Abstract
Systematic genetic studies of a handful of diverse organisms over the past 50 years have transformed our understanding of biology. However, many aspects of primate biology, behavior, and disease are absent or poorly modeled in any of the current genetic model organisms including mice. We surveyed the animal kingdom to find other animals with advantages similar to mice that might better exemplify primate biology, and identified mouse lemurs (Microcebus spp.) as the outstanding candidate. Mouse lemurs are prosimian primates, roughly half the genetic distance between mice and humans. They are the smallest, fastest developing, and among the most prolific and abundant primates in the world, distributed throughout the island of Madagascar, many in separate breeding populations due to habitat destruction. Their physiology, behavior, and phylogeny have been studied for decades in laboratory colonies in Europe and in field studies in Malagasy rainforests, and a high quality reference genome sequence has recently been completed. To initiate a classical genetic approach, we developed a deep phenotyping protocol and have screened hundreds of laboratory and wild mouse lemurs for interesting phenotypes and begun mapping the underlying mutations, in collaboration with leading mouse lemur biologists. We also seek to establish a mouse lemur gene “knockout” library by sequencing the genomes of thousands of mouse lemurs to identify null alleles in most genes from the large pool of natural genetic variants. As part of this effort, we have begun a citizen science project in which students across Madagascar explore the remarkable biology around their schools, including longitudinal studies of the local mouse lemurs. We hope this work spawns a new model organism and cultivates a deep genetic understanding of primate biology and health. We also hope it establishes a new and ethical method of genetics that bridges biological, behavioral, medical, and conservation disciplines, while providing an example of how hands-on science education can help transform developing countries.
Keywords: mouse lemur, model organism, Microcebus, primate genetics, Madagascar
THE pantheon of genetic model organisms (including the bacterium Escherichia coli, yeast Saccharomyces cerevisiae, nematode Caenorhabditis elegans, fruit fly Drosophila melanogaster, zebrafish Danio rerio, mouse Mus musculus, and mustard weed Arabidopsis thaliana) has transformed our understanding of biology (Davis 2004; Fields and Johnston 2005). The focus on a small number of species has been important to this scientific success by providing the critical mass of investigators necessary to tackle complex biological systems, and by creating synergies and economies of scale that enabled systematic, genome-wide approaches. Central among these is the ability to achieve genetic saturation—identifying many or all of the genes involved in a biological process—and then to have elegant tools available to organize the genes into genetic pathways and localize their site of action.
Many of the genes, pathways, and principles elaborated in genetic model organisms have turned out to be broadly conserved, aiding understanding of organisms throughout the tree of life. However, the focus on a small number of organisms has impeded progress in areas of biology not represented in the pantheon. “Boutique” model organisms have sprung up to target some of the neglected areas, such as the flatworm Planaria torva for tissue regeneration (Newmark and Sanchez Alvarado 2002; Reddien et al. 2005), stickleback fish Gasterosteus aculeatus for vertebrate evolution (Peichel et al. 2001; Jones et al. 2012), and killifish Nothobranchius furzeri for vertebrate aging (Harel et al. 2015; Kim et al. 2016). Remarkably, primate biology, which holds some of the most fascinating and important questions in all biology, has been left without its own model, relying on mouse and simpler model organisms and human genetic studies.
Mouse Fails to Mimic Many Aspects of Primate Biology, Behavior, and Disease
The laboratory mouse M. musculus is a nearly perfect mammalian model organism. It has a short generation time (2–3 months) and large litter size (8–12 pups), and is small and easy to maintain in a laboratory setting. Its rise to prominence followed the invention of the technology for introducing specific gene mutations into mice (Kuehn et al. 1987; Thomas and Capecchi 1987). Since that time, gene targeting has been used to elucidate the function of >3000 genes (http://www.mousephenotype.org/), and the nearly $1 billion International Mouse Phenotyping Consortium was established to generate and phenotype knockouts in all protein-coding genes during this decade (Collins et al. 2007; Abbott 2010; Skarnes et al. 2011). This mouse knockout strategy has revolutionized the study of mammalian biology and led to the establishment of thousands of mouse models to explore disease mechanisms and evaluate new diagnostics and therapeutics.
The strategy, however, does not always work. This became clear when Evans’ Nobel Prize-winning knockout of the Lesch–Nyhan (HPRT) gene (Kuehn et al. 1987) was found to have the biochemical defect but not the behavioral manifestations (self-mutilation) of the human syndrome (Engle et al. 1996). Other examples followed, such as failure of mouse CFTR knockouts to model the devastating lung disease of cystic fibrosis patients (Grubb and Boucher 1999), along with similar setbacks for other lung diseases (Baron et al. 2012). Parkinson’s, Huntington’s, and other neurodegenerative diseases have been particularly difficult to model in mice (Schnabel 2008; Beal 2010), as have immune (Mestas and Hughes 2004; Zschaler et al. 2014), infectious (Rittirsch et al. 2007; Kim et al. 2014), and metabolic diseases (Panchal and Brown 2011; Perlman 2016). No organ system has escaped this problem, and reviews now appear with titles like “The mousetrap: what we can learn when the mouse model does not mimic the human disease” (Elsea and Lucas 2002). A systematic comparison of human and mouse genes and knockout phenotypes found that the mouse does not model the critical function, or have an assignable ortholog, of ∼40% of the human genes required for viability (Liao and Zhang 2008); the authors speculate that the fraction is likely greater for nonessential genes, such as most human behavior and disease genes.
Of course, the mouse would not be expected to model every aspect of mammalian biology and behavior, especially those features of primates that distinguish them from other mammals. Rodents have relatively fast life histories with low survivorship, which selects for rapid reproduction and low parental investment, whereas primates have long life spans and few offspring. Hence mice are poor models for human aging and chronic conditions of aging, including atherosclerosis (Bentzon and Falk 2010) and Alzheimer’s disease (Phinney et al. 2003; Geerts 2009), and for parenting and family bonding. Rodents also perceive the world differently. Primates, with their large, forward-facing eyes, are highly reliant on vision and have high acuity and a complex oculomotor system for acquiring and processing visual information, whereas rodents see with low resolution (20/2000 vision) and do not track objects (Baker 2013; Izpisua Belmonte et al. 2015). Primates also have the manual dexterity to grasp objects and create tools. Perhaps the most salient differences are in higher order brain function, especially cognition and the formation of complex social systems that involve elaborate vocal communication and specialized brain structures in primates (e.g., expanded frontal cortex) that are limited or absent in rodents (Izpisua Belmonte et al. 2015). Also, mouse genetic studies have focused on laboratory mice, so they have had little impact on understanding behaviors and biological phenomena that manifest only in natural settings.
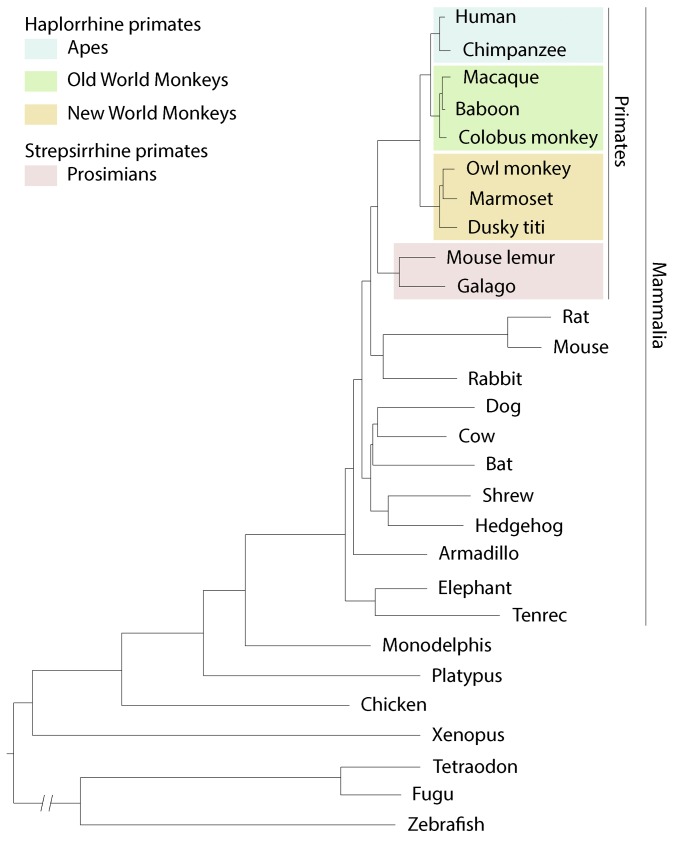
A genetic model organism that better mimics primate biology, behavior, and health is urgently needed. Researchers have begun targeting homologs of human disease genes in other animals including rat (Tong et al. 2010) and pig (Rogers et al. 2008). However, rats and mice are on an evolutionary tangent: although rodents diverged more recently from the human lineage than almost all other animals outside of primates, their mutational clock has run several times faster (Li et al. 1996; Huttley et al. 2007) (Figure 1). Pigs diverged earlier (Groenen et al. 2012) and are large and expensive to maintain in a laboratory. Rats and pigs are therefore unlikely to provide a general solution to the biological limitations of mice. Scientists have therefore begun developing advanced genetic approaches in nonhuman primates.
