Abstract
BACKGROUND. Both seasonal and novel avian influenza viruses can result in severe infections requiring hospitalization. Anti-influenza antibodies (Abs) with Fc-mediated effector functions, such as Ab-dependent cellular cytotoxicity (ADCC), are of growing interest in control of influenza but have not previously been studied during severe human infections. As such, the objective of this study was to examine Fc-mediated Ab functions in humans hospitalized with influenza infection.
METHODS. Serum Ab response was studied in subjects hospitalized with either pandemic H7N9 avian influenza virus in China (n = 18) or circulating seasonal influenza viruses in Melbourne, Australia (n = 16). Recombinant soluble Fc receptor dimer ELISAs, natural killer (NK) cell activation assays, and Ab-dependent killing assays with influenza-infected target cells were used to assess the Fc functionality of anti-influenza hemagglutinin (HA) Abs during severe human influenza infection.
RESULTS. We found that the peak generation of Fc functional HA Abs preceded that of neutralizing Abs for both severe H7N9 and seasonal influenza infections. Subjects who succumbed to complications of H7N9 infection demonstrated reduced HA-specific Fc receptor–binding Abs (in magnitude and breadth) immediately prior to death compared with those who survived. Subjects who recovered from H7N9 and severe seasonal influenza infections demonstrated increased Fc receptor–binding Abs not only against the homologous infecting strain but against HAs from different influenza A subtypes.
CONCLUSION. Collectively, survivors of severe influenza infection rapidly generate a functional Ab response capable of mediating ADCC against divergent influenza viruses. Broadly binding HA Abs with Fc-mediated functions may be a useful component of protective immunity to severe influenza infection.
FUNDING. The National Health and Medical Research Council ([NHMRC] grants 1023294, 1041832, and 1071916), the Australian Department of Health, and the joint University of Melbourne/Fudan University International Research and Research Training Fund provided funding for this study.
Keywords: Immunology, Infectious disease
Fc functional antibodies are decreased in severe avian influenza patients and occur earlier than strain-specific neutralizing antibodies in patients with seasonal and avian influenza.
Introduction
Yearly epidemics of seasonal influenza cause 3–5 million severe illnesses each year, and current vaccines are only partially effective (1). Although less prevalent than seasonal influenza, avian influenza viruses more commonly cause severe influenza illness. Avian influenza viruses of the H7N9 subtype recently emerged in China and have the potential to cause global pandemics if they acquire the ability to transmit human to human (2). As of February 2017, there have been 1,258 reported human cases of H7N9 avian influenza, with a case fatality rate of 41%, primarily due to rapidly progressing pneumonia (3). During the fifth epidemic of human H7N9 avian influenza (beginning on October 1, 2016), there has been a dramatic rise in the number of cases reported (460 cases) compared with the previous 4 epidemics, suggesting that H7N9 avian influenza poses an ongoing threat to human health.
Immune control of severe seasonal and avian influenza infections is likely multifactorial and is not well characterized (4, 5). Neutralizing antibodies (Abs) (NAbs), T cells, and natural killer (NK) cell responses were associated with recovery in a cohort of 18 subjects with severe human H7N9 infection (6, 7). In the absence of vaccination, partial immunity to seasonal influenza is driven by immune responses to repeated infections with related strains. In contrast, most humans lack previous exposure to novel avian influenza viruses and have lower preexisting humoral immunity (8). Furthermore, human H7N9 infection causes serious immune dysregulation (e.g., lymphocytopenia, reduced monocytes, and altered cytokine and chemokine levels) that may limit key functions of the antiviral response (9–11). Comparing severe seasonal and avian influenza infections in humans may yield insight into the immune mechanisms of recovery.
Humoral immunity has long been implicated in protection from influenza through NAbs that block virus entry into cells (12, 13). The narrow specificity of influenza NAbs, particularly those directed against the immunodominant hemagglutinin (HA) head, is problematic for protection against influenza viruses (14, 15). However, in addition to neutralization, Abs can also mediate several functions through their Fc region, including complement-dependent cytotoxicity (CDC) (16, 17), Ab-dependent cellular cytotoxicity (ADCC) (18–21), and Ab-dependent phagocytosis (ADP) (22). ADCC occurs when Abs bind to viral antigens on infected cells and engage receptors for the Fc region of IgG (FcγRs) found on the surface of innate immune effector cells. Multimeric engagement of neighboring FcγRs on effector cells, often referred to as cross-linking, leads to cell activation and the release of antiviral cytokines (e.g., IFN-γ) and cytolytic granules to kill the infected target cell. ADCC is primarily mediated through human FcγRIIIa (CD16a) found on NK cells, monocytes, and macrophages (23).
Several mouse studies, macaque studies, and human cohort studies in recent years have implicated ADCC and other Fc-mediated Ab effector mechanisms with control of influenza (24–29). A recent study suggested that high titers of ADCC-mediating Abs might reduce clinical symptoms and viral replication following an A/Wisconsin/67/131/2005 (H3N2) influenza challenge of humans (30). Human influenza challenge models, however, are necessarily only modestly pathogenic and may not mirror severe clinical infections. As such, we studied ADCC responses in cohorts of hospitalized subjects with H7N9 and seasonal influenza infections.
Results
Hospitalized influenza-infected subjects.
Sera were studied from 18 H7N9-infected patients referred to and hospitalized at the Shanghai Public Health Clinical Centre (SPHCC) (Table 1). The patients were hospitalized at the SPHCC 6–16 days (median 9.5 days) after symptom onset, and the first available samples were generally collected within 48 hours of admission to the SPHCC (median 11 days after symptom onset). The median age of the H7N9-infected patients was 71 years (range 47–88 years). All subjects were treated with oseltamivir starting a median of 6.5 days after symptom onset (range 3–11 days). Six patients succumbed to H7N9 infection, and twelve recovered from the infection. The median sequential organ failure assessment (SOFA) score, calculated shortly after admission to SPHCC, was 3.5 (range 2–9) for the H7N9-infected patients.
Table 1. Characteristics of the H7N9 influenza-infected cohort, the DISI seasonal influenza-infected cohort, and control subjects.
Plasma and sera samples were also studied from 16 hospitalized subjects with confirmed infection with either H1N1 or H3N2 seasonal influenza A viruses or seasonal influenza B viruses in Melbourne, Australia (Table 1), as part of the Dissection of Influenza-Specific Immunity (DISI) cohort recruited from the Alfred Hospital. The median age of the patients hospitalized with seasonal influenza was 57 years (range 22–84 years). Subjects with severe seasonal influenza infection were hospitalized 1–7 days (median 4.5 days) after symptom onset, and the first samples were generally collected within 48 hours of hospital admission (median 6 days after symptom onset). One patient contracted influenza in the hospital and died. Eleven of the sixteen seasonal influenza-infected subjects were treated with oseltamivir starting a median of 3 days after symptom onset (range 1–8 days). The median SOFA score, calculated shortly after hospital admission, was 2 (range 0–6) for subjects with severe seasonal influenza infection, which was significantly lower than that in subjects with H7N9 infection (P = 0.0001).
