Abstract
The goal of the Collaborative Cross (CC) project was to generate and distribute over 1000 independent mouse recombinant inbred strains derived from eight inbred founders. With inbreeding nearly complete, we estimated the extinction rate among CC lines at a remarkable 95%, which is substantially higher than in the derivation of other mouse recombinant inbred populations. Here, we report genome-wide allele frequencies in 347 extinct CC lines. Contrary to expectations, autosomes had equal allelic contributions from the eight founders, but chromosome X had significantly lower allelic contributions from the two inbred founders with underrepresented subspecific origins (PWK/PhJ and CAST/EiJ). By comparing extinct CC lines to living CC strains, we conclude that a complex genetic architecture is driving extinction, and selection pressures are different on the autosomes and chromosome X. Male infertility played a large role in extinction as 47% of extinct lines had males that were infertile. Males from extinct lines had high variability in reproductive organ size, low sperm counts, low sperm motility, and a high rate of vacuolization of seminiferous tubules. We performed QTL mapping and identified nine genomic regions associated with male fertility and reproductive phenotypes. Many of the allelic effects in the QTL were driven by the two founders with underrepresented subspecific origins, including a QTL on chromosome X for infertility that was driven by the PWK/PhJ haplotype. We also performed the first example of cross validation using complementary CC resources to verify the effect of sperm curvilinear velocity from the PWK/PhJ haplotype on chromosome 2 in an independent population across multiple generations. While selection typically constrains the examination of reproductive traits toward the more fertile alleles, the CC extinct lines provided a unique opportunity to study the genetic architecture of fertility in a widely genetically variable population. We hypothesize that incompatibilities between alleles with different subspecific origins is a key driver of infertility. These results help clarify the factors that drove strain extinction in the CC, reveal the genetic regions associated with poor fertility in the CC, and serve as a resource to further study mammalian infertility.
Keywords: QTL mapping, line extinction, sperm motility, infertility, mouse, multiparental populations, MPP
THE Collaborative Cross (CC) is a mouse genetic reference population designed to contain high genetic variation across strains and minimal genetic variation within a strain (Collaborative Cross Consortium 2012). The CC Consortium initiated several thousand inbred funnels with high levels of genetic diversity due to the inclusion of three wild-derived inbred founders from three subspecific origins (WSB/EiJ from Mus musculus domesticus, PWK/PhJ from M. musculus musculus, and CAST/EiJ from M. musculus castaneus), along with five classical inbred strains that have a mostly M. musculus domesticus origin (A/J, C57BL/6J, 129S1/SvImJ, NOD/ShiLtJ, and NZO/HILtJ) (Yang et al. 2011). The design enabled the CC to capture 90% of genetic variation found in common laboratory stocks (Roberts et al. 2007). However, the level and origin of genetic variation may have acted as a double-edged sword, as the CC population has suffered extreme extinction throughout its breeding process. The rate of extinction, previously reported to be 73.04% (Collaborative Cross Consortium 2012), has steadily increased and is considerably larger than other similar research populations and predicted models of inbreeding (Radwan 2003; Peirce et al. 2004; Swindell and Bouzat 2006; Kover et al. 2009; McMullen et al. 2009; Huang et al. 2011; King et al. 2012; Mackay et al. 2012; Pekkala et al. 2012). This is believed to be due in large part to genetic incompatibilities between the three M. musculus subspecies (Chesler et al. 2008; Aylor et al. 2011; Threadgill and Churchill 2012). Other factors such as colony management, reduced fertility, and small litter sizes may have influenced the high extinction rate but remained unidentified, though genetic loci influencing litter size and other reproductive parameters have been detected (Philip et al. 2011). In contrast to the CC white paper, which stated that no “heroic” efforts would be made to rescue uncooperative strains (Churchill et al. 2004), extraordinary efforts were undertaken, including male fertility testing, combining the remaining males and females together in all possible combinations, and creating cousin lines for funnel rescue, which makes the high level of extinction even more remarkable.
The CC and its founders have been used to successfully identify genetic association for several traits, including spontaneous colitis and susceptibility to infection by viruses such as Ebola, West Nile, Severe Acute Respiratory Syndrome, and influenza (Ferris et al. 2013; Chesler 2014; Rasmussen et al. 2014; Gralinski et al. 2015, 2017; Green et al. 2017). The CC is an ideal population to study fertility since it contains a large amount of genetic diversity, has wide variation in fertility, and has complementary resources to validate discoveries (Threadgill and Churchill 2012; Morgan and Welsh 2015; Morgan et al. 2017). The set of lines destined for extinction is a valuable, but transient, population that is only available during the creation of a multi-parental population (MPP). To our knowledge, this is a unique resource as other similar research populations do not retain this level of information during inbreeding (Kover et al. 2009; McMullen et al. 2009; Huang et al. 2011; King et al. 2012; Pekkala et al. 2012; Pool et al. 2012; Bouchet et al. 2017; Cubillos et al. 2017; Mangandi et al. 2017; Raghavan et al. 2017; Tisné et al. 2017). With detailed genotyping and reproductive phenotyping of CC lines that were declared extinct, we can address questions related to survivability and line extinction during the process of inbreeding. The first focus on this resource will be on male fertility and its variability in the extinct CC lines, although implications on fertility expand well beyond the CC.
We focused on male fertility based on a combination of practical and theoretical considerations. One important factor is that measuring fertility in males is much easier than in females due to their ability to undergo concurrent fertility testing with multiple females. Moreover, fertility testing with unrelated mice was not practical until a line was declared extinct, by which time many females were past their reproductive age. Additionally, targeted gene disruptions and large-scale mutagenesis programs have identified many more mutations that selectively affect fertility in males than in females (Handel et al. 2006; Kennedy and O’Bryan 2006; Matzuk and Lamb 2008). It is hypothesized that the sex differences in these studies may be connected to the large number of genes with restricted expression during spermatogenesis (Schultz et al. 2003; Wu et al. 2004; Chalmel et al. 2007). Finally, the “faster-males” theory predicts that male infertility is common in species where males are the heterogametic sex, as males will be disproportionally affected by infertility since they possess a single copy of chromosome X (Haldane 1922; Orr and Turelli 1996; Johnson and Lachance 2012).
Here, we report on the magnitude of extinction in the CC, characterize reproductive traits on males from 347 CC extinct lines, identify genomic regions associated with eight traits, and perform a validation experiment to confirm the association of a region on chromosome 2 with a measure of sperm velocity. We observed a wide range of phenotypic variation for all reproductive traits compared to the CC founders, which is expected due to the genetic diversity within the CC. Through the use of fertility testing, we conclude that male infertility is responsible for nearly half of all extinction. We also compared genomic features between the CC extinct and CC living populations and observed notable differences in allele frequencies, particularly with CAST/EiJ and PWK/PhJ haplotypes. These two strains, which have different subspecific origins from the remaining CC founder strains and were previously observed to be outliers in several reproductive traits (Odet et al. 2015), drive the allelic effects in many of the QTL identified in this study.
Materials and Methods
Nomenclature
We use the following nomenclature with regard to CC animals (Supplemental Material, Figure S1). Funnels are independent iterations of the breeding schema. CC line refers to the generations between G2:F1 and the point they became > 90% homozygous or extinct. Finally, strain denotes a CC line that has reached 90% homozygosity. The CC breeding schema used a design so that each of the eight founder inbred strains had an equal probability to be represented in the autosomes. The inbred founder strains are referred to as G0 and were mated in a specific order known as the funnel code to generate four G1s. The mitochondrial genome is inherited from the strain in the first position of the funnel code, while the Y chromosome is inherited from the founder strain listed in the last position. Founders in positions 4, 7, and 8 do not contribute to chromosome X. Once a CC line reached 90% homozygosity it was renamed and made publicly available as a CC strain (Srivastava et al. 2017).
Throughout the manuscript, we use the following colors and one letter symbols to represent the eight founder strains: A/J, yellow, A; C57BL/6J, gray, B; 129S1/SvImJ, pink, C; NOD/ShiLtJ, dark blue, D; NZO/HlLtJ, light blue, E; CAST/EiJ, green, F; PWK/PhJ, red, G; and WSB/EiJ, purple, H.
Mice
All funnels reported here were initially bred at Oak Ridge National Laboratory (ORNL) in Oak Ridge, Tennessee (Chesler et al. 2008), and then at the University of North Carolina (UNC) at Chapel Hill since 2009 (Threadgill et al. 2011; Collaborative Cross Consortium 2012; Welsh et al. 2012). A single male from each of 347 independent lines, which were declared extinct between 2008 and 2011, was transferred to the laboratory of Fernando Pardo-Manuel de Villena at the UNC at Chapel Hill. A funnel was declared extinct when no combinations of males and females within that strain would produce fertile offspring necessary to maintain a colony. Reproductive phenotypes were measured for adult males, ranging from 179 to 862 days of age. Before declaring a funnel extinct, males were mated in several possible combinations within a funnel while attempting to maintain existing CC lines, a strategy that contributed to the age variation in males from extinct lines.
All breeding mice used for the Vcl1 QTL validation experiment were acquired from the UNC Systems Genetics Core Facility (csbio.unc.edu/CCstatus/; Welsh et al. 2012). Mice from six CC strains (CC008/GeniUnc, CC013/GeniUnc, CC021/Unc, CC034/Unc, CC053/Unc, and CC065/Unc), referred to as “cases,” have the PWK/PhJ allele at the Vcl1 QTL peak on chromosome 2, and mice from six other CC strains (CC001/Unc, CC002/Unc, CC003/Unc, CC010/GeniUnc, CC011/Unc, and CC018/Unc), referred to as “controls,” do not have any PWK/PhJ contribution on chromosome 2. Mice within cases and controls were outcrossed to each other to generate 12 and 11 of the possible 15 F1 hybrids in cases and controls, respectively. Finally, F1 hybrids were outcrossed within cases and control populations so that four different CC strains were represented in each F2 mouse. The number of case and control mice phenotyped from each generation and their pedigrees are provided in Table S1.
