model: P = 0.015).
Figure 4.
Sex ratio (# males/total offspring) produced by Liposcelis sp. females as they aged. Females produce a female biased sex ratio overall, which varied as females aged, with a more male biased sex ratio when females were young compared to when they were older. Black, red, and blue data points indicate the low, medium, and high density treatments, respectively.
Discussion
We explored the mode of reproduction and sex determination in Liposcelis sp., and found that this species exhibits paternal genome elimination. Within males, paternally inherited chromosomes were never transmitted to offspring. Also, immunofluorescence microscopy revealed the presence in males, but not in females, of densely packed chromocenters associated with H3K9me3 (an epigenetic mark associated with heterochromatinization), suggesting that paternal chromosomes are silenced in males. This is an exciting finding as this is the first species in the order Psocodea in which paternal genome elimination has been conclusively demonstrated. An earlier study found that some, but not all, male human body lice, P. humanus, transmit only the genes that they inherit from their mother (McMeniman and Barker 2006), suggesting that paternal genome elimination may be widespread in this order, although in P. humanus it is not clear why all males did not exhibit this chromosome inheritance pattern.
PGE has been documented in five other arthropod orders: mites (Phytoseiidae, Otopheidomenidae, and Ascoidea), flies [it has evolved twice, in Sciaridae (fungus gnats) and Cecidomyiidae (gall midges)], springtails (Symphypleona), beetles [Cryphalini (bark beetles)], and scale insects (Neococcoidea) (Metz 1938; Helle et al. 1978; Nur 1980; Stuart and Hatchett 1988; Brun et al. 1995; Dallai et al. 2000). In all of these lineages, males develop from fertilized eggs, but fail to transmit chromosomes they inherited from their fathers. However, how paternal genome elimination occurs in these lineages is quite different. In sciarid and cecidomyiid flies, and in symphyleonan springtails, paternally inherited sex chromosomes (but not autosomes) are ejected during male development, often in complex combinations (Metz 1938; Stuart and Hatchett 1988; Dallai et al. 2000). On the other hand, mites, bark beetles, and scale insects do not have sex chromosomes at all. Instead, the entire paternal chromosome complement is eliminated or inactivated in males (Nelson-Rees et al. 1980; Nur 1980; Brun et al. 1995). Paternal chromosomes can be heterochromatinized early in development and excluded from viable sperm during spermatogenesis (e.g., Lecanoid and Comstockiella scale insects), or they can be lost entirely in early development in males (Diaspidid scale insects) (Ross et al. 2010a). The lack of consistent molecular features makes it difficult to diagnose paternal genome elimination in species without extensive investigation into male meiosis or crossing experiments that follow alleles in males over several generations. Because of this, it is likely that PGE is present in more species than it has been identified in to date.
Our finding of heterochromatinization occurring throughout male but not female abdominal tissue, and the lack of an obvious sex chromosome in our genomic analysis, suggests that paternal genome elimination in Liposcelis booklice is likely similar to the Lecanoid/Comstockiella systems in scale insects, with paternal chromosomes being heterochromatinized in male body tissues as well as the germline, rather than being eliminated in somatic tissue, or present but not heterochromatinized (Ross et al. 2010a). In many species that exhibit PGE, paternal chromosomes are epigenetically silenced in males through heterochromatinization and form a large chromocenter; this has been best studied in scale insects, particularly the citrus mealybug P. citri (Bongiorni et al. 2004, 2007). Heterochromatinization is thought to occur through imprinting, as paternal chromosome heterochromatinization occurs soon after fertilization, before embryonic genes are highly expressed (Sabour 1972). Paternal genome heterochromatinization in males involves many of the same components that are involved in facultative heterochromatinization in other animals. For instance, H3K9me3 is involved in paternal chromosome heterochromatinization in P. citri, and Liposcelis, and also in X-chromosome inactivation in mammals (Cowell et al. 2002).