Figure 1.
Phylogenetic tree showing evolutionary relationship of primates and other vertebrates. Branch length (horizontal distance) reflects the mutational load in coding and noncoding sequences for a species. Note that rodents (rat, mouse) diverged from the primate lineage more recently than all other animals shown, but their rate of mutation accumulation (mutational clock) is several times greater than that of other vertebrates, as indicated by the long cumulative branch length since separation of mouse and rat from the primate lineage. Genetic distance between species can be estimated from the sum of the branch lengths connecting them, which is about two times greater from mouse to human than from mouse lemur to human. Phylogenetic tree adapted from Margulies et al. (2007).
Most Nonhuman Primates Are Unsuitable for Systematic Genetics
Research on nonhuman primates has been dominated for decades by longitudinal studies of the physiology and behavior of individuals and their communities, both in laboratory and natural settings. Such studies have identified many interesting and important physiological and behavioral traits, as exemplified by Jane Goodall’s pioneering work on chimp social behavior and personality in Tanzania (Goodall 1967, 1968). However, long generation times and the small numbers of individuals examined make it impractical or impossible to map the genes and alleles underlying such traits, as is routine for genetic model organisms. The classical genetic approach is of course just as powerful for primates, as evidenced by myriad human genetic studies that overcome the disadvantage of long generation time by collecting phenotypic and genetic information on many individuals in large family pedigrees (Chong et al. 2015). Application of this approach to other primates would require characterization of large family pedigrees, as is underway for large breeding colonies of baboons and macaques (Cox et al. 2013; Vinson et al. 2013), or study of primates with a much shorter generation time.
Primate geneticists are also exploring the “reverse-genetic” approach, in which a specific gene is targeted for disruption and the phenotypic consequences subsequently determined—the approach that catapulted mouse and yeast to prominence. Viral-mediated gene delivery was demonstrated in macaque Macaca mulatta embryos 16 years ago (Chan et al. 2001), and generation of transgenic marmosets, Callithrix jacchus, with germline transmission of the introduced reporter gene was shown 8 years later (Sasaki et al. 2009). The latter achievement led to hailing of the common marmoset on the cover of Nature as the next biomedical “supermodel.” Efforts toward gene editing in macaques and marmosets have since expanded, and in the last several years the first targeted gene knockouts were achieved by TALEN- and CRISPR-mediated deletions in preimplantation embryos (Liu et al. 2014; Niu et al. 2014; Sato et al. 2016). However, the procedures are still inefficient and complicated by somatic mosaicism, off-target effects, and persistent activity of the introduced nuclease. In addition, germline transmission of the mutation has not been demonstrated (Luo et al. 2016). Other targeting approaches are under consideration, such as genetic modification and selection in vitro of embryonic stem (ES) cells (as in mice), although creation of germline-competent primate ES cells has remained elusive (Tachibana et al. 2012).
Even if the technical limitations are overcome, there are serious ethical issues surrounding germline modification of higher primates (Nature Editors 2009; Olsson and Sandoe 2010; Collins 2015), as well as myriad practical considerations. For example, the macaque has a gestation time of 5.5 months, a generation time of ∼4 years, a litter size of 1, and maintenance costs of ∼$25K per generation (Table 1). The marmoset offers considerable advantages, with a gestation time of 4.5 months and generation time of ∼2 years, but maintenance costs of ∼$5K per generation are still one to two orders of magnitude greater than for mice. On practical considerations alone, it is hard to envision widespread adoption of either of these primates for systematic genetic studies, though they are likely to play increasingly prominent roles in medical and behavioral research. Below we describe a systematic approach to primate genetics that does not require germline modification.
Table 1. Reproductive characteristics and maintenance costs of several small mammals and primates.
| Species | Divergence from humans (MY) | Human sequence [ortholog] identity (%)a | Weight (g)b | Gestation period (mo) | Age of sexual maturity (mo) | Litter size (no.) | Typical [max] life span (yr)c | Cost per generation ($)d |
|---|---|---|---|---|---|---|---|---|
| Nonprimate small mammals | ||||||||
| Mouse (M. musculus) | 90–110 | 84 [74] | 25 | 0.7 | 1.5 | 8–12 | 2 [4] | 100 |
| Rat (Rattus norvegicus) | 90–110 | 84 [74] | 350 | 0.8 | 2 | 6–14 | 3 [4] | 300 |
| Northern tree shrew (T. belangeri) | 80–105 | 88 [71] | 150 | 1.5 | 4 | 1–5 | ND [11] | ND |
| Primates | ||||||||
| Madame Berthe’s mouse lemur (M. berthae) | 60–75 | 91 [86] | 30 | 2.1 | 8 | 1–4 | 7 [18] | 500 |
| Brown mouse lemur (M. rufus) | | | | | 40 | | | | | | | | | | |
| Gray mouse lemur (M. murinus) | | | | | 60 | | | | | | | | | | |
| Bush baby (G. demidovii) | 60–75 | 89 [85] | 60 | 3.7 | 9 | 1–2 | ND [13] | ND |
| Spectral tarsier (Tarsius spectrum) | 55–65 | 90 [77] | 120 | 6.5 | 17 | 1 | ND [12] | ND |
| Pygmy marmoset (C. pygmaea) | 30–45 | ND | 120 | 4.6 | 16 | 1–2 | ND [18] | ND |
| Common marmoset (C. jacchus) | 30–45 | 95 [89] | 300 | 4.8 | 16 | 1–3 | 8 [16] | 5,000 |
| Rhesus macaque (M. mulatta) | 20–30 | 97 [94] | 8,000 | 5.5 | 36 | 1 | 26 [42] | 20,000 |
| Chimpanzee (Pan troglodytes) | 5–8 | 99 [98] | 50,000 | 7.6 | 120 | 1 | 50 [80] | 250,000 |
| Human (Homo sapiens) | — | — | 70,000 | 9.3 | 156 | 1 | 80 [122] | 230,000 |
The Staff of the Jackson Laboratory 1966; Nowak 1999; Townsend 2001; Wrogemann et al. 2001; Glazko and Nei 2003; Tardif et al. 2003; Andrade et al. 2004; Suckow et al. 2006; Gursky 2007; Chatterjee et al. 2009; Fuchs and Corbach-Sohle 2010; Fischer and Austad 2011; Languille et al. 2012; Fan et al. 2013; Rowe and Myers 2016; Ensembl release 88 2017; Human Ageing Genomic Resources 2017; Lino et al. 2017.
Percent DNA sequence identity to human orthologous exon sequences. Brackets, percent of exon sequences with identified human ortholog. Species used for comparison: mouse lemur (M. murinus), galago (Otolemur garnettii), tarsier (T. syrichta).
Average weight of adult male in the wild.
75th percentile for age of mortality in captivity. Brackets, oldest recorded age in captivity.
Generation time (gestation period plus age of sexual maturity in days) multiplied by the 2016 per diem laboratory maintenance rates from National Institutes of Health, Division of Veterinary Resources (https://www.ors.od.nih.gov/sr/dvr/documents/dvrrates.pdf). Mouse lemur value from MHNH colony. Chimpanzee per diem rate from 2015. Human value from 2015 United States Department of Agriculture estimate of child expenditures from birth through age 17.
Search for a Suitable Organism for Systematic Genetic Analysis of Primate-Specific Biology
In the summer of 2009, three high school interns (C.E., J.W., and M.R.K.) surveyed the animal kingdom for a species that might be suitable as a genetic model organism for primate-specific biology. The students first considered the attributes of other genetic model organisms that contributed to their scientific success. The most obvious were rapid reproduction (short gestation and generation times), abundance of offspring, small size, and ease of laboratory maintenance, which enable vast numbers of genetic crosses. Also considered was each species’ effectiveness in modeling primate features, prior use in research, safety, cost of colony maintenance, abundance in the wild and conservation status, plus ethical and legal issues. Through literature searches, the students compiled a database of species and these attributes, from which we selected the most appealing candidates.
Generation time and laboratory maintenance cost generally scale with body size, so the top candidates were all small, similar in size to mice (25 g) and rats (350 g). Because rodents diverged from the primate lineage more recently than almost any other animal (Murphy et al. 2001) (Figure 1), there were few candidates to consider outside of primates. One was the northern tree shrew Tupaia belangeri (Figure 2A), which is small (100–200 g) and has favorable reproductive characteristics (Table 1), and has even been proposed as a model for some human disorders (Zhao et al. 2002; Fan et al. 2013). However, genomic sequence analysis places the tree shrew’s divergence from the primate lineage only ∼5 MY after rodent divergence (Fan et al. 2013), so it may not provide a substantial modeling advantage over the mouse.
Figure 2.
Candidates for a genetic model organism that could better mimic primate biology than mouse. Animals shown are small mammals (<200 g) that diverged more recently from the primate lineage than mouse. (A) Northern tree shrew T. belangeri (∼150 g), a mammal found in southeast Asia. (B) Bush baby G. demidovii (∼60 g), a prosimian (strepsirrhine) primate found in west and central Africa. (C) Spectral tarsier T. spectrum (∼120 g), a prosimian primate (recently grouped with haplorrhines) found in Southeast Asia. (D) Pygmy marmoset C. pygmaea (∼120 g), the world’s smallest monkey (haplorrhine), found in the Amazon basin of South America. Photo credits: (A) http://www.philadelphiazoo.org, (B) http://www.arkive.org, (C) http://www.ecologyasia.com, and (D) http://www.justviral.eu.