Mice were housed in standard 20 × 30-cm ventilated polysulfone cages with laboratory grade Bed-O-Cob bedding. Water and Purina Prolab RMH3000 were provided ad libitum, and a small section of polyvinyl chloride pipe was present in each cage for enrichment. All procedures involving animals were performed according to the Guide for the Care and Use of Laboratory Animals with prior approval by the Institutional Animal Care and Use Committee within the Association for Assessment and Accreditation of Laboratory Animal Care-accredited program at the UNC at Chapel Hill (Animal Welfare Assurance Number: A-3410-01).
Reproductive phenotyping
Each CC male was mated to as many females from the same funnel as possible, as well as to unrelated females, such as Swiss Webster and FVB/NJ. Mate pairs were set up for a minimum of 7 weeks after a line was declared extinct. Males that were unproductive in all matings were considered infertile. Reproductive phenotyping was performed on subsets of the 347 males from extinct CC lines as previously described for the CC founder strains (Odet et al. 2015). Each male was killed using CO2 asphyxiation followed by cervical dislocation and the carcass was weighed. After dissection, the weights for each testis, each epididymis with attached vas deferens, and seminal vesicles were recorded.
Sperm counts were determined after collecting sperm from the right cauda epididymis using a procedure to maximize recovery. The epididymis was stored at 4° overnight in phosphate-buffered saline, clipped with iris scissors in 500 μl of phosphate-buffered saline, and incubated for 10 min in a 37° incubator. Sperm were then extruded from the cauda with fine forceps. The sperm suspension was transferred to a microfuge tube and the collection well was rinsed with an additional 500 μl of phosphate-buffered saline. Sperm were diluted, if necessary, and counted using a hemocytometer.
We measured sperm motility using sperm from the left cauda epididymis. The cauda was clipped with iris scissors and transferred to a 37° incubator (5% CO2 in air), allowing sperm to swim out for 10 min into 1 ml of human tubule fluid (HTF) medium + 5 mg/ml bovine serum albumin (Goodson et al. 2011). HTF medium typically supports the development of hyperactivated motility over a 90-min period (Goodson et al. 2011; Odet et al. 2015). After appropriate dilution with the same medium, sperm were transferred to Leja chambers (100 µm-depth; Leja Products BV, Nieuw-Vennep, The Netherlands) and motility was assessed by computer-assisted sperm analysis (CASA) using a CEROS imaging system (Hamilton Thorne Biosciences; version 12.3H IVOS software). Sperm tracks (90 frames, 1.5 sec) and kinetic parameters for individual sperm were captured at 60 Hz using motility analysis parameters (mouse 2) recommended by Hamilton Thorne Biosciences, except that slow cells were counted as motile (Goodson et al. 2011). Tracks in 10 fields were typically recorded for each mouse, along with the percentage of motile sperm and the average path velocity (VAP, µm/sec), straight-line velocity (VSL, µm/sec), curvilinear velocity (VCL, µm/sec), amplitude of lateral head displacement (ALH, µm), and beat cross frequency (BCF, Hz) for each motile sperm. CASAnova, a support vector machines program based on CASA parameters (Goodson et al. 2011), was used to classify individual sperm as progressive, intermediate, hyperactivated, slow, or weakly motile.
The HTF sperm suspension used for motility analysis was also used to assess sperm morphology. Ten-microliter aliquots of this suspension were spread onto positively charged slides and allowed to air dry briefly until moisture had just evaporated. After fixation with −20° methanol for 10 min, the samples were air dried and stored at −20°. Acrosomes were stained with peanut agglutinin conjugated to a fluorescent tag (Alexa Fluor 488; Invitrogen, Carlsbad, CA) (Lee et al. 2008) before microscopic analysis. Sperm were scored by a trained observer, classifying each sperm as having normal morphology, abnormal head shape, abnormal tail bending (≥ 90°), or broken tails (severed at the head/neck junction or at more distal locations along the length of the flagellum).
Testis histology
After wet weights were determined, the left testis was flash frozen in liquid nitrogen and preserved for future use while the right testis was prepared for histological examination. If there were remarkable differences between the two testes, such as one being unusually large in size, then the abnormal testis was selected for histology. Distinct differences occurred in < 1 out of every 50 samples. Typically, the right testis was fixed in Bouin’s solution, cut in half horizontally, and embedded in paraffin. Testis sections (8 µm) were stained with periodic acid-Schiff reagent and counterstained with hematoxylin (Russell et al. 1990). Multiple images of each section were recorded using an Olympus BX51 microscope equipped with a motorized stage and an Olympus DP72 digital camera. A composite image of each transverse section (20 ×) was generated using MetaMorph automation and image analysis software (Molecular Devices, Sunnyvale, CA). Testis composite images were annotated as previously described using a custom interactive image analysis package (Odet et al. 2015). For each testis, the tubule centers were automatically found and refined by a trained observer. The mean tubule radius and the number of tubules were used to estimate the length of the seminiferous epithelium. An interactive tool was used to denote tubules with few or many vacuoles. We classified vacuolization in six categories ranging from no vacuoles to many vacuoles in > 20 tubules, and further classified the six categories into two that generalized few to no vacuoles (categories 1–4) and many vacuoles (categories 5–6).
Statistical analysis
We used one-way fixed effect ANOVA and Student’s t-tests to evaluate differences in reproductive phenotypes within the extinct lines and in the validation populations. Levene’s test was used to access the equality of variances. Phenotypic measurements from CC founders were obtained from Odet et al. (2015). All statistical analyses were conducted with JMP 12 or R 3.2.3 software.
Genotyping
Genotyping for 347 samples was performed with the Mouse Diversity Array (MDA) (Yang et al. 2009). Mice were selected so that only extinct funnel males with reproductive phenotypes were genotyped for analysis. The MDA contains 623,124 SNP probe sets with an average spacing of 4.3 kb. MDA markers were selected to provide genetic information over a uniform grid based on SNP discovery performed in 17 inbred strains (Yang et al. 2007). MDA is highly informative for the discrimination between inbred strains and between the three main house mouse subspecies, M. musculus domesticus, M. musculus musculus, and M. musculus castaneus (Yang et al. 2011). Marker genotypes from the MDA were called using the MouseDivGeno package (Didion et al. 2012). The marker set was then refined through a set of filters. We first selected informative markers among the CC founder strains. This informative set was further filtered to select markers that yielded consistent genotypes among the majority of multiple technical and biological replicates from each founder strain available from The Jackson Laboratory’s MDA genotype repository of inbred mouse strains (ftp://ftp.jax.org/petrs/MDA/). A final filter removed any genotypes that are inconsistent between predicted and observed genotypes in F1 hybrids derived from crosses of founder inbred strains. A resulting set of 381,351 MDA genotyping markers survived these filtering steps, and only this subset was used in subsequent analyses of the 347 CC lines that we analyzed.
HMM haplotype reconstruction
A hidden Markov model (HMM) was used to infer the founder haplotype mosaics from the filtered MDA genotypes. We used a forward–backward inference to establish probabilities at every marker for each of the 36 founder genotype possibilities. The emission probabilities at each marker were derived from the founder and F1 genotypes described in the previous section. Pseudocounts for each unobserved MDA genotype call were included, sufficient to model a uniform 2% genotyping error rate. A transition model was derived to estimate the probability of transitions between the founder genotypes of adjacent markers by recombination (36 by 36). Transition probabilities were modeled by an (1−e(−Δ/K)) exponential decay where Δ is the separation between adjacent markers and whose damping factor, K, is estimated by fitting a linkage map (Liu et al. 2010) for every nonoverlapping 10 Mb block. A correction factor was added for shared recombination between homozygous inbred states. Forty extinct males were also genotyped using the Mouse Universal Genotyping Array (Morgan et al. 2016a). Visual comparison of samples genotyped on multiple arrays concluded that haplotype reconstruction was virtually identical between the two platforms and HMMs.
A modified eight-state genotype HMM was used to infer the founder likelihoods at each marker of chromosome X to accommodate hemizygous males. A 500-kb region on the end of chromosome X containing the pseudoautosomal region (PAR) was excluded from the HMM. The reasons for exclusion include the variable length of the PAR region in the founder strains (White et al. 2012), the fact that it recombines with chromosome Y, and the presence of de novo duplications or translocations discovered in the related Diversity Outbred (DO) population that can lead to apparent heterozygosity (Chesler et al. 2016), which is disallowed in the eight-state model. We also incorporated a significant penalty against the fourth, seventh, and eighth positions of each line’s funnel code into the HMM’s transition probabilities. This accounts for a constraint imposed by the funnel breeding structure used to develop the CC. The founders in those positions cannot contribute to X; however, a small nonzero probability (1 × 10−4) of such transitions was included to allow for potential breeding errors. Finally, we used a forward–backward inference to establish probabilities at every marker for each of eight possible founder genotype possibilities. A complete list of ORNL funnel codes used for each CC line is provided in Table S2.
QTL mapping
QTL mapping was performed with the R package DOQTL, v1.6.0 (Gatti et al. 2014). DOQTL performs QTL mapping by regressing the phenotypes on the founder haplotype probabilities estimated with our HMM. Average testes size, body weight, and age were conditionally included as covariates for the reproductive traits. QTL significance intervals were defined by the 95% Bayesian credible interval, calculated by normalizing the area under the QTL curve (Sen and Churchill 2001). Log of the odds ratio (LOD) was the reported mapping statistic. The significance thresholds for QTL were calculated independently for autosomes and chromosome X using 1000 permutations. Significant QTL were determined using a genome-wide P-value of 0.05 and suggestive QTL were determined using a P-value of 0.1. The founder allelic effect was identified using a regression of the phenotype on the genotype probabilities at each locus.