Although the lineages in which PGE occurs are taxonomically widespread, they share some striking similarities in their ecology. Species that exhibit PGE are typically small and have limited dispersal throughout their life, resulting in a high degree of mating between close relatives. The reason for the association between PGE and inbreeding remains unclear. Several theoretical studies have proposed that inbreeding promotes the evolution of PGE and other asymmetric genetic systems (Hamilton 1967; Haig 1993; Gardner and Ross 2014; alternatively, see Bull (1979)); however, there has been little empirical work quantifying the level of inbreeding in PGE species and related taxa. Liposcelis exhibit many of the ecological factors that are associated with PGE, being small, wingless, and with limited dispersal, which may result in a high degree of inbreeding. Obtaining estimates of sex ratio and inbreeding in wild Liposcelis may help elucidate why there is an association between inbreeding and PGE.
Additionally, species with PGE often have female biased sex ratios with maternal control over the offspring sex ratio. This has been studied best in mites (Helle et al. 1978, Nagelkerke and Sabelis 1998) and scale insects (Varndell and Godfray 1996; Ross et al. 2010b, 2012). The results from our controlled laboratory experiments point toward maternal control of sex ratio in Liposcelis sp. We found highly female-biased sex ratios in Liposcelis sp., which altered as a female aged, with a more male biased sex ratio produced when females were young. The finding that females produce more males early in reproduction is intriguing, as something similar was found in P. citri (Ross et al. 2012); we speculate that this might be driven by the need to ensure mating in groups with little dispersal. It is unlikely that the sex ratio differences we observed were due to differential mortality, as females produced approximately the same amount of offspring each week in the experiment. To confirm that females are able to control offspring sex ratio, it would be interesting to conduct similar experiments in more natural settings, and alter other ecological factors such as relatedness of individuals and resource availability.
It is likely that paternal genome elimination is widespread in Psocodea, and is perhaps the mode of sex determination for the entire lineage that includes Liposcelididae and Phthiraptera (Yoshizawa and Johnson 2010). A number of features strongly suggest that human parasitic lice, P. humanus (and probably other parasitic lice) exhibit PGE. First, as mentioned earlier, a previous study found that some, but not all, male P. humanus only transmitted their maternal copy of microsatellite markers (McMeniman and Barker 2006). Additionally, human lice do not have sex chromosomes (Tombesi and Papeschi 1993; Golub and Nokkala 2004; Bressa et al. 2015) and exhibit highly female-biased sex ratios (Buxton 1941).
Parasitic lice also have unusual spermatogenesis and sperm morphology that have been suggested to be linked to PGE (Ross and Normark 2015; Blackmon et al. 2017). Spermatogenesis is highly distinctive in parasitic lice, consisting of several mitotic divisions at the end of spermatogenesis, the last one being unequal, and resulting in half the products of the mitotic division forming functional sperm and the other half forming nonfunctional pycnotic nuclei (Hindle and Pontecorvo 1942; Tombesi and Papeschi 1993; Golub and Nokkala 2004). Although little is currently known about spermatogenesis in Liposcelididae, this group is known to have an unusual sperm morphology which is also present in P. humanus and other lice species (Dallai and Afzelius 1991; Ross and Normark 2015) where sperm contain two axonemes rather than the usual single one (King and Ahmed 1989).
Even if PGE is widespread in Psocodea, it is likely to be quite different between parasitic lice and booklice. Cytogenetic studies of parasitic lice (Golub and Nokkala 2004; Bressa et al. 2015) report males and females having the same number of chromosomes, and do not mention any differences in the appearance of chromosomes in males and females, suggesting that male chromosomes may not be heterochromatinized. Thus, Psocodea represents an exciting new model for studying the evolution, ecology, and genetics of paternal genome elimination, an enigmatic and interesting mode of sex determination. The ease with which booklice can be maintained in the laboratory compared to other arthropods with paternal genome elimination makes them especially promising for study.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.199786/-/DC1.