Among the nearly 500 primate species of the world, several stood out. The pygmy marmoset Cebuella pygmaea (100–150 g), less than half the size of the common marmoset (Figure 2D and Table 1), is the world’s smallest monkey. Although their small size would make them less costly to maintain in the laboratory, like all “higher” primates their long gestation period (4.5 months) and sexual maturation (1–2 years) render them impractical as a genetic model organism, as does the somatic chimerism observed among littermates (dizygotic twins) in many marmoset species (and presumably C. pygmaea) (Ross et al. 2007; Sweeney et al. 2012). Several small prosimian primates (50–150 g), such as the bush baby Galagoides demidovii (Figure 2B) and the tarsier Tarsius spectrum (Figure 2C), were likewise attractive candidates because of their relatively short generation times (∼1 and ∼2 years, respectively) (Table 1). However, tarsiers are notoriously difficult to maintain in captivity due to specific dietary and environmental requirements (Wright et al. 2003), and bush babies in captivity have high levels of infanticide and can harbor zoonotic disease (Chang et al. 1980; Tartabini 1991).
Mouse Lemur, an Excellent Candidate for a Primate Genetic Model Organism
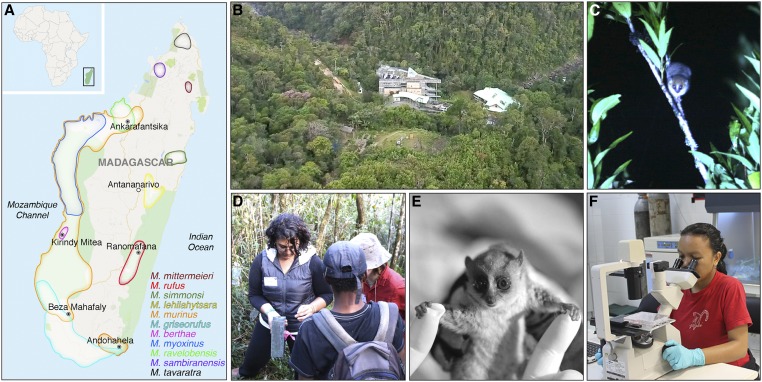
The top candidate was the mouse lemur (Microcebus spp.) (Figure 3E, Figure 4B, and Table 1), prosimian primates roughly half the genetic distance between mouse and human (estimated 91% nucleotide identity between human and mouse lemur orthologs) (Margulies et al. 2007; Ensembl release 88 2017) (Figure 1). They are the smallest (30 g, Microcebus berthae; most other species and subspecies 40–60 g in the wild) and the fastest developing (2 months gestation) and maturing (6–8 months to sexual maturity) primates in the world. They are also among the most fecund (litter size 1–4) and abundant primates: we estimate there are millions to tens of million individuals (Andriaholinirina et al. 2014), distributed in many isolated or overlapping populations throughout Madagascar (Figure 3A) (Weisrock et al. 2010).
Figure 3.
Field study of mouse lemurs. (A) Geographical distribution of mouse lemurs in Madagascar (off east coast of mainland Africa, boxed in inset) and major field study sites (filled ○). Capital city Antananarivo (○) is located centrally, inland from coastal forested areas (green shading). The approximate distribution of 11 of the >20 described species of mouse lemurs is shown by color outlines as indicated in the legend; most species are “microendemic” to a region delimited by rivers, mountains, or elevation gradients. Field study sites include: RNP (rain forest), Kirindy Mitea National Park (dry deciduous forest), Ankarafantsika Nature Reserve (dry tropical forest), Andohahela National Park (spiny forest), and Beza Mahafaly Reserve (gallery forest). (B) Aerial view of CVB research station at RNP, established by Patricia Wright and colleagues in 1991 (RNP) and 2003 (CVB). The state-of-the art interdisciplinary research station (http://www.stonybrook.edu/commcms/centre-valbio/) focuses on the unique flora and fauna of the surrounding rain forest, and includes a full molecular and cell biology laboratory and conference center where ecologists and conservation biologists interact with genetics, health, and engineering experts as well as educators and artists. (C) A brown mouse lemur, M. rufus, foraging at night near CVB. Mouse lemurs are nocturnal so field studies are done by capture and release in the evening when they awaken. (D) Field researchers inspecting an aluminum Sherman live trap baited with banana to attract mouse lemurs. (E) A captured brown mouse lemur brought back to the laboratory. An identifying microchip is implanted and the animal thoroughly examined (“deep phenotyping”) before its release back into the wild at the capture site the same evening. Individuals are studied longitudinally by periodic recapture throughout their up to 10 year life span. (F) Scientist examining mouse lemur fibroblast culture in CVB laboratory. Map data from Google (copyright 2016). Microcebus spp. geographical distributions adapted from International Union for Conservation of Nature Red List of Threatened Species (http://www.iucnredlist.org/). Photo credits: (B) Khen Randriamamonjy, (C–E) Guy Albertelli.
Figure 4.
Laboratory study of mouse lemurs. Mouse lemurs have been studied in the laboratory since establishment of a breeding colony of gray mouse lemur M. murinus by J.-J. Petter and A. Petter-Rousseaux in the 1960s from a dozen individuals imported from southwest Madagascar, like the individual shown foraging in the dry deciduous forest of Kirindy Mitea National Park (A). These tiny strepsirrhine primates are docile (B) and readily maintained in enriched cages with branches or tubes simulating their arboreal habitat, and nest boxes (C) simulating the tree holes where they typically sleep in small groups (D). Photo credits: Guy Albertelli (A, C, and D).
Like all lemurs, mouse lemurs are native only to Madagascar. They inhabit virtually all of the island’s diverse biomes, including primary rainforests (Figure 3, B and C), dry forests (Figure 4A), and even secondary forests and other disturbed habitats (Radespiel 2007). Although there have not been any functional genetic studies, their biology (Atsalis 2008), behavior (Radespiel 2000; Weidt et al. 2004), phylogeny (Yoder et al. 2000; Horvath and Willard 2007; Weisrock et al. 2010), and population structure (Wimmer et al. 2002; Fredsted et al. 2005) are under active investigation at multiple field sites around the country (Figure 3A). At these sites, microchip identification tags are implanted and in some cases radio collars attached and video cameras strategically placed so that individual mouse lemurs can be followed throughout their 5–10 year life span (Zohdy et al. 2014), supplemented by periodic trapping (Figure 3, C and D) for measurements and biological sampling (Figure 3, E and F) (Deppe et al. 2016). This work has shown that mouse lemurs exhibit many primate characteristics of interest, including living in complex social groups (Radespiel 2000; Radespiel et al. 2001), vocal communication (Cherry et al. 1987; Leliveld et al. 2011), and omnivorous foraging behaviors that suggest good learning and memory capabilities (Radespiel 2007; Luhrs et al. 2009).
Gray mouse lemurs (M. murinus) have been maintained continuously in a laboratory colony for nearly 50 years at the Muséum National d’Histoire Naturelle (MNHN) in Brunoy, France (Perret 1982), where Martine Perret, Fabienne Aujard, and their colleagues have investigated many aspects of their physiology including reproduction, metabolism, circadian rhythms, and aging (Andres et al. 2003; Aujard et al. 2006; Marchal et al. 2013; Biggar et al. 2015). Breeding colonies have also been established at several other European and American sites (Wrogemann et al. 2001; Rassoul et al. 2010; Zehr et al. 2014), many originating from the MNHN colony, along with small colonies in zoos around the world. These groups keep extensive records of individual health and behavior. Indeed, mouse lemurs have been proposed as a model of primate aging and Alzheimer’s disease because 5–20% of adults over age 5 in one colony show signs of premature aging, including brain atrophy, amyloid plaques, and cytoskeletal Tau pathology, accompanied by cognitive and social decline (Bons et al. 2006; Languille et al. 2012). Mouse lemurs are also susceptible to a variety of tumors (Remick et al. 2009) and eye diseases (Beltran et al. 2007; Alleaume et al. 2017).
Although genetic model organisms have traditionally been selected for laboratory suitability, we find it advantageous that mouse lemurs are studied in both their natural environment and in the laboratory, so they can be used to genetically dissect processes in most areas of biology and medicine. In the fall of 2009, we proposed mouse lemurs as a potential new genetic model organism to veterinarians in Stanford’s Department of Comparative Medicine, and a primate specialist (M.A.A.) joined as a key collaborator. Over the next year the group traveled to Duke Lemur Center; MNHN in Brunoy, France; and several field sites around Madagascar, guided by a mouse lemur ecologist (S.Z.) to learn about their field biology and husbandry and to meet some of the leaders in the field.
