Abstract
The first β-lactone synthetase enzyme is reported, creating an unexpected link between the biosynthesis of olefinic hydrocarbons and highly functionalized natural products. The enzyme OleC, involved in the microbial biosynthesis of long-chain olefinic hydrocarbons, reacts with syn- and anti-β-hydroxy acid substrates to yield cis- and trans-β-lactones, respectively. Protein sequence comparisons reveal that enzymes homologous to OleC are encoded in natural product gene clusters that generate β-lactone rings, suggesting a common mechanism of biosynthesis.
Table of Contents Artwork
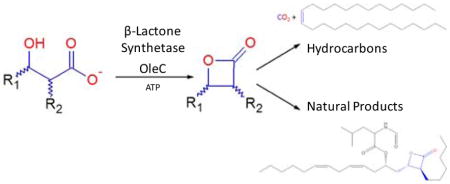
The β-lactone (2-oxetanone) substructure is well known in organic synthesis and microbial natural products, some of which are presently being investigated for anti-obesity, anti-cancer, and antibiotic properties1–5. Although multiple organic synthesis routes exist for β-lactones6, no specific enzyme that catalyzes the formation of this functional group had been identified. While defining the chemistry of a well-known olefinic hydrocarbon biosynthesis pathway, we identified a β-lactone synthetase whose presence extends into natural product biosynthesis.
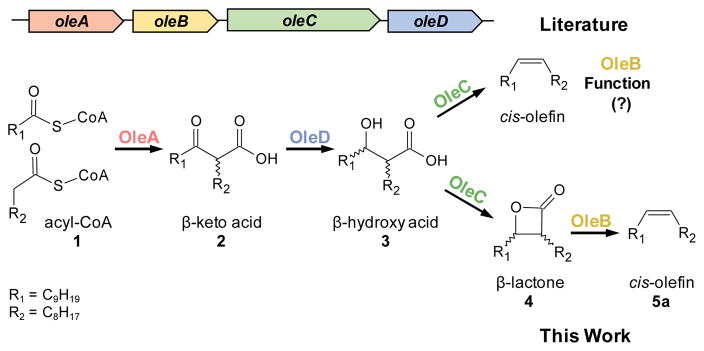
The olefin biosynthesis pathway is encoded by a four-gene cluster, oleABCD, and is found in over 250 divergent bacteria7. Ole enzymes produce long-chain hydrocarbon cis-alkenes from activated fatty acids through the reported chemistry shown in Figure 18,9. OleA, the first enzyme of the pathway, has been studied in Xanthomonas campestris (Xc) and found to catalyze the “head-to-head” coupling of CoA-activated fatty acids (1) to unstable β-keto acids (2)10. The second enzyme, OleD, couples the reduction of 2 with NADPH oxidation to yield stable β-hydroxy acids (3) as defined in Stenotrophomonas maltophilia (Sm)11. Finally, using GC detection methods, we have observed and others have reported that Sm OleC catalyzes an apparent decarboxylative dehydration reaction to generate the final cis-olefin product12. Together, these findings indicate that there is no defined purpose for the ever-present fourth gene in the cluster, oleB.
Figure 1.
ole gene cluster and enzymatic pathway.
We now report that the observation of cis-alkenes as products of OleC is an artifact, a misleading observation caused by GC-analytical methods used to identify the high-boiling point products of OleC. Rather, using 1H NMR, we demonstrate that OleC from four different bacteria produce thermally-labile β-lactones from β-hydroxy acids in an ATP-dependent reaction; no alkenes are observed. Further analyses of gene clusters for β-lactone-containing natural products reveal oleC homologues that likely perform this previously unknown biological β-lactone ring closure reaction.