Subspecific origin
We determined the subspecific origin of the haplotypes that drive the QTL using the Mouse Phylogeny Viewer (http://msub.csbio.unc.edu/) (Wang et al. 2012). The subspecific origin for all eight CC founders has been reported previously (Yang et al. 2011). In the case that the LOD interval contains > 1 subspecific origin, these are listed in decreasing order of contribution with the specific origin at the QTL peak underlined.
Data availability
Table S3 contains QTL mapping phenotypes for the 347 mice that were phenotyped and genotyped. Table S4 contains all raw phenotype data for these mice. Testis histology images and annotations for all samples can be found at http://database.csbio.unc.edu/Infertility. Marker information can be found in Table S5. MDA genotyping data can be found at https://doi.org/10.5281/zenodo.400269. The genotypes of extant strains (including most recent common ancestors and sequenced males) are available to the public linked through the NIH-funded Mutant Mouse Resource and Research Center at (https://www.med.unc.edu/mmrrc/genotypes), (csbio.unc.edu/CCstatus/CCGenomes), and (https://doi.org/10.5281/zenodo.377036).
Results
Over 95% of CC funnels initiated at ORNL are extinct
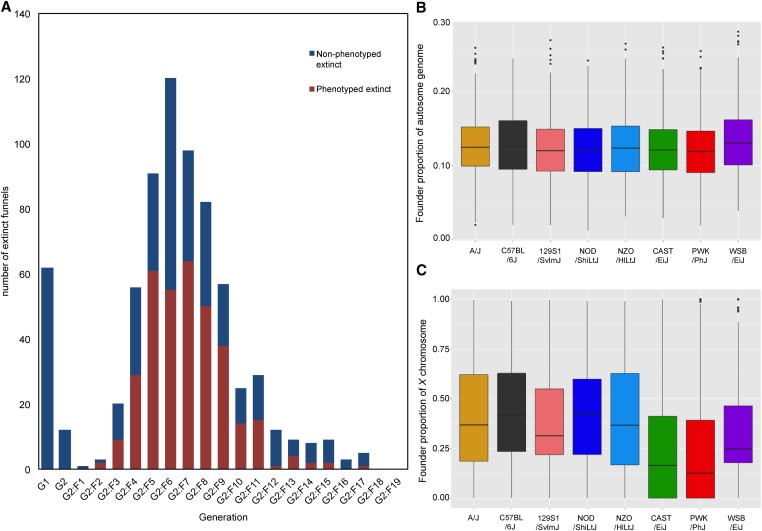
Since the start of the CC project in 2004 at ORNL, 707 out of 738 funnels (95%) have been declared extinct. Extinction follows a bimodal distribution and ranges from G1 to G2:F17 (Figure 1A). Males from 347 funnels that became extinct from 2008 to 2011 were included in this study. These males range from generations G2:F2 to G2:F17, with the bulk of the extinction (86%) taking place between G2:F4 and G2:F9 (Figure 1A).
Figure 1.
CC extinct funnels from ORNL. (A) Distribution of funnel extinction as a function of generation. Extinct phenotyped lines are displayed in red and extinct nonphenotyped lines in blue. (B) Boxplots of allele autosomal frequency for CC phenotyped extinct funnels (C) Boxplots of X chromosome allele frequency for CC phenotyped extinct lines. CC, Collaborative Cross; ORNL, Oak Ridge National Laboratory.
CC extinct lines have balanced founder contributions in the autosomes but not in chromosome X
Inspection of the haplotypes identified six extinct lines in which one or more of the eight founder strains were absent and, therefore, were excluded from the analysis presented in this section. We measured the frequency of the founder haplotypes in the remaining 341 lines that inherited all eight founder haplotypes (Figure 1, B and C, Figure S2, and Table 1). All eight founder strains contribute similarly to the autosomes of extinct lines (Figure 1B) with the average founder haplotype frequencies ranging from 0.1203 from the PWK/PhJ haplotype to 0.1332 from the WSB/EiJ haplotype (Figure 1B and Table 1). The number and width of haplotype blocks is similar to the range previously reported in a cohort of CC lines (Figure S3, A and B). We conclude that extinction is not associated with global selection in favor or against any particular founder strain.
Table 1. Proportion of the genome from each of the eight founder haplotypes in extinct phenotyped and living CC lines.
| Founder Haplotype | CC Extinct Autosomes | CC Living Autosomes | CC Extinct X Chromosome | CC Living X Chromosome |
|---|---|---|---|---|
| A/J | 0.1260 | 0.1226 | 0.1240 | 0.1057 |
| C57BL/6J | 0.1290 | 0.1349 | 0.1332 | 0.1668 |
| 129S1/SvImJ | 0.1229 | 0.1423 | 0.1355 | 0.2041 |
| NOD/ShiLtJ | 0.1220 | 0.1449 | 0.1333 | 0.194 |
| NZO/HlLtJ | 0.1245 | 0.1447 | 0.1454 | 0.1253 |
| CAST/EiJ | 0.1221 | 0.0945 | 0.0961 | 0.0465 |
| PWK/PhJ | 0.1203 | 0.0863 | 0.1174 | 0.0477 |
| WSB/EiJ | 0.1332 | 0.1318 | 0.1150 | 0.11 |
CC, Collaborative Cross.
We measured founder haplotype frequency in chromosome X independently from the autosomes because of the constraints imposed by funnel code (Materials and Methods) (Figure 1C, Figure S2, and Table 1). Allele frequencies of the extinct CC lines in chromosome X are more variable than in the autosomes, ranging from 0.1454 for NZO/HlLtJ to 0.096 for CAST/EiJ. There is an apparent deficit in the contributions of the wild-derived strains, which is not driven by a single locus, but rather is chromosome-wide (Figure S2 and Table 1). These features strongly suggest the presence of negative selection for X-linked variation.
Uniform distribution and unbiased founder contribution to heterozygosity characterized extinct CC lines
Heterozygosity varies widely in the CC extinct mice, ranging from 0.76 to 0.03. This is expected as the level of inbreeding increases with the number of generations. In fact, there is a very good fit between the observed and predicted (Welsh and McMillian 2012) levels of heterozygosity between G2:F2 and G2:F7 (Figure S3C). In later generations, there is apparent excess of heterozygosity among extinct lines. The level of heterozygosity is almost uniform along the autosomes and the contribution from each of the founder strains is unbiased (Figure S3D). Finally, there is no evidence for bias in pairwise contributions to heterozygosity (Figure S3E). We conclude that extinction is not associated with strong selection in favor of or against heterozygosity at one or a few loci.
Males are infertile in nearly half of the extinct CC lines
In the CC extinct population, the mice varied in age (179–862 days old), but only six mice were > 2 years of age. Once a CC line was declared extinct, the remaining male mice from this funnel were made available for phenotyping. These CC male mice were first tested for fertility with unrelated outbred females. A male mouse was designated “genetically fertile” if it was able to produce a litter with either a related female from the same funnel or an outbred female, and “genetically infertile” if it was unable to produce offspring in any mating pairs (Table 2). While this binary definition of fertility is unable to distinguish between highly productive and marginally productive males, it allowed us to rapidly categorize reproductive potential across a wide range of ages. Of the 347 mice measured, 164 (47%) were genetically infertile, while 183 (53%) were genetically fertile (Table 2). This indicates that a large number of the CC lines went extinct due to male infertility. For the 183 males that were fertile, 99 (54%) were only productive with an outbred female, indicating that female infertility and genetic incompatibility could play a role in CC extinction.
Table 2. Fertility testing to determine fertility status in CC extinct phenotyped lines.
| Mated | Fertility Status | ||||
|---|---|---|---|---|---|
| Initially with Females from Same Lines | Later with Unrelated Outbred Females | Funnel Female | Unrelated Female | Number of Lines | |
| + | + | + | + | 28 | 183 Fertile |
| + | + | − | + | 99 | |
| + | + | + | − | 21 | |
| + | − | + | na | 22 | |
| − | + | na | + | 13 | |
| + | + | − | − | 149 | 164 Infertile |
| − | + | na | − | 15 | |
| + | − | − | na | 0 | |
na, not applicable.
We assessed a large number of male reproductive traits in the CC extinct lines, including reproductive organ weights, sperm counts, sperm quality characteristics, and testis histology. Table S4 lists the traits measured in representative males from 347 genotyped lines. Overall, the CC extinct fertile and infertile males have statistically similar means and distributions for the majority of the reproductive traits measured (Table S6). When fertile and infertile males are significantly different, the infertile males tend to have phenotype values associated with reduced fitness. This is illustrated in Figure 2 and Figure S4, which display distributions for each trait compared to the distributions of the same trait in the CC founder strains (Odet et al. 2015). The CC extinct lines also tend to have means that are associated with lower reproductive fitness compared to the founders (Figure 2 and Figure S4). Both the CC extinct infertile and fertile males have a wider phenotypic range than the founders, which is expected because the CC are genetic mosaics of the founders (Threadgill et al. 2011).
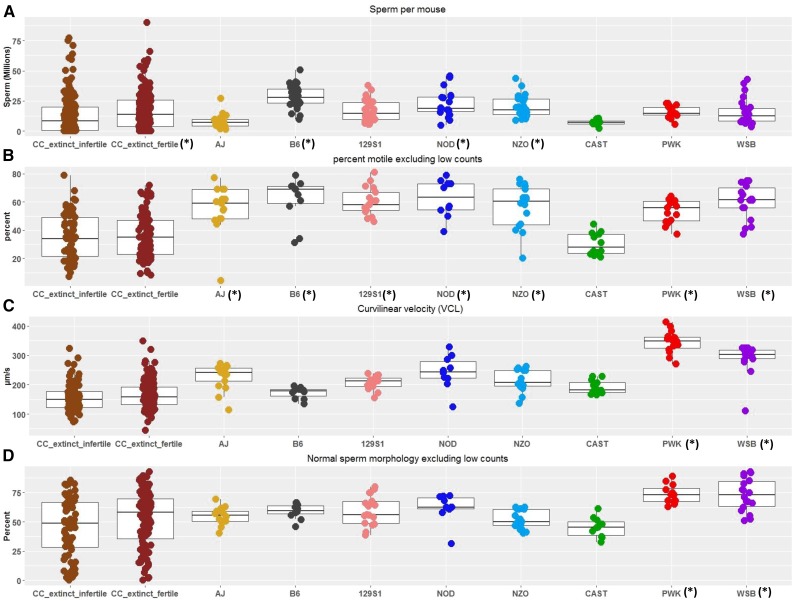
Figure 2.
