Abstract
Meiotic homologous recombination, a critical event for ensuring faithful chromosome segregation and creating genetic diversity, is initiated by programmed DNA double-strand breaks (DSBs) formed at recombination hotspots. Meiotic DSB formation is likely to be influenced by other DNA-templated processes including transcription, but how DSB formation and transcription interact with each other has not been understood well. In this study, we used fission yeast to investigate a possible interplay of these two events. A group of hotspots in fission yeast are associated with sequences similar to the cyclic AMP response element and activated by the ATF/CREB family transcription factor dimer Atf1-Pcr1. We first focused on one of those hotspots, ade6-3049, and Atf1. Our results showed that multiple transcripts, shorter than the ade6 full-length messenger RNA, emanate from a region surrounding the ade6-3049 hotspot. Interestingly, we found that the previously known recombination-activation region of Atf1 is also a transactivation domain, whose deletion affected DSB formation and short transcript production at ade6-3049. These results point to a possibility that the two events may be related to each other at ade6-3049. In fact, comparison of published maps of meiotic transcripts and hotspots suggested that hotspots are very often located close to meiotically transcribed regions. These observations therefore propose that meiotic DSB formation in fission yeast may be connected to transcription of surrounding regions.
Keywords: meiosis, meiotic recombination, chromatin, transcription, DNA double-strand break formation
MEIOTIC recombination ensures proper chromosome segregation during the first meiotic division and creates genetic diversity (Hunter 2015). It is initiated by programmed DNA double-strand breaks (DSBs), introduced predominantly at chromosomal regions called recombination hotspots (de Massy 2013). Since DSBs are potentially lethal DNA insults, the initiation process of meiotic recombination is regulated in many ways (Keeney et al. 2014). Factors that directly promote the reaction, such as the topoisomerase-like protein Spo11 and its partners, and the chromatin structure are central players of meiotic DSB regulation. In addition, DSB formation is known to be influenced by other chromosomal events such as DNA replication (Borde et al. 2000; Murakami and Keeney 2014; Wu and Nurse 2014) and transcription (de Castro et al. 2012; Sun et al. 2015).
Possible connection between meiotic recombination and transcription has been reported in many organisms. For example, early studies on budding and fission yeasts have demonstrated that transcription factors bind to hotspots and activate recombination (White et al. 1991; Kon et al. 1997). It is also shown that locations of hotspots coincide with those of promoters in various species, including budding yeast (Pan et al. 2011) and Arabidopsis thaliana (Choi et al. 2013). While these observations appear to indicate a functional interaction of the two events, the situation is much more enigmatic: recombination activation by hotspot-binding transcription factors in yeasts is not always accompanied by transcription activation, and is rather context dependent (Mieczkowski et al. 2006; Pan et al. 2011; Zhu and Keeney 2015). In species including fission yeast (see below) (Fowler et al. 2014) and mice (Brick et al. 2012), hotspots are located outside of promoters of protein-coding genes. Therefore, whether and how transcription influences initiation of meiotic recombination is still controversial, and analyzing their interplay may shine a new light on the mechanism of meiotic recombination.
Fission yeast Schizosaccharomyces pombe is an excellent model organism for studying meiotic recombination. Its DSB hotspots are preferentially localized in large intergenic regions, but unlike budding yeast they rarely coincide with transcription promoters (Fowler et al. 2014). Historically, recombination studies in this organism has been led by a hotspot called ade6-M26, which is created by a G/T point mutation within the ade6 ORF, and is associated with a cyclic AMP response element (CRE)-related M26-sequence (ATGACGT; M26 mutation is underlined.) (Ponticelli et al. 1988; Szankasi et al. 1988). This hotspot is intriguing because it is bound and activated by Atf1-Pcr1, a heterodimer of sequence-specific DNA-binding transcription factors (Kon et al. 1997). Both Atf1 and Pcr1 belong to the well-conserved activating transcription factor/CRE binding (ATF/CREB) transcription factor family, and are transcription regulators that bind to promoters of various stress-responsible genes through CRE sequences (Chen et al. 2003). Later analyses have shown that Atf1 and Pcr1 also activate other mutation-created M26-sequence-dependent hotspots located in the ade6 ORF (Fox et al. 2000; Steiner and Smith 2005a). More importantly, Atf1, Pcr1, and the M26 sequence are likely to activate meiotic recombination at authentic hotspots in the fission yeast genome (Steiner and Smith 2005a; Wahls and Davidson 2010; Fowler et al. 2014).
We have been interested in how meiotic DSB formation occurs at fission yeast hotspots, including M26-sequence-dependent hotspots. Our previous studies have proposed that Atf1-Pcr1 facilitates DSB formation at ade6-M26 at least partly by modulating the surrounding chromatin structure (Mizuno et al. 1997; Yamada et al. 2004, 2013). Although recombination at this hotspot is not caused by increased transcription of ade6 messenger RNA (mRNA) (Grimm et al. 1991; Kon et al. 1997), a later study found a short transcript specific to and emanating from around ade6-M26 (Hirota et al. 2008). These previous reports led us to wonder whether Atf1-Pcr1 involves transcription to activate meiotic recombination. In this study, based on the analyses of another M26-sequence-dependent hotspot, ade6-3049, as well as the Atf1 protein, we discuss a possibility that Atf1 promotes DSB formation through inducing transcription from its binding site. Moreover, comparison of previously published genome-wide data sets unveiled proximity of hotspots and meiotically transcribed regions. These observations may suggest that, at least at a subset of fission yeast hotspots, transcription participates in regulation of meiotic recombination initiation.
Materials and Methods
Yeast strains
Yeast strains used in this study are listed in Supplemental Material, Table S1 in File S1. To construct an atf1-HRAΔ strain, atf1 DNA fragment lacking the homologous recombination activation (HRA) region was transformed into an atf1Δ::ura4+ cell.
Northern blotting and RACE analyses
h+/h− diploid cells were induced to enter meiosis by transferring them into nitrogen-free medium, and collected at indicated times after induction. Total RNAs were analyzed by Northern blotting with 32P-labeled DNA probes. Signals were quantified using a Fuji BAS 2000 Bio-Imaging Analyzer (Fujifilm). RACE was performed with SMARTer RACE 5′/3′ Kit (Clontech Laboratories) according to the manufacturer’s instructions.