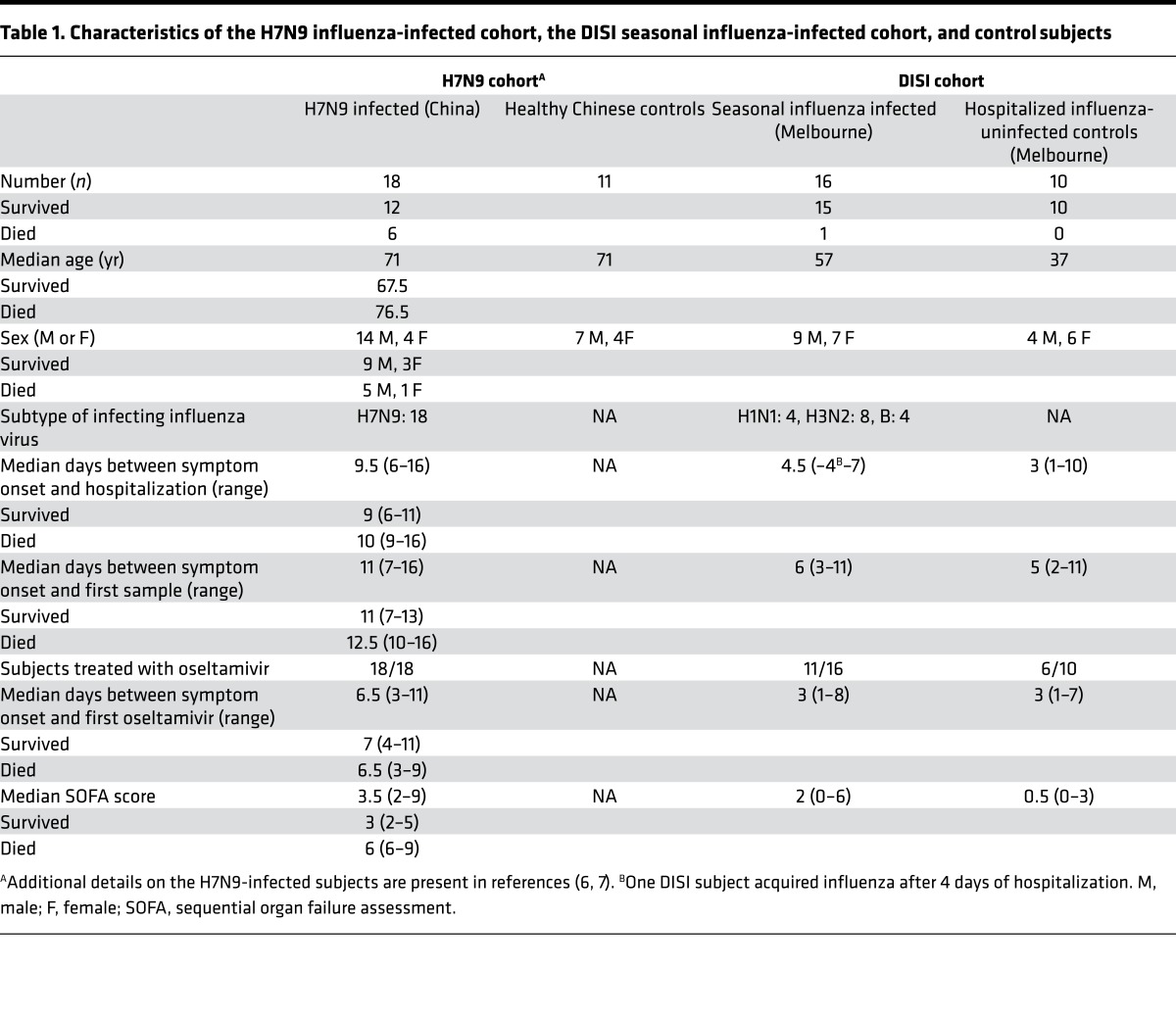
FcγR-binding HA-specific Abs during severe influenza. Nonneutralizing Abs have been shown to assist in the clearance of influenza infections in animal models and may be important in the control of severe influenza infections (24–30). We first studied serial samples from subjects infected with H7N9 and seasonal influenza using a recently developed recombinant soluble FcγR (rsFcγR) dimer ELISA (31). At hospital admission and hospital release/death, sera from H7N9-infected subjects showed significantly increased rsFcγRIIIa and rsFcγRIIa dimer binding by anti-H7 Abs compared with uninfected controls (Figure 1, A and C). Binding by H7 Abs to the rsFcγRIIa dimer (P = 0.003), but not the rsFcγRIIIa dimer (P = 0.49), increased modestly by the time of hospital release/death compared with hospital admission (Figure 1, A and C). The absence of a large rise in FcγR-binding Abs during hospitalization likely reflects the relatively long 9.5-day median duration of influenza-like illness (ILI) at the time of hospital admission — the subjects had already developed FcγR-binding Abs by the time the first serum sample was available.
Figure 1. Increased HA-specific rsFcγR dimer–binding Abs after severe influenza infections.
Subjects infected with H7N9 influenza (A and C) and seasonal influenza (B and D) were examined for dimeric rsFcγRIIIa (A and B) and rsFcγRIIa (C and D) binding to Ab-opsonized HA at hospital admission, hospital release/death, and approximately 30 days after hospitalization (seasonal infection only). Dimeric rsFcγR binding by Abs to the HA protein of A/Shanghai/1/2013 (H7N9) is shown for 18 H7N9-infected (black circles) and 11 healthy age-matched control (black diamonds) subjects (A and C). For seasonal influenza-infected subjects (B and D), dimeric rsFcγR engagement by Abs was measured against either (a) H1 protein from A/California/04/2009 (H1N1) virus for subjects infected with seasonal H1N1 influenza (4 subjects, gray squares), (b) H3 protein from A/Switzerland/9715293/2013 (H3N2) virus for subjects infected with seasonal H3N2 influenza (8 subjects, black circles), or (c) HA protein from B/Phuket/3073/2013 virus for seasonal influenza B–infected subjects (4 subjects, white triangles). As controls for seasonal influenza, 10 hospitalized influenza-negative subjects (B and D, black diamonds) were tested for dimeric rsFcγR binding by Abs against all 3 seasonal HA proteins. All influenza-infected and control subjects were also tested for dimeric rsFcγR binding by Abs against an irrelevant HIV-1 protein gp140, and representative data are shown for a single time point (all control gp140-specific responses are shown in Supplemental Figure 1). A Kruskal-Wallis test was used to compare healthy age-matched controls to H7N9-infected subjects at hospital admission and hospital release/death, while a Wilcoxon matched-pairs signed-rank test was used to compare rsFcγR dimer binding between hospital admission and hospital release/death samples in the H7N9-infected subjects (A and C). A Mann Whitney U test was used to compare hospitalized seasonal influenza-infected subjects with hospitalized influenza-negative subjects at the same time point of collection (B and D). **P < 0.01, ***P < 0.001.
Subjects with severe seasonal influenza infection demonstrated detectable baseline levels of serum HA-specific rsFcγRIIIa and rsFcγRIIa dimer binding at the time of hospital admission (Figure 1, B and D), a median of only 4.5 days after ILI onset. This likely reflects common prior exposure to influenza viruses in the adult population (26, 27, 32), as the levels were similar to those in noninfluenza-infected controls (rsFcγRIIIa: P = 0.36 and rsFcγRIIa: P = 0.40, Figure 1, B and D). A progressive increase in rsFcγR dimer binding by HA-specific Abs was observed in subjects with confirmed seasonal influenza infection, but not in the influenza-uninfected controls (Figure 1, B and D). Negligible levels of FcγR dimer binding were observed against a control immobilized HIV-1 gp140, since all subjects were HIV negative (Figure 1 and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.92750DS1). Collectively, these results suggest that HA Abs capable of forming complexes with FcγRs develop early in the sera of individuals hospitalized with pandemic H7N9 and seasonal influenza infections.
HA Abs induced during severe influenza infections activate primary NK cells.
FcγRIIIa cross-linking activates NK cells to produce cytokines and to degranulate releasing cytotoxic granules. To validate that the FcγRIIIa dimer ELISA data had functional relevance, we studied serial samples from H7N9- and seasonal influenza-infected cohorts for their capacity to induce NK cell activation. Serum HA Abs from H7N9-infected subjects, but not healthy age-matched controls, demonstrated H7-specific primary NK cell IFN-γ and/or CD107a expression (Supplemental Figure 2A). The H7N9-infected cohort did not show a significant difference in NK cell activation between the hospital admission samples and the hospital release/death samples (P = 0.07, Figure 2A), consistent with the delay prior to the first sample being available and the results from the rsFcγRIIIa dimer ELISA. Subjects with seasonal influenza developed HA-specific Ab-dependent NK cell activation during the course of their illness, unlike control hospitalized patients, supporting the increased HA-specific FcγRIIIa cross-linking Ab observed (Figure 2B). Indeed, a significant correlation between dimeric rsFcγRIIIa binding and NK cell activation was observed for both the H7N9-infected and seasonal influenza-infected cohorts (Figure 2, C and D). As with the FcγR dimer ELISA, control HIV-1 gp140-specific NK cell–activating Abs were low (Figure 2A and Supplemental Figures 2 and 3). Overall, severe H7N9 and seasonal influenza infections generate a humoral anti-HA response that can cross-link FcγRs and activate primary NK cells.
Figure 2. Increased NK cell–activating HA-specific Abs after severe influenza infection.