The Mouse Lemur Research Community
Mouse lemurs were first described in the 18th century by English illustrator John Frederick Miller (Miller and Shaw 1796), and the field’s early history includes one of the most thorough compendia of developmental anatomy, carried out by Swiss explorer Hans Bluntschli in the 1930s (Bluntschli collection, Department of Mammalogy, American Museum of Natural History, New York). Today there are ∼20 mouse lemur research groups around the world, roughly equally divided among laboratory scientists and field biologists. In June 2011, we organized the first meeting ever devoted to mouse lemurs—a mouse lemur genetics workshop—at the Howard Hughes Medical Institute’s Janelia Farm research campus. Among the several dozen participants were leading mouse lemur conservation, evolutionary, and ecology researchers; laboratory scientists studying mouse lemur physiology, behavior, and disease; prominent primate biologists; model organism geneticists; and genomics experts. It was a diverse teach-in with lively discussion, brainstorming, and socializing that fostered interactions among laboratory and field biologists from around the world, many of whom had never met before. The workshop consolidated the field and spurred completion of a high quality mouse lemur reference genome sequence [Jeffrey Rogers, Anne Yoder, Kim Worley (https://www.hgsc.bcm.edu/non-human-primates/mouse-lemur-genome-project); Assembly Mmur_3.0 (accession number GCF_000165445.2) (NCBI annotation release 101 2017)]. The potential of the mouse lemur as a genetic model organism was discussed, and the ethical use of lemurs in research debated. These discussions, and those at a second workshop 4 years later (October 2015) at the MNHN in Paris, shaped the approaches described below.
A Classical Approach to Mouse Lemur Genetics
The initial workshop fueled the idea of creating a genetic model using noninvasive or minimally invasive techniques, like those used by field researchers, that leverages the large standing genetic diversity of mouse lemurs across Madagascar. The idea is that with millions of mouse lemurs, and assuming a de novo mutation rate of ∼1.2 × 10−8/bp per generation (∼50 new mutations in each individual) as shown for other primates (The 1000 Genomes Project Consortium 2010), genetic saturation could be achieved by screening the large pool of existing variants. This is similar to the way human genetics is done, with a focus on thorough phenotyping and genotyping of many individuals and existing mutations, rather than induction of new mutations (Kaiser 2014). We thus began exploring the feasibility of two parallel approaches, a classical genetic approach and a reverse-genetic approach, both of which take advantage of the rich genetic diversity of mouse lemurs.
To initiate a classical genetic approach, we set out to identify mouse lemurs with distinctive traits, the way that Drosophila, mouse, and human genetics began. We developed a deep phenotyping protocol that measures >50 morphological, physiological, and behavioral characteristics of each individual. Many of the assays are based on those developed by the International Mouse Phenotyping Consortium for comprehensive phenotyping of mouse knockout lines, in their effort to create “the first truly comprehensive functional catalogue” of a mammalian genome (Koscielny et al. 2014) (http://www.mousephenotype.org). Our assessment includes a complete veterinary physical exam and blood chemistry panel, with a small amount of blood archived along with cultured skin fibroblasts derived from a 2-mm ear punch biopsy to provide a renewable source of cells and genomic DNA. So far we have screened several hundred mouse lemurs in Malagasy rain forests and laboratory colonies in Europe and the United States, in collaborations with P. Wright and colleagues [Ranomafana National Park (RNP), Madagascar] and F. Aujard, M. Perret, and colleagues (MNHN), and created an extensive mouse lemur phenotype database. Wild mouse lemurs are released back into the forest after phenotyping, and captured again periodically throughout their lives for additional testing, as P. Wright and colleagues have done to follow M. rufus mouse lemurs longitudinally around the Centre ValBio (CVB) field station at RNP for the past 15 years (Figure 3, B–F) (Atsalis 2008; Wright et al. 2012). In 2013 we installed a modern genetics and molecular biology laboratory at the field station to facilitate phenotyping, biological sampling, cell culture, and DNA isolation and amplification (Figure 3F).
From the phenotyping of several hundred M. murinus and M. rufus mouse lemurs we and our collaborators have identified >20 distinct traits, including eye color variants, a progressive eye disease, morbid obesity, hypercholesterolemia, hyperlipidemia, hyperglycemia, cardiac arrhythmias, as well as behavioral and vocalization variants. We have begun genomic sequencing of the phenotyped individuals to define family pedigrees and map the genetic loci underlying each trait.
A Mouse Lemur “Knockout” Library by Sequencing Naturally Occurring Variants
We are also exploring a reverse-genetic approach to establish a mouse lemur knockout collection, equivalent to the one being created for mice by the international consortium. Although targeted gene knockouts can now be generated in primates using engineered site-specific nucleases such as CRISPR, important technical obstacles and ethical issues remain (see above). We propose instead to identify, rather than create, null mutations in mouse lemur genes by screening the large pool of existing genetic variants for naturally occurring null alleles and thereby establish a living library of knockouts in mouse lemur genes, as has begun for humans (MacArthur et al. 2012; Saleheen et al. 2017).
This strategy is predicated on the discovery from genomic sequence analysis of thousands of humans, that each individual carries the surprisingly high mutational load of ∼100–200 putative null (protein-truncating) alleles in different genes, many of them common (>5% allele frequency) but ∼15% rare or private alleles (<1% allele frequency) (MacArthur et al. 2012; Lek et al. 2016). If the same holds true for mouse lemurs, as it appears to for macaques (Fawcett et al. 2011), then sequencing 1000 individuals should identify over 100,000 null alleles. Although these are unlikely to be randomly distributed across the genome, such a large collection should include null alleles in many of the estimated 20,000 protein coding genes, especially if sequenced individuals are from reproductively isolated groups like most mouse lemur populations. With genome sequencing costs down to ∼$1000 and dropping (http://www.genome.gov/sequencingcosts), it is now feasible to sequence genomic DNA from thousands of living mouse lemurs. Most of the putative null alleles found in the human studies are heterozygous, but ∼30% (35 per individual) are homozygous (Lek et al. 2016). If the same is true for mouse lemurs, then for the remaining 70% it would be necessary to search among the relatives in their community to find homozygous individuals, or cross heterozygotes to determine the gene’s full loss-of-function phenotype.
These classical and reverse-genetic approaches set the stage for systematic genetic analysis of a nonhuman primate. The major obstacle for this ambitious plan is to conduct these approaches at a scale to achieve genetic saturation. Because neither approach requires technical sophistication, we have embarked on a citizen science effort involving Malagasy high school students across the country, as described below. This effort could achieve the equivalent of the $1-billion mouse knockout project at a fraction of the cost, in a fraction of the time, and with minimal maintenance costs because animals are “maintained” in the wild.
Malagasy Science Education, Citizen Science, and Mouse Lemur Genetics
Science education is in the midst of a transformation, from traditional classroom lectures, rote learning, and cookbook-style science laboratories, to curricula focused on how scientific concepts are experimentally ascertained and with laboratories that engage students with opportunities for scientific inquiry and exploration (Wood 2009; Shah and Martinez 2016). The transformation has begun in some industrialized countries but has yet to reach developing nations. This is a missed opportunity, especially for countries like Madagascar that have some of the most fascinating and unexplored biology in the world literally outside their school doors (Figure 5, A and B). Students live and play near the same remarkable forests that scientists come from around the world to study, yet few Malagasy students are aware of the opportunity this biological treasure trove presents.
Figure 5.
High school students exploring the unique biology outside their school. (A) Lycée Kelilalina, a new high school in a small rural village near CVB field station. (B) Like most schools in Madagascar, it has a “living laboratory” right outside the door, ripe for exploration. (C) Students initializing a catch-and-release field study on mouse lemurs around the school. (D–F) Students assembling powerful paper microscopes (Foldscopes, http://www.foldscope.com) to explore the microscale biology surrounding the school.
Biology education in Madagascar has the potential to become an active, hands-on discovery curriculum in which students are citizen scientists exploring the unique but largely uncharted biology around their schools. We are therefore designing lecture and laboratory units, each centered on a major biological concept, that use the surrounding environment as a living laboratory the students explore with frugal science tools, such as the powerful $1 paper microscopes (“Foldscopes”) invented by our Stanford colleague Manu Prakash (Cybulski et al. 2014) (Figure 5, D–F). We have begun piloting these units at local high schools near the CVB field station (Figure 5). We are also conducting annual workshops for students of Hantanirina Rasamimanana at L’École Normale Supérieure (ENS), University of Antananarivo, who are training to become high school biology teachers. Each year we host several classes of ENS students and faculty at the field station, and provide them with an introductory experience in field biology, molecular biology, cell biology, and genetics using mouse lemurs in the wild to exemplify the topics. We hope to expand this active science discovery curriculum throughout the country, starting with high schools staffed by teachers trained at our rain forest workshops. Our goal is to invigorate Malagasy biology education, and at the same time identify highly motivated students who love exploration and could partner with local and international scientists exploring Madagascar’s biology, such as our genetic studies of the biology, behavior, health, and conservation of local mouse lemurs.
Another goal of the education plan is to develop the university genetics and molecular biology curriculum. Currently, there is no laboratory course and little equipment in Madagascar to learn genetics and molecular biology approaches, even at the premier universities. We are seeking funds to establish a basic molecular biology laboratory like the one we installed at the field station, and use it to develop a laboratory component for biology courses at University of Antananarivo. Sadly, virtually all of the biodiversity and population genetics samples collected in Madagascar are exported out of the country for analysis. This is a big administrative burden and cost for researchers, and it limits engagement of Malagasy students in genetics research and development of scientific capacity in Madagascar. We envision that samples collected throughout the country by international and domestic researchers, as well as by high school students doing citizen science projects, could be analyzed by university students in the new molecular biology laboratory.