Tiling array and high throughput sequencing data analysis
The meiotic transcript map and the map of oligonucleotides attached to Rec12 (Rec12-oligo) were previously described in de Castro et al. (2012) and Fowler et al. (2014), respectively. The fission yeast genome annotation (version of May 12, 2016) was downloaded from PomBase (http://www.pombase.org/). The data were processed using Perl (http://www.perl.org/) and R (http://www.r-project.org/).
Chromatin analyses
The chromatin structure around ade6-3049/3057 was examined as previously described in Mizuno et al. (1997), except for the following modifications. Genomic DNA was treated with 0, 20, or 30 units of micrococcal nuclease (MNase) (Takara), digested with Sac I, and separated on a 1.2% agarose gel. The probe was amplified by PCR (see below), and labeled with 32P using the Random Primer DNA Labeling Kit version 2 (Takara).
Other methods and information
Recombination frequency was measured by random-spore analyses. Chromatin immunoprecipitation (ChIP) and budding yeast one-hybrid analyses were performed as described in Yamada et al. (2013) and Morita et al. (2011), respectively. For ChIP of Rec12-DNA linkage, cross-linking was omitted. Antibodies used were anti-H3K9ac (Millipore, Bedford, MA), M2 anti-FLAG (Sigma Chemical, St. Louis, MO), and anti-Atf1 (Takemata et al. 2016). Meiotic DSB fragments were detected as previously described in Steiner et al. (2002). For all statistical analyses, the one-tailed Student’s t-test was used. Primers used for PCR are listed in Table S2 in File S1, or previously described (Yamada et al. 2013).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Reagents are available upon request.
Results and Discussion
Short transcripts associated with the ade6-3049 recombination hotspot
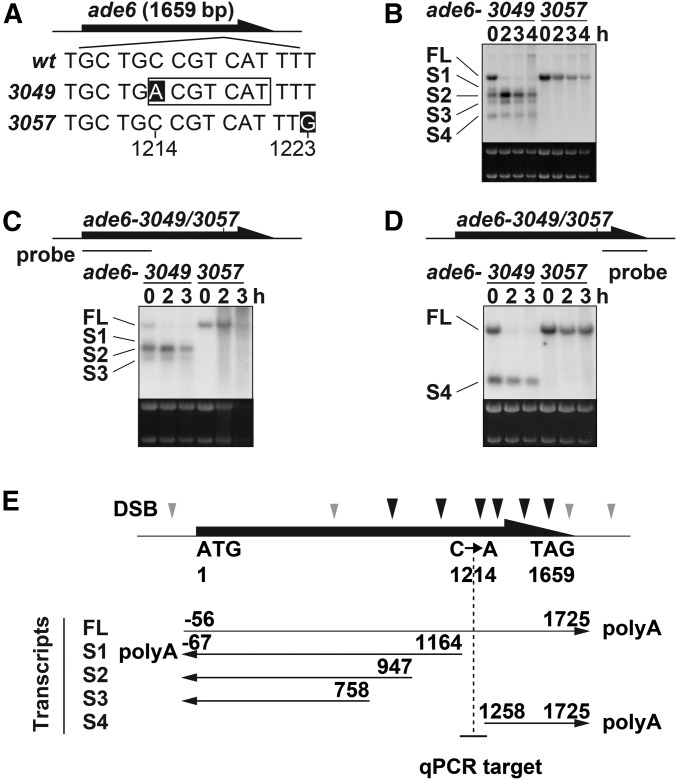
We were intrigued by an ectopic transcript starting from around the ade6-M26 hotspot (Hirota et al. 2008). Wondering whether other M26-sequence-dependent hotspots are also associated with similar transcripts, another M26-sequence-dependent hotspot, ade6-3049, was tested. This hotspot, activated by Atf1-Pcr1, is located near the 3′ end of the ade6 gene and its activity is stronger than that of ade6-M26 (Figure 1A) (Steiner et al. 2002; Steiner and Smith 2005b). h+/h− diploid cells carrying the ade6-3049 hotspot, along with those carrying the nonhotspot control allele ade6-3057, were induced to enter meiosis by nitrogen withdrawal, and their total RNA was analyzed by Northern blotting with a probe recognizing the entire ade6 ORF. The results revealed dramatically different transcript patterns between ade6-3049 and ade6-3057 (Figure 1B), as explained in the next paragraph.
Figure 1.
ade6-3049 mutation induces ectopic transcripts at the ade6-3049 meiotic recombination hotspot. (A) Positions of ade6-3049 and ade6-3057 mutations (white letters). The numbers below the sequences indicate the positions relative to the first A of the ade6 ORF. (B) Transcripts around the ade6-3049/3057. Cells were induced to meiosis by nitrogen depletion and harvested at the indicated time after meiosis induction. Their total RNA was analyzed by Northern blotting using the probe recognizing the full-length ade6 ORF. Ribosomal RNA stained by ethidium bromide is shown as a loading control. (C) Same analyses as (B), but using a probe recognizing 5′ end of the ade6 ORF. (D) Same analyses as (B), but using a probe recognizing 3′ end of the ade6 ORF. (E) Estimated 5′ and 3′ end of ade6 mRNA based on Northern blotting and RACE analyses. Previously identified break sites are indicated by arrowheads, with stronger break sites by black ones (Steiner et al. 2002). The vertical dotted line and the horizontal short line indicate the position of the 3049 mutation and of the quantitative PCR (qPCR) fragment analyzed in Figure 2C, Figure 5B, and Figure S1, respectively. wt, wild type.
In the ade6-3049 cells, multiple RNAs [at least five: full length (FL), and S1–S4] containing ade6 sequence were detected both before and after meiosis induction. In stark contrast to 3049, ade6-3057 cells produced only one ade6 RNA species (FL), which gradually declined through meiosis progression. Therefore, the M26 sequence at ade6-3049 was associated with unique transcripts. These results, together with a previous one on ade6-M26, prompted us to infer that spurious transcription initiation may be a common feature among at least a part of M26-sequence-dependent hotspots.