The first suggestions of β-lactone synthetase activity arose when monitoring reactions of Xc OleC with ATP, MgCl2, and a synthetic, diastereomeric mixture of 3 by GC. Full 1H NMR spectra of all synthesized compounds from this study are shown in Supplemental Figures S1–S6. Two peaks were observed by GC, coupled to both a mass spectrometer (MS) and flame ionization detector (FID), with mass spectra and retention times identical to synthetic cis- and trans-olefin standards (5a and 5b, respectively). However, the GC/FID peak area of the enzymatically-generated olefin varied significantly with GC inlet temperature and inlet liner purity, while synthetic standards were unaffected, suggesting that the observed olefin may be a thermal decomposition product of the actual OleC product.
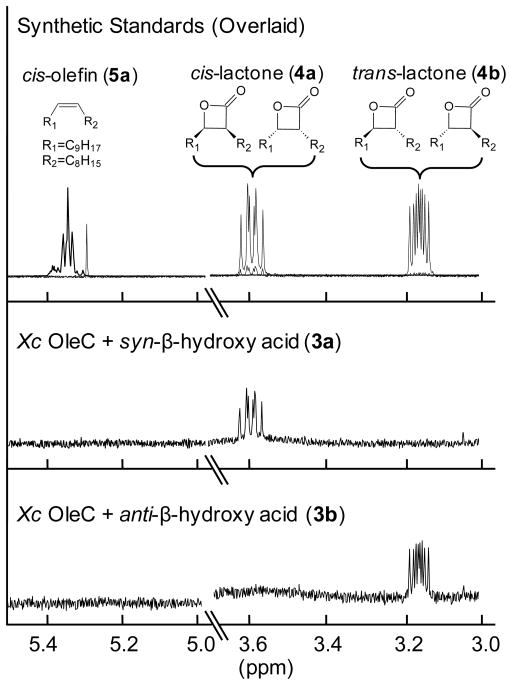
To test this hypothesis, Xc OleC reactions with 3 were scaled to generate sufficient quantities for 1H NMR. No resonances consistent with the prepared olefin standards (Figures S1 and S2) (5a, 5b) were observed; rather, four distinct multiplets, appearing as doublet of doublets of doublets, arose (Figures S7 and S8). These resonances were consistent with the two hydrogens of cis- and trans-β-lactone rings and perfectly matched our authentic standards of cis- and trans-3-octyl-4-nonyloxetan-2-one (4a, 4b) (Figures S3 and S4). Furthermore, when 4a and 4b were analyzed by GC, retention times and mass spectra matched the olefin standards 5a and 5b, with sensitivity to inlet conditions being observed. The thermal decarboxylation of cis- and trans-β-lactone to cis- and trans-olefin, respectively, is well known13,14. We believe that thermal decomposition during GC/MS analysis caused the product of OleC catalysis to be misidentified. Additionally, when reviewing supplemental NMR data from the literature report of Sm OleC characterization, resonances of the cis- and trans-β-lactones, consistent with those synthesized here, are visible12.
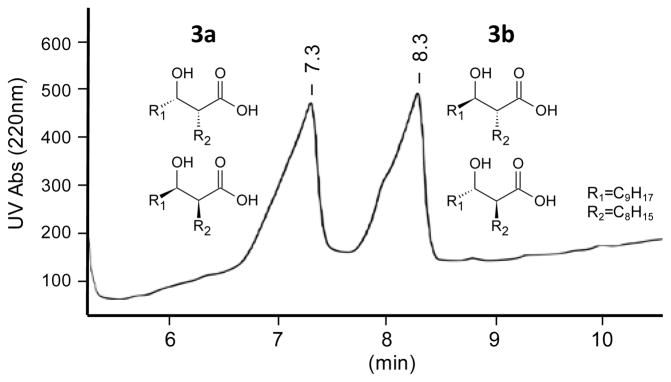
The stereochemical origins of 4a and 4b were then investigated by reacting Xc OleC with syn- and anti-β-hydroxy acids (Figures S9 and S10). HPLC was used to separate 3 into its syn- and anti-diastereomeric pairs (3a and 3b) (Figure 2). Examining 3a and 3b by 1H NMR and GC/MS, post-methylation, demonstrated each contained <10% of the opposite racemic diastereomer (Figures S5 and S6). When reacting with Xc OleC, 3a produced 4a, while 3b gener-ated 4b (Figure 3). GC/MS analysis supported this conclusion, as OleC reactions with 3a and 3b yielded the β-lactone breakdown products, 5a and 5b, respectively. OleC consumed greater than 90% of substrates 3a and 3b by GC/MS, supporting the conclusions of Kancharla, et al. that all four β-hydroxy acid isomers are utilized by OleC12. Taken together, Xc OleC represents the first reported β-lactone synthetase, converting β-hydroxy acid substrates to β-lactones in the presence of ATP and MgCl2. MgATP is likely required to activate the hydroxyl or carboxyl group and promote β-lactone ring formation.