Selected reproductive traits in the Collaborative Cross (CC) extinct population and the CC founders. Symbols beside strain labels (*) reflect simplified categorical significance differences using a Student’s t-test. (A) Mean sperm count per mouse (millions of sperm). (B) Percent of sperm that were motile. (C) Mean value for curvilinear velocity (VCL, µm/sec). (D) Percent of sperm with normal morphology.
CC extinct lines show large variation in reproductive organ weights
Wider phenotypic range for males from the extinct lines was apparent in the distributions of testis weight, epididymis + vas deferens weight, and seminal vesicle weight (Figure S4) (Levene’s test, P-value < 0.0001). Sixty-seven mice (19.3%) in the CC extinct population had small testes, with a mean size < 0.04 g, and 64 of those 67 mice had testes with extensive germ cell loss. Three males had one very large testis and were excluded from the comparison of means. Histological analysis identified one testis with expanded seminiferous tubules (OR1400m135), as well as a large tumor (OR5253m147) and a large cyst (OR1626m161) that filled most of each testis. Enlarged seminal vesicles, which have been associated with aging in mice (Finch and Girgis 1974; Yamate et al. 1990), were also observed. These features were absent in the previous reproductive phenotyping study in the CC founders. Body weight was highly positively correlated with testis, epididymis + vas deferens weight, and seminal vesicle weight (Table S7). Seminal vesicle weight was correlated with age, but other reproductive organ weights were not.
Many CC extinct lines have low sperm counts
Mean sperm counts in CC extinct infertile males (13.4 × 106 sperm/mouse) were significantly lower than counts for CC extinct fertile males (16.7 × 106 sperm/mouse) and C57BL/6J, NOD/ShiLtJ, and NZO/HlLtJ males (F Ratio = 5.4218, P-value < 0.0001) (Figure 2A). The distributions of sperm counts in the CC extinct population and in the founders were dramatically different (Figure 2A). Sperm counts have a positively skewed distribution with a long tail in the CC extinct mice. Ninety mice (26%) in our study population had sperm counts ≤ 1 × 106/mouse, including 41 mice with no sperm recovered and 33 with < 0.5 × 106 sperm/mouse. These low counts likely contribute to lowered reproductive fitness. Sperm counts are negatively correlated with age (r = −0.1328, P-value = 0.0143) (Table S7) and positively correlated with testis weight and other reproductive traits measured.
CC extinct lines have low sperm motility
CASA was used to measure several quantitative parameters of motility immediately after sperm were isolated from the cauda epididymis. CASA tracks were recorded for 1–1761 motile sperm/mouse. We restricted our analysis to individuals with at least 40 motile sperm to mitigate artifacts associated with small sample size. This removed 58 samples, leaving 151 for the analysis (Table S3). The distribution of the percentage of motile sperm is wider in the extinct CC lines than in the eight founder strains and the mean percentage is significantly lower in CC extinct lines compared to the majority of the founder strains (F Ratio = 27.0528, P-value < 0.0001). The exception is in CAST/EiJ mice, where the mean percentage of motile sperm was 30% (Figure 2B). As in the CC founders (Odet et al. 2015), age was not significantly correlated with the percentage of motile sperm in CC extinct mice. We measured other kinematic parameters of sperm motility, including VAP (µm/sec), VSL (µm/sec), VCL (µm/sec), ALH (µm), and BCF (Hz) (Figure S4). Mean VCL values for the CC extinct mice were lower than the means for the eight founder strains (F Ratio = 42.7656, P-value < 0.0001) (Figure 2C and Table S6).
We also assessed sperm motility in the extinct lines using CASAnova, a support vector machine program that identifies sperm motility patterns (Goodson et al. 2011). This automated program classifies each CASA sperm track into one of five motility categories: progressive, intermediate, hyperactivated, slow, or weakly motile. We again included only samples that had ≥ 40 motile sperm. For progressive motility, the extinct lines had lower means compared to the eight founders (Figure S4). Hyperactivated motility has a wider distribution for extinct lines (Levene’s test, P-value < 0.0001), although the average percent hyperactivated is close to the average found in the majority of the founder strains. Age of the mouse was not correlated with any motility category, which was previously observed (Odet et al. 2015).
Sperm morphology varies widely among CC extinct lines
Sperm morphology was measured on 205 males from extinct lines. The distribution of sperm assessed ranged from 1 to 395 sperm per mouse. We included a cutoff that required a minimum of 40 sperm classified for morphological features. This removed 36 samples, leaving 169 for the analysis (Table S3). Sperm morphology was scored in four categories: percent of sperm with normal morphology (Figure 2D), percent with abnormal head shape, percent with abnormal tail bending, and percent with broken tails (Figure S4). For all categories, we see wide distributions in both the fertile and infertile CC extinct mice compared to the eight founder strains. For the percent of sperm with normal morphology, the means for both CC extinct fertile and infertile (50.1 and 45.5%, respectively) are significantly lower than two founder strains, PWK/PhJ and WSB/EiJ (73.7 and 72.9%, respectively) (F Ratio = 4.4363, P-value < 0.0001). Age of the mouse was not significantly correlated with any of the morphology traits. In the CC founders, age was correlated with abnormal head shape, abnormal tail bending, and broken tails, but not normal morphology (Odet et al. 2015).
Vacuolization of seminiferous tubules is prevalent in infertile males
To evaluate testis histology, we generated a composite image of a complete transverse testis section for each mouse (http://database.csbio.unc.edu/Infertility). The number and mean radius of the seminiferous tubules were determined as previously described (Odet et al. 2015). Figure S5 provides an example of the tools used for annotation. Distribution of the tubule number and the length of the seminiferous epithelium in each testis section are shown in Figure S4. Both CC extinct fertile and infertile males have approximately the same mean and distribution for tubule count and seminiferous epithelium length, but the variance is greater compared to the founder strains (Table S6) (Levene’s test, P-value < 0.0001). Age was not correlated with tubule number but was significantly negatively correlated with seminiferous epithelium length (r = −0.1201, P-value = 0.0259).
Testis histology in the CC extinct lines was highly variable, ranging from normal spermatogenesis to partial or complete loss of germ cells. Additional abnormal features observed include vacuoles in the seminiferous epithelium, germ cell sloughing into the tubule lumen, abnormal germ cells, expanded rete testis, abnormal blood vessels, tumors or cysts, and accumulations of material intensely stained with periodic acid-Schiff reagent. Since vacuolization was commonly observed, we examined the frequency of tubules with vacuoles and classified each testis as either having no/few vacuoles or many vacuoles (Table S8). Compared to CC fertile males, CC infertile males are much more likely to have many vacuoles as opposed to having only a few or no vacuoles (χ2 = 5.6099, P-value = 0.017859). This level of vacuolization was rarely observed in the CC founders (Odet et al. 2015), and is strongly associated with infertility in the CC extinct population.
QTL mapping
Using MDA genotyping and a specialized HMM, we performed QTL analysis using 347 male mice representing 347 independent extinct CC lines. For each trait, the number of mice with phenotypic information ranged from 151 to 347. Of the 33 phenotypes measured, we identified significant QTL (P-value < 0.05) for seven traits, including genetic infertility, reproductive organ weights, and measures of sperm quality, and a suggestive QTL (P-value < 0.1) for testis weight (Table 3).
Table 3. QTL identified for fertility traits in males of CC extinct lines.
| Trait | Locus | Chr | Peak (Mbp) | LOD interval (Mbp) | QTL LOD score | Haplotype | Subspecies | Allele Direction |
|---|---|---|---|---|---|---|---|---|
| Genetic fertility | Fer1 | X | 101 | 55–150 | 6.191 | PWK/PhJ | mus | Decreased |
| Epididymis + vas deferens weight | Epid1 | 4 | 95 | 90–100 | 10.06 | NZO/HILtJ PWK/PhJ | dom, mus | Increased |
| mus | Decreased | |||||||
| Seminal vesicle weight | Svw1 | 4 | 117 | 90–119 | 8.486 | NZO/HILtJ PWK/PhJ | dom, mus | Increased |
| mus | Decreased | |||||||
| Seminal vesicle weight | Svw2 | X | 134 | 130–155 | 6.68 | A/J | dom | Increased |
| Testis weight | Tw1 | 1 | 170 | 3–185 | 6.728 | C57BL/6J | dom, mus | Increased |
| VCL | Vcl1 | 2 | 72 | 62–137 | 7.779 | PWK/PhJ | mus | Increased |
| A/J | dom, mus | Decreased | ||||||
| ALH | Alh1 | 6 | 91 | 78–97 | 7.93 | A/J; | dom | Increased |
| NZO/HILtJ WSB/EiJ; | dom, mus | Increased | ||||||
| C57BL/6J | dom | Decreased | ||||||
| dom | Decreased | |||||||
| Percent hyperactivated sperm | Phs1 | 14 | 19 | 3–21 | 7.652 | PWK/PhJ | mus | Increased |
| Percent broken sperm | Pbs1 | 13 | 109 | 102–112 | 8.299 | CAST/EiJ | cast | Increased |
In the case that the LOD interval contains > 1 subspecific origin, these are listed in decreasing order of contribution with the specific origin at the QTL peak underlined. Chr, chromosome; mus, M. musculus musculus; dom, M. musculus domesticus; VCL, curvilinear velocity; ALH, amplitude of lateral head displacement; cast, M. musculus castaneus.
