Meiotic recombination ensures the faithful segregation of chromosomes and influences patterns of genetic diversity. Morgan et al. used genotype data..
Keywords: meiotic recombination, recombination hotspots, copy-number variation, genetic mapping, multiparental populations, MPP
Abstract
Meiotic recombination is an essential feature of sexual reproduction that ensures faithful segregation of chromosomes and redistributes genetic variants in populations. Multiparent populations such as the Diversity Outbred (DO) mouse stock accumulate large numbers of crossover (CO) events between founder haplotypes, and thus present a unique opportunity to study the role of genetic variation in shaping the recombination landscape. We obtained high-density genotype data from DO mice, and localized 2.2 million CO events to intervals with a median size of 28 kb. The resulting sex-averaged genetic map of the DO population is highly concordant with large-scale (order 10 Mb) features of previously reported genetic maps for mouse. To examine fine-scale (order 10 kb) patterns of recombination in the DO, we overlaid putative recombination hotspots onto our CO intervals. We found that CO intervals are enriched in hotspots compared to the genomic background. However, as many as of CO intervals do not overlap any putative hotspots, suggesting that our understanding of hotspots is incomplete. We also identified coldspots encompassing 329 Mb, or of observable genome, in which there is little or no recombination. In contrast to hotspots, which are a few kilobases in size, and widely scattered throughout the genome, coldspots have a median size of 2.1 Mb and are spatially clustered. Coldspots are strongly associated with copy-number variant (CNV) regions, especially multi-allelic clusters, identified from whole-genome sequencing of 228 DO mice. Genes in these regions have reduced expression, and epigenetic features of closed chromatin in male germ cells, which suggests that CNVs may repress recombination by altering chromatin structure in meiosis. Our findings demonstrate how multiparent populations, by bridging the gap between large-scale and fine-scale genetic mapping, can reveal new features of the recombination landscape.
MEIOTIC recombination is a process that exchanges genetic material between homologous chromosomes during gametogenesis in eukaryotes. It serves important roles in individual fitness and the maintenance of genetic diversity in populations (Otto and Lenormand 2002). The formation of crossovers (COs) between homologous chromosomes provides mechanical support to allow pairing, and to orient chromosomes on the meiotic spindle for proper segregation to daughter cells at meiosis I. Failure to form COs or to distribute them properly is associated with aneuploidies that are invariably deleterious (Hassold et al. 2007; Fledel-Alon et al. 2009). Recombination facilitates the maintenance of genetic diversity in populations by allowing beneficial mutations to dissociate from linked deleterious mutations, and by limiting the loss of genetic variation in the vicinity of loci undergoing selective sweeps (Maynard Smith and Haigh 1974; Hudson and Kaplan 1995).
Meiotic recombination in mammals begins early in the first meiotic prophase with the programmed introduction of double-strand breaks (DSBs) (Sun et al. 1989). In primates and in mice, the location of most DSBs is determined by the trimethylation of histone 3 lysine 4 (H3K4me3) by the DNA binding enzyme PRDM9 (Hayashi et al. 2005; Borde et al. 2009; Parvanov et al. 2010; Baudat et al. 2010; Brick et al. 2012). The sequence specificity of PRDM9 is determined by a highly polymorphic tandem array of zinc fingers that recognizes a degenerate 13 bp motif (Myers et al. 2008). As a result, recombination occurs almost exclusively at discrete hotspots that are ∼1 kb in length, and are centered on a PRDM9 binding motif. DSBs are repaired using the homologous chromosome as a template, which results in either a CO—if the repair involves exchange of flanking regions of the chromosome—or a noncrossover (NCO) if it does not. In mouse, ∼200 DSBs occur per meiosis, of which ∼15 to 35 will lead to COs, and the remainder to NCO products (Handel and Schimenti 2010).
The local rate of recombination—defined as the frequency of COs per unit of physical distance—varies along the genome. At large scales (of order 10 Mb), CO events have characteristic distributions that are similar across a wide range of mammalian species. These include suppression of CO events near centromeres (Beadle 1932) and increased recombination rates in the distal portion of chromosome arms in male meiosis (Dunn and Bennett 1967; Broman et al. 1998; Cox et al. 2009; Wong et al. 2010; Liu et al. 2014). Recombination is suppressed in specific megabase-sized regions variously described as “‘coldspots,” “recombination deserts” (Smagulova et al. 2011), “cold zones” (Paigen et al. 2008) or “cold regions” (Liu et al. 2014). At fine scales (of order 10 kb), variation in recombination rates is a direct consequence of the locations of hotspots, which are highly variable within, and between, populations due to the dynamic coevolution of hotspots, and functional polymorphism in PRDM9. Recombination rates differ between the sexes in most mammals at both the large and fine scales of spatial distribution. The mechanisms that resolve DSBs are reasonably well understood (Gray and Cohen 2016), but the factors influencing which DSBs will become COs remain largely unknown.
Multi-parent populations (MPPs) are derived from interbreeding of multiple founder individuals or inbred strains. MPPs can accumulate large numbers of CO events between the founder haplotypes over successive generations, and high-density genotyping, or genome-wide sequencing, can be used to obtain precise localization of these events. Thus MPPs present a unique opportunity to study the recombination landscape at multiple levels of resolution.
The Diversity Outbred (DO) population is a mouse MPP derived from the Collaborative Cross (CC), which was, in turn, derived from eight inbred founder strains (A/J, C57BL/6J, 129S1/SvImJ, NOD/ShiLtJ, NZO/HlLtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ) (Svenson et al. 2012). Here, we analyze high-density genotyping data from DO mice spanning 16 breeding generations. We use the accumulated CO events to construct a dense sex-averaged recombination map, and address two outstanding questions regarding the effect of genetic variation on recombination. What is the relationship between hotspots and the distribution of CO events in the DO? What genomic features are associated with recombination coldspots? We complement our genotype data with whole-genome sequence data from 228 DO and 69 CC mice. We use published data to propose an epigenetic model of coldspots and to show that these features of the murine recombination landscape are not unique to rodents.
Materials and Methods
Mice
Breeding and maintenance of the DO at the Jackson Laboratory is described in detail elsewhere (Svenson et al. 2012; Chesler et al. 2016). Mice included in this study represent the “distribution” branch of the DO breeding program, except for the G21 cohort, which includes some breeders in the “production” branch. Data were contributed to this study by a large group of investigators (see Chesler et al. 2016); all animal work was performed in accordance with regulations set out by the animal care and use committees of their respective institutions.
Whole-genome sequencing in the DO
Whole-genome sequencing of 228 male DO mice from generations was performed at the Wellcome Trust Sanger Institute (Hinxton, Cambridge, UK). Barcoded libraries were prepared from fragmented genomic DNA using the Illumina TruSeq kit and pooled. Paired-end reads ( bp) were generated using 14 lanes of an Illumina HiSeq 2500 instrument, for an approximate coverage of per sample. Integrity of raw reads was confirmed using FastQC. Reads for each sample were realigned to the mm10 reference using bwa-mem v0.7.12 with default parameters (Li 2013). Optical duplicates were removed with samblaster (Faust and Hall 2014).
Genotyping and haplotype inference
DO mice were genotyped on either the MegaMUGA ( markers) or GigaMUGA ( markers) arrays (Morgan et al. 2016a) by the commercial service of Neogen/Geneseek (Linclon, NE). Genotypes were generously contributed by many investigators, and curated at the Jackson Laboratory as described in Chesler et al. (2016). Quality control checks were performed using the argyle package for R (Morgan 2016); samples with missing calls were excluded from subsequent analysis.
Haplotypes were reconstructed from array genotypes for all samples using the hidden Markov model (HMM) implemented in DOQTL (Gatti et al. 2014), with filtered genotypes from MegaMUGA ( QC-passing markers) or GigaMUGA ( QC-passing markers) as input. Diplotypes were “pseudophased” using a greedy algorithm: moving left to right along each chromosome, choose the configuration that minimizes the total number of COs. In sibships identified based on kinship estimates from SNP genotypes, we attempted to improve phasing using a dynamic programming algorithm. For a given chromosome pair (e.g., chromosome 1), there are chromosomes in a group of k siblings. An individuals chromosomes can have at most two phasing configurations (only one when homozygous), so there are possible configurations in the sibship. We used a scoring function that gives equal weight to every CO, and used dynamic programming to choose the state path that minimizes the total number of COs in a chromosome pair across the k siblings. Only more COs were shared between siblings after phasing improvement. We concluded that greedy phasing is an acceptable heuristic for our purposes.
Whole-genome sequencing and haplotype reconstruction for CC mice are described in detail elsewhere (Srivastava et al. 2017, this issue). The position of each CO was refined to the nearest flanking sequence variants in the parental strains using variant calls from the Sanger Mouse Genomes Project REL-1505 set (ftp://ftp-mouse.sanger.ac.uk/REL-1505-SNPs_Indels/mgp.v5.merged.snps_all.dbSNP142.vcf.gz; Keane et al. (2011)). Briefly, variants within 100 kb of the putative CO and informative between the flanking haplotypes were identified, and the number of reads supporting each allele counted. The refined CO interval was defined as the interval between the first pair of consecutive variants such that the previous five sites were consistent with the left founder haplotype and the next five sites were consistent with the right founder haplotype.
Pedigree reconstruction
Kinship coefficients were estimated for all pairs of individuals within generations from SNP genotypes using KING v1.4 (Manichaikul et al. 2010). Markers with missing data or minor allele frequency were removed. Unlike some other kinship estimators, the estimator implemented in KING does not require that markers be in linkage equilibrium, and its sampling variance decreases as the number of markers increases. Approximately autosomal SNPs were used at each generation. We used as a cutoff for siblings (relationship degree 2), and as a cutoff for cousins (relationship degree 3), based on inspection of the distribution of pairwise kinship coefficients across all generations.
Estimation of genetic map
Estimation of the genetic map in the DO is challenging. Traditional approaches to the construction of linkage maps in pedigrees assume that every CO is distinct, and can be attributed to, at most, one of two specific meioses (in the case of unknown phase). In the DO, however, COs cannot be uniquely assigned to a specific meiosis, the number of effective meiosis is unknown, and the same CO may be observed multiple times if it is shared identical by descent (IBD) between two or more individuals. We first sought to reduce our total dataset of 2.2 million COs to a set of distinct COs. To do so, we identified COs with the same haplotypes at the junction (e.g., HF), and overlapping coordinates in overlapping windows of two generations For each overlap, we identified the individual chromosomes on which the COs were identified, and tested whether the next CO or the previous CO on the same chromosomes were also shared. If at least one other neighboring CO was shared between the chromosomes, we considered the entire set of COs—both the focal pair and the neighboring pair(s)—as shared. (See Supplemental Material, Figure S2 for an example.) After performing this analysis in adjacent generations, we constructed a graph of shared COs across all generations Nodes in the graph are individual COs, and edges represent sharing between chromosomes. Connected components in the graph correspond to distinct COs transmitted over multiple generations; nodes with no incoming or outgoing edges correspond to singletons that are, by definition, distinct.