Serum HA Abs in subjects infected with H7N9 (A) and seasonal influenza (B) were analyzed for their ability to activate primary NK cells to express IFN-γ and/or CD107a. The time course, control subjects, statistical analysis, and HA and gp140 proteins studied were as described for Figure 1. Spearman correlations between primary NK cell activation and dimeric rsFcγRIIIa ELISA for H7N9 (C) and seasonal influenza (D) infected cohorts are shown across all times points studied. **P < 0.01, ***P < 0.001.
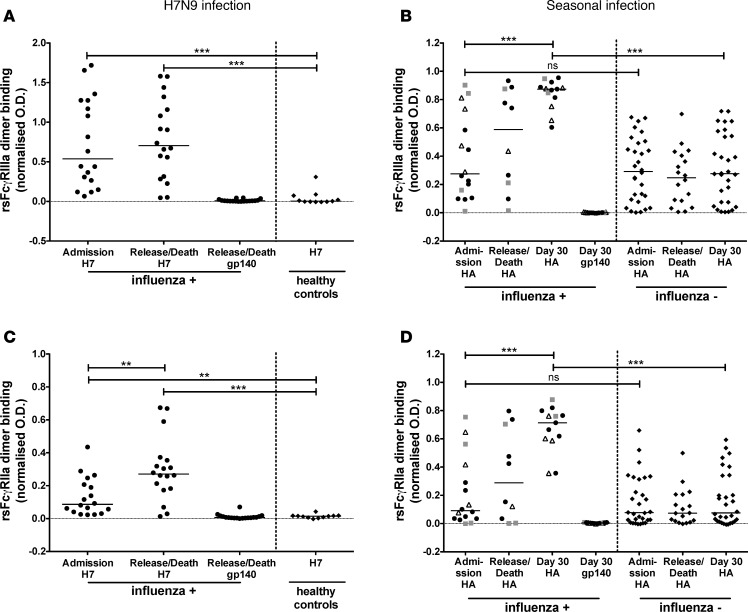
Recovery from H7N9 infection associated with enhanced ADCC Abs.
The evolution of the ADCC Ab response may mirror or influence the course of severe human influenza infection. To examine the relationship between severe influenza infection and ADCC-mediating Abs, we compared serum HA Abs from H7N9-infected subjects grouped based on infection outcome. At admission, there were comparable levels of rsFcγRIIIa and rsFcγRIIa dimer binding by anti-H7 Abs in sera from the 12 subjects who recovered and the 6 subjects who succumbed to H7N9 infection (Figure 3, A and B). At discharge or death, however, Abs collected from the recovered subjects at hospital release showed significantly higher dimeric rsFcγR binding when incubated with H7 protein than serum Abs collected prior to death from the 6 subjects who succumbed to H7N9 infection (Figure 3, A and B). Ab-dependent NK activation assays also demonstrated a reduction in H7-specific ADCC Abs in those that succumbed to infection, but the difference was not statistically significant (Figure 3C).
Figure 3. Elevated rsFcγR dimer binding and NK cell activation by serum HA Abs in subjects who recovered from H7N9 infection.
The H7N9-infected cohort was divided into subjects who survived (n = 12, circles) and died (n = 6, diamonds). Dimeric rsFcγRIIIa binding (A), dimeric rsFcγRIIa binding (B), and primary NK cell activation (C) by serum Abs was analyzed against the immobilized H7 protein at hospital admission (black symbols) and hospital release/death (white symbols). Background rsFcγR dimer binding or NK cell activation by serum Abs to an irrelevant HIV-1 protein gp140. A Mann Whitney U test was used to compare the subjects who survived H7N9 infection to the subjects who died from H7N9 infection. * P < 0.05.
Severe influenza infections generate cross-reactive Fc functional Ab responses.
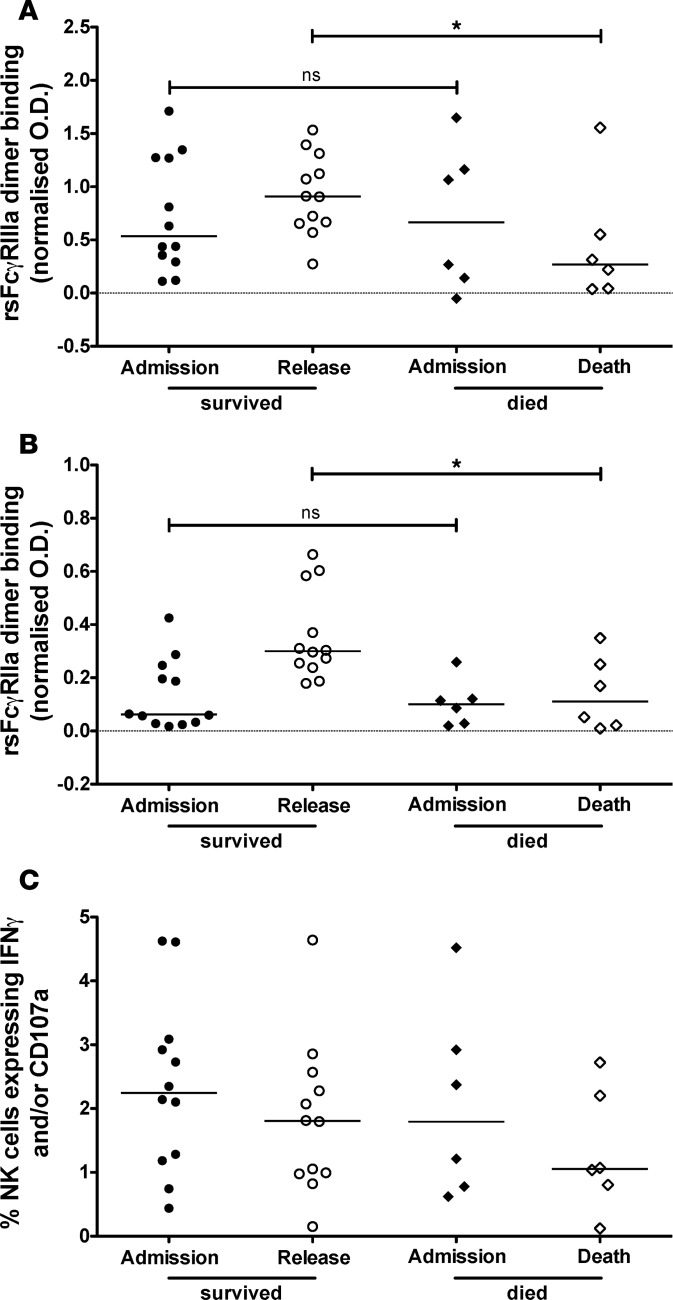
ADCC Abs need not bind variable sites on the HA head and can have broad reactivity to multiple strains of influenza (24–26, 28, 33). We found that FcγRIIIa- and FcγRIIa-binding Abs in the sera of H7N9-infected patients demonstrated significant cross-reactivity to HAs from other group 2 influenza viruses, including the HAs from H3N2 and H4N6 viruses (Figure 4, A and C). FcγR dimer binding was also detected against HAs from group 1 influenza viruses, such as the H5 HA from an avian H5N1 virus and the H2 HA from a noncirculating H2N2 virus, in the H7N9-infected cohort (Figure 4, A and C). Interestingly, in H7N9-infected sera, cross-reactivity to the HA of A/California/04/2009 (H1N1) pandemic virus (H1 Cali/09) was lower than any other subtypes tested in this elderly Chinese cohort (Figure 4, A and C).
Figure 4. Breadth of Fc functional Abs generated during severe influenza infection.