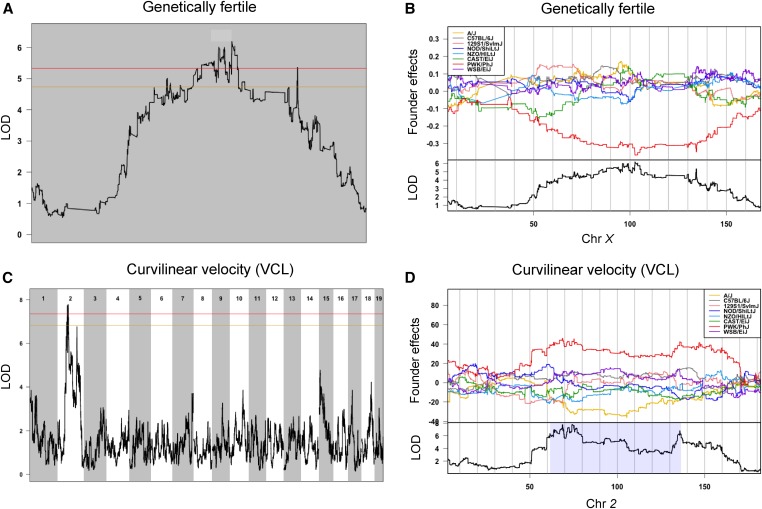
We categorized “genetic fertility” as a binary trait, with fertile males producing offspring with a related or unrelated outbred female, and infertile males producing no offspring with related and unrelated females. In the CC extinct population, males in 53% of lines were genetically fertile, while males in 47% of lines were infertile. We identified a QTL for this binary measure of genetic fertility on chromosome X (Figure 3A). The PWK/PhJ haplotype at the QTL is associated with lower fertility (Figure 3B). PWK/PhJ is the only founder strain with a M. musculus musculus haplotype at the peak.
Figure 3.
Selected QTL in CC extinct lines. Chromosomes are ordered across the x-axis with chromosome number labeled at the top. The y-axis is the LOD value. Autosomes and X chromosome were analyzed separately. The red line indicates a genome-wide LOD significance threshold at P = 0.05, the gold line indicates a genome-wide LOD significance threshold at P = 0.10. Light blue boxes indicated the 95% C.I. for QTL. (A) QTL mapping on chromosome X for genetic fertility. (B) Coefficients for CC founder allelic effects of genetic fertility in chromosome X. (C) Genome-wide scan for VCL. (D) Coefficients for CC founder allelic effects of VCL in chromosome 2. CC, Collaborative Cross; VCL, curvilinear velocity.
We identified QTL for combined testis weight, combined epididymis + vas deferens weight, and weight of seminal vesicles. For epididymis + vas deferens and seminal vesicle weights, we observe a highly significant QTL on chromosome 4 (Figure S6). For both of these traits, the NZO/HlLtJ haplotype is associated with an increase in organ weights while the PWK/PhJ haplotype is associated with lower organ weights. These traits are significantly positively correlated (r = 0.2555, P-value = < 0.0001) (Table S7). We hypothesize that this region on chromosome 4 is involved in reproductive organ development for both traits. We also mapped a significant QTL for seminal vesicle weight, but not epididymis weight, on chromosome X (Figure S6). This suggests that loci controlling seminal vesicle and epididymis weight may be shared, but may be modified differently by loci on chromosome X. For testis weight, we observe two sharp peaks in LOD score on chromosome 1 (Figure S6), with the second peak reaching a genome-wide adjusted P < 0.1.
We identified QTL for two kinetic parameters of sperm motility, VCL, and ALH. For VCL, we observe two peaks on chromosome 2, the first one exceeding the genome-wide significance threshold of P < 0.05 (Figure 3, C and D). The direction of the allelic effect for VCL, which is driven by the PWK/PhJ haplotype, is consistent with a previous report that sperm from PWK/PhJ mice were significantly faster than sperm from other CC founder strains (Odet et al. 2015). Validation of the first QTL on chromosome 2 is described in the next section. We identified a QTL for ALH on chromosome 6 (Figure S6). The allele effects at this QTL are mostly complex (Figure S6 and Table 3). Based on CASAnova analysis of motility patterns, we identified a significant QTL for the percent of hyperactivated sperm on chromosome 14 that exceeded the genome-wide threshold level of P < 0.05 (Figure S6). This hyperactivation trait is driven by the PWK/PhJ haplotype and is consistent with its founder effect (Figure S4).
Finally, we identified a QTL for the percentage of broken tails, a sperm morphology trait. This QTL on chromosome 13 exceeded the significance threshold of P < 0.05 (Figure S6). The CAST/EiJ haplotype has the largest allelic effect, associated with an increase of broken tails (Figure S6).
QTL validation
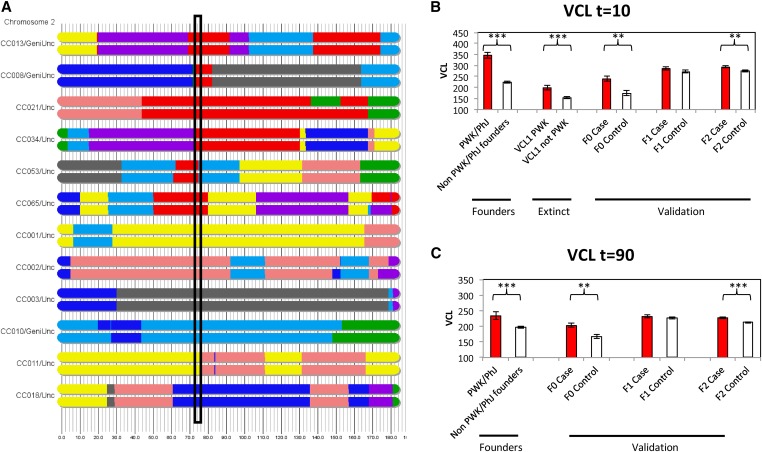
To validate the QTL for VCL on chromosome 2 (Vcl1), we conducted an experiment across three generations of mice in two populations designed to test the effect of the PWK/PhJ allele at the QTL peak. The initial F0 generation consisted of 12 CC living strains (six for the case population and six for the control population, see Materials and Methods), the F1 generation were crosses of the 12 F0 CC strains, and the F2 generation was an outbred population derived from four-way crosses of F1 hybrid mice. The case population is homozygous for the PWK/PhJ allele at Vcl1 (73–75 Mb) while this allele is absent in the control population (Figure 4A). We also verified that there is no long-range linkage disequilibrium between Vcl1 and the rest of the genome (Figure S7). To further verify the effect of Vcl1, we created F1 and F2 experimental populations of cases and controls. The number of mice for each population and generation is provided in Table S1 and phenotypes are provided in Table S9.
Figure 4.
The validation of Vcl1 on chromosome 2. “Case” and “Control” refer to cohorts that contained the PWK/PhJ allele or an alternative allele, in the region denoted by the box (73–75 Mb). (A) The haplotype visualization of chromosome 2 for the six CC strains to generate the cases and the six CC strains to generate the controls. (B) VCL (μm/sec) at 10 min and (C) VCL at 90 min. We measured across CC founders, extinct CC lines, F0 living CC strains, F1 crosses of living CC strains, and F2 crosses of living CC strains. *** P < 0.001, ** P < 0.01, * P < 0.05. CC, Collaborative Cross; VCL, curvilinear velocity.
We measured VCL at t = 0 and t = 90 for the F0, F1, and F2 case and control males (Figure 4, B and C). At both time points, VCL was significantly higher for the cases in the F0 (t = 10, t-ratio = 3.06, P < t = 0.0021; t = 90, t-ratio = 2.95, P < t = 0.0027) and F2 crosses (t = 10, t-ratio = 2.81, P < t = 0.0027; t = 90, t-ratio = 4.17, P < t <0.0001), reproducing the PWK/PhJ haplotype effect in the CC founders and CC extinct populations. Although VCL is not significantly different between case and control F1 hybrid males, the direction of the effect is consistent with our predictions. Failure to reach significance may be due to smaller sample size (t = 10, t-ratio = 1.30, P < t = 0.09; t = 90, t-ratio = 0.69, P < t = 0.24).
Discussion
High levels of line extinction occurred during the inbreeding of the CC that surpassed levels seen in comparable MPP and standard mouse recombinant inbred lines. At the time of this publication, 95% of funnels initiated at ORNL have become extinct, compared to < 50% extinction observed in a recent expansion of the BXD panel (Peirce et al. 2004; R. W. Williams, personal communication). Early reports suggested that genomic incompatibilities and infertility were the likely causes of this high level of extinction (Chesler et al. 2008; Collaborative Cross Consortium 2012). We identified a high frequency of male infertility in the CC extinct lines, where nearly 50% of males were unable to produce offspring with either related or unrelated outbred females. Using males from CC extinct lines, we performed the largest mapping study for reproductive traits in the CC, and identified several genetic associations with sperm morphology, sperm motility, and reproductive organ weights. Infertility was associated with a large region on chromosome X known to contain loci previously associated with hybrid incompatibility, sterility, and speciation (Payseur et al. 2004; Storchova et al. 2004; Good et al. 2008, 2010; Mihola et al. 2009; Wang et al. 2015; Balcova et al. 2016). QTL allelic effects for most reproductive traits were driven by PWK/PhJ or CAST/EiJ haplotypes (Table 3). This finding is significant because these two strains are derived from M. musculus musculus and M. musculus castaneus, while the other six founder strains have mostly a M. musculus domesticus origin. Overall, the results from several experiments suggest that poor fertility traits and hybrid incompatibilities involving the CAST/EiJ and PWK/PhJ haplotypes contribute to breeding difficulties and strain extinction in the CC. Our experiment allows us to pinpoint the genomic regions and founder haplotypes that are associated with extinction and poor fertility in the CC. This information can be used to improve colony management in the CC.