Next we constructed a cumulative map by integrating across all distinct COs on each chromosome. We noticed that the shape of this cumulative map was remarkably similar to the shape of the sex-averaged cumulative map in the CC (Liu et al. 2014). The two populations share the same eight founders at the same expected allele frequencies, so we reasoned that the CC map could be used as a scaffold for approximating the relationship between centimorgans and distinct CO counts in the DO. To obtain this approximation, we fit polynomial regressions of degree k (using least squares) each chromosome as follows:
where is cumulative CO count in the DO. (Note that the model lacks an intercept term because the genetic map must begin at zero.) The fitted values from this regression were taken as the centimorgan positions on the DO map. We found that rescaling with polynomials of even k, or , overestimated map length on every chromosome. Polynomials of odd degree could better accommodate the enrichment of COs in subtelomeric regions. For parsimony, we chose Note that it is possible to obtain a nonmonotone function from these regressions, which violates a fundamental property of the genetic map, that it be nondecreasing. We confirmed that none of the fitted models showed any evidence for violation of this property. Strain-specific maps were estimated similarly. Every distinct CO contributes to two strain-specific maps, corresponding to the two haplotypes at the junction.
If all COs in our data were truly distinct, then the appropriate scaling would be linear, and the slope would provide an estimate of the number of effective meioses we can observe in our cross-section of the DO. We view the higher-order terms in the polynomial as correction factors for the inclusion of cryptic duplicate COs in the map. In practice, however, the linear term dominates: its median value across chromosomes is while the quadratic and cubic terms are and , respectively.
Identification of recombination coldspots
We identified coldspots using a one-dimensional dynamic programming (DP) algorithm to identify regions with 10-fold reduction in frequency of COs via a generic scoring scheme (Karlin and Altschul 1990). Briefly, we first compute local CO density in windows of 500 kb, with 100 kb offset between adjacent windows. Those densities are converted to an excursion score, , as follows:
where λ is the mean CO density per chromosome, and θ is a prespecified enrichment or depletion factor. (Tenfold reduction corresponds to ) Then, a forward pass is made over the excursion scores to calculate the final score,
with Coldspots are finally extracted by performing a traceback over the This method avoids the need for a fixed-size sliding window.
Haplotype-specific coldspots were identified in similar fashion. First, we computed, in 1 Mb windows with 500 kb overlap, the density of COs involving the focal haplotype () and the density of all other COs (R). Define the enrichment score
with a pseuocount to avoid taking logarithms of zero. We then applied the one-dimensional DP algorithm to this score.
Discovery and genotyping of copy-number variant (CNVs)
Multisample CNV discovery and genotyping was performed with GenomeSTRiP (Handsaker et al. 2011). Briefly, this software uses depth of coverage and paired-end mapping patterns in multiple samples to identify candidate CNV regions and infer alternate copy-number allele(s) that are then genotyped in each individual sample. We used the CNVDiscoveryPipeline module with default settings, except as follows: window size 10 kb for initial discovery of candidate variants; minimum mapping quality (MQ) = 0 at both the discovery and genotyping stages; and MQ > 10 at for refining candidate CNV boundaries. These MQ settings are much more permissive than the defaults, and permit discovery of CNVs over SDs with high pairwise identity, over which few or no reads align with MQ > 0.
CNV discovery in a natural population is complicated by the fact that most novel alleles are rare. The only means of control of the false discovery rate is to apply strict filters for genotype quality—filters that naturally create bias against novel alleles overlying the regions of the genome most prone to structural mutation. We therefore took, as our initial CNV callset, the output of the penultimate stage of the pipeline, prior to the application of filters for missingness and genotype quality.
We sought to assign copy numbers to the eight founder alleles of the DO, not to individual samples. (De novo CNVs were beyond the scope of the present investigation.) Using the copy number assigned to each sample by GenomeSTRiP as a quantitative trait, we genetically mapped all candidate CNVs using R/qtl2, and estimated the founder strain copy numbers as the best unbiased linear predictors (BLUPs) at the QTL peak. CNVs mapping with LOD score or minor allele count were excluded as likely false positives or rare variants. Next, we merged CNVs with overlapping coordinates and identical strain distribution pattern into single loci. This yielded a final set of CNVs segregating in the DO.
Analyses of ChIP-seq data
ChIP-seq data for the H3K4me3 mark of active recombination hotspots in spermatocytes was obtained from the NCBI Short Read Archive, accession SRP045879 (Baker et al. 2015), and for the H3K9me2 mark of heterochromatin in spermatocytes, accession SRP059590. Reads for each sample were realigned to the mm10 reference using bwa-mem v0.7.12 with default parameters (Li 2013). Optical duplicates were removed with samblaster (Faust and Hall 2014). Enrichment in regions of interest was calculated by computing normalized read depth per kilobase in the target region(s) and in random intervals having the same size distribution [obtained using bedtools random (Quinlan and Hall 2010)].
Analyses of gene expression
Larson et al. (2016a) measured gene expression in isolated spermatids of three males from each of four F crosses—CZECHII/EiJ PWK/PhJ; LEWES/EiJ PWK/PhJ; PWK/PhJ LEWES/EiJ; and WSB/EiJ LEWES/EiJ—using RNA-seq. Reads were retrieved from the NCBI Short Read Archive (SRP065082). Transcript-level expression was estimated using kallisto (Bray et al. 2016), using the Ensembl 85 transcript catalog (Yates et al. 2016). In the presence of redundant transcripts (i.e., from multiple copies of a coamplified gene family), kallisto uses an expectation-maximization algorithm to distribute the “weight” of each read across transcripts without double-counting. Transcript-level expression estimates were aggregated to the gene level for differential expression testing using the R package tximport.
Gene-level expression estimates were transformed to log scale, and gene-wise dispersion parameters estimated using the voom() function in the R package limma. Genes with total normalized abundance (length-scaled transcripts per million, TPM) in aggregate across all samples were excluded, as were genes with TPM in fewer than three samples. Expression contrasts were estimated using the empirical Bayes procedure implemented in the R package limma (Ritchie et al. 2015).
Test for enrichment of sequence features
We used the GenomicAssociationTester package for Python (Ponjavic et al. 2007) to test for enrichment of various sequence features (i.e., genes, conserved elements, repeat elements) in defined intervals (i.e., coldspots.) GAT estimates enrichment by comparing observed overlap to overlap between defined intervals, and randomly sampled intervals from the genome having the same size distribution as the query intervals. Annotations for repeat elements (LINE, SINE, and LTR) were obtained from the UCSC Genome Browser (http://genome.ucsc.edu/). Gene annotations were obtained from Ensembl v86 (ftp://ftp.ensembl.org/pub/release-86/gff3/mus_musculus/), and evolutionarily constrained elements from the alignment of 40 eutherian mammal genomes generated by the Ensembl Compara team (ftp://ftp.ensembl.org/pub/release-86/maf/ensembl-compara/). Tracts of IBD in classical inbred strains were obtained from the UNC Mouse Phylogeny Viewer (http://msub.csbio.unc.edu).
For gene ontology enrichment tests we used GOrilla (http://cbl-gorilla.cs.technion.ac.il/GOrilla).
Data availability
Sample information is provided in Table S1; coordinates and haplotypes of all COs and distinct COs in Tables S2 and S3 respectively; coordinates of COs from CC strains in Table S5; coordinates of coldspots in Tables S6 and S8; and coordinates and founder strain copy numbers of CNVs in Table S7. File S2 contains genotype and marker information for all samples, in PLINK binary format. The sex-averaged map rescaled to centimorgan units is provided in File S3. Raw CNV calls are provided in File S4. (Note that File S2, File S3, and File S4 have been deposited at Zenodo; doi:10.5281/zenodo.227105). Raw sequence reads for 228 mice have been deposited in the European Nucleotide Archive (accession PRJEB8871).
Results
A catalog of COs in the DO
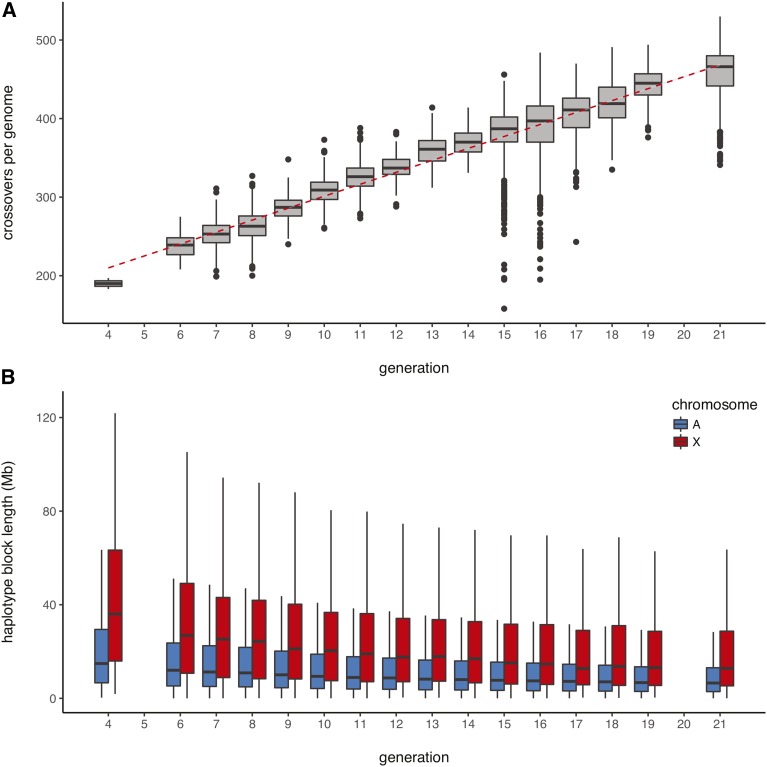
We analyzed genotype data from DO mice spanning 16 breeding generations (Table 1) and identified CO events arising in ∼ meioses (see File S1 for derivation). The number of COs per individual increases linearly over time at a rate of 15.2 COs per genome per generation ( on 6762 d.f., Figure 1), and the size of haplotype blocks decays approximately exponentially (rate constant 0.959; CI ). CO events are localized to a median resolution of 28.5 kb [median absolute deviation (MAD) 29.8 kb] on the autosomes and 58.1 kb (67.2 kb) on the X chromosome (Table 2). The rate of accumulation of CO events in the DO population is substantially lower than an early prediction (23.9 COs/genome/generation) based on the DO breeding design, and estimates of the global recombination rates obtained in a different genetic background (Cox et al. 2009; Broman 2012; Gatti et al. 2014). The discrepancy could be due to sparsity of markers with sufficient information content to trigger a haplotype transition in our HMM in some regions of the genome, or to a lower average rate of recombination in the DO background vs. classical inbred strains.