Subjects infected with H7N9 influenza (A and C) and seasonal influenza A (B and D) were examined over time for breadth of dimeric rsFcγRIIIa (A and B) and rsFcγRIIa (C and D) against a range of HA proteins. Median rsFcγRIIIa (A) and rsFcγRIIa (C) dimer binding by serum Abs from 12 subjects who survived and 6 subjects who died from H7N9 infection is shown against HA proteins from the A/Shanghai/1/2013 (H7N9) virus (H7 Shanghai), the A/California/04/2009 (H1N1) virus (H1 Cali/09), the A/Japan/305/1957 (H2N2) virus (H2 Japan), the A/Perth/16/2009 (H3N2) virus (H3 Perth), the A/Swine/Ontario/01911-1/1999 (H4N6) virus (H4 Ontario), and the A/Vietnam/1194/2004 (H5N1) virus (H5 Vietnam). Median rsFcγRIIIa (B) and rsFcγRIIa (D) dimer binding by serum Abs from 12 subjects infected with seasonal influenza A viruses (of the H1N1 or H3N2 subtypes) and 10 influenza-negative subjects is shown for H1 Cali/09, H2 Japan, H4 Ontario, H5 Vietnam, and H7 Shanghai as well as for H3 proteins from the A/Switzerland/9715293/2013 (H3N2) and X-31 (H3N2) viruses. All influenza-infected and control subjects were background subtracted for dimeric rsFcγR binding by Abs against an irrelevant HIV-1 protein gp140. Data from individual subjects are shown in Supplemental Figures 4 and 5. All 6 HAs were analyzed concurrently with a Wilcoxon matched-pairs signed-rank test to compare hospital admission to hospital release/death in the H7N9-infected cohort (A and C). Similarly for the seasonal influenza-infected and uninfected subjects, all 7 HAs were analyzed concurrently with a Kruskal-Wallis test to compare hospital admission, hospital release/death, and 30 days after hospitalization (B and D). *P < 0.05, ***P < 0.001, ****P < 0.0001.
In subjects recovering from H7N9 infection, dimeric FcγRIIIa binding by anti-HA Abs remained constant from hospital admission to release. In contrast, subjects who succumbed to H7N9 infection showed decreased median FcγRIIIa dimer binding by HA-specific Abs from the time of hospital admission to death across all subtypes tested (Figure 4A and Supplemental Figure 4). Median binding by anti-HA Abs to the FcγRIIa (CD32a) dimer increased across all subtypes from hospital admission to release in the recovered subjects but remained constant in subjects who died from H7N9 infection (Figure 4C and Supplemental Figure 5).
Subjects infected with seasonal strains of influenza A (either H1N1 or H3N2) also demonstrated significant increases in median cross-reactive FcγR dimer–binding Abs against HAs from different subtypes of influenza, including H2 Japan, H4 Ontario, H5 Vietnam, and H7 Shanghai (Figure 4, B and D, and Supplemental Figures 4 and 5). In subjects with confirmed cases of seasonal influenza A infection, HA-specific Abs showed increased FcγRIIIa and FcγRIIa dimer binding from hospital admission to 30 days after hospitalization for all group 1 (H1N1, H2N2, and H5N1) and group 2 (H3N2, H4N6, and H7N9) HAs tested (Figure 4, B and D, and Supplemental Figures 4 and 5). In contrast, hospitalized influenza-negative subjects did not show an increase in FcγR cross-linking Abs against HAs from the 6 different influenza subtypes over time.
Kinetics of ADCC and NAb responses during severe influenza infection.
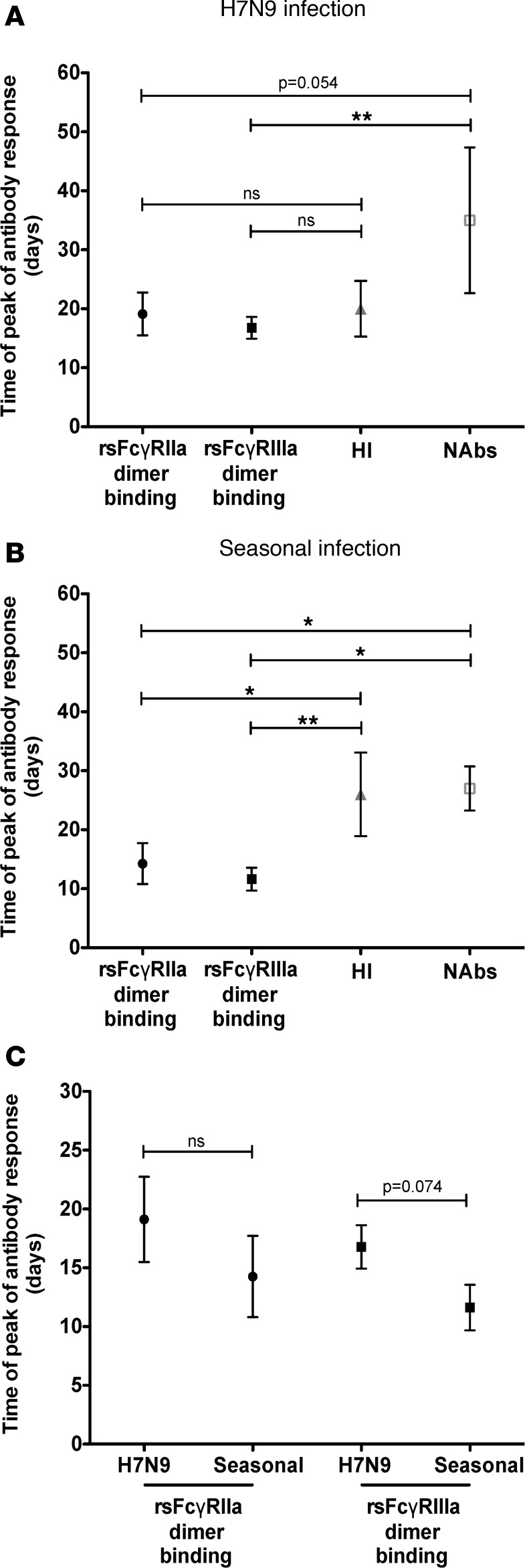
ADCC responses appear relatively early in the course of severe influenza infection and may provide some immunity as the virus-specific NAb response is developing. Whether the peak ADCC response precedes that of the hemagglutination inhibiting or NAb response is not known. Our series of Fc-binding, hemagglutination inhibition (HI), and focus reduction assays (FRA) across the severe influenza infection cohorts allowed us to analyze the kinetics of the responses. To account for the limited series of samples and variable times after infection when the samples were taken from each subject, we modeled the peak of ADCC or NAb responses across data collected from all subjects at all time points tested (2–5 longitudinal samples per subject) based on standard rise and decay characteristics of Abs as previously described (34). The primary data used for modeling is shown in Supplemental Figures 6 and 7.
We first analyzed the H7N9 cohort, which reflects the exposure to a virus with little or no specific Abs prior to infection. We found that, in H7N9 infection, the HA-specific rsFcγRIIIa dimer–binding Ab response reached its peak nearly 3 weeks earlier (mean 17 days, 95% CI 14–20 days) than the NAb response (mean 35 days, 95% CI 13–56 days, P = 0.001, Figure 5A). The HA-specific rsFcγRIIa dimer–binding Ab response also trends toward reaching its peak earlier (mean 19 days, 95% CI 13–25 days) than the NAb response during H7N9 infection, but the peak response was more variable and the difference was marginally significant (P = 0.054, Figure 5A). The time to peak of response between H7-specific FcγR cross-linking Ab responses and the H7-specific HI response was similar (Figure 5A).
Figure 5. Kinetics of Fc functional and NAb responses in severe influenza infection.
Ab kinetics were modeled for the estimated peak generation of responses for rsFcγRIIIa binding, rsFcγRIIa binding, hemagglutination inhibiting (HI), and neutralizing Abs (NAbs) for H7N9 influenza (A) and seasonal influenza (B). Viruses and HA proteins used for each cohort are described in the Methods. Data represent mean and 95% confidence intervals. Individual data points for all subjects are shown in Supplemental Figures 6 and 7. (C) Comparison of peak generation of rsFcγRIIIa-binding and rsFcγRIIa-binding Abs in H7N9 and severe seasonal influenza. *P < 0.05, **P < 0.01 by Wald’s test (described in Methods).
We then analyzed the seasonal influenza-infected cohort, in which some Ab responses, such as nonneutralizing responses, may be preprimed and increase earlier than highly specific NAb responses. We found that both rsFcγRIIa (mean 14 days, 95% CI 8–20 days, P = 0.025, Figure 5B) and rsFcγRIIIa (mean 12 days, 95% CI 8–15 days, P = 0.0092) dimer–binding Ab responses had significantly shorter times to peak than the HI response (mean 26 days, 95% CI 14–38 days, Figure 5B). Moreover, we also found a later peak of NAb response (mean 27 days, 95% CI 20–33 days) compared with both rsFcγRIIIa (P = 0.033, Figure 5B) and rsFcγRIIa (P = 0.043, Figure 5B) dimer–binding Ab responses. Despite its relatively short time to peak, the FcγR cross-linking Ab response correlated with NAb responses for subjects infected with severe H7N9 and seasonal influenza (Supplemental Figure 8). Consistent with previous studies using Abs from intravenous immunoglobulin preparations (31) and influenza vaccinees (35), we found that FcγR cross-linking Ab responses correlate with HI in subjects infected with seasonal influenza but not H7N9-infected subjects (Supplemental Figure 9).