To gain further insights, the hotspot-associated transcripts around ade6-3049 were thoroughly analyzed. Northern blot analyses using two probes hybridizing to either side (5′ or 3′) of 3049/3057 mutations suggested that the longest RNA present both in 3049 and 3057 cells would be the full-length ade6 mRNA (Figure 1, C and D; transcript FL). Also, all shorter RNAs, specifically those detected in the 3049 cells, were hybridized by either upstream (Figure 1C; transcripts S1, S2, and S3) or downstream (transcript S4) probes (Figure 1D). Transcription start and termination sites were subsequently determined by RACE analyses. Remarkably, four shorter transcripts started from a 500-bp region containing the 3049 site, with three of them being transcribed to upstream of the ade6 gene, and the other one, downstream (Figure 1E and Figure S1A). Moreover, ChIP using anti-Atf1 antibody confirmed that Atf1 is indeed localized around ade6-3049, but not ade6-3057 (Figure S1B). These results may imply that Atf1, which binds to the M26 sequence at ade6-3049, functions as a transcriptional activator to instigate transcription.
Transactivation domain of Atf1 is the HRA region
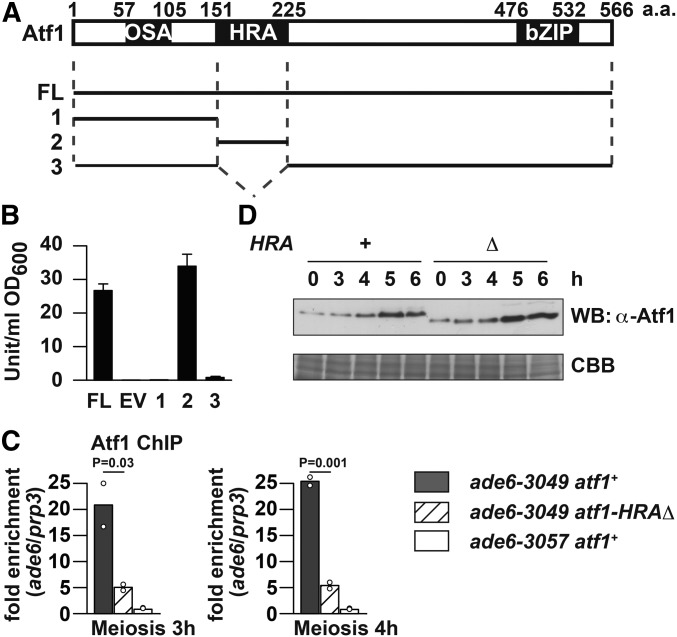
We then wished to understand how Atf1 is involved in generation of short transcripts at hotspots. In terms of the protein structure, a transcription factor can be often divided into functional domains such as a DNA-binding domain and a transactivation domain. This is also the case with Atf1, as its C-terminal bZIP domain serves as a DNA-binding/protein-dimerization domain (Takeda et al. 1995; Kanoh et al. 1996; Shiozaki and Russell 1996; Wilkinson et al. 1996). Gao et al. (2008) later used plasmid-borne, truncated-atf1 genes to identify additional functional domains: the HRA region, which is necessary and sufficient for recombination activation at ade6-M26; the homologous recombination repression region, which represses recombination at the same hotspot; and the osmotic stress activation (OSA) region, which is essential for cell growth in a hyperosmotic environment (Figure 2A). It has not been known, however, whether Atf1 possesses a transactivation domain, and to address this point we performed budding yeast one-hybrid system experiments (Horie et al. 1998; Morita et al. 2011). In this assay, a protein of interest is fused to the DNA-binding domain of LexA (LexA BD) and expressed in the budding yeast L40 strain, carrying a lacZ reporter gene under control of a LexA operator. The fusion protein with transactivation activity can drive transcription of lacZ to produce β-galactosidase, an enzyme hydrolyzing o-Nitrophenyl-β-galactoside to galactose and o-nitrophenol. Colorimetric quantification of o-nitrophenol allows us to estimate transactivation activity of the protein fused to LexA BD. As expected, full-length Atf1 fused to LexA BD, but not LexA BD alone, could activate transcription of lacZ (Figure 2B), which suggested that Atf1 is indeed a transcription activator.
Figure 2.
Transactivation domain of Atf1 is the previously identified HRA region, which is dispensable for Atf1 protein stability. (A) Previously known functional domains of Atf1 (Gao et al. 2008) and Atf1 fragments tested for transactivation activity. FL, full length (of Atf1). (B) Quantification of β-galactosidase activity detected in cells expressing LexA BD-Atf1 fusion protein. Results of three independent experiments with SDs are shown. EV, empty vector. (C) ChIP of Atf1 and Atf1-HRAΔ. Indicated cells of pat1-114 background were induced to enter meiosis by temperature-shift method, and harvested at indicated hours after the induction. ChIP using anti-Atf1 antibody was performed, and DNA was isolated from immunoprecipitates as well as whole-cell extracts. Obtained DNA was analyzed by real-time qPCR, where fragments corresponding to ade6-3049/3057 and the prp3+ promoter were amplified. Relative enrichment at ade6-3049/3057 over prp3 is shown. Bar graphs are created based on mean values of two independent experiments (shown by ○). (D) Western blotting of Atf1 and Atf1-HRAΔ during meiosis. pat1-114 atf1+ and pat1-114 atf1-HRAΔ cells were induced to enter meiosis by temperature-shift method, and harvested at indicated hours after the induction. Whole-cell extract was analyzed by Western blotting using anti-Atf1 antibody (WB: α-Atf1). An SDS-PAGE gel in which the same amount of whole-cell extract was fractionated was stained by Coomassie brilliant blue and is shown as a loading control (CBB).
Three Atf1 fragments depicted in Figure 2A were similarly analyzed. Among them, the HRA region per se was able to activate transcription of the reporter gene, while fragments lacking the HRA region were not. This finding shows that the transactivation domain of Atf1 coincides with the previously identified HRA region. In other words, Atf1 can activate meiotic recombination at ade6-M26 and transcription through the same domain. In contrast to HRA, the OSA region did not confer transactivation activity to Atf1 fragments. This observation may mean that the OSA region functions independently of transcription activation or specifically in response to osmotic stress, or may be obtained because our assay just failed to detect its transcription-stimulating function. In any case, the HRA region is likely to be a key domain responsible for transactivation activity of Atf1.