Table 1. Sample size by generation and sex-chromosome karyotype.
| Sex chromosomes | |||
|---|---|---|---|
| Generation | XX | XY | XO |
| 4 | 0 | 2 | 0 |
| 6 | 117 | 0 | 0 |
| 7 | 1026 | 61 | 8 |
| 8 | 1039 | 301 | 2 |
| 9 | 211 | 238 | 0 |
| 10 | 369 | 274 | 6 |
| 11 | 484 | 388 | 4 |
| 12 | 53 | 136 | 1 |
| 13 | 104 | 89 | 1 |
| 14 | 81 | 130 | 0 |
| 15 | 290 | 127 | 1 |
| 16 | 256 | 319 | 4 |
| 17 | 0 | 142 | 0 |
| 18 | 48 | 160 | 1 |
| 19 | 49 | 96 | 0 |
| 21 | 190 | 55 | 2 |
| Total | 4317 | 2518 | 30 |
Figure 1.
Accumulation of COs in the genomes of DO mice. (A) Distribution of the number of observed COs per genome as a function of generation number. The accumulation of COs is linear (regression line shown in red), with rate 15.2 ( CI ) additional autosomal COs per genome per generation. (B) Distribution of haplotype block lengths by generation.
Table 2. Number and widths of CO intervals identified. Interval widths are given as medians median absolute deviation.
| Chromosome | Total | Distinct | Width (kb) |
|---|---|---|---|
| A | 2,169,584 | 730,367 | 28.5 (29.8) |
| X | 71,104 | 19,193 | 58.1 (67.2) |
Individual DO mice are related to one another, both within and between generations. Thus some CO events may be shared by multiple individuals. In order to accurately estimate a genetic map, we need to distinguish between shared CO events and recurrent events that localize to the same marker interval. To make this distinction, we tried to identify groups of related individuals and infer regions of the genome that are shared between them and thus IBD. This exercise also afforded the opportunity to investigate the degree of population structure in the DO. We estimated kinship coefficients () between all pairs of individuals within each generation. (Kinship coefficients between generations are expected to be small since our sample does not capture parent-offspring pairs; see Materials and Methods). Only a small fraction of these pairs within any generation ( of ) can be detected as relatives at a threshold of (Table S4). Below this threshold, which corresponds to the chance frequency of allele sharing among eight founders, pairs of animals are effectively unrelated. We detected pairs with high kinship coefficients () that represent possible sibships. The distribution of estimated kinship coefficients by generation is shown in Figure S1. For the generations with the largest sample sizes (G7, G8, and G11), the distribution clearly has two nonzero modes corresponding to relationships of degree 3 (cousins) and 2 (siblings and double-cousins).
We estimated kinship between individuals in consecutive generations, and identified clusters of individuals corresponding to local pedigrees. We then identified chromosome segments shared IBD among related individual as illustrated in Figure S2. This exercise reduced our set of 2.2 million observed haplotype junctions to a set of “unique” CO events, defined as the union of the events observed exactly once, and those shared IBD across multiple individuals (Table 2). Among the unique CO events, were singletons—observed exactly once—and were observed in multiple related individuals. In order to confirm the robustness of this classification, we compared kinship coefficients estimated from SNP genotype sharing to the proportion of CO events that we inferred to be shared, and obtained reasonably close agreement (Figure S3). For example, among mice in 508 sibships, we observed an average sharing of which is below the expected value of The degree of underestimation is related to the proportion of singleton CO events, which decreases with increasing number of related individuals. This suggests that our power to detect a CO event as shared between any two individuals improves when more and larger related groups are present, but our ability to detect shared events between a particular pair does not. Consequently, our set of unique COs likely contains some shared events that we have misclassified as recurrent events. The misclassification rate is < Although it may distort the scaling of the DO genetic map, if we assume that misclassifications are randomly distributed among CO events, the “shape” of the map will be unaffected.
The DO provides a dense recombination map
When estimating a genetic map from an intercross or backcross experiment, the per-generation probability of a CO event between two markers can be directly related to the count of observed COs r via a map function. The map function tends to (where M is the number of informative meioses) when the physical distance between markers and the probability of multiple recombination events in the marker interval become negligible. Standard approaches to estimating genetic maps from population data, as opposed to crosses, require computationally intensive inference on the ancestral recombination graph. The DO presents a problem of intermediate complexity. We have described the process of counting the unique CO events (r), but the (effective) number of meioses M is not known, and in fact M will vary across the genome.
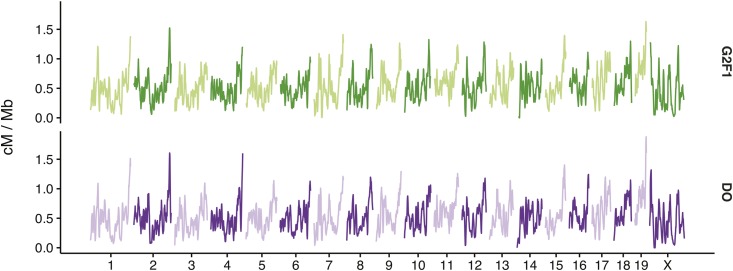
In order to scale CO counts to units of genetic distance (centimorgans) in our DO map, we took advantage of a previously reported genetic map derived from the CC population (Liu et al. 2014). We observed that the cumulative CO counts along each chromosome are highly concordant and can be nearly brought into register by a simple linear scaling of the chromosome lengths (Figure S4). Having established this correspondence, we applied a polynomial regression to estimate the relationship between cumulative CO count and centimorgan position, and used this fitted model to predict the centimorgan position of each CO event in the DO genetic map (see Materials and Methods). We use the rescaled map (Figure 2) for the remainder of the analyses presented here. The map in the DO spans ∼1291 cM on the autosomes (0.52 cM/Mb) and 71 cM (0.41 cM/Mb) on the X chromosome, shorter than length of the standard map for mouse ( cM) obtained from an outbred population derived from eight classical inbred strains (Cox et al. 2009). The DO map was estimated on a marker grid with greater physical extent than the standard map, and we speculate that the difference in map length is due to alleles derived from M. m. musculus and M. m. castaneus that segregate in the DO but not classical inbred strains (Dumont and Payseur 2011b). The unique CO events observed in the DO equate to an average of 11 CO events between any two of the ∼ consecutive marker pairs on the genotyping array. The absence of COs in an interval is therefore, in most cases, strong evidence for true local variation in recombination rate. Although local recombination rates (expressed as cM/Mb) are highly correlated (Spearman’s ) between the DO and CC at broad scales, the correlation decays at finer scales (Figure S5).
Figure 2.
Local sex-averaged recombination rates (centimorgan/megabase), calculated in 5 Mb windows, with 1 Mb offset between adjacent windows, for the CC and DO.
Most COs are associated with putative hotspots
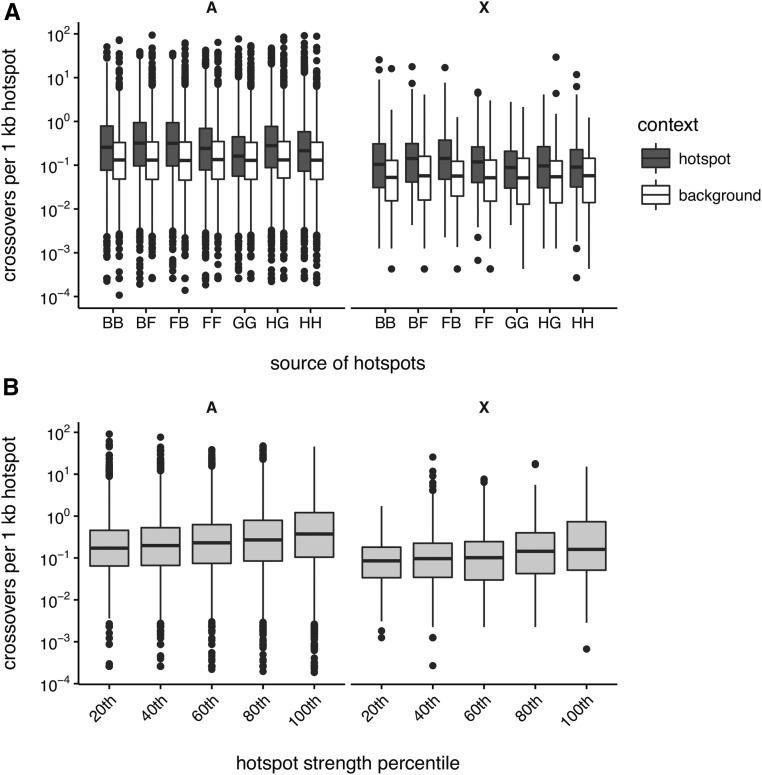
Our genotyping marker set, with an average intermarker spacing of 35 kb, does not offer enough precision to localize CO events to individual hotspots, which are on the order of 1 kb in size. However, the high density of unique CO events provides a powerful test for (sex-averaged) usage of hotspots. We obtained a list of putative hotspots, defined here as PRDM9-dependent H3K4me3 peaks ascertained by ChIP-seq in spermatocytes of male offspring of crosses between C57BL/6J, WSB/EiJ, CAST/EiJ, and PWD/PhJ (closely related to PWK/PhJ) (Baker et al. 2015). We calculated the cumulative density of COs (accounting for uncertainty in CO position) within putative hotspots, and found that they overlap a median 0.18 COs per kilobase of hotspot vs. 0.09 COs per kilobase in random genomic intervals of equal size ( Wilcoxon rank-sum test). The enrichment of putative hotspots for COs is similar regardless of the genetic background in which hotspots were ascertained (Figure 3A). Hotspot strength, as defined by the density of the H3K4me3 signal, is positively correlated (Spearman’s ) with CO density (Figure 3B; hotspots binned by strength for clarity of visualization).
Figure 3.
Recombination hotspot usage in the DO. (A) Distribution of CO density in hotspots defined by H3K4me3 ChIP-seq in testes from several genotypes, vs. random genomic intervals of equal size. Crosses are denoted as (maternal strain) × (paternal strain), and strains denoted by their one-letter codes: B = C57BL/6J, F = CAST/EiJ, G = PWD/PhJ, H = WSB/EiJ. (B) Distribution of within-hotspot CO density by bins of hotspot strength, denoted by their right endpoint. Overlap is computed separately for the autosomes (A, right) and X chromosome (X, left).