The pattern of overall extinction at ORNL has a bimodal distribution, with the first peak occurring at the G1 generation and the second peak occurring between the G2:F4 to G2:F9 generation (Figure 1A). The lines phenotyped in this study do not capture the entire pattern of extinction at ORNL because they miss the first peak, as those funnels became extinct before the collection start date for this study. Funnels classified as extinct in G1 and G2 represent cases of hybrid sterility affecting males (Mihola et al. 2009) and outcrosses that were fully unproductive (Chesler et al. 2008). With the exception of the first peak, our experimental cohort is a good representation of the overall extinction at ORNL.
Interestingly, the observed frequency of founder haplotypes in the autosomes of the extinct lines is close to the expected frequency of 12.5% (Figure 1, Figure S2, and Table 1). This observation is consistent with those reported for G2:F1 (Liu et al. 2014) and multiple reports in CC lines before reaching the 90% inbreeding status (Aylor et al. 2011; Collaborative Cross Consortium 2012; Ferris et al. 2013; Gralinski et al. 2015). We conclude that extinction is not associated with grossly distorted haplotype frequencies. However, there is a striking difference between the global haplotype frequency of the CC extinct lines and CC living strains. CC living strains have significantly lower than expected genetic contribution from CAST/EiJ and PWK/PhJ founders in the autosomes (Srivastava et al. 2017). For chromosome X, both extinct CC lines and living CC strains have a deficit in the contribution of CAST/EiJ and PWK/PhJ haplotypes (Figure 1B, Figure S2, and Table 1), which cannot be explained by the funnel code (i.e., differential contribution to the X chromosome from founders depending on their position in the funnel) (Table S2). It should be also noted that neither the presence of distortion nor the differences between extinct lines and living CC strains can be ascribed to funnels with < 8 founders (Collaborative Cross Consortium 2012) as they were excluded in this comparison. Taken together, these observations suggest that there has been ongoing selection against X-linked variation from CAST/EiJ and PWK/PhJ starting in the earliest stages of the funnels. In contrast, selection in the autosomes against haplotypes from non-domesticus origin arose at generations when most of the genome was becoming homozygous in individual CC mice. Lines that overcame this bottleneck had either lower contribution of CAST/EiJ and PWK/PhJ genome-wide by chance, or were able to purge these alleles before the line became inbred. Selection in the autosomes was not restricted to a few loci with major effects because the only consistent transmission ration distortion (TRD) observed was the overrepresentation of WSB/EiJ on chromosome 2, now known to be due to responder to meiotic drive 2 (R2d2) (Didion et al. 2015, 2016; Morgan et al. 2016b). TRD at R2d2 was present in the DO and drove an almost complete selfish sweep without an increase in infertility (Chesler et al. 2016). In addition, homozygosity for the WSB haplotype for females precludes meiotic drive and is not associated with reduced litter size (Didion et al. 2016), and meiotic drive does not operate through males (Didion et al. 2015). Thus, R2d2 is unlikely to be associated with extinction in the CC. We conclude that selection acting on the autosomes was qualitatively different from the one acting on chromosome X, likely due to different constraints imposed by selection (Haldane 1922; Orr and Turelli 1996; Johnson and Lachance 2012). Based on the presence of similar high levels of extinction, the same patterns of global distortion among living CC strains, and similar patterns of distortion at R2d2 irrespective of the site of origin, we hypothesize that our conclusions on extinction in the ORNL population extend to the other two CC populations created at Tel Aviv University and Geniad in Western Australia.
Male infertility was a major feature of the CC extinct lines, affecting males from nearly half of the 347 lines tested. Although the classification of males as infertile if they fail to produce offspring with both related and unrelated females is straightforward, age is an important factor to consider. If a male was able to successfully breed with related females, it was much more likely to be retained for a longer period of time at ORNL. Therefore, these males tended to be older when they arrived at UNC. This complicates the analysis of reproductive traits that vary with age (i.e., sperm counts and testis histology). In addition, this can also explain why males were fertile with a related female but not with an unrelated female (Table 2). Nevertheless, these mice were still considered genetically fertile because they produced offspring. This factor was accounted for in the genetic mapping experiments by using age as a covariate when appropriate.
We measured 33 different male reproductive traits (Figure 2 and Figure S4). The common feature among many of the measured traits is that the CC extinct lines display wider variation compared to the founders, which is expected because they are mosaics of the eight founders (Threadgill et al. 2011). Traits related to fitness are expected to be under strong directional selection, as selection tends to only favor the direction that leads to increased fitness. This is what makes the CC extinct population so dynamic and unique in this regard. The genetic diversity between the eight founder strains of the CC has created an abundance of allelic combinations that have not occurred in any existing inbred mouse strains (Rogala et al. 2014). These new allelic arrangements produce CC strains that are more reproductively fit than the founder strains, as well as strains that are reproductively unfit and contain abnormal phenotypes. This is an important distinction between the living and extinct CC populations, and makes the CC extinct population superior for reproductive trait mapping because it contains more variation for reproductive ability. However, the drawback is that we cannot go back and add new experiments to the CC extinct population.
We identified several genomic regions associated with eight fertility-related traits (Table 3). Genetic fertility was defined as a male’s ability to produce offspring with either a related or unrelated female. We observed a large region on the X chromosome that was significantly associated with infertility, and was driven by the PWK/PhJ haplotype (Figure 3). Individuals in the CC extinct population that had a PWK/PhJ haplotype on the X chromosome were much more likely to be infertile. PWK/PhJ is the only strain from the M. musculus subspecies and it is likely that incompatibilities arise from crossing together mice from different subspecific origins. The X chromosome is particularly enriched in loci previously identified in hybrid incompatibility, sterility, and speciation (Payseur et al. 2004; Storchova et al. 2004; Good et al. 2008, 2010; Mihola et al. 2009; Wang et al. 2015; Balcova et al. 2016), so new combinations of loci may drive deleterious effects in males.
For seminal vesicles and epididymis weight, we identified novel overlapping QTL on chromosome 4, which has similar allelic effects from the same founder strains. Previous research on the weight of seminal vesicles using 13 inbred strains identified a QTL at a different location on chromosome 4 (Le Roy et al. 2001). We also identified a novel QTL for testis weight on chromosome 1 (Figure S6). Previous QTL mapping studies in other populations identified regions on chromosomes 6, 11, and X that were associated with testis size (Oka et al. 2004; L’Hote et al. 2007; Good et al. 2008). These results demonstrate that the CC contains unique genetic variation that can be used to identify loci associated with variation in reproductive organs.
Defects in sperm motility are common in infertile human males and have been identified in > 40 mouse knockout models (Matzuk and Lamb 2008; Hwang et al. 2011). In a previous study on founder strains, it was reported that the CAST/EiJ strain contained high levels of abnormal sperm with poor motility (Odet et al. 2015). We identified a QTL for broken sperm tail morphology on chromosome 13 that is driven by the CAST/EiJ haplotype. This association may contribute to the previous observation of poor sperm motility in CAST/EiJ mice. We also identified QTL for VCL (Vcl1) and ALH, two quantitative parameters of sperm motility determined by CASA. Prior to our study, a QTL for BCF on chromosome 7 (Golas et al. 2004) was the only QTL that has been identified for any of the CASA motility parameters.
High VCL values are characteristic of progressive motility, which predominates immediately after sperm are released from the epididymis, and hyperactivated motility, which develops in the female reproductive tract and is required for fertility (Turner 2006; Goodson et al. 2011). We identified a significant QTL on chromosome 14 for the percentage of hyperactivated sperm just after isolation from the epididymis. This region of chromosome 14 is enriched with genes associated with sperm development (Moretti et al. 2016). Although intermediate and hyperactivated patterns of motility are typically low at initial time points, both are elevated at time 0 in PWK/PhJ mice, perhaps related to the high VCL values seen in this strain (Odet et al. 2015). Alternatively, premature hyperactivation has been reported as a motility defect in mice with t haplotypes (Olds-Clarke and Johnson 1993), suggesting that this QTL for hyperactivity could have negative effects on fertility by interfering with the movement of sperm to the site of fertilization.
We performed validation testing for Vcl1, the QTL identified for VCL. Validation testing of a QTL is independent from replication, as it can offer new biological understanding of the trait. Validation testing can confirm the location of a region associated with a trait and the direction of the trait, demonstrate that the region is not an artifact of a particular population, and possibly narrow the region into a smaller interval. We chose to validate Vcl1 instead of other traits for several reasons. First, the allele effects under the QTL are clearly driven by the PWK/PhJ haplotype. This is consistent with VCL being significantly higher in PWK/PhJ than the other seven founders at two time points (Figure 4, B and C). There were also enough living CC strains with and without the PWK/PhJ haplotype at Vcl1 that were devoid of long-range linkage disequilibrium to validate the initial observation. Figure 4 shows the consistent pattern of higher VCL values for mice with a PWK/PhJ haplotype at Vcl1 in the CC founders, CC extinct lines, and validation populations. This experiment provides the first successful example of cross validation using complementary CC resources. Cross validation can be difficult because differences in allele frequency between the two populations, epistatic interactions, and the “Bevis effect” can prevent the replication of a QTL effect (Shorter et al. 2015; King and Long et al. 2017; Najarro et al. 2017); however, our carefully designed approach with clear and consistent haplotype effects tested over multiple generations overcame those previously known limitations. Analysis of whole-genome sequences of the eight CC founders (Keane et al. 2011) identified 708 high-quality SNP variants within 26 genes that are unique to the PWK/PhJ haplotype in the 73–75 Mb window under the peak of the QTL (Table S10). This is consistent with the hypothesis that there is a PWK/PhJ exclusive genomic feature at Vcl1 associated with increased VCL activity.