Deletion of the HRA region decreases 3049-bound Atf1, but does not affect protein stability
To understand how the HRA region functions in fission yeast cells, we constructed a strain in which HRA-coding sequence was deleted from the native atf1+ locus (atf1-HRAΔ), and compared atf1-HRAΔ cells to atf1+ cells. Before analyzing recombination and transcription, binding to the ade6-3049 hotspot and protein stability of Atf1 were tested. To achieve highly synchronous meiosis progression, pat1-114 haploid cells of both atf1 backgrounds were induced to enter meiosis by the temperature-shift method. ChIP using anti-Atf1 antibody was performed to compare Atf1 levels at the ade6-3049 hotspot and at the control site prp3+. As shown in Figure 2C, although both wild-type Atf1 and HRA-lacking Atf1 (Atf1-HRAΔ) were substantially enriched more at 3049 than at prp3, the enrichment of Atf1-HRAΔ was reduced to about a fourth to fifth of that of wild-type Atf1. However, by Western blotting using the same antibody, we observed that Atf1 protein levels in atf1+ and atf1-HRAΔ cells were comparable to each other as far as we tested (Figure 2D).
The reduced binding of Atf1-HRAΔ to ade6-3049 can be interpreted in several ways. First, HRA deletion may perturb the protein structure of Atf1 to interfere with binding to the 3049 hotspot. A second interpretation is related to a recent report on transcription activation of the fbp1+ gene, whose promoter is bound by Atf1: binding of Atf1 to the fbp1+ promoter is strengthened through downstream (i.e., fbp1+) transcription by Atf1 (Takemata et al. 2016). If this is the case with ade6-3049, Atf1 may self-enforce its 3049 binding by producing 3049-associated short transcripts; while Atf1-HRAΔ may not generate the short transcripts well enough to enhance its 3049 binding. Third, there is a caveat that the antibody could not equally recognize Atf1 and Atf1-HRAΔ unless they are denatured by SDS and/or not cross-linked with formaldehyde. Therefore at this stage, we cannot determine what is attributable to decreased ChIP signals of Atf1-HRAΔ at ade6-3049. Nevertheless, as HRA deletion appeared not to compromise protein abundance of Atf1, we decided to explore its effects on recombination and transcription.
Deletion of the HRA region reduces meiotic DSB formation and recombination at ade6-3049
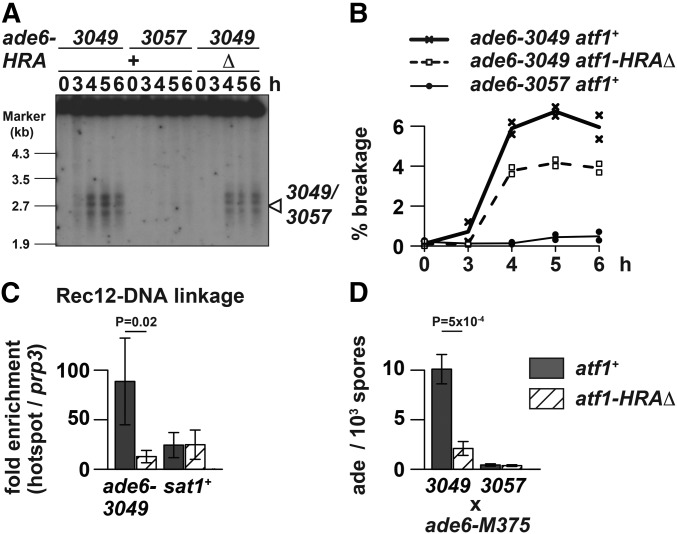
DSB formation at the ade6-3049 hotspot was examined by monitoring two products: DSB fragments and Rec12 (the fission yeast homolog of Spo11) covalently attached to broken ends (Rec12-DNA linkage). For effective detection of these two molecules, which are transient and otherwise difficult to observe, analyses were performed on pat1-114 rad50S haploid cells, as the rad50S mutation prevents DSB repair and accumulates Rec12-attached DSB ends. Also, the rec12+ locus was engineered to express C-terminally FLAG-tagged Rec12 in the analyzed cells, which are fully adept at DSB formation (Cromie et al. 2007; Yamada et al. 2013).
To observe DSB fragments at ade6-3049, genomic DNA; isolated from atf1+ ade6-3049, atf1+ ade6-3057, and atf1-HRAΔ ade6-3049 cells; was digested with the restriction enzyme Afl II and analyzed by Southern blotting, as described previously (Figure 3A) (Steiner et al. 2002). The results showed that 3049-associated DSB signals, which are at the background level around ade6-3057, are weaker in atf1-HRAΔ mutants than in atf1+ cells. Although distribution of break sites was not dramatically altered, quantification of two independent experiments showed that DSB frequency at 5 hr is 6.7 and 4.2% in atf1+ and atf1-HRAΔ (P-value = 0.01), respectively (Figure 3B). Therefore, DSB formation at 3049 was compromised by deletion of the HRA region.
Figure 3.
Deletion of the Atf1 HRA region affected meiotic recombination at ade6-3049. (A) Effects of HRA deletion on DSB formation at the ade6-3049 hotspot. Indicated cells of the pat1-114 rad50S background were induced to enter meiosis by temperature-shift method, and harvested at the indicated hours after the induction. Genomic DNA was isolated, digested with Afl II, and analyzed by Southern blotting as previously described (Steiner et al. 2002). The position of the 3049 or 3057 mutations is marked by the arrowhead. (B) Quantitative analyses of DSB formation presented in (A). The DSB (%) values were obtained by dividing the signal intensity of broken DNA fragments over that of the unbroken parental fragment. Mean values of two independent experiments are plotted. (C) Effects of HRA deletion on Rec12-DNA linkage production at the ade6-3049 hotspot. Rec12-FLAG-expressing pat1-114 rad50S strains with or without HRA were induced to enter meiosis by temperature-shift method, and harvested 5 hr after the induction. Rec12-FLAG-DNA linkage was chromatin immunoprecipitated by anti-FLAG antibody. DNA isolated from immunoprecipitates and whole-cell extracts was analyzed by real-time qPCR, where fragments corresponding to the hotspots (ade6-3049 and sat1) and the prp3+ promoter (prp3) were amplified. Relative enrichment at hotspots over prp3 was calculated and mean values of three independent experiments as well as their SD are shown. (D) Effects of HRA deletion on meiotic gene conversion at the ade6-3049 hotspot. Recombination frequency was measured between ade6-M375 and either ade6-3049 or ade6-3057 by counting the number of ade+ spores. Mean values of three independent experiments and their SD are shown.