When we compared the intervals representing putative hotspots with our CO event intervals, we found that [ CI by nonparametric bootstrap) of CO intervals overlap a putative hotspot. This is substantially lower than estimates obtained from human pedigrees (∼ Baudat et al. (2010)]. The overlap is reduced among CO events between classical inbred strains (A/J, C57BL/6J, 129S1/SvImJ, NOD/ShiLtJ, and NZO/HlLtJ) compared to CO events for which one of the junction haplotypes is from a wild-derived strain (CAST/EiJ, PWK/PhJ, and WSB/EiJ) (Figure S6A). This effect is further exaggerated in regions where the classical founder strains share an ancestral haplotype IBD (Figure S7). We reasoned that the overlap between CO intervals and hotspots might be underestimated when the number of informative markers precludes accurate localization of CO events. To test this hypothesis, we examined the subset of COs ( of the total) that could be resolved to 100 kb or better. To further mitigate bias arising from the arbitrary spacing of markers on the genotyping array, we expanded each CO interval by 17 kb (half the median distance between markers) to the left and to the right. We find that () of these “padded” COs on autosomes and () of padded COs on the X chromosome overlap a putative hotspot, in good agreement with expectations (Figure S6B).
To further evaluate the influence of uncertainty in localizing CO events on our estimates of hotspot usage, we turned to whole-genome sequencing data ( coverage) from 69 recombinant inbred lines from the CC (Srivastava et al. 2017, this issue). COs arising during the breeding of CC were refined to the nearest flanking variants in the parental genomes (Keane et al. 2011). We restricted our analysis to the subset of autosomal COs that could be localized to kb resolution (Table S5). Of these, () overlap a putative hotspot. Furthermore, this degree of hotspot usage does not depend on the founder strain haplotypes at the junction ( on 27 d.f., Figure S8A). CO intervals are enriched for putative hotspots relative to random genomic intervals of equal size, and the degree of enrichment increases with hotspot strength (Figure S8B). Based on these analyses, we conclude that the majority (∼) of CO events in the DO are likely associated with a putative hotspot(s) predicted from epigenetic assays.
COs are suppressed near large CNVs
In a previous study of recombination in the CC, we identified 59 regions (spanning 129 Mb) that are at least 500 kb in size, and show at least 10-fold reduction in recombination rates relative to the genome-wide average. These coldspots were frequently associated with segmental duplications (SDs) in the reference genome. Because SDs in the reference genome are associated with CNV across individuals (Sharp et al. 2005; Perry et al. 2006; Bailey and Eichler 2006; She et al. 2008; Sudmant et al. 2015), we hypothesized that heterozygosity for CNVs might suppress normal crossing-over, and give rise to coldspots.
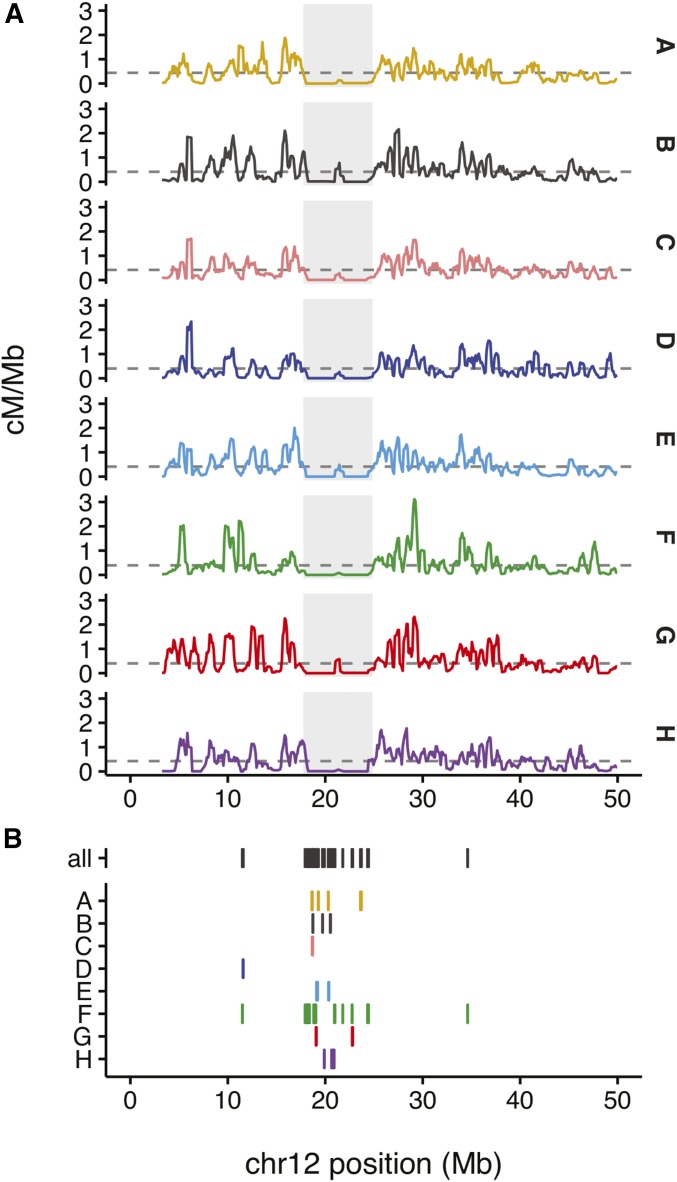
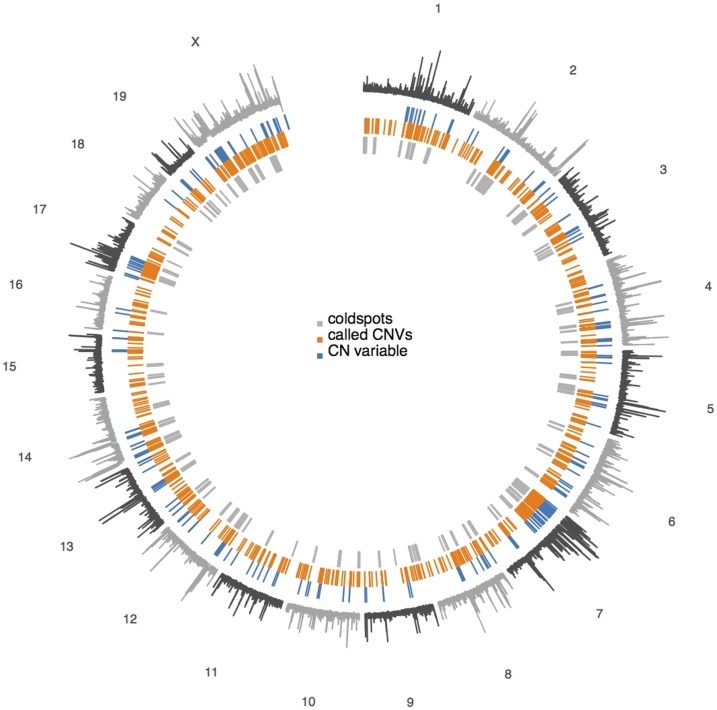
Using our DO genetic map, we identified 105 regions that are at least 600 kb in length, and show at least a 100-fold reduction in local recombination rate relative to the background rate (estimated separately for autosomes and the X chromosome) (Figure S9 and Table S6). These coldspots are not simply the complement of hotspots. We note that hotspots have median spacing 14.6 kb (MAD 15.9 kb), but coldspots span 600 kb to 13.6 Mb. COs accumulate at an approximately constant rate outside of coldspots but not inside of coldspots (Figure S10). Coldspots are found on all chromosomes, but are particularly abundant on the X chromosome (accounting for of its length; see Discussion). An example coldspot on chromosome 12 is illustrated in Figure 4.
Figure 4.
Example of a recombination coldspot on proximal chromosome 12. (A) Strain-specific local recombination rates (in centimorgan/megabase) across the proximal 50 Mb of chromosome 12. Median chromosome-wide recombination rates for each strain are marked with gray dashed lines. The coldspot is indicated by the gray shaded region. (B) CNVs ascertained in the DO. Top line, all CNVs irrespective of strain distribution pattern; remaining lines, CNVs with an allele private to a single strain.
In order to identify the sequence, structural, or epigenetic features that could be responsible for the apparent suppression of recombination in coldspots, we examined several reference genome annotations (Table 3). Coldspots contain fewer protein-coding genes but more pseudogenes than random genomic regions of similar sizes. Among the protein-coding genes present in coldspots, genes encoding olfactory receptors (4.1-fold; ), odorant peptides (10.1-fold; ), and vomeronasal receptors (3.8-fold; ) are highly overrepresented. Coldspots are enriched for some classes of transposable elements (LINEs and LTRs) but not others (SINEs). They lie in evolutionary labile regions of the genome: they are half as likely as a random genomic interval to contain a conserved element in a multiple sequence alignment of 40 eutherian mammals. However, the distinguishing feature of coldspots is a 3.6-fold enrichment of SDs, defined as duplications in the reference sequence >1 kb with mutual sequence identity.
Table 3. Enrichment of various genomic annotations in coldspots vs. genome background.
| Feature | Enrichment | q-Value |
|---|---|---|
| Reference genome annotations | ||
| Protein-coding gene | 0.60 | |
| Pseudogene | 1.98 | |
| LINE | 1.69 | |
| SINE | 0.69 | |
| LTR | 1.67 | |
| Segmental duplications | 3.66 | |
| Variation between species | ||
| GERP constrained elements | 0.46 | |
| Variation within species | ||
| IBD among classical strains | 0.69 | |
| CNV regions in DO | 3.84 | |
| Called CNVs in DO | 3.46 | |
| Biallelic | 2.46 | |
| Multiallelic | 4.34 | |
Significance was computed over 1000 shuffles and is expressed as the q-value, proportion of tests expected to represent false discoveries. GERP constrained elements are sequences conserved across 40 eutherian mammals as defined by the Ensembl Compara pipeline Yates et al. (2016). CNVs are defined on the basis of whole-genome sequencing of 228 DO mice as described in the main text.
We next turned to low-coverage whole-genome sequencing data that we obtained on 228 individual DO mice to identify CNV regions, and examine their overlap with coldspots. We calculated normalized read depth (an estimator of copy number) in nonoverlapping 25 kb windows across the genome for each individual, and then calculated the coefficient of variation (median/median absolute deviation) across the 228 mice for each window. Contiguous regions with high coefficient of variation were deemed “CNV regions.” We found that of these CNV regions overlap coldspots, an enrichment of 3.8-fold relative to random genomic intervals of similar size (Figure 5 and Table 3). We note that coldspots encompass the CNV regions: only of the span of all coldspots is occupied by CNVs. Thus, the suppression of recombination can extend beyond the CNV region, in some cases for several megabases (e.g., central chromosome 2, distal chromosome X).
Figure 5.
Genome-wide view of CNV in the DO. Outer track shows coefficient of variation (median/MAD) of normalized read depth across the population. Inner tracks show CNV regions identified by the HMM (blue); CNVs genotyped in individual samples (orange); and recombination coldspots (gray).