Acknowledgments
We thank Laura Ross, members of the Ross laboratory, and members of the Perlman laboratory for useful discussion on this work. This work was supported by a National Sciences and Engineering Council of Canada Discovery grant to S.J.P. S.J.P. acknowledges support from the Integrated Microbial Biodiversity Program of the Canadian Institute for Advanced Research.
Footnotes
Communicating editor: K. Peichel
Literature Cited
- Bachtrog D., Mank J. E., Peichel C. L., Kirkpatrick M., Otto S. P., et al. , 2014. Sex determination: why so many ways of doing it? PLoS Biol. 12: e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell L. R., Maine E. M., Schedl P., Cline T. W., 1988. Sex-lethal, a Drosophila sex determination switch gene, exhibits sex-specific RNA splicing and sequence similarity to RNA binding proteins. Cell 55: 1037–1046. [DOI] [PubMed] [Google Scholar]
- Beukeboom L. W., Perrin N., 2014. The Evolution of Sex Determination. Oxford University Press, Oxford, UK. [Google Scholar]
- Beye M., Hasselmann M., Fondrk M. K., Page R. E., Omholt S. W., 2003. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell 114: 419–429. [DOI] [PubMed] [Google Scholar]
- Blackmon H., Ross L., Bachtrog D., 2017. Sex determination, sex chromosomes, and karyotype evolution in insects. J. Hered. 108(1): 78–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert S., Raymond F., Godzaridis É., Laviolette F., Corbeil J., 2012. Ray Meta: scalable de novo metagenome assembly and profiling. Genome Biol. 13: R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongiorni S., Fiorenzo P., Pippoletti D., Prantera G., 2004. Inverted meiosis and meiotic drive in mealybugs. Chromosoma 112: 331–341. [DOI] [PubMed] [Google Scholar]
- Bongiorni S., Pasqualini B., Taranta M., Singh P. B., Prantera G., 2007. Epigenetic regulation of facultative heterochromatinisation in Planococcus citri via the Me(3)K9H3–HP1-Me(3)K20H4 pathway. J. Cell Sci. 120: 1072–1080. [DOI] [PubMed] [Google Scholar]
- Bressa M. J., Papeschi A. G., Toloza A. C., 2015. Cytogenetic features of human head and body lice (Phthiraptera: Pediculidae). J. Med. Entomol. 52: 918–924. [DOI] [PubMed] [Google Scholar]
- Brun L. O., Stuart J., Gaudichon V., Aronstein K., French-Constant R. H., 1995. Functional haplodiploidy: a mechanism for the spread of insecticide resistance in an important international insect pest. Proc. Natl. Acad. Sci. USA 92: 9861–9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull J. J., 1979. An advantage for the evolution of male haploidy and systems with similar genetic transmission. Heredity 43: 361–381. [Google Scholar]
- Buxton P. A., 1941. Studies on populations of head-lice (Pediculus humanus capitis: Anoplura). Parasitology 33: 224–242. [Google Scholar]
- Cowell I. G., Aucott R., Mahadevaiah S. K., Burgoyne P. S., Huskisson N., et al. , 2002. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma 111: 22–36. [DOI] [PubMed] [Google Scholar]