The above observation was further verified by measuring Rec12 covalently attached to DSB ends. Cells 5 hr after meiosis induction were harvested without formaldehyde-cross-linking and analyzed by ChIP of Rec12-FLAG (Figure 3C). At ade6-3049, Rec12-DNA linkage was less precipitated in atf1-HRAΔ cells than in atf1+ cells, which suggests that HRA deletion impaired DSB formation at this hotspot. In contrast, at the sat1 hotspot (Miyoshi et al. 2012), which does not harbor an M26 sequence, atf1+ cells and atf1-HRAΔ cells produced similar amounts of Rec12-DNA linkage. Based on these results, we concluded that HRA would promote DSB formation at the ade6-3049 hotspot. It should be pointed out that effects of HRA deletion were less overtly manifested by Southern blotting than by Rec12-DNA linkage ChIP. The reason for this difference is currently unknown, but we emphasize that a same conclusion was drawn from two distinct procedures.
Consistently, lack of HRA reduced recombination frequency between ade6-3049 and ade6-M375, but not between ade6-3057 and ade6-M375 (Figure 3D). These experiments suggest that HRA is specifically involved in DSB formation as well as subsequent recombination at ade6-3049, and probably other M26-sequence-dependent hotspots. They were in agreement with a previous observation that expression of HRA-lacking Atf1 from a multi-copy plasmid does not fully restore meiotic recombination defects at M26 in atf1Δ cells (Gao et al. 2008).
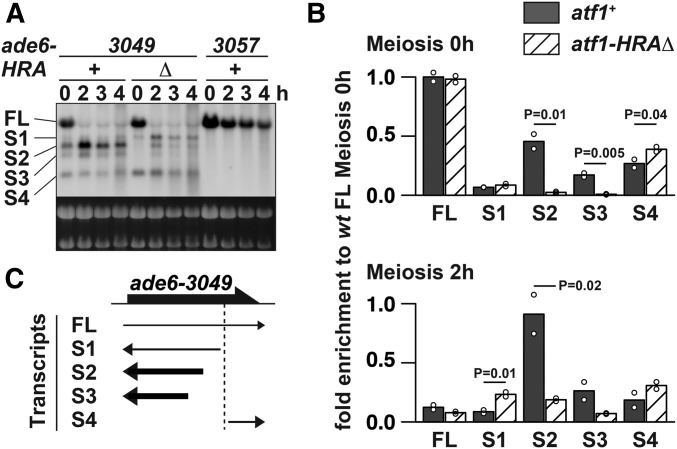
Deletion of the HRA region reduces several short transcripts at ade6-3049
We next compared 3049-associated short transcripts in atf1+ and atf1-HRAΔ cells. Northern hybridization probing the entire ade6 ORF demonstrated that the transcripts’ pattern is disturbed in the atf1-HRAΔ mutant (Figure 4A). In particular, detailed quantification of blots showed that HRA deletion severely decreases the transcripts S2 and S3, both of which were antisense transcripts starting from around the M26 sequence (Figure 4, B and C). These results would indicate that the lack of HRA not only impairs recombination, as was originally proposed, but also affects transcription of 3049-associated short transcripts.
Figure 4.
Deletion of the Atf1 HRA region affected transcription at ade6-3049. (A) Effects of HRA deletion on transcripts production around ade6-3049. Total RNA was analyzed by Northern blotting as explained in Figure 1B. (B) Quantitative analyses of ade6 transcripts shown in (A). Abundance of transcripts, detected in atf1+ (filled bars) and atf1-HRAΔ (hatched bars) cells at 0 h (top) and 2 hr (bottom) after meiosis induction, were estimated by quantifying signal intensities of each transcript and normalizing them to the mean value of full-length ade6 mRNA present in atf1+ cells at 0 hr. The numbers of transcripts are the same as those indicated in (A). Bar graphs are created based on mean values of two independent experiments (shown by ○). (C) Graphic summary of results shown in (A) and (B). RNAs strongly affected (more than twofold for both time points) by the atf1-HRAΔ mutation were shown as bold black arrows. The vertical dotted line indicates the position of the 3049 mutation.
It is intriguing that small amounts of short transcripts were still present even in the absence of HRA. This may be because HRA-lacking Atf1 can, albeit at a reduced level, bind to ade6-3049 and modify surrounding chromatin to produce short transcripts, as the Atf1 bZIP domain itself has been recently reported to influence nucleosome phasing around its binding site (Garcia et al. 2014).
ade6-3049, but not ade6-3057, is associated with open chromatin structure, which is at least partly mediated by HRA
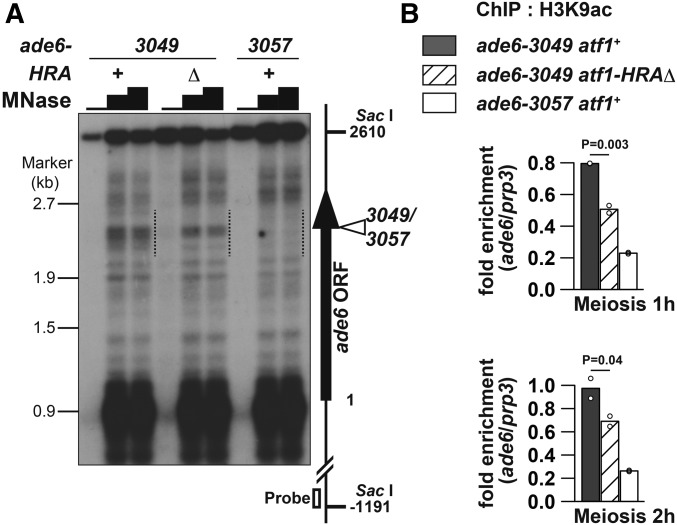
The impact of HRA deletion on both transcription and recombination around ade6-3049 may suggest that lack of HRA affects the local chromatin environment, since chromatin influences all DNA-templated events. To address this point, chromatin structure around ade6-3049 was compared between atf1+ and atf1-HRAΔ cells by a method using MNase. The crude chromatin fraction, isolated from meiotic pat1-114 haploid cells, was treated with varying amounts of the nuclease, and genomic DNA was subsequently purified. The obtained DNA was digested with the restriction endonuclease Sac I and analyzed by Southern blotting using a probe hybridizing to one end of the 3049-containing Sac I fragment (Figure 5A).
Figure 5.