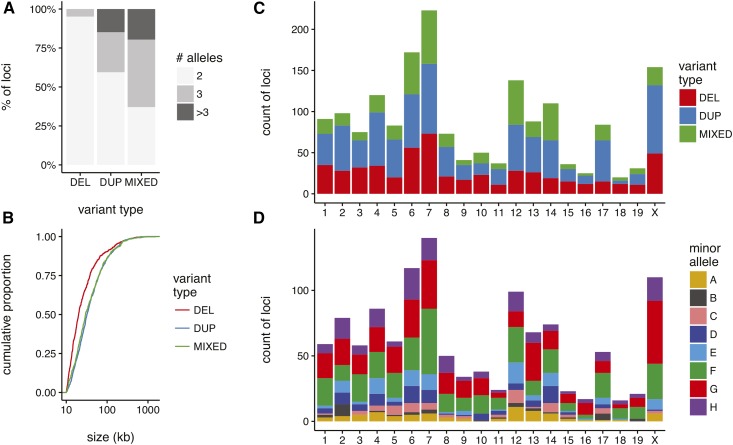
In order to investigate the relationship between copy number and COs, we examined haplotypes at CNV loci ( on the autosomes, and 154 on the X chromosome) of at least 10 kb in size (File S4 and Table S7). We further restricted our attention to common CNV alleles with minor-allele count among the 228 sequenced individuals. We excluded CVNs whose position could not be confirmed by genetic mapping (Figure S11). Lastly, overlapping CNVs with identical strain distribution patterns were merged. A majority of filtered CNV loci ( ) have a minor allele private to a single founder strain, and most of these private alleles (862; ) are contributed by the wild-derived strains CAST/EiJ, PWK/PhJ, and WSB/EiJ. As expected, CNVs cluster near SDs: CNV loci () overlap SDs in the reference genome, and the great majority of these () are multiallelic. Coldspots are enriched in CNVs, as expected, and the enrichment is stronger for multiallelic CNVs (Table 3). Figure 6 summarizes the properties of CNVs ascertained in the DO.
Figure 6.
Properties of CNVs ascertained in the DO. (A) Proportion of CNV loci according to variant type (DEL, deletion; DUP, duplication; MIXED, complex variants) and number of alleles in the DO. (B) Cumulative distribution of nominal size of CNV loci by variant type, on log10 scale. (C) Count of variants per chromosome, by type. (D) Count of private variants (minor allele found in exactly one founder strain) per chromosome.
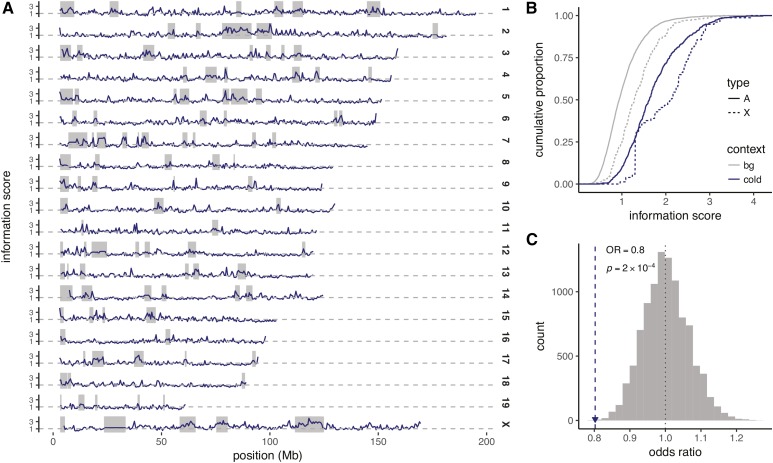
Our working hypothesis is that, in CNV regions, CO events should be biased in favor of junction between founder haplotypes with equal or similar copy number, and depleted for haplotypes with different copy number. To measure this bias we calculated an “information score,” the Kullback-Leibler divergence, between the observed and expected frequency of junctions across the possible pairs of founder haplotypes. In a randomly mating population such as the DO, the two chromosomes paired at a particular meiosis represent independent random draws from the population. The expected frequency of a CO between a given pair of founder haplotypes is equal to the product of the marginal frequency of those haplotypes. Our information score has expected value 1; larger values indicate more extreme departures from random joining of haplotypes. The score clearly tends to take larger values within coldspots on both the autosomes and the X chromosome ( Wilcoxon rank-sum test) (Figure 7, A and B). We next asked whether, within CNV regions, haplotypes with the same copy number were more likely to recombine than haplotypes with different copy numbers. For each CNV region, we computed the similarity between haplotypes using genotypes at underlying called CNVs (orange track in Figure 5), and then calculated the odds ratio for the association between copy-number identity and incidence of COs overlapping each region. The observed value () was compared to the distribution of permutations (within loci) of founder-haplotype copy numbers (Figure 7C). Consistent with our working model, haplotypes with different copy number profiles are significantly less likely to form COs near CNV regions than those with the same copy number It follows from our model that the most variable loci are predicted to be least permissive for COs between any pair of haplotypes, and, therefore, most likely to be coldspots, and we note that coldspots are nearly twice as enriched for CNVs with more than two alleles (4.3-fold) vs. biallelic CNVs (2.5-fold, Table 3).
Figure 7.
Biased distribution of COs in the vicinity of coldspots is explained by copy number differences. (A) CO information score, measuring departure from expected frequency of COs with respect to founder strain pairs, in 500 kb windows across the genome. (B) Cumulative distribution of the information score in cold regions (blue) vs. the remainder of the genome (gray), calculated separately for the autosomes (A) and X chromosome (X). (C) Odds ratio (OR) for association between copy-number difference and haplotype-specific CO incidence at CNV regions (blue track in Figure 5), vs. permutations (histogram).
Haplotype-specific coldspots are relatively rare
To obtain more concrete evidence of the effect of copy number on crossing-over, we focused on a more narrow question: could we identify “haplotype-specific” coldspots, that is, regions depleted for COs involving one founder haplotype but with normal levels of recombination among the remaining haplotypes? This analysis was motivated by a previous observation that COs involving the WSB/EiJ haplotype are completely suppressed in the middle of chromosome 2, presumably due to a 4 Mb insertion at the R2d2 locus in WSB/EiJ (Morgan et al. 2016b). We calculated an enrichment score (see Materials and Methods) across the autosomes and X chromosome, and identified 27 putative haplotype-specific coldspots. These regions have median size 2.9 Mb (MAD 2.4 Mb) and their union spans 155 Mb (Table S8). The highest-scoring region is the WSB/EiJ-specific coldspot on chromosome 2 described in detail elsewhere. We note that the haplotype-specificity of coldspots is a population-level property. If a haplotype-specific coldspot exists, and the “cold” haplotype is present at sufficient frequency, the region will also be a nonspecific coldspot. In the DO, of the span of nonspecific coldspots also lies in haplotype-specific coldspots, while of the span of haplotype-specific coldspots also lies in nonspecific coldspots.
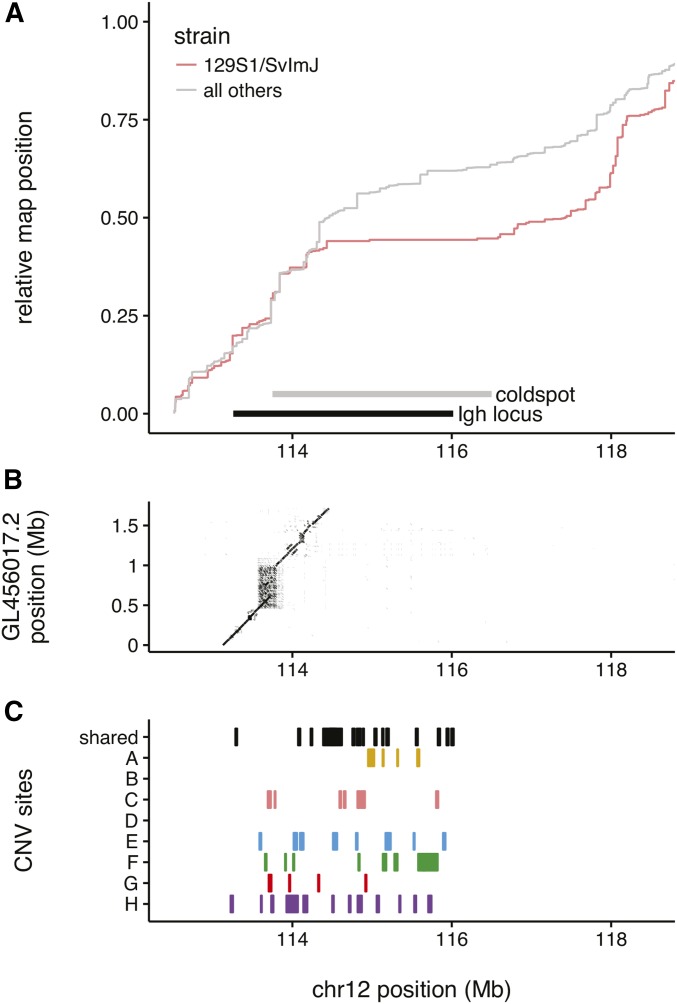
A representative example is the 129S1/SvImJ-specific coldspot on distal chromosome 12 (Figure 8A). This coldspot spans the Igh locus, a cluster of duplicated genes encoding immunoglobulin heavy-chain peptides, which is known to be highly polymorphic in wild mice (Tutter and Riblet 1989) and classical inbred strains (Retter et al. 2007). At least three classical Igh haplotypes are segregating in the DO: Ighb (C57BL/6J, NOD/ShiLtJ); Ighe (A/J) and Igha (129S1/SvImJ). A portion of the Igh locus in 129S1/SvImJ has been sequenced and assembled from BACs, and is included as an ALT sequence (GenBank ID GL456017.2) in the current mouse reference genome assembly (https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.25/). Alignment of GL456017.2 to the reference genome reveals a 300 kb insertion in 129S1/SvImJ just proximal to the haplotype-specific coldspot (Figure 8B). The region is tiled with additional CNVs identified by whole-genome sequencing in the DO (Figure 8C).
Figure 8.
Example of a haplotype-specific recombination coldspot at the Igh locus. (A) Cumulative recombination map for COs involving (pink) or not involving (gray) the 129S1/SvImJ haplotype in the vicinity of the Igh locus on distal chromosome 12. Maps are scaled to arbitrary units so that both start at zero and end at one. Extent of 129S1/SvImJ-specific coldspot is indicated by gray bar. (B) Dotplot representation of alignment of GL456017.2 (alternate sequence derived from 129S1/SvImJ BACs) to the mouse reference genome. (C) CNVs in the vicinity of the Igh locus. Alleles private to each strain are shown in individual tracks; alleles shared by two or more strains are shown in top track.