Figure 2.
HPLC separation of syn- and anti-β-hydroxy acids (3a and 3b, respectively) for use as Xc OleC substrates. The rise in baseline is caused by impurities from the MTBE gradient that absorb at 220 nm.
Figure 3.
1H NMR (400 MHz; CDCl3) analyses of Xc OleC products with β-hydroxy acid substrates.
To determine if β-lactone synthetase activity is a common enzymatic step in long-chain olefin biosynthesis, we obtained four oleC genes from oleABCD gene clusters in divergent microorganisms (Table 1). Purified OleC enzyme from the four organisms was reacted overnight with ATP and 3, then analyzed by 1H NMR and GC/MS. The products of OleC proteins from the bacteria Stenotrophomonas maltophilia, Arenimonas malthae, and Lysobacter dokdonensis were both 4a and 4b β-lactones (Figures S11–S13), with no 5a or 5b olefins being observed, indicating that OleC enzymes from diverse sources are β-lactone synthetases. The Gram-positive bacterium Micrococcus luteus (Ml) was specifically chosen because its sequence is very divergent from Xc OleC, and it contains a natural fusion of the oleB and oleC genes. This natural oleBC fusion is found in all actinobacteria, which comprise ~30% of the microorganisms that contain identifiable oleABCD genes. Reaction of purified Ml OleBC fusion with MgCl2, ATP, and 3 produced β-lactones 4a and 4b as well as small amounts of cis-olefin, 5a (Figure S14). No trace of trans-olefin, 5b, was detected. Further characterization is ongoing, but we believe that OleB performs a syn-elimination of cis-β-lactone to form the final cis-olefin product. This is consistent with previous studies of microorganisms expressing ole genes that contain ole-fins with a cis-relative configuration exclusively7–9. These data also demonstrate that an enzyme domain with only 35% amino acid identity to the Xc OleC generates β-lactones, indicating that this activity is likely common among all olefinic hydrocarbon biosynthesis OleC homologues.
Table 1.
Other OleC enzymes make β-lactones
| Organism | Ascension # | % IDa |
|---|---|---|
| Xanthomonas campestris | WP_011035474.1 | 100 |
| Stenotrophomonas maltophilia | AFC01244.1 | 77 |
| Arenimonas malthae | WP_043804215.1 | 73 |
| Lysobacter dokdonensis | WP_036166093.1 | 70 |
| Micrococcus luteusb | WP_010078536.1 | 35 |
% identity based on amino acid sequence
OleC and OleB are a natural fusion in Ml.
Establishing the widespread nature of lactone synthetase activity within olefinic hydrocarbon biosynthesis led us to search sequence databases for OleC homologues in other biosynthetic pathways. OleC is a member of the ubiquitous AMP-dependent ligase/synthetase enzyme superfamily; as such, homologues are found in all organisms15. As of November 2016, a BLAST search of NCBI’s non-redundant protein sequence database identified over 900 sequences with greater than 35% sequence identity and over 16,000 with greater than 25% sequence identity to Xc OleC.