The results from this study on CC extinct males have several implications for mouse and non-mouse MPP efforts. The inclusion of three subspecies was motivated by the desire to increase genetic diversity in the CC. The five classical founder strains in the CC, as well as the majority of the classical mouse strains commonly used in research, are derived from the same few fancy mice and have limited and nonrandom haplotype diversity across the genome (Yang et al. 2011). An MPP using only these strains would have lower overall genetic diversity and regions of identity by descent shared by all founder strains would leave “blind spots” covering a substantial fraction of the genome, reducing its value as a mapping resource. A future design that contains several unrelated or wild-derived inbred strains from the same subspecies would likely contain fewer incompatibilities and still maintain a high level of genetic diversity. Reproductive performance of the founder strains is another factor essential to a successful MPP. Many of the CC founders have combinations of poor sperm morphology, motility, and production traits (Odet et al. 2015). Future MPP efforts should select inbred strains with the best reproductive potential through extensive reproductive phenotyping.
The systematic archiving of samples from each funnel, coupled with deep reproductive phenotyping and genotyping, provides a unique opportunity to study the genetic underpinning of fertility and genetic incompatibility. The CC extinct resource contains unique allele combinations that are absent in any other live laboratory mouse stocks. Although extinction endangered the CC project, the living CC inbred strains provide a genetic reference population with unique and powerful characteristics and associated resources (Morgan and Welsh 2015; Oreper et al. 2017; Srivastava et al. 2017; Tyler et al. 2017).
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.199596/-/DC1.
Acknowledgments
We would like to thank Lisa Branstetter, T. Justin Gooch, Stephanie Hansen, Mark Calaway, Jennifer Shockley, and Jason Spence for mouse work. We wish to note that the scope of the work presented here required substantial efforts from a diversity of very skilled junior scientists and that just authorship order cannot capture these contributions properly. This work was supported by National Institutes of Health (NIH) grant R01HD065024 to D.A.O. and F.P.-M.d.V. from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. Additional support for personnel and resources was provided by NIH grant U19AI100625 to F.P.-M.d.V. from the National Institute of Allergy and Infectious Diseases.
Footnotes
Communicating editor: K. M. Nichols
Literature Cited
- Aylor D. L., Valdar W., Foulds-Mathes W., Buus R. J., Verdugo R. A., et al. , 2011. Genetic analysis of complex traits in the emerging collaborative cross. Genome Res. 21: 1213–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcova M., Faltusova B., Gergelits V., Bhattacharyya T., Mihola O., et al. , 2016. Hybrid sterility locus on chromosome X controls meiotic recombination rate in mouse. PLoS Genet. 12(4): e1005906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet S., Olatoye M. O., Marla S. R., Perumal R., Tesso T., et al. , 2017. Increased power to dissect adaptive traits in global sorghum diversity using a nested association mapping population. Genetics 206: 573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmel F., Rolland A. D., Niederhauser-Wiederkehr C., Chung S. S. W., Demougin P., et al. , 2007. The conserved transcriptome in human and rodent male gametogenesis. Proc. Natl. Acad. Sci. USA 104: 8346–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler E. J., 2014. Out of the bottleneck: the diversity outcross and collaborative cross mouse populations in behavioral genetics research. Mamm. Genome 25: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler E. J., Miller D. R., Branstetter L. R., Galloway L. D., Jackson B. L., et al. , 2008. The collaborative cross at oak ridge national laboratory: developing a powerful resource for systems genetics. Mamm. Genome 19: 382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler E. J., Gatti D. M., Morgan A. P., Strobel M., Trepanier L., et al. , 2016. Diversity outbred mice at 21: maintaining allelic variation in the face of selection. G3 (Bethesda) 6: 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Airey D. C., Allayee H., Angel J. M., Attie A. D., et al. , 2004. The collaborative cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137. [DOI] [PubMed] [Google Scholar]
- Collaborative Cross Consortium , 2012. The genome architecture of the collaborative cross mouse genetic reference population. Genetics 190: 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos F. A., Molinet J., Brice C., Tisné S., Abarca V., Tapia S. M., et al. , 2017. Identification of nitrogen consumption genetic variants in yeast through QTL mapping and bulk segregant RNA-seq analyses. G3 (Bethesda) 7: 1693–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didion J. P., Yang H. N., Sheppard K., Fu C. P., McMillan L., et al. , 2012. Discovery of novel variants in genotyping arrays improves genotype retention and reduces ascertainment bias. BMC Genomics 13: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didion J. P., Morgan A. P., Clayshulte A. M. F., McMullan R. C., Yadgary L., et al. , 2015. A multi-megabase copy number gain causes maternal transmission ratio distortion on mouse Chromosome 2. PLoS Genet. 11(2): e1004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didion J. P., Morgan A. P., Yadgary L., Bell T. A., McMullan R. C., et al. , 2016. R2d2 drives selfish sweeps in the house mouse. Mol. Biol. Evol. 33: 1381–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris M. T., Aylor D. L., Bottomly D., Whitmore A. C., Aicher L. D., et al. , 2013. Modeling host genetic regulation of influenza pathogenesis in the Collaborative Cross. PLoS Pathog. 9: e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch C. E., Girgis F. G., 1974. Enlarged seminal-vesicles of senescent C57BL/6J mice. J. Gerontol. 29: 134–138. [DOI] [PubMed] [Google Scholar]
- Gatti D. M., Svenson K. L., Shabalin A., Wu L. Y., Valdar W., et al. , 2014. Quantitative trait locus mapping methods for diversity outbred mice. G3 (Bethesda) 4: 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golas A., Grzmil P., Muller C., Styrna J., 2004. Chromosome 7q11 controls sperm beat cross frequency (BCF) in mice. Folia Biol. (Krakow) 52: 211–217. [DOI] [PubMed] [Google Scholar]
- Good J. M., Dean M. D., Nachman M. W., 2008. A complex genetic basis to X-linked hybrid male sterility between two species of house mice. Genetics 179: 2213–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good J. M., Giger T., Dean M. D., Nachman M. W., 2010. Widespread over-expression of the X chromosome in sterile f-1 hybrid mice. PLoS Genet. 6(9): e1001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson S. G., Zhang Z., Tsuruta J. K., Wang W., O’Brien D. A., 2011. Classification of mouse sperm motility patterns using an automated multiclass support vector machines model. Biol. Reprod. 84: 1207–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L. E., Ferris M. T., Aylor D. L., Whitmore A. C., Green R., et al. , 2015. Genome wide identification of SARS-CoV susceptibility loci using the collaborative cross. PLoS Genet. 11(10): e1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L. E., Menachery V. D., Morgan A. P., Totura A., Beall A., et al. , 2017. Allelic variation in the toll-like receptor adaptor protein ticam2 contributes to SARS-coronavirus pathogenesis in mice. G3 (Bethesda) 7: 1653–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R., Wilkins C., Thomas S., Sekine A., Hendrick D. M., et al. , 2017. Oas1b-dependent immune transcriptional profiles of West Nile virus infection in the collaborative cross. G3 (Bethesda) 7: 1665–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J. B. S., 1922. Sex ratio and unisexual sterility in hybrid animals. J. Genet. 12: 101–109. [Google Scholar]
- Handel M. A., Lessard C., Reinholdt L., Schimenti J., Eppig J. J., 2006. Mutagenesis as an unbiased approach to identify novel contraceptive targets. Mol. Cell. Endocrinol. 250: 201–205. [DOI] [PubMed] [Google Scholar]
- Huang X., Paulo M. J., Boer M., Effgen S., Keizer P., et al. , 2011. Analysis of natural allelic variation in Arabidopsis using a multiparent recombinant inbred line population. Proc. Natl. Acad. Sci. USA 108: 4488–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang K., Walters R. C., Lipshultz L. I., 2011. Contemporary concepts in the evaluation and management of male infertility. Nat. Rev. Urol. 8: 86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N. A., Lachance J., 2012. The genetics of sex chromosomes: evolution and implications for hybrid incompatibility. Ann. N. Y. Acad. Sci. 1256: E1–E22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane T. M., Goodstadt L., Danecek P., White M. A., Wong K., et al. , 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy C. L., O’Bryan M. K., 2006. N-ethyl-N-nitrosourea (ENU) mutagenesis and male fertility research. Hum. Reprod. Update 12: 293–301. [DOI] [PubMed] [Google Scholar]
- King E. G., Long A. D., 2017. The Beavis effect in next-generation mapping panels in Drosophila melanogaster. G3 (Bethesda) 7: 1643–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Macdonald S. J., Long A. D., 2012. Properties and power of the Drosophila synthetic population resource for the routine dissection of complex traits. Genetics 191: 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M., et al. , 2009. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5(7): e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roy I., Tordjman S., Migliore-Samour D., Degrelle H., Roubertoux P. L., 2001. Genetic architecture of testis and seminal vesicle weights in mice. Genetics 158: 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.-F., Tam Y.-T., Zuo Y., Cheong A. W. Y., Pang R. T. K., et al. , 2008. Characterization of an acrosome protein VAD1.2/AEP2 which is differentially expressed in spermatogenesis. Mol. Hum. Reprod. 14: 465–474. [DOI] [PubMed] [Google Scholar]