Genes in coldspots have epigenetic features of repressed chromatin
Epigenetic marks play an important role in defining the location of CO events. In primates and in mouse, the H3K4me3 mark established by PRDM9 designates hotspots for DSBs. In taxa without a functional PRDM9 homolog, such as birds (Singhal et al. 2015), dogs (Auton et al. 2013), and some yeast (Lam and Keeney 2015), recombination is directed toward gene promoters and CpG islands. (The mechanism for this PRDM9-free targeting may be related to DNA methylation, or to features of chromatin architecture such as nucleosome spacing.) We asked whether a complementary suite of epigenetic features could be defined for coldspots, with a view toward identifying properties that could explain the decoupling of the spatial distribution of DSBs from COs. Our prototype is the sex chromosomes in male meiosis, where DSBs occur in the heterologous regions of the X and Y chromosomes (Mahadevaiah et al. 2001), but do not, under normal circumstances, resolve as CO events.
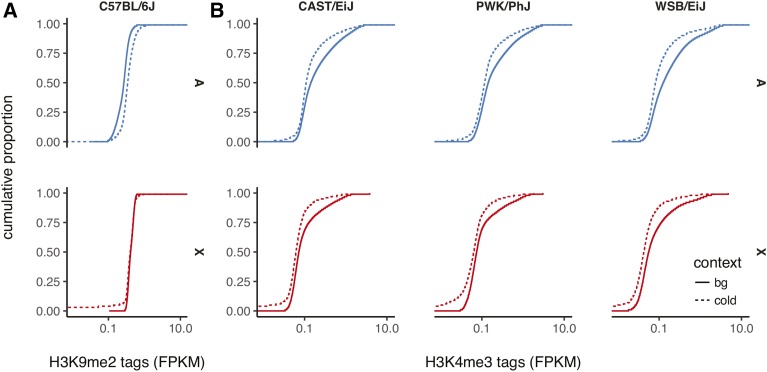
We examined the density of two histone modifications, histone 3 lysine 9 dimethylation (H3K9me2) and H3K4me3, in spermatocytes. H3K9me2 is associated with heterochromatin, and is a characteristic mark of the silenced X chromosome in male meiosis (Khalil et al. 2004). H3K4me3, by contrast, is associated not only with recombination hotspots, but also the transcription start sites of actively transcribed genes (Liu et al. 2005; Barski et al. 2007). Coldspots on the autosomes are enriched, relative to randomly drawn genomic intervals of equal size, for H3K9me2 in pachytene spermatocytes (Walker et al. (2015); Figure 9A). The enrichment is absent on the X chromosome, as expected, because H3K9me2 is associated with meiotic sex-chromosome inactivation (MSCI) and the onset of MSCI is in pachytene. However, coldspots are depleted for H3K4me3 on both the autosomes and the X chromosome in mixed-stage spermatocytes of wild-derived inbred strains from all three mouse subspecies (Baker et al. (2015); Figure 9B). Together, these patterns point to repression of coldspot chromatin during male meiosis.
Figure 9.
Epigenetic features of coldspots in male germ cells. (A) Distribution of tag read density (as fragments per million mapped, FPKM) for the H3K9me2 mark associated with heterochromatin in pachytene spermatocytes from C57BL/6J males. Distributions are plotted separately for autosomes (A, blue) and the X chromosome (X, red), and for coldspots (dashed lines) vs. randomly drawn regions of equal size (solid lines). (B) Distribution of tag read density for the H3K4me3 mark associated with recombination hotspots and actively transcribed genes in mixed-stage spermatocytes from three inbred strains.
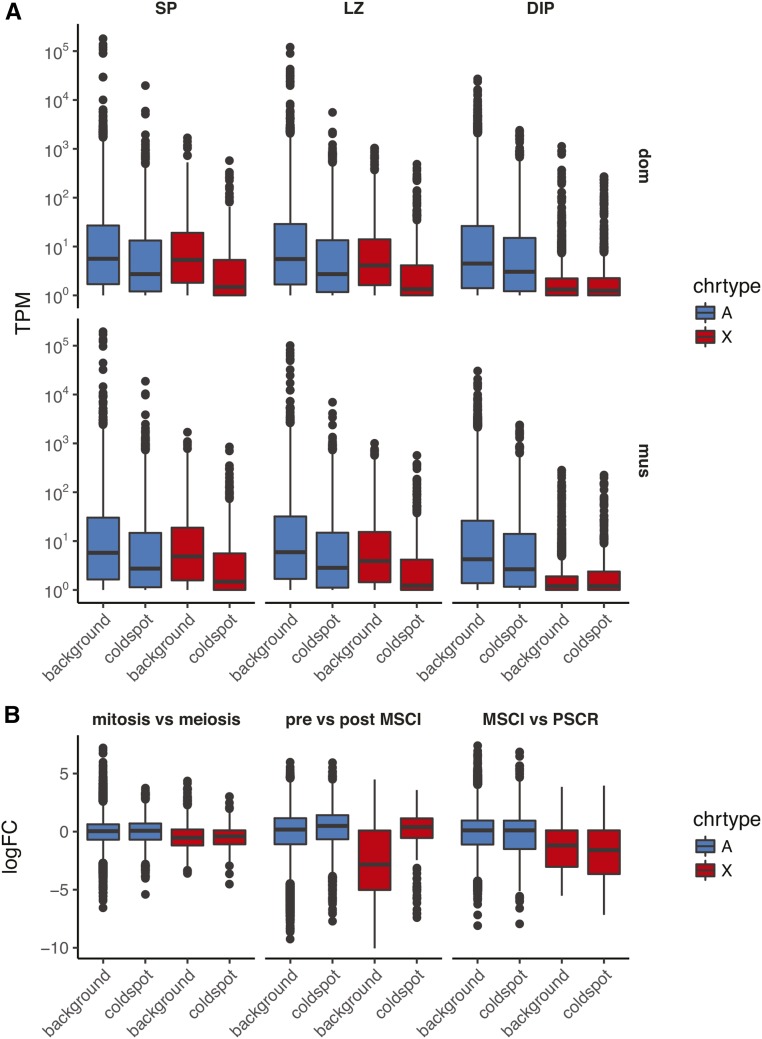
We next examined the association between gene expression and coldspot status in male germ cells. We reanalyzed a published RNA-seq experiment (Larson et al. 2016a,b) on germ cell populations isolated by fluorescence-activated cell sorting. Median expression of genes in coldspots is ∼10-fold lower than genes outside coldspots for all cell types examined ( Wilcoxon rank-sum test, for all comparisons), and the effect holds for both the autosomes and the X chromosome (Figure 10A).
Figure 10.
Transcriptional control of coldspots in male germ cells. (A) Distribution of normalized expression levels (transcripts per million mapped reads, TPM) according to chromosome type (autosomes, A; X chromosome, X) and coldspot status in WSB/EiJ×LEWES/EiJ F1 (dom) or PWK/PhJ×CZECHII/EiJ F1 (mus) males. SP, spermatogonia; LZ, leptotene/zygotene spermatocytes; DIP, diplotene spermatocytes. (B) Distribution of log2 fold-difference (logFC) values in comparisons between stages of spermatognesis. Horizontal axis and color scheme follow (A). MSCI, meiotic sex chromosome inactivation; PSCR, postmeiotic sex chromosome repression.
If COs are suppressed in coldspots because of defects in pairing or synapsis, we would expect that genes in coldspots would be subject to meiotic silencing of unpaired chromatin (MSUC). Onset of MSUC should be concomitant with MSCI and occur by early diplotene. To test this prediction, we estimated expression contrasts between pre- and post-MSCI cell types, using difference between mitotic (spermatogonia) and meiotic cells (spermatocytes) as a negative control (Figure 10B). Contrary to our prediction, we observed no difference in expression change between coldspot genes and background genes across the mitosis–meiosis transition. The expression of coldspot genes on both the autosomes and the X chromosome increases relative to background with the onset of MSCI, and the difference persists after meiosis (during postmeiotic sex chromatin repression, PSCR).
Coldspots are not unique to the rodent lineage
Our findings support the hypothesis that coldspots in mouse are associated with megabase-scale structural properties of genomic DNA. However, both the global rate and the fine-scale spatial distribution of recombination are rapidly evolving in murid rodents (Dumont and Payseur 2011a; Dumont et al. 2011; Baker et al. 2015), and coldspots might be a byproduct of this process rather than a common feature of mammalian meiosis.
To test the generality of coldspots, we sought a second mammal for which there exists both a high-quality reference genome assembly and a dense pedigree-based genetic map. (Genetic maps derived from patterns of LD in populations are prone to artifacts in SDs and other repetitive sequences where it is difficult to ascertain variants using short-read sequencing.) We chose the domestic dog (Canis lupus familiaris): large pedigrees are available (Campbell et al. 2016), and, like the mouse, the dog has an all-acrocentric karyotype, mitigating the possibility of confounding of coldspots with the centromere effect (Beadle 1932). More interestingly, domestic dogs, and other canids, apparently lack a functional PRDM9 ortholog (Axelsson et al. 2012), providing an opportunity to test whether coldspots are independent of PRDM9, and therefore independent of the lineage-specific spatial distribution of recombination hotspots.
We reanalyzed a published genetic map derived from a golden retriever pedigree spanning ∼408 effective meioses (Campbell et al. 2016). Local sex-specific recombination rates across the 38 dog autosomes are shown in Figure S12. The dog map recapitulates the major feature of the mouse map: elevated recombination rate in the distal portion of chromosomes in males but not females. Applying the same strategy as we used for the sex-averaged mouse map from the DO, we identified 66 coldspots on 13 chromosomes in the sex-averaged dog map. They are larger than coldspots in mouse—ranging in size from 400 kb to 11.4 Mb—but cover a slightly smaller fraction of the autosomes () as a consequence of the lower density of the dog map, and therefore lower power to discriminate true coldspots from random variation in the background recombination rate. As with mouse, coldspots in the dog genome are 2.5-fold enriched for SDs Example coldspots on dog chromosomes 9 and 22 are shown in Figure S13.
Discussion
Here, we describe the most extensive study to date of recombination rate variation along the mouse genome (Rowe et al. 1994; Cox et al. 2009; Paigen et al. 2008; Liu et al. 2014). The large number of accumulated CO events and high density of genotyped markers allow us to examine features of the recombination landscape across scales spanning several orders of magnitude. The DO affords the power to test relationships between local sequence features and recombination rate.