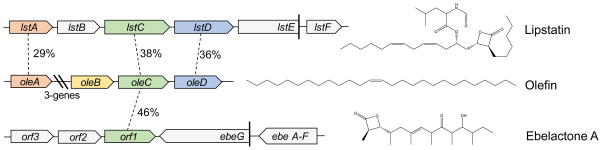
Of the sequences examined, two Xc OleC homologues were clearly encoded in gene clusters known to produce β-lactone natural products (Figure 4). The first, LstC, is an uncharacterized enzyme found in the lipstatin biosynthesis pathway from Streptomyces toxytricini. Lipstatin is the precursor to Orlistat, the only over-the-counter, FDA-approved anti-obesity drug. LstC is a member of the AMP-dependent ligase/synthetase superfamily, and its protein sequence is 38% similar to Xc OleC, more similar than the sequence of the β-lactone synthetase domain of Ml OleBC (35%). Surprisingly, further investigation revealed homologues to OleA and OleD encoded by the lipstatin gene cluster, suggesting that the two gene clusters have a common ancestry. The synthesis of both lipstatin and olefinic hydrocarbons are initiated by the condensation of two fatty acyl-CoAs to form a β-keto acid. In the case of lipstatin, the two fatty acids are 3-hydroxy-linoleic and octanoic acid1. The hydroxyl group of linoleic acid is later functionalized by LstE and LstF with a modified valine1. LstD and OleD likely perform the same NADPH-dependent reduction of the β-keto group to a hydroxyl group. Formation of the trans-β-lactone is likely accomplished by the OleC homologue LstC, to generate the final product in lipstatin biosynthesis. Olefin biosynthesis is completed by the putative OleB-dependent elimination of CO2 to generate the final olefin product. The lipstatin gene cluster lacks any gene product that shows homology to OleB, consistent with the accumulation of the β-lactone natural product and further supporting our hypothesis that OleB performs the final step in the biosynthetic pathway to olefins.
Figure 4.
Homologous OleC enzymes encoded in β-lactone biosynthesis gene clusters. Percent iden-tity is based on amino acid sequences. The E values for OleC to LstC and Orf1 are 2e-72 and 1e-143, respectively. Lipstatin is the precursor to the anti-obesity drug Orlistat. Ebelactone A is a commercially available general esterase inhibitor.
The gene cluster responsible for the biosynthesis of ebelactone A, a commercially available esterase inhibitor, in Streptomyces aburaviensis shows a gene, orf1, with 46% amino acid sequence identity to Xc OleC and is directly adjacent to ebeA-G (Figure 4). Unlike lipstatin, ebelactone A is formed partly by a polyketide synthase multi-domain protein rather than fatty acid condensation; as such, OleA and OleD homologues are not encoded in the surrounding gene cluster. Literature reports suggest that the β-lactone ring of ebelactone A is formed spontaneously from the final, enzyme-linked, β-hydroxy-thioester intermediate5. While a spontaneous β-hydroxy-thioester cyclization is mechanistically plausible, β-lactone formation from β-hydroxy-thioesters in ubiquitous pathways such as fatty acid oxidation or synthesis has not been reported to our knowledge16. Additionally, β-hydroxy-thioester intermediates are extremely common in polyketide synthesis pathways, while β-lactone formation is comparatively rare. An Orf1-independent cyclization would require a unique property of ebelactone A precursors or a novel polyketide domain architecture to promote β-lactone ring cyclization. However, in favor of an Orf1-independent mechanism is that no thioesterase domain exists in the final polyketide synthesis domain, suggesting that no free β-hydroxy acid is released for the putative ATP-dependent Orf1 to act on. Other polyketide-type β-lactone gene clusters, such as those for salinosporamide A, cinnabaramide, and oxazolomycin, do not encode an OleC homologue with high sequence similarity (>35%). Polyketide-derived β-lactones are thought to form by the cyclization of the final thioester, enzyme-linked intermediate, but this has never been characterized2,17–19. It is reasonable to hypothesize that specialized polyketide synthase domains represent a second mechanism of β-lactone formation. Regardless, the discovery of a stand-alone β-lactone synthetase here creates new opportunities for the natural product field. Preliminary screening of Streptomyces and Nocardia genomes suggests that β-lactone natural products may be more widespread than currently realized.