Chromatin structure around ade6-3049 is more open than around ade6-3057, and HRA is involved in the chromatin regulation. (A) MNase sensitivity of chromatin around ade6-3049/3057. Indicated cells of the pat1-114 background were induced to enter meiosis by temperature-shift method, and harvested 3 hr after the induction. MNase-digested DNA was prepared by MNase (0, 20, or 30 units) treatment of chromatin fraction and subsequent purification. DNA was further cut with Sac I, fractionated on 1.2% agaraose gel, and analyzed by Southern blotting using the probe indicated by the open box. The vertical arrow and the open arrowhead show the ade6 ORF and the 3049/3057 site, respectively. The numbers on the left and the right side of the panel indicate the positions of λ/EcoT14 I fragments used for electrophoresis marker and the positions from the first A of the ade6 ORF, respectively. The dotted lines indicate a region in which MNase-sensitive sites are clustered around ade6-3049. Presented is an example from two independent experiments, whose results were similar to each other. (B) HRA deletion reduced acetylation level of H3K9 around ade6-3049. Indicated cells of the pat1-114 background were induced to enter meiosis by temperature-shift method, and harvested 1 or 2 hr after the induction. ChIP using anti-H3K9ac antibody was performed as described previously (Yamada et al. 2013). DNA isolated from immunoprecipitates and whole-cell extracts was analyzed by real-time qPCR, where fragments corresponding to ade6-3049/3057 and the prp3+ promoter were amplified. Relative enrichment at ade6-3049/3057 over prp3 is shown. Bar graphs are created based on mean values of two independent experiments (shown by ○).
The analyses on atf1+ cells revealed that an ∼600-bp region (Figure 5A, dotted lines) surrounding ade6-3049 contains multiple MNase-hypersensitive sites, while the corresponding region around ade6-3057 does not; suggesting that chromatin is more accessible around 3049 than 3057. This difference, similar to the case of the ade6-M26 hotspot and its negative control locus ade6-M375 (Mizuno et al. 1997), is consistent with the idea that hotspots are located in open chromatin to help DSB machinery function at hotspots.
Remarkably, deletion of the HRA region eliminated several but not all of the 3049-surrounding MNase-hypersensitive sites (Figure 5A). Therefore, this domain is involved in formation of an accessible chromatin environment around ade6-3049, and by doing so, would promote transcription and recombination.
HRA deletion partially reduces hotspot-associated H3Lys9 acetylation
Open chromatin is associated with acetylated histones and we have previously reported that H3Lys9 acetylation (H3K9ac) is a major mark of hotspots in fission yeast (Yamada et al. 2013). We hence asked whether deletion of the HRA region may influence the high level of H3K9ac at ade6-3049. Since we have observed that histone-acetylation levels decline through meiosis progression (Yamada et al. 2004), cells 1 and 2 hr after meiosis induction were examined. As shown in Figure 5B, HRA deletion decreased this modification to an intermediate level between wild type (atf1+ ade6-3049) and its negative control (atf1+ ade6-3057) strains. This result indicates that HRA partly but significantly promotes H3K9ac at the ade6-3049 hotspot, and the histone modification may be involved in transcription, recombination, or both at 3049.
Possibility of functional connection between 3049-associated transcription and recombination
How are our present results integrated to describe what occurs around ade6-3049 in meiosis? Several clues are available from this and previous studies. First, transcription preceded DSB formation; short transcripts were already present in premeiotic cells, and in particular, transcript S2 was induced 2 hr after meiosis induction, when proteins involved in DSB formation are barely expressed (Mata et al. 2002). Second, in atf1-HRAΔ mutants, recombination frequency was reduced with concomitant diminishment of open chromatin hallmarks, namely MNase sensitivity and H3K9ac, around the hotspot. Third, at a CRE site in the promoter of fbp1+, transcription activated by Atf1 increases H3K9ac to further stimulate downstream transcription (Takemata et al. 2016).
One of possible models deduced from the above three points is as follows. Atf1 that is bound to 3049 stimulates transcription (of short transcripts) to promote H3K9ac and locally decondense surrounding chromatin, and thereby would facilitate DSB formation near the transcription sites. This scenario may be related to a previous report that artificial activation of transcription creates a DSB hotspot by reducing nucleosome occupancy (de Castro et al. 2012), in that transcription activates DSB formation. Since other possibilities are also possible and we cannot exclude a possibility that correlation of the two events is merely a coincidence, further analyses are necessary.
Pervasive correlation between transcription and recombination initiation sites
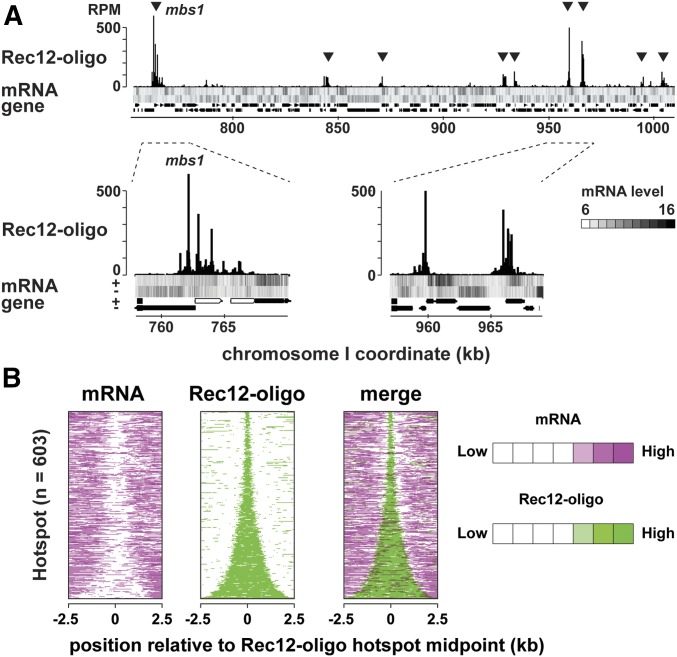
Having shown that transcribed regions of short transcripts and meiotic DSB sites are closely located to each other at the ade6-3049 hotspot, we wondered whether this possible connection could be extended to other hotspots. This notion appears to be incompatible with a known feature that promoters of protein-coding genes are not major sites for DSB hotspots in fission yeast. However, high-resolution mapping of Rec12-oligo, byproducts of DSB processing, proved that hotspots often encompass a large region with multiple Rec12-oligo sites scattered (Fowler et al. 2014). In addition, hotspots are directed to loci expressing noncoding RNAs (Wahls et al. 2008). These observations led us to consider that at least a part (or “edge”) of hotspots may be adjoined to transcribed regions. We therefore compared meiotic transcriptome data at 3 hr after meiosis induction (de Castro et al. 2012), instead of gene positions, and a Rec12-oligo hotspot map (Fowler et al. 2014). As shown in Figure 6A, most hotspots are found in intergenic regions, as previously reported (compare rows “Rec12-oligo” and “gene”) (Cromie et al. 2007; Fowler et al. 2014). Remarkably, regions associated with Rec12-oligo are in many cases located near those where at least one strand is transcribed (compare rows “Rec12-oligo” and “mRNA”), suggesting that a part of a hotspot is frequently juxtaposed to meiotically transcribed regions.