Next, we asked whether there is a difference in the kinetics between severe H7N9 and seasonal infections within the same FcγR cross-linking Ab response. We hypothesized that prior exposure to seasonal viruses might prime memory B cell responses for a more rapid induction after infection. We observed a trend toward later peak of rsFcγRIIIa dimer–binding H7-specific Abs in the H7N9-infected cohort compared with the seasonal HA-specific rsFcγRIIIa dimer–binding Abs in the seasonal influenza-infected cohort (P = 0.074, Figure 5C). There was no significant difference in the time of peak for the HA-specific dimeric rsFcγRIIa-binding Ab response in H7N9 infection compared with seasonal influenza infection (P = 0.11, Figure 5C).
Ab-mediated killing of target cells infected with a divergent, homosubtypic influenza virus is increased following severe seasonal influenza infection.
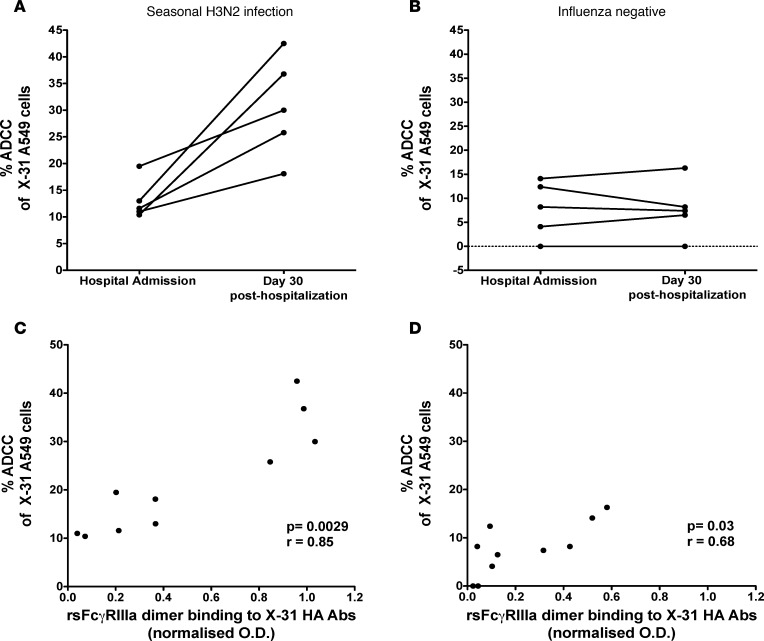
A key potential advantage of cross-reactive ADCC Abs is their ability to kill respiratory cells infected with antigenically divergent influenza viruses. To assess whether the HA-specific rsFcγRIIIa dimer binding and NK cell–activating Abs boosted following severe influenza infection are capable of mediating ADCC against heterologous strains of influenza, we infected a respiratory cell line (A549 cells) with the X-31 virus, which expresses H3 from the pandemic A/Aichi/2/1968 (H3N2) virus, and added effector NK cells to assess ADCC by lactate dehydrogenase (LDH) release. We chose the X-31 virus, since the 1968 HA is distinct from the HA expressed by 2014/2015 seasonal H3N2 viruses. We studied serum Abs from 5 subjects infected with seasonal H3N2 influenza because H3N2 infection resulted in uniform increases in rsFcγR dimer binding and NK cell–activating Abs against H3N2 HAs, and we had adequate sera samples from these subjects. All 5 subjects showed a marked increase in ADCC of X-31-infected cells 30 days after hospitalization compared with hospital admission (Figure 6A). In contrast, the 5 influenza-negative subjects tested showed no difference in ADCC at hospital admission and at 30 days after hospitalization (Figure 6B). We observed significant correlations between the rsFcγRIIIa dimer binding ELISA to X-31 HA protein and ADCC against X-31–infected A549 cells for both control and influenza-infected subjects, suggesting that HA-specific Abs play a role in killing influenza-infected cells (Figure 6, C and D). Recent studies have also shown that neuraminidase and nucleoprotein Abs may be partially responsible for influenza-specific ADCC (24, 36). Interestingly, ADCC against X-31–infected A549 cells also correlated with the rsFcγRIIIa dimer binding ELISA (P = 0.01, r = 0.78) and with the NK cell activation assay (P = 0.05, r = 0.64) performed using a heterologous HA protein from the seasonal A/Switzerland/9715293/2014 (H3N2) influenza virus.
Figure 6. Generation of ADCC Abs after severe seasonal H3N2 infection.
Sera were tested at hospital admission and 30 days later for their ability to kill H3N2-infected (X-31) respiratory epithelial cells (A549 cells) in the presence of an NK cell line (NK-92 cells). Killing was measured by the specific release of LDH from target cells. Sera from 5 subjects with confirmed seasonal H3N2 infection (see Supplemental Table 1 for strains) (A) and 5 subjects hospitalized with noninfluenza respiratory infections (B) were tested. Spearman correlation between ADCC activity and rsFcγRIIIa-binding Abs in the influenza-infected (C) and influenza-uninfected (D) subjects across two time points studied.
Discussion
There has been renewed interest in the nonneutralizing functions of Abs during clinically important human infections, such as those involving HIV (37–41), dengue (42), and influenza viruses (24–30). In both animal models and mild-to-moderate human influenza infections, Fc-FcγR interactions aid in the clearance of influenza infection (24–30). Herein, we performed the first comprehensive investigation to our knowledge of the Fc functional Ab response in serial samples across two cohorts of subjects hospitalized with severe H7N9 or seasonal influenza infections.
A key finding was that subjects who succumbed to H7N9 infection showed significantly lower FcγR dimer binding, reduced NK cell activation, and reduced HA breadth in their Fc functional Abs just prior to death compared with subjects who recovered. Severe human H7N9 infection has a high mortality rate (3) and leads to immune dysfunction on many levels, so we cannot assert that this observed reduction in HA-specific Fc functional Abs was causally related to their death (9–11). Previous work on this cohort showed that the CD8+ T cell response was also impaired in people who died from H7N9 infection (6) and that these subjects commonly had other comorbidities (7). There is a growing interest in passive transfer of anti-influenza Abs in severe influenza, with some efficacy observed in small human studies (43, 44) and animal models (45). Both polyclonal immunoglobulin preparations (28) and some monoclonal Abs can have significant Fc-mediated functions across a range of influenza strains (24, 25, 33). Future interventional studies complementing anti-influenza ADCC in subjects with severe influenza are warranted.
A second key finding was the relatively rapid peak generation of Fc functional Abs in both cohorts of severe influenza infection in different geographical regions compared with NAbs. The average time gap between the peak generation of Fc functional Abs and NAbs was in the order of 13–18 days — this is potentially highly clinically relevant in the setting of a rapid progressive infection. For the seasonal influenza-infected cohort, we speculate that this reflects the presence of cross-reactive memory B cells prior to infection (26, 27, 29, 30), which are more rapidly recalled to produce broadly binding Fc functional Abs compared with the generation of de novo strain-specific NAbs. For the H7N9-infected cohort, Fc functional Ab responses trended toward slower development than observed in the seasonal influenza-infected cohort but were still more rapid than the H7-specific NAb response. Differences in the kinetics of the Ab response, between seasonal and H7N9 influenza, may reflect lower levels of and/or more weakly cross-reactive preprimed memory B cells against H7 prior to infection. We acknowledge that our analyses are limited both by the time taken for the subjects to present to hospital and by the modest number of samples able to be collected from these very ill cohorts that are relatively difficult to recruit, thus necessitating a modeling approach to determine the peak responses. Nonetheless, our work represents the largest analysis of Fc functional Abs in clinically relevant influenza to date, and a large time gap between Fc functional Abs and NAbs was observed across two independent cohorts. Similar patterns of more rapid generation of Fc functional Abs than NAbs have been reported in other infectious diseases, such as HIV (46).