- Dallai R., Afzelius B. A., 1991. Sperm flagellum of insects belonging to orders Psocoptera, Mallophaga and Anoplura. Ultrastructural and phylogenetic aspects. Bolletino di Zool. 58: 211–216. [Google Scholar]
- Dallai R., Fanciulli P. P., Frati F., 2000. Aberrant spermatogenesis and the peculiar mechanism of sex determination in symphypleonan Collembola (Insecta). J. Hered. 91: 351–358. [DOI] [PubMed] [Google Scholar]
- Dübendorfer A., Hediger M., Burghardt G., Bopp D., 2002. Musca domestica, a window on the evolution of sex-determining mechanisms in insects. Int. J. Dev. Biol. 46: 75–79. [PubMed] [Google Scholar]
- Gardner A., Ross L., 2014. Mating ecology explains patterns of genome elimination. Ecol. Lett. 17: 1602–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub N. V., Nokkala S., 2001. The karyotypes of two bark-lice species (Psocoptera, Psocomorpha, Amphipsocidae): the first description of neo-XY sex chromosome system in Psocoptera. Folia Biol. 49: 153–156. [PubMed] [Google Scholar]
- Golub N. V., Nokkala S., 2004. Chromosome numbers of two sucking louse species (Insecta, Phthiraptera, Anoplura). Hereditas 141: 94–96. [DOI] [PubMed] [Google Scholar]
- Golub N. V., Nokkala S., 2009. Chromosome numbers in eight species of Palaearctic Psocoptera (Insecta). Comp. Cytogenet. 3: 33–41. [Google Scholar]
- Haig D., 1993. The evolution of unusual chromosomal systems in coccoids: extraordinary sex ratios revisited. J. Evol. Biol. 6: 69–77. [Google Scholar]
- Hall A. B., Basu S., Jiang X., Qi Y., Timoshevskiy V. A., et al. , 2015. A male-determining factor in the mosquito Aedes aegypti. Science 348: 1268–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton W. D., 1967. Extraordinary sex ratios. Science 156: 477–488. [DOI] [PubMed] [Google Scholar]
- Helle W., Bolland H. R., Van Arendonk R., De Boer R., Schulten G. G. M., et al. , 1978. Genetic evidence for biparental males in haplo-diploid predator mites (Acarina: Phytoseiidae). Genetica 49: 165–171. [Google Scholar]
- Herrick G., Seger J., 1999. Imprinting and paternal genome elimination in insects. Results Probl. Cell Differ. 25: 41–71. [DOI] [PubMed] [Google Scholar]
- Hindle E., Pontecorvo G., 1942. Mitotic divisions following meiosis in Pediculus corporis males. Nature 149: 668. [Google Scholar]
- King P. E., Ahmed K. S., 1989. Sperm structure in the Psocoptera. Acta Zoologica 70: 57–61. [Google Scholar]
- Kiuchi T., Koga H., Kawamoto M., Shoji K., Sakai H., et al. , 2014. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 509: 633–636. [DOI] [PubMed] [Google Scholar]
- Kozielska M., Weissing F. J., Beukeboom L. W., Pen I., 2010. Segregation distortion and the evolution of sex-determining mechanisms. Heredity 104: 100–112. [DOI] [PubMed] [Google Scholar]
- Krzywinska E., Dennison N. J., Lycett G. J., Krzywinski J., 2016. A maleness gene in the malaria mosquito Anopheles gambiae. Science 353: 67–69. [DOI] [PubMed] [Google Scholar]
- Li, H., 2013 Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Available at: https://arxiv.org/abs/1303.3997.