Figure 6.
Correlation between transcription and recombination initiation sites around fission yeast hotspots. (A) An example of uniquely mapped Rec12-oligo and complementary DNA of meiotic transcripts hybridized to genome tiling array. The x-axis shows the chromosomal coordinates in base pairs, and the y-axis shows reads per million base pairs (RPM). The heat map shows the log2 signal strength of forward and reverse transcripts. Black and white represent up- or downregulation of gene expression. Protein-coding genes and RNA genes are shown as filled black and gray boxes at the bottom of the figure, respectively. Two regions are zoomed to show the locations of Rec12-oligo and transcripts within hotspots. (B) Heat maps of transcripts (left) and Rec12-oligo (middle) around hotspots. All hotspots on the genome (n = 603) were ranked by width and their midpoints were aligned. Data of transcripts (magenta) and Rec12-oligo (green) were overlaid to show on a color scale (right). Magenta and green represent high signals, while white represents low signals.
To gain more comprehensive insights, the same data sets were exploited to map meiotic transcripts around all hotspots, which were aligned across the y-axis according to the hotspot width (Figure 6B). The comparison showed that most hotspots are juxtaposed to transcribed regions (Figure 6B), reminiscent of the cases explained in Figure 1E (ade6-3049) and Figure 6A. Therefore, spatial proximity between transcription and DSB formation can be widely observed in fission yeast.
Finally, we also asked how chromatin structure is related to the above relationship by comparing a heat map of nucleosome depletion to those presented in Figure 6B (Figure S2). Consistent with previous reports that fission yeast hotspots, including ade6-M26 and ade6-3049, generally have low nucleosome occupancy (de Castro et al. 2012; Yamada et al. 2013), our analysis found that hotspots can be generally superimposed on low-level nucleosome regions. Furthermore, it is noteworthy that the width of hotspots and NDRs correlated with each other, implying functional relation between the two regions. It should be noted, however, that a more recent and higher resolution analysis unveiled that the extent of nucleosome depletion is much milder than budding yeast hotspots (Fowler et al. 2014). One possibility to account for all these observations would be that transcription adjacent to hotspots set up a moderately accessible chromatin region, which would be sufficient for fission yeast DSB machinery to initiate meiotic recombination.
Conclusion
Contribution of transcription (and transcription factors) to meiotic DSB formation has been less appreciated partly due to the fact that its (their) roles are context dependent. In this study, however, we demonstrate that the HRA region of Atf1, which had been shown to activate recombination at ade6-M26, is also a transactivation domain and that it directly or indirectly promotes both production of short transcripts and meiotic recombination at the ade6-3049 hotspot. In addition, revisiting genome-wide data of meiotic transcripts and Rec12-oligo raise a possibility that DSB formation occurs in close proximity to transcribed regions.
Recent studies on various organisms reveal that mechanisms of meiotic DSB formation vary among species, and that, even in a single species, there are several ways to accomplish this event (Brick et al. 2012; Borde and de Massy 2013; Ismail et al. 2014). Considering such diversity, exploring possible connection between transcription and meiotic recombination may be necessary to thoroughly understand the initiation mechanism of meiotic recombination.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.197954/-/DC1.
Acknowledgments
We thank Wayne P. Wahls for sharing information on the M26-associated short transcript before launching this study. We also thank Gerald Smith for strains, and Emi Takaya and Tomohiko Morita for technical assistance. The authors are grateful to members in Ohta’s laboratory for discussion. This work was supported by the Grant-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) (T.Y.); the Grant-in-Aid for Scientific Research on Innovative Areas from MEXT (K.O.; 3114003, 212414946, and 26291018); and the Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science (S.Y.). It was also supported by the Platform Project for Supporting Drug Discovery and Life Science Research (Platform for Dynamic approaches to Living Systems) from MEXT and the Japan Agency for Medical Research and Development (K.O.).