A third key finding was the remarkable breadth of Fc functional Abs generated by survivors of severe influenza infection. Nonneutralizing Abs recognizing conserved HA structures (such as the HA stem) may be more cross-reactive than NAbs, which predominantly target the globular head of HA. Recent data suggest that broadly reactive anti-HA mAbs may require FcγR engagement for protection regardless of neutralization or HA domain specificity (25). Influenza vaccination and infection studies with pandemic and seasonal H1N1 viruses have been shown to recall memory B cell responses that cross-react with avian H5N1 viruses (47–49). While the breadth of anti-HA Abs has been thoroughly examined for group 1 viruses, it had not been previously studied during severe H7N9 and seasonal influenza in humans. During H7N9 and seasonal infection, dimeric FcγR-binding Abs cross-reacted with HAs from group 1 and 2 influenza viruses. H7N9-infected serum Abs appeared to preferentially bind HAs from H3N2 and H4N6 subtypes, which likely reflects increased homology and epitope conservation within group 2 influenza viruses. Conversely, seasonal influenza A infection induced Abs capable of binding HAs from avian viruses to which previous exposure is unlikely. In subjects who survived severe influenza infection, HA-specific FcγR dimer–binding Abs showed increased breadth, suggesting the expansion of both strain-specific and cross-reactive B cell subsets over time.
We studied both FcγRIIIa-binding Abs, which typically mediate ADCC, as well as FcγRIIa-binding Abs, which mediate ADP. ADP has recently been suggested as the primary mechanism of Fc-mediated protection by nonneutralizing Abs in mouse models of H7N9 infection (50). Although binding to the FcγRIIa dimer generally aligned with responses mediated through FcγRIIIa in our analyses, future studies should consider performing functional influenza-specific ADP assays. Recent work on HA-specific ADP in sera from nonsevere human and macaque influenza infections suggests that functional ADP responses may contribute to the limitation of influenza spread in vitro (32).
Collectively, we show that FcγR cross-linking Abs are present in humans hospitalized with severe influenza infection, for both pandemic H7N9 and seasonal influenza viruses. We found that FcγR cross-linking Abs demonstrate significant breadth against HA proteins from many different subtypes of influenza virus and that their time to peak Ab response is earlier than NAbs. Plasma from patients infected with severe seasonal influenza contained FcγRIIIa cross-linking Abs that were capable of inducing ADCC against target cells infected with a heterologous influenza virus in vitro. These data highlight a potential role for ADCC Abs in the control of severe human influenza infection.
Methods
Hospitalized influenza-infected cohorts.
Longitudinal humoral responses from 4 groups were compared in this study: 18 patients with PCR-confirmed H7N9 influenza hospitalized at SPHCC in 2013; 11 age-matched healthy control subjects from China; 16 patients with PCR-confirmed seasonal influenza hospitalized at the Alfred Hospital in 2014–15; and 10 patients hospitalized with acute respiratory illness at the Alfred Hospital in 2014–15 but who were influenza-negative by PCR.
The cohort of H7N9-infected subjects was previously described (either partially or fully) in publications by Wang et al. (6) and Zhang et al. (7), and their characteristics are summarized in Table 1. Sera were collected from 18 subjects infected with H7N9 influenza (median age of 71 years, range 47–88) and 11 healthy control subjects from China (also with a median age of 71 years). Of the 18 H7N9-infected subjects, 17 were referred to SPHCC from other hospitals a median of 5 days (range 2–9 days) after initial hospitalization.
Plasma or sera samples were also collected from 16 subjects (median age of 57 years, range 22–84 years) hospitalized with seasonal influenza as part of the DISI cohort (Table 1). Plasma/sera samples were generally taken within 48 hours of hospital admission, 3 to 5 days later, at the time of hospital discharge (or just prior to death), and approximately 30 days after hospitalization. Nasal swabs taken within 3 days of hospitalization were sequenced by standard Sanger sequencing methods. Based on the sequences of HA, neuraminidase, nucleoprotein, and matrix 1 proteins, the influenza strain of infection was determined. As controls for the seasonal cohort, we studied 10 subjects hospitalized during the same time frame with acute respiratory infection who were PCR negative for influenza.
Clinical data collection for hospitalized patients included the SOFA score, a clinical score that reflects the severity of sepsis-related illness (51).
HA proteins and influenza viruses.
All 18 subjects from the H7N9-infected cohort were infected with the same influenza virus subtype (avian H7N9), and HA protein based assays were performed with a recombinant HA from the A/Shanghai/1/2013 (H7N9) virus (Sinobiological). The A/Shanghai/4664T/2013 (H7N9) strain was used for the HI assay, and a pseudovirus-based neutralization assay was performed with a nonreplicative HIV backbone expressing influenza H7 and N9 proteins.
Of the 16 subjects hospitalized with seasonal influenza infection, 4 were infected with H1N1 influenza viruses, 8 were infected with H3N2 influenza viruses, and 4 were infected with influenza B viruses. The 4 subjects hospitalized with H1N1 influenza were all infected with A/California/07/2009 (H1N1) pandemic-like viruses, and HA protein-based assays, HI and FRA, were performed with A/California/07/2009 (H1N1) HA protein or virus. Of the 8 subjects hospitalized with H3N2 influenza, 7 were infected with strains classified as clade 3C.3 and 1 was infected with a 3C.2 virus (Supplemental Table 1). We performed HA protein-based assays, HI and FRA, against the 3C.3 strain A/Switzerland/9715293/2014 (H3N2) HA protein or virus (the 2014-15 seasonal vaccine strain). The HA proteins of the 8 infecting H3N2 viruses had greater than 97% amino acid identity with the HA protein of A/Switzerland/9715293/2013. Of the 4 subjects hospitalized with influenza B, 2 were infected with viruses from the Yamagata lineage, 1 was infected with a virus from the Victoria lineage, and 1 virus could not be sequenced (Supplemental Table 1). We performed HA protein-based assays, HI and FRA assays, against the Yamagata lineage B/Phuket/3073/2013 HA protein or virus (the 2014-15 seasonal vaccine strain).
The recombinant HA proteins from A/Shanghai/1/2013 (H7N9), A/Switzerland/9715293/2013 (H3N2) virus, and B/Phuket/3073/2013 virus were expressed in a baculovirus-insect expression system, whereas all other recombinant HA proteins (H1 A/California/04/2009, H2 A/Japan/305/1957, H3 A/Perth/16/2009, H3 A/X-31, H4 A/Swine/Ontario/01911-1/99 and H5 A/Vietnam/1194/2004) used in the FcγR dimer ELISAs and NK cell activation assays were expressed in human cells.
HI and FRA.
HI and luciferase-based pseudovirus neutralization (with an HIV backbone carrying H7 and N9) assays with H7N9-infected sera were performed as previously described (6, 7). HI assays with plasma/sera from subjects with seasonal influenza infections were carried out as described in Kristensen et al. (35). FRA were performed as described in ref. 52, with modification to the determination of virus neutralization titers. Inhibition of infectious virus ≥50% was considered positive for neutralization.
Dimeric rsFcγRIIIa and rsFcγRIIa binding ELISA.
A rsFcγR dimer ELISA was employed to model the need for ADCC-inducing Abs to cross-link FcγRs as previously described (31, 35). This assay demonstrated a stronger correlation with Ab-dependent NK cell activation than a standard IgG ELISA (31). In subjects from the Australian DISI cohort infected with H3N2 influenza, we confirmed that, while the H3-specific FcγRIIIa dimer ELISA correlated with both Ab-mediated NK cell activation and an H3-specific IgG ELISA, there was no significant correlation between primary NK cell activation and an IgG ELISA (data not shown). Briefly, 96-well ELISA plates were coated with 50 ng purified influenza HA protein (Sinobiological) overnight, washed, blocked, and washed again, and then 1:40 plasma dilutions (based on previous work, refs. 31, 32, 35, and limited titration) were added and incubated at 37°C for 1 hour. After a further 5 washes, 50 μl of either 0.1 μg/ml rsFcγRIIIa biotinylated dimer (V176 high-affinity variant) or 0.2 μg/ml rsFcγRIIa biotinylated dimer (H131 high-affinity variant) was added for 1 hour at 37°C. HRP-streptavidin (1:10,000, ThermoFisher Scientific) was added to all wells for 1 hour; the plates were washed 8 times and blotted dry, and then 50 μl of TMB substrate was added and the plate was developed in the dark. The reaction was stopped with 1 M HCl and absorbance read at 450 nm. Intragam 5 (5 μg/ml, bioCSL) was used as a positive control and allowed for normalization between plates as previously described (31, 35).
Ab-mediated NK cell activation assay.