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Shao R., Song N., Song F., Jiang P., et al. , 2015. Higher-level phylogeny of paraneopteran insects inferred from mitochondrial genome sequences. Sci. Rep. 5: 8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMeniman C. J., Barker S. C., 2006. Transmission ratio distortion in the human body louse, Pediculus humanus (Insecta: Phthiraptera). Heredity 96: 63–68. [DOI] [PubMed] [Google Scholar]
- Metz C. W., 1938. Chromosome behavior, inheritance and sex determination in Sciara. Am. Nat. 743: 485–520. [Google Scholar]
- Nagelkerke C. J., Sabelis M. W., 1998. Precise control of sex allocation in pseudo-arrhenotokous phytoseiid mites. J. Evol. Biol. 11: 649–684. [Google Scholar]
- Nelson-Rees W. A., Hoy M. A., Roush R. T., 1980. Heterochromatinization, chromatin elimination and haploidization in the parahaploid mite Metaseiulus occidentalis (Nesbitt) (Acarina: Phytoseiidae). Chromosoma 77: 263–276. [DOI] [PubMed] [Google Scholar]
- Normark B. B., 2003. The evolution of alternative genetic systems in insects. Annu. Rev. Entomol. 48: 397–423. [DOI] [PubMed] [Google Scholar]
- Nur U., 1980. Evolution of unusual chromosome systems in scale insects (Coccoidea: Homoptera), pp. 97–117 in Insect Cytogenetics, edited by Blackman R. L., Hewitt G. M., Ashburner M. Blackwell, Oxford. [Google Scholar]
- Perlman S. J., Hodson C. N., Hamilton P. T., Opit G. P., Gowen B. E. 2015. Maternal transmission, sex ratio distortion, and mitochondria. Proc. Natl. Acad. Sci. USA 112: 10162–10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2014. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Ross L., Normark B. B., 2015. Evolutionary problems in centrosome and centriole biology. J. Evol. Biol. 28: 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross L., Pen I., Shuker D. M., 2010a Genomic conflict in scale insects: the causes and consequences of bizarre genetic systems. Biol. Rev. Camb. Philos. Soc. 85: 807–828. [DOI] [PubMed] [Google Scholar]
- Ross L., Langenhof M. B. W., Pen I., Beukeboom L. W., West S. A., et al. , 2010b Sex allocation in a species with paternal genome elimination: the roles of crowding and female age in the mealybug Planococcus citri. Evol. Ecol. Res. 12: 89–104. [Google Scholar]
- Ross L., Langenhof M. B. W., Pen I., Shuker D. M., 2012. Temporal variation in sex allocation in the mealybug Planococcus citri: adaptation, constraint, or both? Evol. Ecol. 26: 1481–1496. [Google Scholar]
- Sabour M., 1972. RNA synthesis and heterochromatization in early development of a mealybug. Genetics 70: 291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez L., 2010. Sciara as an experimental model for studies on the evolutionary relationships between the zygotic, maternal and environ- mental primary signals for sexual development. J. Genet. 89: 325–331. [DOI] [PubMed] [Google Scholar]
- Stuart J. J., Hatchett J. H., 1988. Cytogenetics of the Hessian fly: II. Inheritance and behavior of somatic and germ-line-limited chromosomes. J. Hered. 79: 190–199. [DOI] [PubMed] [Google Scholar]
- Tombesi M. L., Papeschi A. G., 1993. Meiosis in Haematopinus suis and Menacanthus stramineus (Phthiraptera, Insecta). Hereditas 119: 31–38. [Google Scholar]
- Varndell N. P., Godfray H. C. J., 1996. Facultative adjustment of the sex ratio in an insect (Plannococcus citri, Pseudococcidae) with paternal genome loss. Evolution 50: 2100–2105. [DOI] [PubMed] [Google Scholar]
- Vicoso B., Bachtrog D., 2015. Numerous transitions of sex chromosomes in Diptera. PLoS Biol. 13: e1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S. K., Thornton I. W. B., 1966. Chromosome numbers of some psocid genera (Psocoptera). Nature 211: 214–215. [DOI] [PubMed] [Google Scholar]
- Yoshizawa K., Johnson K. P., 2003. Phylogenetic position of Phthiraptera (Insecta: Paraneoptera) and elevated rate of evolution in mitochondrial 12S and 16S rDNA. Mol. Phylogenet. Evol. 29: 102–114. [DOI] [PubMed] [Google Scholar]
- Yoshizawa K., Johnson K. P., 2010. How stable is the “Polyphyly of Lice” hypothesis (Insecta: Psocodea)?: a comparison of phylogenetic signal in multiple genes. Mol. Phylogenet. Evol. 55: 939–951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
File S1 contains supplementary information including information on genotypes in the inheritance study, and additional results from the immunohistochemistry staining. Illumina sequence data are deposited in GenBank (NCBI) under BioProject ID PRJNA355858, and allele inheritance study sequence data under accessions KY454577 and KY454578.