Supplementary Material
Acknowledgments
Funding
This was supported by U of M Biotechnology Institute Microbial Factories Award (LPW and CMW), NIH Training for Future Biotechnology Development (2T32GM008347-22) (JKC), NIH Chemical-Biology Interface Training Grant (GM-008700) (MRJ), an NSF-LSSURP grant (JYN).
We thank Anna Kim, Tiffany Engel, and Shintaro Nakayama for their practical support in the lab.
Footnotes
Notes
The authors declare no competing financial interest.
References
- 1.Bai T, Zhang D, Lin S, Long Q, Wang Y, Ou H, Kang Q, Deng Z, Liu W, Tao M. Appl Environ Microbiol. 2014;80:7473–7483. doi: 10.1128/AEM.01765-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Angew Chem Int Ed. 2003;42:355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- 3.Lee M, Gwak HS, Park BD, Lee ST. J Am Oil Chem Soc. 2005;82:181–188. [Google Scholar]
- 4.Masamune S, Hayase Y, Chan WK, Sobczak RL. J Am Chem Soc. 1976;98:7874–7875. doi: 10.1021/ja00440a096. [DOI] [PubMed] [Google Scholar]
- 5.Wyatt MA, Ahilan Y, Argyropoulos P, Boddy CN, Magarvey NA, Harrison PH. J Anti-biot (Tokyo) 2013;66:421–431. doi: 10.1038/ja.2013.48. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Tennyson RL, Romo D. Heterocycles. 2004;64:605–658. [Google Scholar]
- 7.Sukovich DJ, Seffernick JL, Richman JE, Gralnick JA, Wackett LP. Appl Environ Micro-biol. 2010;76:3850–3862. doi: 10.1128/AEM.00436-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albro PW, Dittmer JC. Biochemistry. 1969;8:394–405. doi: 10.1021/bi00829a055. [DOI] [PubMed] [Google Scholar]
- 9.Frias JA, Richman JE, Wackett LP. Appl Environ Microbiol. 2009;75:1774–1777. doi: 10.1128/AEM.02547-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frias JA, Richman JE, Erickson JS, Wackett LP. J Biol Chem. 2010;286:10930–10938. doi: 10.1074/jbc.M110.216127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonnett SA, Papireddy K, Higgins S, del Cardayre S, Renyolds KA. Biochemistry. 2011;50:9633–9640. doi: 10.1021/bi201096w. [DOI] [PubMed] [Google Scholar]
- 12.Kancharla P, Bonnett SA, Reynolds KA. Chembiochem. 2016;17:1426–1429. doi: 10.1002/cbic.201600063. [DOI] [PubMed] [Google Scholar]
- 13.Noyce DS, Banitt EH. J Org Chem. 1966;31:4043–4047. [Google Scholar]
- 14.Mulzer J, Zippel M, Bruentrup G. Angew Chem. 1980:469–470. [Google Scholar]
- 15.Conti E, Franks NP, Brick P. Structure. 1996;4:287–298. doi: 10.1016/s0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- 16.Dick LR, Cruikshank AA, Grenier L, Melandri FD, Nunes SL, Stein RL. J Biol Chem. 1996;271:7273–7276. doi: 10.1074/jbc.271.13.7273. [DOI] [PubMed] [Google Scholar]
- 17.Rachid S, Huo L, Herrmann J, Stadler M, Kopcke B, Bitzer J, Muller R. ChemBioChem. 2011;12:922–931. doi: 10.1002/cbic.201100024. [DOI] [PubMed] [Google Scholar]
- 18.Zhao C, Coughlin JM, Ju J, Zhu D, Wendt-Pienkowski E, Zhou X, Wang Z, Shen B, Deng Z. J Biol Chem. 2010;285:20097–20108. doi: 10.1074/jbc.M109.090092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hemmerling F, Hahn F. J Org Chem. 2016;12:1512–1550. doi: 10.3762/bjoc.12.148. Beilstein. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.