Potential Impact on the Science of Evolution, Ecology, and Conservation
The canonical genetic model organisms were originally selected for laboratory studies. Some of these, as well as some newer model organisms, are now studied in the wild, broadening the impact of genetic model organisms to areas of biology previously outside their purview, such as evolution and ecology. An important advantage of mouse lemurs is that they are already studied in the wild as well as in the laboratory, so their establishment as a genetic model organism could rapidly affect almost every area of primate biology and health.
One of the most urgent applications is conservation biology. Madagascar is a land of exceptional beauty and the world’s premier hotspot of biodiversity (Myers et al. 2000), home to over 700 vertebrate, 5000 invertebrate, and 8000 plant species found nowhere else (Goodman and Benstead 2005; Callmander et al. 2011). Yet much of this biodiversity is threatened by ravaging of its rain forests for logging, agriculture, and mining (Mittermeier et al. 2004). Although conservationists are working fervently to stave off the crisis, a thorough understanding of how specific genes and mutations influence the health, survival, and adaptability of each ecological community, species, and individual in the face of such rapid environmental change would inform conservation strategies.
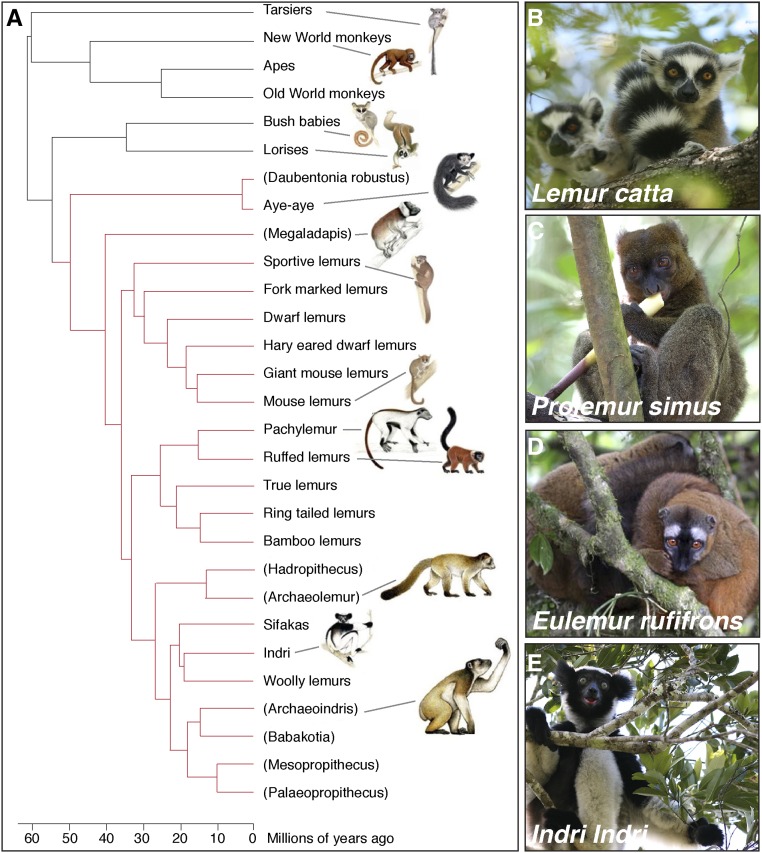
An especially visible aspect of this crisis is the threat to the island’s lemur species (Schwitzer et al. 2014). Lemurs are “Darwin’s finches” of the primates (Martin 1972; Yoder 2013). The island was colonized some 60 MYA by an ancestral primate thought to have rafted across the Mozambique Channel from mainland Africa (Simpson 1940; Poux et al. 2005; Ali and Huber 2010). Lemurs subsequently radiated into >100 species from the gorilla-size sloth lemurs (Archaeoindris fontoynontii) that once roamed the island, to the common ring tail lemur (Lemur catta), the popular Coquerel’s sifaka (Propithecus coquereli), the exotic aye-aye (Daubentonia madagascariensis), and the tiny mouse lemurs featured here (Mittermeier et al. 2010) (Figure 6). Today lemurs are the most threatened mammalian group on earth (Schwitzer et al. 2014), with at least 17 species already extinct, 24 species critically endangered, and all extant species threatened due to habitat destruction (Schwitzer et al. 2013). Although the ecology, behavior, and phylogeny of lemurs have received significant research attention, little is known about their genes, physiology, and diseases and how these impact their evolution and conservation. Establishment of mouse lemurs as a genetic model organism could bring these questions to the fore.
Figure 6.
“Darwin’s finches” of the primates. (A) Phylogenetic tree of the ∼100 species of lemurs, all endemic only to Madagascar. Parentheses, extinct species. In the ∼60 MY since their common ancestor colonized the island, lemurs radiated into diverse forms and functions adapted to nearly every ecological niche, like Darwin’s finches did in the Galapagos Islands. (B–E) The lemur clade includes (B) the iconic ring-tailed lemur L. catta featured as King Julien XIII in the Dreamworks movie Madagascar, (C) the greater bamboo lemur Prolemur simus that can metabolize the cyanide in ingested bamboo, (D) the red-fronted brown lemur Eulemur rufifrons with its distinctive sexually dichromatic coat, and (E) the indri Indri indri with its bellowing song that can be heard a kilometer away. Like most of Madagascar’s legendary endemic flora and fauna, all lemurs are critically threatened by deforestation, although gray mouse lemurs have International Union for Conservation of Nature “least concern” status because of their abundance. Phylogenetic tree adapted from Herrera and Davalos (2016). Photo credits: (B–E) Guy Albertelli.
Impact on Development in Madagascar
Madagascar is among the richest countries in the word in its biodiversity, natural resources, and cultural diversity. But Madagascar is also one of the economically poorest countries (average daily income ∼70 cents) (International Monetary Fund 2016), facing the environmental crisis noted above along with equally pressing problems in food insecurity, sanitation, health, education, economics, and politics. These interacting conditions keep Madagascar and other developing nations stuck in a “poverty trap,” and it will likely require sustained effort addressing each of them to effect change (Bowles et al. 2006; Ngonghala et al. 2014). We hope the proposal described here spawns a new model organism and deep understanding of primate biology, while establishing a new and ethical way of doing genetics that bridges biological, behavioral, medical, and conservation research. We also fervently hope it shows how hands-on science education can help transform a developing country by creating scientific and economic opportunities that pave the way to health and prosperity.
Acknowledgments
We thank the many people who contributed to the ideas and development of this work. In particular, we thank our collaborators Martine Perret, Fabienne Aujard, Jérémy Terrien, Fathia Djelti, and Fabien Pifferi, and colleagues at Muséum National D’Histoire Naturelle in France, who pioneered the laboratory study of mouse lemurs and graciously welcomed us into their research world and generously shared their expertise. We thank Anne Yoder and colleagues at Duke Lemur Center; Elke Zimmerman and Ute Radespiel at University of Hannover; Jean-Michel Verdier, Corinne Lautier, and colleagues at Université de Montpellier; and Martin Bauert at Zoo Zurich for hosting us at their facilities and introducing us to their research and their mouse lemur colonies and methods. We are grateful to our collaborator Patricia Wright of Stony Brook University, who has dedicated her career to the study and preservation of Madagascar’s biodiversity. Her success in establishing the Ranomafana National Park and building the Centre ValBio (CVB) research station has opened the way for generations of students, conservationists, and researchers (including us), and her enthusiastic collaboration has been invaluable in our entry into the field. We thank our partners in Madagascar, including: Josiane Rakotonirina and colleagues at Ranomafana National Park and the Madagascar National Park leadership for their interest and support of research; Benjamin Andriamihaja and his team at the Madagascar Institute for the Conservation of Tropical Environments (MICET) for masterfully handling the logistical aspects of our research in Madagascar; Jean-Claude Razafimahaimodison, Pascal Rabeson, John Cadle, and the rest of the CVB staff for help installing and managing the laboratory, and for facilitating and graciously hosting our research and stay each year; James and Robin Hernnstein for their vision, guidance, and generous support of CVB; Hantanirina Rasamimanana and her colleagues and students at the École Normale Supérieur of University of Antananarivo; and Mbola Rajeriniaina and Rence Randrianindrina at Lycée Kelilalina for their devotion to improving science education in Madagascar, and their collaboration and support of the workshops. A special thanks to our CVB field guides Pierre Lahitsara, Victor Rasendry, Justin Rakotonjatovo, Dina Andrianoely, and Francois Zakamanana, and to our student colleagues Hajanirina Razafindrakoto and Tojo Razanaparany for their exceptional collaboration in phenotyping mouse lemurs. We thank Jeffrey Rogers and Kim Worley at Baylor College of Medicine, and Anne Yoder for their leadership in completing and sharing the mouse lemur reference genome sequence. We appreciate the contributions of all the participants of the mouse lemur genetics workshops, and the valuable early discussions about the project with Evan Eichler, Ross Metzger, David Kingsley, and Gail Martin. We thank interns Emily Willick, Nira Krasnow, Marie Ezran, and Kate Apostolou for help designing the biology curriculum for the education workshops; Guy Albertelli for photography; and other laboratory members and Stanford University colleagues, especially Matthew Footer, Hernan Espinoza, and Jonathan James, for their guidance and help establishing the CVB laboratory. It has been a pleasure working with Stanford professors Michelle Barry, Matthew Bonds, Manu Prakash, and their colleagues in bridging research, science education, and health in Ranomafana. We greatly appreciate the administrative support of Maria Petersen, Jessica Metzger, John Kennedy, and Kristine Kerivan. This work was supported by the Howard Hughes Medical Institute and the Vera Moulton Wall Center of Stanford University.