Footnotes
Communicating editor: J. Surtees
Literature Cited
- Borde V., de Massy B., 2013. Programmed induction of DNA double strand breaks during meiosis: setting up communication between DNA and the chromosome structure. Curr. Opin. Genet. Dev. 23: 147–155. [DOI] [PubMed] [Google Scholar]
- Borde V., Goldman A. S., Lichten M., 2000. Direct coupling between meiotic DNA replication and recombination initiation. Science 290: 806–809. [DOI] [PubMed] [Google Scholar]
- Brick K., Smagulova F., Khil P., Camerini-Otero R. D., Petukhova G. V., 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature 485: 642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Toone W. M., Mata J., Lyne R., Burns G., et al. , 2003. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14: 214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Zhao X., Kelly K. A., Venn O., Higgins J. D., et al. , 2013. Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 45: 1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie G. A., Hyppa R. W., Cam H. P., Farah J. A., Grewal S. I., et al. , 2007. A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLoS Genet. 3: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro E., Soriano I., Marin L., Serrano R., Quintales L., et al. , 2012. Nucleosomal organization of replication origins and meiotic recombination hotspots in fission yeast. EMBO J. 31: 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B., 2013. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet. 47: 563–599. [DOI] [PubMed] [Google Scholar]
- Fowler K. R., Sasaki M., Milman N., Keeney S., Smith G. R., 2014. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res. 24: 1650–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox M. E., Yamada T., Ohta K., Smith G. R., 2000. A family of cAMP-response-element-related DNA sequences with meiotic recombination hotspot activity in Schizosaccharomyces pombe. Genetics 156: 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Davidson M. K., Wahls W. P., 2008. Distinct regions of ATF/CREB proteins Atf1 and Pcr1 control recombination hotspot ade6–M26 and the osmotic stress response. Nucleic Acids Res. 36: 2838–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P., Paulo E., Gao J., Wahls W. P., Ayte J., et al. , 2014. Binding of the transcription factor Atf1 to promoters serves as a barrier to phase nucleosome arrays and avoid cryptic transcription. Nucleic Acids Res. 42: 10351–10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C., Schaer P., Munz P., Kohli J., 1991. The strong ADH1 promoter stimulates mitotic and meiotic recombination at the ADE6 gene of Schizosaccharomyces pombe. Mol. Cell. Biol. 11: 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K., Mizuno K., Shibata T., Ohta K., 2008. Distinct chromatin modulators regulate the formation of accessible and repressive chromatin at the fission yeast recombination hotspot ade6–M26. Mol. Biol. Cell 19: 1162–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie S., Watanabe Y., Tanaka K., Nishiwaki S., Fujioka H., et al. , 1998. The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol. Cell. Biol. 18: 2118–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N., 2015. Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 7: a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail M. B., Shinohara M., Shinohara A., 2014. Dot1-dependent histone H3K79 methylation promotes the formation of meiotic double-strand breaks in the absence of histone H3K4 methylation in budding yeast. PLoS One 9: e96648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J., Watanabe Y., Ohsugi M., Iino Y., Yamamoto M., 1996. Schizosaccharomyces pombe gad7+ encodes a phosphoprotein with a bZIP domain, which is required for proper G1 arrest and gene expression under nitrogen starvation. Genes Cells 1: 391–408. [DOI] [PubMed] [Google Scholar]
- Keeney S., Lange J., Mohibullah N., 2014. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu. Rev. Genet. 48: 187–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon N., Krawchuk M. D., Warren B. G., Smith G. R., Wahls W. P., 1997. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 94: 13765–13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J., Lyne R., Burns G., Bahler J., 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32: 143–147. [DOI] [PubMed] [Google Scholar]
- Mieczkowski P. A., Dominska M., Buck M. J., Gerton J. L., Lieb J. D., et al. , 2006. Global analysis of the relationship between the binding of the Bas1p transcription factor and meiosis-specific double-strand DNA breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 1014–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T., Ito M., Kugou K., Yamada S., Furuichi M., et al. , 2012. A central coupler for recombination initiation linking chromosome architecture to S phase checkpoint. Mol. Cell 47: 722–733. [DOI] [PubMed] [Google Scholar]
- Mizuno K., Emura Y., Baur M., Kohli J., Ohta K., et al. , 1997. The meiotic recombination hot spot created by the single-base substitution ade6–M26 results in remodeling of chromatin structure in fission yeast. Genes Dev. 11: 876–886. [DOI] [PubMed] [Google Scholar]
- Morita T., Yamada T., Yamada S., Matsumoto K., Ohta K., 2011. Fission yeast ATF/CREB family protein Atf21 plays important roles in production of normal spores. Genes Cells 16: 217–230. [DOI] [PubMed] [Google Scholar]
- Murakami H., Keeney S., 2014. Temporospatial coordination of meiotic DNA replication and recombination via DDK recruitment to replisomes. Cell 158: 861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Sasaki M., Kniewel R., Murakami H., Blitzblau H. G., et al. , 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144: 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli A. S., Sena E. P., Smith G. R., 1988. Genetic and physical analysis of the M26 recombination hotspot of Schizosaccharomyces pombe. Genetics 119: 491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K., Russell P., 1996. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 10: 2276–2288. [DOI] [PubMed] [Google Scholar]
- Steiner W. W., Smith G. R., 2005a Natural meiotic recombination hot spots in the Schizosaccharomyces pombe genome successfully predicted from the simple sequence motif M26. Mol. Cell. Biol. 25: 9054–9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner W. W., Smith G. R., 2005b Optimizing the nucleotide sequence of a meiotic recombination hotspot in Schizosaccharomyces pombe. Genetics 169: 1973–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner W. W., Schreckhise R. W., Smith G. R., 2002. Meiotic DNA breaks at the S. pombe recombination hot spot M26. Mol. Cell 9: 847–855. [DOI] [PubMed] [Google Scholar]
- Sun X., Huang L., Markowitz T. E., Blitzblau H. G., Chen D., et al. , 2015. Transcription dynamically patterns the meiotic chromosome-axis interface. Elife 4: e07424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szankasi P., Heyer W. D., Schuchert P., Kohli J., 1988. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe. Wild-type and mutant alleles including the recombination host spot allele ade6–M26. J. Mol. Biol. 204: 917–925. [DOI] [PubMed] [Google Scholar]
- Takeda T., Toda T., Kominami K., Kohnosu A., Yanagida M., et al. , 1995. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 14: 6193–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemata N., Oda A., Yamada T., Galipon J., Miyoshi T., et al. , 2016. Local potentiation of stress-responsive genes by upstream noncoding transcription. Nucleic Acids Res. 44: 5174–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls W. P., Davidson M. K., 2010. Discrete DNA sites regulate global distribution of meiotic recombination. Trends Genet. 26: 202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls W. P., Siegel E. R., Davidson M. K., 2008. Meiotic recombination hotspots of fission yeast are directed to loci that express non-coding RNA. PLoS One 3: e2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. A., Wierdl M., Detloff P., Petes T. D., 1991. DNA-binding protein RAP1 stimulates meiotic recombination at the HIS4 locus in yeast. Proc. Natl. Acad. Sci. USA 88: 9755–9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M. G., Samuels M., Takeda T., Toone W. M., Shieh J. C., et al. , 1996. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 10: 2289–2301. [DOI] [PubMed] [Google Scholar]
- Wu P. Y., Nurse P., 2014. Replication origin selection regulates the distribution of meiotic recombination. Mol. Cell 53: 655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S., Ohta K., Yamada T., 2013. Acetylated histone H3K9 is associated with meiotic recombination hotspots, and plays a role in recombination redundantly with other factors including the H3K4 methylase Set1 in fission yeast. Nucleic Acids Res. 41: 3504–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Mizuno K., Hirota K., Kon N., Wahls W. P., et al. , 2004. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 23: 1792–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Keeney S., 2015. High-resolution global analysis of the influences of Bas1 and Ino4 transcription factors on meiotic DNA break distributions in Saccharomyces cerevisiae. Genetics 201: 525–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Reagents are available upon request.