- L’Hote D., Serres C., Laissue P., Oulmouden A., Rogel-Gaillard C., et al. , 2007. Centimorgan-range one-step mapping of fertility traits using interspecific recombinant congenic mice. Genetics 176: 1907–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E. Y., Zhang Q., McMillan L., Pardo-Manuel de Villena F., Wang W., 2010. Efficient genome ancestry inference in complex pedigrees with inbreeding. Bioinformatics 26: i199–i207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E. Y., Morgan A. P., Chesler E. J., Wang W., Churchill G. A., et al. , 2014. High-resolution sex-specific linkage maps of the mouse reveal polarized distribution of crossovers in male germline. Genetics 197: 91–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster genetic reference panel. Nature 482: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangandi J., Verma S., Osorio L. F., Peres N. A., van de Weg E., et al. , 2017. Pedigree-based analysis in a multiparental population of octoploid strawberry reveals QTL alleles conferring resistance to Phytophthora cactorum. G3 (Bethesda) 7: 1707–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk M. M., Lamb D. J., 2008. The biology of infertility: research advances and clinical challenges. Nat. Med. 14: 1197–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen M. D., Kresovich S., Villeda H. S., Bradbury P., Li H., et al. , 2009. Genetic properties of the maize nested association mapping population. Science 325: 737–740. [DOI] [PubMed] [Google Scholar]
- Mihola O., Trachtulec Z., Vlcek C., Schimenti J. C., Forejt J., 2009. A mouse speciation gene encodes a meiotic histone h3 methyltransferase. Science 323: 373–375. [DOI] [PubMed] [Google Scholar]
- Morgan A. P., Welsh C. E., 2015. Informatics resources for the collaborative cross and related mouse populations. Mamm. Genome 26: 521–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. P., Fu C. P., Kao C. Y., Welsh C. E., Didion J. P., et al. , 2016a The mouse universal genotyping array: from substrains to subspecies. G3 (Bethesda) 6: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. P., Holt J. M., McMullan R. C., Bell T. A., Clayshulte A. M. F., et al. , 2016b The evolutionary fates of a large segmental duplication in mouse. Genetics 204: 043687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. P., Gatti D. M., Keane T. M., Galante R. J., Pack A. I., et al. , 2017. Structural variation shapes the landscape of recombination in mouse. Genetics 206: 603–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti C., Vaiman D., Tores F., Cocquet J., 2016. Expression and epigenomic landscape of the sex chromosomes in mouse post-meiotic male germ cells. Epigenetics Chromatin 9: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najarro M. A., Hackett J. L., Macdonald S., 2017. Loci contributing to boric acid toxicity in two reference populations of Drosophila melanogaster. G3 (Bethesda) 7: 1631–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odet F., Pan W. Q., Bell T. A., Goodson S. G., Stevans A. M., et al. , 2015. The founder strains of the collaborative cross express a complex combination of advantageous and deleterious traits for male reproduction. G3 (Bethesda) 5: 2671–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Mita A., Sakurai-Yamatani N., Yamamoto H., Takagi N., et al. , 2004. Hybrid breakdown caused by substitution of the X chromosome between two mouse subspecies. Genetics 166: 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olds-Clarke P., Johnson L. R., 1993. T-haplotypes in the mouse compromise sperm flagellar function. Dev. Biol. 155: 14–25. [DOI] [PubMed] [Google Scholar]
- Oreper D. G., Cai Y., Tarantino L. M., Pardo-Manuel de Villena F., Valdar W., 2017. Inbred strain variant database (ISVDB): a repository for probabilistically informed sequence differences among the collaborative cross strains and their founders. G3 (Bethesda) 7: 1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., Turelli M., 1996. Dominance and Haldane’s rule. Genetics 143: 613–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur B. A., Krenz J. G., Nachman M. W., 2004. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution 58: 2064–2078. [DOI] [PubMed] [Google Scholar]
- Peirce J. L., Lu L., Gu J., Silver L. M., Williams R. W., 2004. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekkala N., Knott K. E., Kotiaho J. S., Puurtinen M., 2012. Inbreeding rate modifies the dynamics of genetic load in small populations. Ecol. Evol. 2: 1791–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip V. M., Sokoloff G., Ackert-Bicknell C. L., Striz M., Branstetter L., et al. , 2011. Genetic analysis in the collaborative cross breeding population. Genome Res. 21: 1223–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool J. E., Corbett-Detig R. B., Sugino R. P., Stevens K. A., Cardeno C. M., et al. , 2012. Population genomics of sub-saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 8: e1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwan J., 2003. Inbreeding depression in fecundity and inbred line extinction in the bulb mite, Rhizoglyphus robini. Heredity 90: 371–376. [DOI] [PubMed] [Google Scholar]
- Raghavan C., Mauleon R. P., Apostol V. L., Jubay M. L. S., Zaw H., Bonifacio J. B., et al. , 2017. Approaches in characterizing genetic structure and mapping in a rice multi-parental population. G3 (Bethesda) 7: 1721–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A. L., Okumura A., Ferris M. T., Green R., Feldmann F., et al. , 2014. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science 346: 987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Pardo-Manuel de Villena F., Wang W., McMillan L., Threadgill D. W., 2007. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm. Genome 18: 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogala A. R., Morgan A. P., Christensen A. M., Gooch T. J., Bell T. A., et al. , 2014. The collaborative cross as a resource for modeling human disease: CC011/Unc, a new mouse model for spontaneous colitis. Mamm. Genome 25: 95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell L. D., Ettlin R. A., Sinha Hikim A. P., Clegg E. D., 1990. Histological and histopathological evaluation of the testis. Cache River Press, Clearwater. [Google Scholar]
- Schultz N., Hamra F. K., Garbers D. L., 2003. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc. Natl. Acad. Sci. USA 100: 12201–12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S., Churchill G. A., 2001. A statistical framework for quantitative trait mapping. Genetics 159: 371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J. R., Couch C., Huang W., Carbone M. A., Peiffer J., et al. , 2015. Genetic architecture of natural variation in Drosophila melanogaster aggressive behavior. Proc. Natl. Acad. Sci. USA 112: E3555–E3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Morgan A. P., Najarian M., Sarsani V. K., Sigmon J. S., et al. , 2017. Genomes of the mouse collaborative cross. Genetics 206: 537–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova R., Gregorova S., Buckiova D., Kyselova V., Divina P., et al. , 2004. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm. Genome 15: 515–524. [DOI] [PubMed] [Google Scholar]
- Swindell W. R., Bouzat J. L., 2006. Reduced inbreeding depression due to historical inbreeding in Drosophila melanogaster: evidence for purging. J. Evol. Biol. 19: 1257–1264. [DOI] [PubMed] [Google Scholar]
- Threadgill D. W., Churchill G. A., 2012. Ten years of the collaborative cross. G3 (Bethesda) 2: 153–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill D. W., Miller D. R., Churchill G. A., de Villena F. P. M., 2011. The collaborative cross: a recombinant inbred mouse population for the systems genetic era. ILAR J. 52: 24–31. [DOI] [PubMed] [Google Scholar]
- Tisné S., Pomiès V., Riou V., Syahputra I., Cochard B., et al. , 2017. Identification of Ganoderma disease resistance loci using natural field infection of an oil palm multiparental population. G3 (Bethesda) 7: 1683–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner R. M., 2006. Moving to the beat: a review of mammalian sperm motility regulation. Reprod. Fertil. Dev. 18: 25–38. [DOI] [PubMed] [Google Scholar]
- Tyler A. L., Ji B., Gatti D. M., Munger S. C., Churchill G. A., et al. , 2017. Epistatic networks jointly influence phenotypes related to metabolic disease and gene expression in diversity outbred mice. Genetics 206: 621–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. R., de Villena F. P. M., McMillan L., 2012. Comparative analysis and visualization of multiple collinear genomes. BMC Bioinformatics 13: S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. J., White M. A., Payseur B. A., 2015. The pace of hybrid incompatibility evolution in house mice. Genetics 201: 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh C. E., McMillan L., 2012. Accelerating the inbreeding of multi-parental recombinant inbred lines generated by sibling matings. G3 (Bethesda) 2: 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh C. E., Miller D. R., Manly K. F., Wang J., McMillan L., et al. , 2012. Status and access to the collaborative cross population. Mamm. Genome 23: 706–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. A., Ikeda A., Payseur B. A., 2012. A pronounced evolutionary shift of the pseudoautosomal region boundary in house mice. Mamm. Genome 23: 454–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. M., Baxendale V., Chen Y. L., Pang A. L. Y., Stitely T., et al. , 2004. Analysis of mouse germ-cell transcriptome at different stages of spermatogenesis by SAGE: biological significance. Genomics 84: 971–981. [DOI] [PubMed] [Google Scholar]
- Yamate J., Tajima M., Kudow S., Sannai S., 1990. Background pathology in BDF1 mice allowed to live out their life-span. Lab. Anim. 24: 332–340. [DOI] [PubMed] [Google Scholar]
- Yang H., Bell T. A., Churchill G. A., Pardo-Manuel de Villena F., 2007. On the origin of the laboratory mouse. Nat. Genet. 39: 1100–1107. [DOI] [PubMed] [Google Scholar]
- Yang H., Ding Y. M., Hutchins L. N., Szatkiewicz J., Bell T. A., et al. , 2009. A customized and versatile high-density genotyping array for the mouse. Nat. Methods 6: 663–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. N., Wang J. R., Didion J. P., Buus R. J., Bell T. A., et al. , 2011. Subspecific origin and haplotype diversity in the laboratory mouse. Nat. Genet. 43: 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Table S3 contains QTL mapping phenotypes for the 347 mice that were phenotyped and genotyped. Table S4 contains all raw phenotype data for these mice. Testis histology images and annotations for all samples can be found at http://database.csbio.unc.edu/Infertility. Marker information can be found in Table S5. MDA genotyping data can be found at https://doi.org/10.5281/zenodo.400269. The genotypes of extant strains (including most recent common ancestors and sequenced males) are available to the public linked through the NIH-funded Mutant Mouse Resource and Research Center at (https://www.med.unc.edu/mmrrc/genotypes), (csbio.unc.edu/CCstatus/CCGenomes), and (https://doi.org/10.5281/zenodo.377036).