NK cell activation was measured by the ability of Abs bound to a plate coated with influenza proteins to induce NK cell expression of IFN-γ and CD107a as previously described (26). Briefly, 96-well ELISA plates (Nunc) were coated with 600 ng purified influenza HA protein overnight and then incubated with a 1:10 dilution of heat-inactivated sera or plasma for 2 hours at 37°C. After washing, 106 peripheral blood mononuclear cells (PBMCs) were added to each well along with anti-human CD107a allophycocyanin-H7 Ab (clone H4A3; BD Biosciences), 5 μg/ml brefeldin A (Sigma-Aldrich), and 5 μg/ml monensin (Golgi Stop; BD Biosciences) for 5 hours at 37°C with 5% CO2. PBMCs were then incubated with 1 mM EDTA to minimize cell adherence to the plates, anti-human CD3 Pacific Blue or PerCP (clone SP34-2), and anti-human CD56 allophycocyanin (clone B159; both from BD Biosciences) for 30 minutes at room temperature in the dark. Cells were fixed with 1% formaldehyde (Sigma-Aldrich) for 10 minutes and permeabilized with FACS permeabilizing solution 2 (BD Biosciences) for 10 minutes. PBMCs were then incubated at room temperature for 1 hour with IFN-γ AF700 (clone B27; BD Biosciences) in the dark. Cells were again fixed with 1% formaldehyde and acquired on a LSR Fortessa flow cytometer (BD Biosciences). Gating was performed as previously described (32) using FlowJo X 10.0.7r2 software (FlowJo LLC). The proportion of gated CD3–CD56+dim NK cells activated by sera Abs to express IFN-γ and/or CD107a was analyzed (Supplemental Figure 2).
Modeling the kinetics of influenza-specific Ab responses.
To analyze differences between influenza-specific Ab kinetics, we used a log linear regression as previously described (34). Ab responses increased exponentially until a certain time (Tpeak), after which they decrease exponentially. In line with general Ab responses to other acute viral infections, we assume influenza-specific Ab levels increase in the first few weeks with a rate r until Tpeak and will then decline with a rate d (i.e., constraining the model to have negative or 0 slope after Tpeak). This simplified model can be written as follows:
(Equation 1)
where A0 is the initial antibody level, A(t) represents the concentration of influenza-specific Ab response at time t, r represents the expansion rate of the influenza-specific Ab response, d represents the decay of the influenza-specific Ab response after Tpeak, and Tpeak represents the time of the peak influenza-specific Ab response.
A nonlinear mixed effect regression was used to fit the OD and titer data. To enable comparisons between OD readings of the rsFcγR dimer assays and the titers of HI and FRA (NAb) assays, the variability in the titer level was removed by dividing the value by the mean, so the scale between OD and titer was homogenous. We used a binary covariate (representing seasonal vs. H7N9 infection, or rsFcγRIIIa/rsFcγRIIa dimer–binding Ab vs. NAb or HI response) to determine whether the timing of the peak (see Equation 1) is different. Significance was determined based on the value of this covariate (whether it was significantly different from 0), calculated using the Wald test from the standard errors calculated in package nlme (v 3.1-112) in R (v 3.2.2).
ADCC killing assay with influenza-infected A549 cells.
To study ADCC-mediated killing, we adapted a LDH release assay as previously described (53, 54). Briefly, A549 respiratory cell line (ATCC CRL-185) targets were infected with X-31 influenza virus (a reassortant influenza virus expressing the HA and neuraminidase from A/Aichi/2/1968) at an MOI of 10 for 5 hours at 37°C. Experimental wells contained 2 × 104 NK92 cells expressing the high-affinity (V176) variant of FcγRIIIa (provided by K. Campbell, Institute for Cancer Research, Philadelphia, Pennsylvania, USA) combined with 2 × 104 X-31–infected A549 cells in an effector-to-target ratio of 1:1 and a 1:100 dilution of heat-inactivated plasma from influenza-infected donors in triplicate. LDH release was measured with the Cytotox 96 kit (Promega). Percentage cytotoxicity was calculated as follows: [(experimental – effector spontaneous – target spontaneous)/(maximum LDH – target spontaneous)]. Any above background killing in no-Ab control wells was subtracted from the percentage cytotoxicity to give the percentage ADCC. Killing in the no-Ab control wells was minimal and was <3% for all LDH assays performed.
Statistics.
Statistical analysis was performed with GraphPad Prism version 5.0 (GraphPad Software). Data presented in Figures 1, B and D; Figure 2B and Figure 3 were analyzed by Mann Whitney U test. Wilcoxon matched-pairs signed-rank tests were performed to analyze data presented in Figure 1A (paired admission and release/death samples), Figure 1C (paired admission and release/death samples), Figure 2A (paired admission and release/death samples), and Figure 4, A and C. Data presented in Figure 1A (unpaired H7N9-infected samples and healthy controls), Figure 1C (unpaired H7N9-infected samples and healthy controls), Figure 2A (unpaired H7N9-infected samples and healthy controls), and Figure 4, B and D were analyzed by a Kruskal-Wallis test. Spearman correlations were performed in Figure 2, C and D; Figure 6, C and D; Supplemental Figure 8, A–D and Supplemental Figure 9, A–D. A P value of 0.05 was considered statistically significant unless otherwise specified in the figure legend.
Study approval.
Written informed consent was obtained from all subjects prior to inclusion in these studies. The study of the H7N9 cohort was reviewed and approved by the SPHCC Ethics Committee. The study of the DISI cohort was reviewed and approved by the Alfred Health Ethics Committee.
Author contributions
HAV performed the Fc functional Ab assays, with assistance and advice from FASB, LL, MSP, and SJK. LL, YW, XZ, and JX recruited and organized samples from and performed neutralization assays on the H7N9 cohort. TK, ACC, THON, and KK recruited and organized samples from the seasonal influenza-infected cohort. ACH and DT performed neutralization assays and genotyping of the seasonal influenza cohort. AR and MPD performed the mathematical modeling. BW and PMH developed the FcγR dimer ELISA, provided reagents, and provided expert guidance on the use of this assay. HAV and SJK conceived the study and wrote the manuscript. All authors contributed to the preparation and discussion of the manuscript.
Supplementary Material
Acknowledgments
We thank Janine Roney, Leah Christie, and Jill Garlick for patient recruitment (Alfred Hospital). We thank Malet Aban and Louise Carolan for conducting HI assays and Yi-Mo Deng for sequencing analysis (WHO Collaborating Centre for Reference and Research on Influenza). We thank Brad Gilbertson and Lorena Brown for generously providing influenza virus. We thank Zhongfang Wang for graciously facilitating the collaboration between the University of Melbourne and SPHCC/Fudan University. This work was supported by a joint University of Melbourne/Fudan University International Research and Research Training Fund and an NHMRC project grants (1023294 to SJK, 1041832 to SJK, and 1071916 to KK). The Australian Department of Health provides financial support to conduct influenza surveillance (including clinical data collection) at the Alfred Hospital as part of the Australian FluCAN study. SJK and KK are NHMRC Research Fellows and ACC is a NHMRC Career Development Fellow. The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(13):e92750. https://doi.org/10.1172/jci.insight.92750.
Contributor Information
Lu Liu, Email: liulu11n@163.com.
Fernanda Ana-Sosa-Batiz, Email: fernandaa@student.unimelb.edu.au.
Yanmin Wan, Email: wanyanmin_2004@163.com.
Bruce Wines, Email: bwines@burnet.edu.au.
Danielle Tilmanis, Email: Danni.Tilmanis@influenzacentre.org.
Arnold Reynaldi, Email: areynaldi@kirby.unsw.edu.au.
Aeron C. Hurt, Email: aeron.hurt@influenzacentre.org.
Tom Kotsimbos, Email: tom.kotsimbos@monash.edu.
Katherine Kedzierska, Email: kkedz@unimelb.edu.au.
Xiaoyan Zhang, Email: zhangxiaoyan@shaphc.org.
Jianqing Xu, Email: xujianqing@shphc.org.cn.
References
- 1. Influenza (Seasonal): Fact Sheet. World Health Organization. http://www.who.int/mediacentre/factsheets/fs211/en/ Accessed May 23, 2017.
- 2. Influenza at the human-animal interface. World Health Organization. http://www.who.int/influenza/human_animal_interface/Influenza_Summary_IRA_HA_interface_10_03_2016.pdf Accessed May 23, 2017.