Estimation of a genetic map from the DO in the absence of pedigree information involves detection of CO events and determination of their IBD status, i.e., sharing of CO events across individuals; it further requires imputation of pedigree relationships among the genotyped individuals; and, finally, it requires a means to scale the map to standard centimorgan units. In order to check the accuracy of our map, we performed several ancillary analyses. Inferred sharing of CO events occurs at the expected frequency between siblings (Figure S3). The accumulation of CO events in individual genomes across generations likewise matches our expectations for this population (Figure 1). There is close correspondence between (unscaled) cumulative counts of CO events in the DO with cumulative counts of CO events in the CC an independently derived population with a genetic background similar to the DO (Figure S4). This observation supports our rescaling of the DO recombination rates against the CC genetic map. The resulting map of the DO recapitulates established large-scale properties of the recombination landscape in mouse (Figure 2), while offering the precision needed to investigate variation in the recombination rate at much finer scales.
Hotspot usage, Prdm9 alleles, and the evolution of fine-scale recombination rates
Most, perhaps all, CO events occur within epigenetically defined hotspots. This hypothesis is consistent with patterns of linkage disequilibrium in humans and great apes (Coop and Przeworski 2007), as well as high-resolution maps of DSBs during meiotic prophase in male mice (Smagulova et al. 2011). CO locations are constrained by positioning of DSBs, which are, in turn, determined by the strength of interaction of sequence motifs with DNA binding domains of the histone methyltransferase PRDM9 (Walker et al. 2015). Tens of thousands of putative hotspots have been experimentally predicted in male mice using inbred strains and F1 hybrids. Ours is the first study to estimate the distribution of realized COs with respect to putative hotspots in a genetically diverse and outbred population of mice. We find that ∼ of CO events are associated with a hotspot. The remaining of CO events may be associated with as yet uncharacterized PRDM9 binding sites, such as those with female-specific activity (Paigen et al. 2008), or with hotspots that fell below the limit of detection in epigenetic assays.
Prdm9 is highly polymorphic in natural populations, and variation is concentrated in its DNA-binding domains (Buard et al. 2014). Four Prdm9 alleles are segregating in the DO— dom2 (A/J, C57BL/6J, 129S1/SvImJ, NZO/HlLtJ) and dom3 (NOD/ShiLtJ, WSB/EiJ) from M. m. domesticus; msc (PWK/PhJ, PWD/PhJ) from M. m. musculus; and cst (CAST/EiJ) from M. m. castaneus. Thus, most DO mice will carry two functionally distinct alleles of Prdm9. There is a hierarchy among the alleles such that the stronger variant typically acts in a dominant fashion to activate its own cognate hotspots. In addition, as many as of the hotspots that are observed in F1 hybrids are not active in either parental strain. In admixed populations like the DO and CC, one or the other Prdm9 allele in each individual will be mismatched with the local ancestry across most of the genome. In this case, we expect many COs to be initiated at ancestral hotspots whose activity is lower than hotspots specific to the founder strains (Baker et al. 2015; Davies et al. 2016). We obtained qualitative support for this hypothesis by comparing the enrichment of precisely localized CO events in the CC to hotspots ascertained in the context of different combinations of Prdm9 alleles (Figure S14). For COs between haplotypes of different local ancestry (Yang et al. 2011), enrichment is strongest for hotspots ascertained in F1s with at least one conspecific Prdm9 allele.
Structural variation and CO suppression
We identified 105 megabase-sized coldspots in which few or no COs have been observed in > effective meioses spanning 16 generations of DO breeding (Figure S10). We find that coldspots are strongly associated with regions of SDs in the reference genome that correspond to multi-allelic CNVs in the founder strains of the DO (Table 3). The proximity to repetitive DNA raises the concern that coldspots might be technical artifacts associated with genotyping of repetitive sequences. The SNP markers on our genotyping arrays are biased away from repetitive sequences (Morgan et al. 2016a). However, CO events are readily identified by markers flanking the repetitive regions, provided that consecutive CO events are at least a few megabases apart. This is true for the majority of CO events, at least through generation 21 (Figure 1), and the likelihood of missing more than a handful of CO events is negligible. Alternatively, one might imagine that coldspots are due to systematic genotyping errors in repetitive regions. But genotyping error tends to increase, not decrease, the number of inferred CO events (Gatti et al. 2014). We have observed coldspots at concordant locations in other mouse crosses and conclude that they are a genuine feature of the recombination landscape.
The longitudinal nature of the DO provides strong evidence for a causal link between standing CNV and reduction in recombination. We have restricted our analysis to common CNVs—sites that vary between founder haplotypes, not more recent mutations arising de novo in the DO—and to COs that occurred during breeding of the DO. In this sense, the absence of COs cannot have caused the associated CNVs. It may still be the case that coldspots in the DO are simply those regions that would have been cold in the (unobserved) ancestral populations giving rise to the founder strains, and that “coldness” and structural mutation have a shared basis related to the underlying chromatin state. Two lines of evidence suggest that this explanation alone is not sufficient. First, copy-number profiles in CNV regions predict which haplotypes will recombine (Figure 7). Most CNVs in the DO—like any other sequence variants—segregate between subspecies (Figure 6). This implies that at least some of the reduction in recombination is due to heterozygosity for haplotypes of different ancestry, beyond any “coldness” in the ancestral population. Second, in the case of two loci examined in detail (R2d2 and Igh), a copy-number allele private to one founder strain is associated with nearly complete suppression of only COs involving that haplotype. In these cases, we have no reason to believe that the locus was inherently “cold” before or after the divergence of the M. musculus subspecies except in the context of heterozygosity for the CNV.
A classical explanation for regional absence of CO events in pedigrees is inversion (Sturtevant 1921). The reciprocal products of a CO in an inversion heterozygote are acentric and dicentric, respectively, and generally cannot be properly segregated to gametes. COs between inverted and noninverted haplotypes are therefore rare or absent in progeny. Although we were unable to find evidence for inversion at either R2d2 or Igh based on whole-genome sequence reads from 228 DO or 69 CC mice (Srivastava et al. 2017, this issue), we hypothesize that large structural variants behave in a manner qualitatively similar to inversions. If recent studies of structural variants in humans (Usher et al. 2015; Huddleston and Eichler 2016) and great apes (Gordon et al. 2016) are any guide, the loci we have identified as CNVs may be a proxy for more complex rearrangements. We hypothesize that, when different structural alleles meet at meiosis, pairing and synapsis are disrupted.
To test this prediction, we used expression data from testes of F1 hybrids between wild-derived strains of M. m. domesticus and M. m. musculus origin that are expected to carry different structural alleles at many coldspots. We find no evidence that genes in coldspots are subject to MSUC, as we would expect if coldspots are excluded from the synaptonemal complex (Turner 2007). However, this is probably due to the fact that coldspot genes already have very low expression levels throughout spermatogenesis (Figure 10). In C57L/6J spermatocytes, coldspots are enriched for H3K9me2, a mark of heterochromatin and the inactivated X chromosome, and depleted for H3K4me3, the mark of active genes, and of recombination hotspots (Figure 9). To exclude the possibility that coldspots are dependent on the fine-scale distribution of DSBs, we investigated their presence in the recombination map for a second mammal, the domestic dog. Canids lack a functional PRDM9 ortholog and fine-scale recombination patterns are driven by CpG methylation rather than H3K4me3 (Auton et al. 2013). We identified coldspots in the dog map (Figure S13) that are conspicuously associated with SDs in the reference genome and with CNVs among dog breeds (Nicholas et al. 2011). Similarity in the size and sequence features of coldspots between dog and mouse, whose last common ancestor lived ∼55 MYA (Meredith et al. 2011), suggests that coldspots are a general feature of meiosis in mammals. Together, these findings are consistent with our hypothesis that absence of COs in coldspots is a consequence of structural heterozygosity at the megabase scale.
Independent of the causal relationship between coldspots and CNVs, reduction of recombination facilitates the accumulation of mutations and exacerbates the effects of selective sweeps and genetic draft. Nonrecombining regions may facilitate phenotypic divergence (of which the sex chromosomes are and extreme case), or the rise of selfish elements such as the t-haplotype (Shin et al. 1983; Lyon 1984). Although we assume that most structural alleles at the coldspots we have identified are neutral, enrichment of coldspots for fast-evolving genes related to immunity, olfaction, and kin recognition raises the possibility that coldspots may play a role in local adaptation.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.197988/-/DC1.
Acknowledgments
We thank Leonard McMillan for helpful discussions on identifying crossover events in the Collaborative Cross. We thank Alan Attie, Gillian Beamer, Carol Bult, Leah Rae Donahue, Alison Harrill, David Harrison, Clarissa Parker, Daniel Pomp, Luanne Peters, and Karen Svenson for generously providing genotyping data. This work was supported in part by National Institutes of Health grants P50GM076468, R01GM070683 (G.A.C.); U19AI100625 (F.P.M.dV); P01AG017628, R01HL134015, R01HL111725 (A.I.P.); and F30MH103925 (A.P.M.).