Footnotes
These authors contributed equally to this work.
Communicating editor: O. Hobert
Literature Cited
- Abbott A., 2010. Mouse project to find each gene’s role. Nature 465: 410. [DOI] [PubMed] [Google Scholar]
- Ali J. R., Huber M., 2010. Mammalian biodiversity on Madagascar controlled by ocean currents. Nature 463: 653–656. [DOI] [PubMed] [Google Scholar]
- Alleaume C., Mrini M. E., Laloy E., Marchal J., Aujard F., et al. , 2017. Scleral and corneal xanthomatous inflammation in a gray mouse lemur (Microcebus murinus). Vet. Ophthalmol. 20: 177–180. [DOI] [PubMed] [Google Scholar]
- Andrade M. C., Ribeiro C. T., Silva V. F., Molinaro E. M., Goncalves M. A., et al. , 2004. Biologic data of Macaca mulatta, Macaca fascicularis, and Saimiri sciureus used for research at the Fiocruz primate center. Mem. Inst. Oswaldo Cruz 99: 581–589. [PubMed] [Google Scholar]
- Andres M., Solignac M., Perret M., 2003. Mating system in mouse lemurs: theories and facts, using analysis of paternity. Folia Primatol. (Basel) 74: 355–366. [DOI] [PubMed] [Google Scholar]
- Andriaholinirina N., Baden A., Blanco M., Chikhi L., Cooke A., et al. , 2014. Microcebus murinus. The IUCN Red List of Threatened Species. International Union for Conservation of Nature and Natural Resources, Gland, Switzerland. [Google Scholar]
- Atsalis S., 2008. A Natural History of the Brown Mouse Lemur. Pearson/Prentice Hall, Upper Saddle River, NJ. [Google Scholar]
- Aujard F., Cayetanot F., Bentivoglio M., Perret M., 2006. Age-related effects on the biological clock and its behavioral output in a primate. Chronobiol. Int. 23: 451–460. [DOI] [PubMed] [Google Scholar]
- Baker M., 2013. Neuroscience: through the eyes of a mouse. Nature 502: 156–158. [DOI] [PubMed] [Google Scholar]
- Baron R. M., Choi A. J., Owen C. A., Choi A. M., 2012. Genetically manipulated mouse models of lung disease: potential and pitfalls. Am. J. Physiol. Lung Cell. Mol. Physiol. 302: L485–L497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal M. F., 2010. Parkinson’s disease: a model dilemma. Nature 466: S8–S10. [DOI] [PubMed] [Google Scholar]
- Beltran W. A., Vanore M., Ollivet F., Nemoz-Bertholet F., Aujard F., et al. , 2007. Ocular findings in two colonies of gray mouse lemurs (Microcebus murinus). Vet. Ophthalmol. 10: 43–49. [DOI] [PubMed] [Google Scholar]
- Bentzon J. F., Falk E., 2010. Atherosclerotic lesions in mouse and man: is it the same disease? Curr. Opin. Lipidol. 21: 434–440. [DOI] [PubMed] [Google Scholar]
- Biggar K. K., Wu C. W., Tessier S. N., Zhang J., Pifferi F., et al. , 2015. Primate torpor: regulation of stress-activated protein kinases during daily torpor in the gray mouse lemur, Microcebus murinus. Genomics Proteomics Bioinformatics 13: 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bons N., Rieger F., Prudhomme D., Fisher A., Krause K. H., 2006. Microcebus murinus: a useful primate model for human cerebral aging and Alzheimer’s disease? Genes Brain Behav. 5: 120–130. [DOI] [PubMed] [Google Scholar]
- Bowles S., Durlauf S. N., Hoff K., 2006. Poverty Traps. Princeton University Press, Princeton, NJ. [Google Scholar]
- Callmander M. W., Phillipson P. B., Schatz G. E., Andriambololonera S., Rabarimanarivo M., et al. , 2011. The endemic and non-endemic vascular flora of Madagascar updated. Plant Ecol. Evol. 144: 121–125. [Google Scholar]
- Chan A. W., Chong K. Y., Martinovich C., Simerly C., Schatten G., 2001. Transgenic monkeys produced by retroviral gene transfer into mature oocytes. Science 291: 309–312. [DOI] [PubMed] [Google Scholar]
- Chang J., Wagner J. L., Kornegay R. W., 1980. Fatal Yersinia pseudotuberculosis infection in captive bushbabies. J. Am. Vet. Med. Assoc. 177: 820–821. [PubMed] [Google Scholar]
- Chatterjee H. J., Ho S. Y., Barnes I., Groves C., 2009. Estimating the phylogeny and divergence times of primates using a supermatrix approach. BMC Evol. Biol. 9: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J. A., Izard M. K., Simons E. L., 1987. Description of ultrasonic vocalizations of the mouse lemur (Microcebus murinus) and the fat-tailed dwarf lemur (Cheirogaleus medius). Am. J. Primatol. 13: 181–185. [DOI] [PubMed] [Google Scholar]
- Chong J. X., Buckingham K. J., Jhangiani S. N., Boehm C., Sobreira N., et al. , 2015. The genetic basis of Mendelian phenotypes: discoveries, challenges, and opportunities. Am. J. Hum. Genet. 97: 199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, F. S., 2015 NIH Will No Longer Support Biomedical Research on Chimpanzees National Institutes of Health. Available at: https://www.nih.gov/about-nih/who-we-are/nih-director/statements/nih-will-no-longer-support-biomedical-research-chimpanzees. [Google Scholar]
- Collins F. S., Rossant J., Wurst W., 2007. A mouse for all reasons. Cell 128: 9–13. [DOI] [PubMed] [Google Scholar]
- Cox L. A., Comuzzie A. G., Havill L. M., Karere G. M., Spradling K. D., et al. , 2013. Baboons as a model to study genetics and epigenetics of human disease. ILAR J. 54: 106–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski J. S., Clements J., Prakash M., 2014. Foldscope: origami-based paper microscope. PLoS One 9: e98781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. H., 2004. The age of model organisms. Nat. Rev. Genet. 5: 69–76. [DOI] [PubMed] [Google Scholar]
- Deppe A. M., Baden A., Wright P. C., 2016. The Dwarf and Mouse Lemurs of Madagascar: Biology, Behavior and Conservation Biogeography of the Cheirogaleidae. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- Elsea S. H., Lucas R. E., 2002. The mousetrap: what we can learn when the mouse model does not mimic the human disease. ILAR J. 43: 66–79. [DOI] [PubMed] [Google Scholar]
- Engle S. J., Womer D. E., Davies P. M., Boivin G., Sahota A., et al. , 1996. HPRT-APRT-deficient mice are not a model for lesch-nyhan syndrome. Hum. Mol. Genet. 5: 1607–1610. [DOI] [PubMed] [Google Scholar]
- Fan Y., Huang Z. Y., Cao C. C., Chen C. S., Chen Y. X., et al. , 2013. Genome of the Chinese tree shrew. Nat. Commun. 4: 1426. [DOI] [PubMed] [Google Scholar]
- Fawcett G. L., Raveendran M., Deiros D. R., Chen D., Yu F., et al. , 2011. Characterization of single-nucleotide variation in Indian-origin rhesus macaques (Macaca mulatta). BMC Genomics 12: 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S., Johnston M., 2005. Cell biology. Whither model organism research? Science 307: 1885–1886. [DOI] [PubMed] [Google Scholar]
- Fischer K. E., Austad S. N., 2011. The development of small primate models for aging research. ILAR J. 52: 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredsted T., Pertoldi C., Schierup M. H., Kappeler P. M., 2005. Microsatellite analyses reveal fine-scale genetic structure in grey mouse lemurs (Microcebus murinus). Mol. Ecol. 14: 2363–2372. [DOI] [PubMed] [Google Scholar]
- Fuchs, E., and S. Corbach-Sohle, 2010 Tree Shrews, pp. 262–275 in The UFAW Handbook on the Care and Management of Laboratory and Other Research Animals, Ed. 8, edited by R. Hubrecht and J. Kirkwood. Universities Federation for Animal Welfare, St. Albans, United Kingdom. [Google Scholar]
- Geerts H., 2009. Of mice and men: bridging the translational disconnect in CNS drug discovery. CNS Drugs 23: 915–926. [DOI] [PubMed] [Google Scholar]
- Glazko G. V., Nei M., 2003. Estimation of divergence times for major lineages of primate species. Mol. Biol. Evol. 20: 424–434. [DOI] [PubMed] [Google Scholar]
- Goodall J., 1967. My Friends the Wild Chimpanzees. National Geographic Society, Washington, DC. [Google Scholar]
- Goodall J., 1968. Behaviour of free-living chimpanzees of the Gombe Stream Area. Animal Behaviour Monographs 1: 161–311. [Google Scholar]
- Goodman S. M., Benstead J. P., 2005. Updated estimates of biotic diversity and endemism for Madagascar. Oryx 39: 73–77. [Google Scholar]
- Groenen M. A., Archibald A. L., Uenishi H., Tuggle C. K., Takeuchi Y., et al. , 2012. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491: 393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb B. R., Boucher R. C., 1999. Pathophysiology of gene-targeted mouse models for cystic fibrosis. Physiol. Rev. 79: S193–S214. [DOI] [PubMed] [Google Scholar]
- Gursky S., 2007. The Spectral Tarsier. Taylor & Francis Group, Abingdon, United Kingdom. [Google Scholar]
- Harel I., Benayoun B. A., Machado B., Singh P. P., Hu C. K., et al. , 2015. A platform for rapid exploration of aging and diseases in a naturally short-lived vertebrate. Cell 160: 1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera J. P., Davalos L. M., 2016. Phylogeny and divergence times of lemurs inferred with recent and ancient fossils in the tree. Syst. Biol. 65: 772–791. [DOI] [PubMed] [Google Scholar]
- Herrero J., Muffato M., Beal K., Fitzgerald S., Gordon L., et al. , 2016. Ensembl comparative genomics resources. Database: bav096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath J. E., Willard H. F., 2007. Primate comparative genomics: lemur biology and evolution. Trends Genet. 23: 173–182. [DOI] [PubMed] [Google Scholar]
- Human Ageing Genomic Resources , 2017. AnAge database of animal ageing and longevity. Available at: http://genomics.senescence.info/species/.
- Huttley G. A., Wakefield M. J., Easteal S., 2007. Rates of genome evolution and branching order from whole genome analysis. Mol. Biol. Evol. 24: 1722–1730. [DOI] [PubMed] [Google Scholar]
- International Monetary Fund , 2016. World economic outlook database. Available at: https://www.imf.org/external/pubs/ft/weo/2016/01/weodata/index.aspx.
- Izpisua Belmonte J. C., Callaway E. M., Caddick S. J., Churchland P., Feng G., et al. , 2015. Brains, genes, and primates. Neuron 86: 617–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones F. C., Grabherr M. G., Chan Y. F., Russell P., Mauceli E., et al. , 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484: 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser J., 2014. The hunt for missing genes. Science 344: 687–689. [DOI] [PubMed] [Google Scholar]
- Kim H. K., Missiakas D., Schneewind O., 2014. Mouse models for infectious diseases caused by Staphylococcus aureus. J. Immunol. Methods 410: 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Nam H. G., Valenzano D. R., 2016. The short-lived African turquoise killifish: an emerging experimental model for ageing. Dis. Model. Mech. 9: 115–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koscielny G., Yaikhom G., Iyer V., Meehan T. F., Morgan H., et al. , 2014. The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Res. 42: D802–D809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn M. R., Bradley A., Robertson E. J., Evans M. J., 1987. A potential animal model for Lesch-Nyhan syndrome through introduction of HPRT mutations into mice. Nature 326: 295–298. [DOI] [PubMed] [Google Scholar]
- Languille S., Blanc S., Blin O., Canale C. I., Dal-Pan A., et al. , 2012. The grey mouse lemur: a non-human primate model for ageing studies. Ageing Res. Rev. 11: 150–162. [DOI] [PubMed] [Google Scholar]
- Lek M., Karczewski K. J., Minikel E. V., Samocha K. E., Banks E., et al. , 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leliveld L. M., Scheumann M., Zimmermann E., 2011. Acoustic correlates of individuality in the vocal repertoire of a nocturnal primate (Microcebus murinus). J. Acoust. Soc. Am. 129: 2278–2288. [DOI] [PubMed] [Google Scholar]
- Li W. H., Ellsworth D. L., Krushkal J., Chang B. H., Hewett-Emmett D., 1996. Rates of nucleotide substitution in primates and rodents and the generation-time effect hypothesis. Mol. Phylogenet. Evol. 5: 182–187. [DOI] [PubMed] [Google Scholar]
- Liao B. Y., Zhang J., 2008. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc. Natl. Acad. Sci. USA 105: 6987–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino, M., K. Kuczynski, N. Rodriguez, and T. Schap, 2017 Expenditures on Children by Families, 2015. Miscellaneous Publication No. 1528-2016. U.S. Department of Agriculture, Center for Nutrition Policy and Promotion, Alexandria, VA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Chen Y., Niu Y., Zhang K., Kang Y., et al. , 2014. TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell Stem Cell 14: 323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhrs M. L., Dammhahn M., Kappeler P. M., Fichtel C., 2009. Spatial memory in the grey mouse lemur (Microcebus murinus). Anim. Cogn. 12: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Li M., Su B., 2016. Application of the genome editing tool CRISPR/Cas9 in non-human primates. Dongwuxue Yanjiu 37: 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur D. G., Balasubramanian S., Frankish A., Huang N., Morris J., et al. , 2012. A systematic survey of loss-of-function variants in human protein-coding genes. Science 335: 823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal J., Dal-Pan A., Epelbaum J., Blanc S., Mueller S., et al. , 2013. Calorie restriction and resveratrol supplementation prevent age-related DNA and RNA oxidative damage in a non-human primate. Exp. Gerontol. 48: 992–1000. [DOI] [PubMed] [Google Scholar]
- Margulies E. H., Cooper G. M., Asimenos G., Thomas D. J., Dewey C. N., et al. , 2007. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 17: 760–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. D., 1972. Adaptive radiation and behaviour of the Malagasy lemurs. Proc. R. Soc. Lond. B Biol. Sci. 264: 295–352. [DOI] [PubMed] [Google Scholar]
- Mestas J., Hughes C. C., 2004. Of mice and not men: differences between mouse and human immunology. J. Immunol. 172: 2731–2738. [DOI] [PubMed] [Google Scholar]
- Miller J. F., Shaw G., 1796. Cimelia Physica: Figures of Rare and Curious Quadrupeds, Birds, etc. Together With Several of the Most Elegant Plants. Thomas Bensley, London. [Google Scholar]
- Mittermeier R. A., Gil P. R., Hoffmann M., Pilgrim J., Brooks T., et al. , 2004. Hotspots Revisited: Earth’s Biologically Richest and Most Endangered Terrestrial Ecoregions. Conservation International and CEMEX, Mexico City. [Google Scholar]
- Mittermeier R. A., Edward L. E., Richardson M., Schwitzer C., Langrand O., et al. , 2010. Lemurs of Madagascar, Ed. 3. Conservation International, Arlington, VA. [Google Scholar]
- Murphy W. J., Eizirik E., Johnson W. E., Zhang Y. P., Ryder O. A., et al. , 2001. Molecular phylogenetics and the origins of placental mammals. Nature 409: 614–618. [DOI] [PubMed] [Google Scholar]
- Myers N., Mittermeier R. A., Mittermeier C. G., da Fonseca G. A., Kent J., 2000. Biodiversity hotspots for conservation priorities. Nature 403: 853–858. [DOI] [PubMed] [Google Scholar]
- Nature Editors , 2009. Time to connect. Nature 459: 483. [DOI] [PubMed] [Google Scholar]
- NCBI annotation release 101 , 2017. Microcebus murinus Annotation Release 101. National Center for Biotechnology Information, Bethesda MD. [Google Scholar]
- Newmark P. A., Sanchez Alvarado A., 2002. Not your father’s planarian: a classic model enters the era of functional genomics. Nat. Rev. Genet. 3: 210–219. [DOI] [PubMed] [Google Scholar]
- Ngonghala C. N., Plucinski M. M., Murray M. B., Farmer P. E., Barrett C. B., et al. , 2014. Poverty, disease, and the ecology of complex systems. PLoS Biol. 12: e1001827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y., Shen B., Cui Y., Chen Y., Wang J., et al. , 2014. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156: 836–843. [DOI] [PubMed] [Google Scholar]
- Nowak R. M., 1999. Walker’s Primates of the World. Johns Hopkins University Press, Baltimore. [Google Scholar]
- Olsson I. A., Sandoe P., 2010. “What’s wrong with my monkey?” Ethical perspectives on germline transgenesis in marmosets. Transgenic Res. 19: 181–186. [DOI] [PubMed] [Google Scholar]
- Panchal S. K., Brown L., 2011. Rodent models for metabolic syndrome research. J. Biomed. Biotechnol. 2011: 351982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel C. L., Nereng K. S., Ohgi K. A., Cole B. L., Colosimo P. F., et al. , 2001. The genetic architecture of divergence between threespine stickleback species. Nature 414: 901–905. [DOI] [PubMed] [Google Scholar]
- Perlman R. L., 2016. Mouse models of human disease: an evolutionary perspective. Evol. Med. Public Health 2016: 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret M., 1982. Influence du groupement social sur la reproduction de la femelle de Microcebus murinus (Miller, 1777). Z. Tierpsychol. 60: 47–65. [PubMed] [Google Scholar]
- Phinney A. L., Horne P., Yang J., Janus C., Bergeron C., et al. , 2003. Mouse models of Alzheimer’s disease: the long and filamentous road. Neurol. Res. 25: 590–600. [DOI] [PubMed] [Google Scholar]
- Poux C., Madsen O., Marquard E., Vieites D. R., de Jong W. W., et al. , 2005. Asynchronous colonization of Madagascar by the four endemic clades of primates, tenrecs, carnivores, and rodents as inferred from nuclear genes. Syst. Biol. 54: 719–730. [DOI] [PubMed] [Google Scholar]
- Radespiel U., 2000. Sociality in the gray mouse lemur (Microcebus murinus) in northwestern Madagascar. Am. J. Primatol. 51: 21–40. [DOI] [PubMed] [Google Scholar]
- Radespiel U., 2007. Ecological diversity and seasonal adaptations of mouse lemurs (Microcebus spp.), pp. 211–234 in Lemurs: Ecology and Adaptation, edited by Gould L., Sauther M. L. Springer, Boston. [Google Scholar]