Footnotes
Communicating editor: E. Eskin
Literature Cited
- Auton A., Li Y. R., Kidd J., Oliveira K., Nadel J., et al. , 2013. Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet. 9: e1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E., Webster M. T., Ratnakumar A., Consortium T. L., Ponting C. P., et al. , 2012. Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res. 22: 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. A., Eichler E. E., 2006. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat. Rev. Genet. 7: 552–564. [DOI] [PubMed] [Google Scholar]
- Baker C. L., Kajita S., Walker M., Saxl R. L., Raghupathy N., et al. , 2015. PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination. PLoS Genet. 11: e1004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.-Y., Schones D. E., et al. , 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. [DOI] [PubMed] [Google Scholar]
- Baudat F., Buard J., Grey C., Fledel-Alon A., Ober C., et al. , 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle G. W., 1932. A possible influence of the spindle fibre on crossing-over in Drosophila. Proc. Natl. Acad. Sci. USA 18: 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V., Robine N., Lin W., Bonfils S., Géli V., et al. , 2009. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 28: 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N. L., Pimentel H., Melsted P., Pachter L., 2016. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34: 525–527. [DOI] [PubMed] [Google Scholar]
- Brick K., Smagulova F., Khil P., Camerini-Otero R. D., Petukhova G. V., 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature 485: 642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., 2012. Haplotype probabilities in advanced intercross populations. G3 (Bethesda) 2: 199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Murray J. C., Sheffield V. C., White R. L., Weber J. L., 1998. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am. J. Hum. Genet. 63: 861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J., Rivals E., Segonzac D. D. d., Garres C., Caminade P., et al. , 2014. Diversity of Prdm9 zinc finger array in wild mice unravels new facets of the evolutionary turnover of this coding minisatellite. PLoS One 9: e85021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C. L., Bhérer C., Morrow B. E., Boyko A. R., Auton A., 2016. A pedigree-based map of recombination in the domestic dog genome. G3 (Bethesda) 6: 3517–3524. [DOI] [PMC free article] [PubMed]
- Chesler E. J., Gatti D. M., Morgan A. P., Strobel M., Trepanier L., et al. , 2016. Diversity outbred mice at 21: maintaining allelic variation in the face of selection. G3 6: 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G., Przeworski M., 2007. An evolutionary view of human recombination. Nat. Rev. Genet. 8: 23–34. [DOI] [PubMed] [Google Scholar]
- Cox A., Ackert-Bicknell C. L., Dumont B. L., Ding Y., Bell J. T., et al. , 2009. A new standard genetic map for the laboratory mouse. Genetics 182: 1335–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B., Hatton E., Altemose N., Hussin J. G., Pratto F., et al. , 2016. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature 530: 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont B. L., Payseur B. A., 2011a Evolution of the genomic recombination rate in murid rodents. Genetics 187: 643–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont B. L., Payseur B. A., 2011b Genetic analysis of genome-scale recombination rate evolution in house mice. PLoS Genet. 7: e1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont B. L., White M. A., Steffy B., Wiltshire T., Payseur B. A., 2011. Extensive recombination rate variation in the house mouse species complex inferred from genetic linkage maps. Genome Res. 21: 114–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn L. C., Bennett D., 1967. Sex differences in recombination of linked genes in animals. Genet. Res. 9: 211–220. [DOI] [PubMed] [Google Scholar]
- Faust G. G., Hall I. M., 2014. SAMBLASTER fast duplicate marking and structural variant read extraction. Bioinformatics 30: 2503–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fledel-Alon A., Wilson D. J., Broman K., Wen X., Ober C., et al. , 2009. Broad-scale recombination patterns underlying proper disjunction in humans. PLoS Genet. 5: e1000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti D. M., Svenson K. L., Shabalin A., Wu L.-Y., Valdar W., et al. , 2014. Quantitative trait locus mapping methods for diversity outbred mice. G3 4: 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D., Huddleston J., Chaisson M. J. P., Hill C. M., Kronenberg Z. N., et al. , 2016. Long-read sequence assembly of the gorilla genome. Science 352: aae0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S., Cohen P. E., 2016. Control of meiotic crossovers: from double-strand break formation to designation. Annu. Rev. Genet. 50: 175–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel M. A., Schimenti J. C., 2010. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 11: 124–136. [DOI] [PubMed] [Google Scholar]
- Handsaker R. E., Korn J. M., Nemesh J., McCarroll S. A., 2011. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat. Genet. 43: 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T., Hall H., Hunt P., 2007. The origin of human aneuploidy: where we have been, where we are going. Hum. Mol. Genet. 16: R203–R208. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Yoshida K., Matsui Y., 2005. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438: 374–378. [DOI] [PubMed] [Google Scholar]
- Huddleston J., Eichler E. E., 2016. An incomplete understanding of human genetic variation. Genetics 202: 1251–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Kaplan N. L., 1995. The coalescent process and background selection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 349: 19–23. [DOI] [PubMed] [Google Scholar]
- Karlin S., Altschul S. F., 1990. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl. Acad. Sci. USA 87: 2264–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane T. M., Goodstadt L., Danecek P., White M. A., Wong K., et al. , 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A. M., Boyar F. Z., Driscoll D. J., 2004. Dynamic histone modifications mark sex chromosome inactivation and reactivation during mammalian spermatogenesis. Proc. Natl. Acad. Sci. USA 101: 16583–16587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam I., Keeney S., 2015. Nonparadoxical evolutionary stability of the recombination initiation landscape in yeast. Science 350: 932–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson E. L., Keeble S., Vanderpool D., Dean M. D., Good J. M., 2016a The composite regulatory basis of the large X-effect in mouse speciation. Mol. Biol. Evol. 34: 282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson E. L., Vanderpool D., Keeble S., Zhou M., Sarver B. A. J., et al. , 2016b Contrasting levels of molecular evolution on the mouse X chromosome. Genetics 203: 1841–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., 2013 Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN].
- Liu C. L., Kaplan T., Kim M., Buratowski S., Schreiber S. L., et al. , 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 3: e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E. Y., Morgan A. P., Chesler E. J., Wang W., Churchill G. A., et al. , 2014. High-resolution sex-specific linkage maps of the mouse reveal polarized distribution of crossovers in male germline. Genetics 197: 91–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M. F., 1984. Transmission ratio distortion in mouse t-haplotypes is due to multiple distorter genes acting on a responder locus. Cell 37: 621–628. [DOI] [PubMed] [Google Scholar]
- Mahadevaiah S. K., Turner J. M. A., Baudat F., Rogakou E. P., de Boer P., et al. , 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27: 271–276. [DOI] [PubMed] [Google Scholar]
- Manichaikul A., Mychaleckyj J. C., Rich S. S., Daly K., Sale M., et al. , 2010. Robust relationship inference in genome-wide association studies. Bioinformatics 26: 2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- Meredith R. W., Janečka J. E., Gatesy J., Ryder O. A., Fisher C. A., et al. , 2011. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334: 521–524. [DOI] [PubMed] [Google Scholar]
- Morgan A. P., 2016. argyle: An R package for analysis of Illumina genotyping arrays. G3 (Bethesda) 6: 281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. P., Fu C.-P., Kao C.-Y., Welsh C. E., Didion J. P., et al. , 2016a The mouse universal genotyping array: from substrains to subspecies. G3 (Bethesda) 6: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. P., Holt J. M., McMullan R. C., Bell T. A., Clayshulte A. M.-F., et al. , 2016b The evolutionary fates of a large segmental duplication in mouse. Genetics 204: 267–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S., Freeman C., Auton A., Donnelly P., McVean G., 2008. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat. Genet. 40: 1124–1129. [DOI] [PubMed] [Google Scholar]
- Nicholas T. J., Baker C., Eichler E. E., Akey J. M., 2011. A high-resolution integrated map of copy number polymorphisms within and between breeds of the modern domesticated dog. BMC Genomics 12: 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S. P., Lenormand T., 2002. Resolving the paradox of sex and recombination. Nat. Rev. Genet. 3: 252–261. [DOI] [PubMed] [Google Scholar]
- Paigen K., Szatkiewicz J. P., Sawyer K., Leahy N., Parvanov E. D., et al. , 2008. The recombinational anatomy of a mouse chromosome. PLoS Genet. 4: e1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov E. D., Petkov P. M., Paigen K., 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327: 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G. H., Tchinda J., McGrath S. D., Zhang J., Picker S. R., et al. , 2006. Hotspots for copy number variation in chimpanzees and humans. Proc. Natl. Acad. Sci. USA 103: 8006–8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J., Ponting C. P., Lunter G., 2007. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 17: 556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retter I., Chevillard C., Scharfe M., Conrad A., Hafner M., et al. , 2007. Sequence and characterization of the Ig heavy chain constant and partial variable region of the mouse strain 129S1. J. Immunol. 179: 2419–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., et al. , 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe L. B., Nadeau J. H., Turner R., Frankel W. N., Letts V. A., et al. , 1994. Maps from two interspecific backcross DNA panels available as a community genetic mapping resource. Mamm. Genome 5: 253–274. [DOI] [PubMed] [Google Scholar]
- Sharp A. J., Locke D. P., McGrath S. D., Cheng Z., Bailey J. A., et al. , 2005. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 77: 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X., Cheng Z., Zöllner S., Church D. M., Eichler E. E., 2008. Mouse segmental duplication and copy-number variation. Nat. Genet. 40: 909–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H. S., Flaherty L., Artzt K., Bennett D., Ravetch J., 1983. Inversion in the H-2 complex of t-haplotypes in mice. Nature 306: 380–383. [DOI] [PubMed] [Google Scholar]
- Singhal S., Leffler E. M., Sannareddy K., Turner I., Venn O., et al. , 2015. Stable recombination hotspots in birds. Science 350: 928–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F., Gregoretti I. V., Brick K., Khil P., Camerini-Otero R. D., et al. , 2011. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472: 375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Morgan A. P., Najarian M. L., Sarsani V. K., Sigmon J. S., et al. , 2017. Genomes of the mouse collaborative cross. Genetics 206: 537–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1921. A case of rearrangement of genes in Drosophila. Proc. Natl. Acad. Sci. USA 7: 235–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant P. H., Mallick S., Nelson B. J., Hormozdiari F., Krumm N., et al. , 2015. Global diversity, population stratification, and selection of human copy-number variation. Science 349: aab3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Treco D., Schultes N. P., Szostak J. W., 1989. Double-strand breaks at an initiation site for meiotic gene conversion. Nature 338: 87–90. [DOI] [PubMed] [Google Scholar]
- Svenson K. L., Gatti D. M., Valdar W., Welsh C. E., Cheng R., et al. , 2012. High-resolution genetic mapping using the mouse diversity outbred population. Genetics 190: 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J. M. A., 2007. Meiotic sex chromosome inactivation. Development 134: 1823–1831. [DOI] [PubMed] [Google Scholar]
- Tutter A., Riblet R., 1989. Evolution of the immunoglobulin heavy chain variable region (Igh-V) locus in the genusMus. Immunogenetics 30: 315. [DOI] [PubMed] [Google Scholar]
- Usher C. L., Handsaker R. E., Esko T., Tuke M. A., Weedon M. N., et al. , 2015. Structural forms of the human amylase locus and their relationships to SNPs, haplotypes and obesity. Nat. Genet. 47: 921–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M., Billings T., Baker C. L., Powers N., Tian H., et al. , 2015. Affinity-seq detects genome-wide PRDM9 binding sites and reveals the impact of prior chromatin modifications on mammalian recombination hotspot usage. Epigenetics Chromatin 8: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. K., Ruhe A. L., Dumont B. L., Robertson K. R., Guerrero G., et al. , 2010. A comprehensive linkage map of the dog genome. Genetics 184: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang J. R., Didion J. P., Buus R. J., Bell T. A., et al. , 2011. Subspecific origin and haplotype diversity in the laboratory mouse. Nat. Genet. 43: 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A., Akanni W., Amode M. R., Barrell D., Billis K., et al. , 2016. Ensembl 2016. Nucleic Acids Res. 44: D710–D716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sample information is provided in Table S1; coordinates and haplotypes of all COs and distinct COs in Tables S2 and S3 respectively; coordinates of COs from CC strains in Table S5; coordinates of coldspots in Tables S6 and S8; and coordinates and founder strain copy numbers of CNVs in Table S7. File S2 contains genotype and marker information for all samples, in PLINK binary format. The sex-averaged map rescaled to centimorgan units is provided in File S3. Raw CNV calls are provided in File S4. (Note that File S2, File S3, and File S4 have been deposited at Zenodo; doi:10.5281/zenodo.227105). Raw sequence reads for 228 mice have been deposited in the European Nucleotide Archive (accession PRJEB8871).