- Radespiel U., Sarikaya Z., Zimmermann E., 2001. Sociogenetic structure in a free-living nocturnal primate population: sex-specific differences in the grey mouse lemur (Microcebus murinus). Behav. Ecol. Sociobiol. 50: 493–502. [Google Scholar]
- Rassoul R. A., Alves S., Pantesco V., De Vos J., Michel B., et al. , 2010. Distinct transcriptome expression of the temporal cortex of the primate Microcebus murinus during brain aging vs. Alzheimer’s disease-like pathology. PLoS One 5: e12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W., Bermange A. L., Murfitt K. J., Jennings J. R., Sanchez Alvarado A., 2005. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell 8: 635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remick A. K., Van Wettere A. J., Williams C. V., 2009. Neoplasia in prosimians: case series from a captive prosimian population and literature review. Vet. Pathol. 46: 746–772. [DOI] [PubMed] [Google Scholar]
- Rittirsch D., Hoesel L. M., Ward P. A., 2007. The disconnect between animal models of sepsis and human sepsis. J. Leukoc. Biol. 81: 137–143. [DOI] [PubMed] [Google Scholar]
- Rogers C. S., Stoltz D. A., Meyerholz D. K., Ostedgaard L. S., Rokhlina T., et al. , 2008. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science 321: 1837–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C. N., French J. A., Orti G., 2007. Germ-line chimerism and paternal care in marmosets (Callithrix kuhlii). Proc. Natl. Acad. Sci. USA 104: 6278–6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe N., Myers M., 2016. All the World’s Primates. Pogonias Press, Charlestown, RI. [Google Scholar]
- Saleheen D., Natarajan P., Armean I., Zhao W., Rasheed A., et al. , 2017. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature 544: 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E., Suemizu H., Shimada A., Hanazawa K., Oiwa R., et al. , 2009. Generation of transgenic non-human primates with germline transmission. Nature 459: 523–527. [DOI] [PubMed] [Google Scholar]
- Sato K., Oiwa R., Kumita W., Henry R., Sakuma T., et al. , 2016. Generation of a nonhuman primate model of severe combined immunodeficiency using highly efficient genome editing. Cell Stem Cell 19: 127–138. [DOI] [PubMed] [Google Scholar]
- Schnabel J., 2008. Neuroscience: standard model. Nature 454: 682–685. [DOI] [PubMed] [Google Scholar]
- Schwitzer C., Mittermeier R. A., Davies N., Johnson S., Ratsimbazafy J., et al. , 2013. Lemurs of Madagascar: A Strategy for Their Conservation 2013–2016. IUCN SSC Primate Specialist Group, Bristol Conservation and Science Foundation, and Conservation International, Bristol, United Kingdom. [Google Scholar]
- Schwitzer C., Mittermeier R. A., Johnson S. E., Donati G., Irwin M., et al. , 2014. Conservation. Averting lemur extinctions amid Madagascar’s political crisis. Science 343: 842–843. [DOI] [PubMed] [Google Scholar]
- Shah H. R., Martinez L. R., 2016. Current approaches in implementing citizen science in the classroom. J. Microbiol. Biol. Educ. 17: 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G. G., 1940. Mammals and land bridges. J. Wash. Acad. Sci. 30: 137–163. [Google Scholar]
- Skarnes W. C., Rosen B., West A. P., Koutsourakis M., Bushell W., et al. , 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474: 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckow, M., S. H. Weisbroth, and C. L. Franklin (Editors), 2006 The Laboratory Rat, Ed. 2 (American College of Laboratory Animal Medicine Series). Elsevier, Amsterdam. [Google Scholar]
- Sweeney C. G., Curran E., Westmoreland S. V., Mansfield K. G., Vallender E. J., 2012. Quantitative molecular assessment of chimerism across tissues in marmosets and tamarins. BMC Genomics 13: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Sparman M., Ramsey C., Ma H., Lee H. S., et al. , 2012. Generation of chimeric rhesus monkeys. Cell 148: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif S. D., Smucny D. A., Abbott D. H., Mansfield K., Schultz-Darken N., et al. , 2003. Reproduction in captive common marmosets (Callithrix jacchus). Comp. Med. 53: 364–368. [PubMed] [Google Scholar]
- Tartabini A., 1991. Mother-infant cannibalism in thick-tailed bushbabies (Galago crassicaudatus umbrosus). Primates 32: 379–383. [Google Scholar]
- The 1000 Genomes Project Consortium , 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Staff of the Jackson Laboratory , 1966. Biology of the Laboratory Mouse. Dover Publications, Mineola, NY. [Google Scholar]
- Thomas K. R., Capecchi M. R., 1987. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51: 503–512. [DOI] [PubMed] [Google Scholar]
- Tong C., Li P., Wu N. L., Yan Y., Ying Q. L., 2010. Production of p53 gene knockout rats by homologous recombination in embryonic stem cells. Nature 467: 211–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend W. R., 2001. Mammalian species: Callithrix pygmaea. Am. Soc. Mammal. 665: 1–6. [Google Scholar]
- Vinson A., Prongay K., Ferguson B., 2013. The value of extended pedigrees for next-generation analysis of complex disease in the rhesus macaque. ILAR J. 54: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidt A., Hagenah N., Randrianambinina B., Radespiel U., Zimmermann E., 2004. Social organization of the golden brown mouse lemur (Microcebus ravelobensis). Am. J. Phys. Anthropol. 123: 40–51. [DOI] [PubMed] [Google Scholar]
- Weisrock D. W., Rasoloarison R. M., Fiorentino I., Ralison J. M., Goodman S. M., et al. , 2010. Delimiting species without nuclear monophyly in Madagascar’s mouse lemurs. PLoS One 5: e9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer B., Diethard T., Kappeler P. M., 2002. The genetic population structure of the gray mouse lemur (Microcebus murinus), a basal primate from Madagascar. Behav. Ecol. Sociobiol. 52: 166–175. [Google Scholar]
- Wood W. B., 2009. Innovations in teaching undergraduate biology and why we need them. Annu. Rev. Cell Dev. Biol. 25: 93–112. [DOI] [PubMed] [Google Scholar]
- Wright P. C., Simons E. L., Gursky S. (Editors), 2003. Tarsiers: Past, Present, and Future. Rutgers University Press, New Brunswick, NJ. [Google Scholar]
- Wright P. C., Erhart E. M., Tecot S., Baden A. L., Arrigo-Nelson S. J., et al. , 2012. Long-term lemur research at Centre ValBio, Ranomafana National Park, pp. 67–100 in Long-Term Field Studies of Primates, edited by Kappeler P. M., Watts D. P. Springer-Verlag, Berlin. [Google Scholar]
- Wrogemann D., Radespiel U., Zimmermann E., 2001. Comparison of reproductive characteristics and changes in body weight between captive populations of rufous and gray mouse lemurs. Int. J. Primatol. 22: 91–108. [Google Scholar]
- Yoder A. D., 2013. The lemur revolution starts now: the genomic coming of age for a non-model organism. Mol. Phylogenet. Evol. 66: 442–452. [DOI] [PubMed] [Google Scholar]
- Yoder A. D., Rasoloarison R. M., Goodman S. M., Irwin J. A., Atsalis S., et al. , 2000. Remarkable species diversity in Malagasy mouse lemurs (primates, Microcebus). Proc. Natl. Acad. Sci. USA 97: 11325–11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr S. M., Roach R. G., Haring D., Taylor J., Cameron F. H., et al. , 2014. Life history profiles for 27 strepsirrhine primate taxa generated using captive data from the Duke Lemur Center. Sci. Data 1: 140019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Tang Z. Y., Klumpp B., Wolff-Vorbeck G., Barth H., et al. , 2002. Primary hepatocytes of Tupaia belangeri as a potential model for hepatitis C virus infection. J. Clin. Invest. 109: 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohdy S., Gerber B. D., Tecot S., Blanco M. B., Winchester J. M., et al. , 2014. Teeth, sex, and testosterone: aging in the world’s smallest primate. PLoS One 9: e109528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zschaler J., Schlorke D., Arnhold J., 2014. Differences in innate immune response between man and mouse. Crit. Rev. Immunol. 34: 433–454. [PubMed] [Google Scholar]