- 3.Iuliano AD, et al. Increase in human infections with avian influenza A(H7N9) virus during the fifth epidemic - China, October 2016-February 2017. MMWR Morb Mortal Wkly Rep. 2017;66(9):254–255. doi: 10.15585/mmwr.mm6609e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan GS, et al. Broadly-reactive neutralizing and non-neutralizing antibodies directed against the h7 influenza virus hemagglutinin reveal divergent mechanisms of protection. PLoS Pathog. 2016;12(4):e1005578. doi: 10.1371/journal.ppat.1005578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkinson TM, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;18(2):274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, et al. Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8+ T cells. Nat Commun. 2015;6:6833. doi: 10.1038/ncomms7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang A, et al. Kinetics of serological responses in influenza A(H7N9)-infected patients correlate with clinical outcome in China, 2013. Euro Surveill. 2013;18(50):20657. doi: 10.2807/1560-7917.ES2013.18.50.20657. [DOI] [PubMed] [Google Scholar]
- 8.Wang W, et al. Serum samples from middle-aged adults vaccinated annually with seasonal influenza vaccines cross-neutralize some potential pandemic influenza viruses. J Infect Dis. 2016;213(3):403–406. doi: 10.1093/infdis/jiv407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diao H, et al. Severe H7N9 infection is associated with decreased antigen-presenting capacity of CD14+ cells. PLoS One. 2014;9(3):e92823. doi: 10.1371/journal.pone.0092823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji H, et al. Epidemiological and clinical characteristics and risk factors for death of patients with avian influenza A H7N9 virus infection from Jiangsu Province, Eastern China. PLoS ONE. 2014;9(3):e89581. doi: 10.1371/journal.pone.0089581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu W, Shi Y, Gao H, Liang W, Sheng J, Li L. Immune derangement occurs in patients with H7N9 avian influenza. Crit Care. 2014;18(2):R43. doi: 10.1186/cc13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, Crowe JE. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol. 2011;85(20):10905–10908. doi: 10.1128/JVI.00700-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang TT, et al. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 2010;6(2):e1000796. doi: 10.1371/journal.ppat.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandbulte MR, et al. Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses. Proc Natl Acad Sci USA. 2011;108(51):20748–20753. doi: 10.1073/pnas.1113801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shil P, Chavan S, Cherian S. Molecular basis of antigenic drift in Influenza A/H3N2 strains (1968-2007) in the light of antigenantibody interactions. Bioinformation. 2011;6(7):266–270. doi: 10.6026/97320630006266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Brien KB, Morrison TE, Dundore DY, Heise MT, Schultz-Cherry S. A protective role for complement C3 protein during pandemic 2009 H1N1 and H5N1 influenza A virus infection. PLoS ONE. 2011;6(3):e17377. doi: 10.1371/journal.pone.0017377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohta R, Torii Y, Imai M, Kimura H, Okada N, Ito Y. Serum concentrations of complement anaphylatoxins and proinflammatory mediators in patients with 2009 H1N1 influenza. Microbiol Immunol. 2011;55(3):191–198. doi: 10.1111/j.1348-0421.2011.00309.x. [DOI] [PubMed] [Google Scholar]
- 18.Greenberg SB, Criswell BS, Six HR, Couch RB. Lymphocyte cytotoxicity to influenza virus-infected cells: response to vaccination and virus infection. Infect Immun. 1978;20(3):640–645. doi: 10.1128/iai.20.3.640-645.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashimoto G, Wright PF, Karzon DT. Antibody-dependent cell-mediated cytotoxicity against influenza virus-infected cells. J Infect Dis. 1983;148(5):785–794. doi: 10.1093/infdis/148.5.785. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto G, Wright PF, Karzon DT. Ability of human cord blood lymphocytes to mediate antibody-dependent cellular cytotoxicity against influenza virus-infected cells. Infect Immun. 1983;42(1):214–218. doi: 10.1128/iai.42.1.214-218.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vella S, Rocchi G, Resta S, Marcelli M, De Felici A. Antibody reactive in antibody-dependent cell-mediated cytotoxicity following influenza virus vaccination. J Med Virol. 1980;6(3):203–211. doi: 10.1002/jmv.1890060303. [DOI] [PubMed] [Google Scholar]
- 22.Huber VC, Lynch JM, Bucher DJ, Le J, Metzger DW. Fc receptor-mediated phagocytosis makes a significant contribution to clearance of influenza virus infections. J Immunol. 2001;166(12):7381–7388. doi: 10.4049/jimmunol.166.12.7381. [DOI] [PubMed] [Google Scholar]
- 23.Bruhns P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood. 2012;119(24):5640–5649. doi: 10.1182/blood-2012-01-380121. [DOI] [PubMed] [Google Scholar]
- 24.DiLillo DJ, Palese P, Wilson PC, Ravetch JV. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J Clin Invest. 2016;126(2):605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiLillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat Med. 2014;20(2):143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jegaskanda S, et al. Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies. J Immunol. 2013;190(4):1837–1848. doi: 10.4049/jimmunol.1201574. [DOI] [PubMed] [Google Scholar]
- 27.Jegaskanda S, et al. Age-associated cross-reactive antibody-dependent cellular cytotoxicity toward 2009 pandemic influenza A virus subtype H1N1. J Infect Dis. 2013;208(7):1051–1061. doi: 10.1093/infdis/jit294. [DOI] [PubMed] [Google Scholar]
- 28.Jegaskanda S, et al. Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity in intravenous immunoglobulin as a potential therapeutic against emerging influenza viruses. J Infect Dis. 2014;210(11):1811–1822. doi: 10.1093/infdis/jiu334. [DOI] [PubMed] [Google Scholar]
- 29.Jegaskanda S, Weinfurter JT, Friedrich TC, Kent SJ. Antibody-dependent cellular cytotoxicity is associated with control of pandemic H1N1 influenza virus infection of macaques. J Virol. 2013;87(10):5512–5522. doi: 10.1128/JVI.03030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jegaskanda S, et al. Generation and protective ability of influenza virus-specific antibody-dependent cellular cytotoxicity in humans elicited by vaccination, natural infection, and experimental challenge. J Infect Dis. 2016;214(6):945–952. doi: 10.1093/infdis/jiw262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wines BD, Vanderven HA, Esparon SE, Kristensen AB, Kent SJ, Hogarth PM. Dimeric FcγR ectodomains as probes of the fc receptor function of anti-influenza virus IgG. J Immunol. 2016;197(4):1507–1516. doi: 10.4049/jimmunol.1502551. [DOI] [PubMed] [Google Scholar]
- 32.Vanderven HA, et al. What lies beneath: antibody dependent natural killer cell activation by antibodies to internal influenza virus proteins. EBioMedicine. 2016;8:277–290. doi: 10.1016/j.ebiom.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333(6044):850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 34.Reynaldi A, et al. Modeling of EBV infection and antibody responses in kenyan infants with different levels of malaria exposure shows maternal antibody decay is a major determinant of early EBV infection. J Infect Dis. 2016;214(9):1390–1398. doi: 10.1093/infdis/jiw396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kristensen AB, et al. Antibody responses with Fc-mediated functions after vaccination of HIV-infected subjects with trivalent influenza vaccine. J Virol. 2016;90(12):5724–5734. doi: 10.1128/JVI.00285-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jegaskanda S, Co MDT, Cruz J, Subbarao K, Ennis FA, Terajima M. Induction of H7N9-cross-reactive antibody-dependent cellular cytotoxicity antibodies by human seasonal influenza A viruses that are directed toward the nucleoprotein. J Infect Dis. 2017;215(5):818–823. doi: 10.1093/infdis/jiw629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ackerman ME, et al. Polyfunctional HIV-specific antibody responses are associated with spontaneous HIV control. PLoS Pathog. 2016;12(1):e1005315. doi: 10.1371/journal.ppat.1005315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonsignori M, et al. Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J Virol. 2012;86(21):11521–11532. doi: 10.1128/JVI.01023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haynes BF, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366(14):1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lambotte O, et al. Heterogeneous neutralizing antibody and antibody-dependent cell cytotoxicity responses in HIV-1 elite controllers. AIDS. 2009;23(8):897–906. doi: 10.1097/QAD.0b013e328329f97d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wren LH, et al. Specific antibody-dependent cellular cytotoxicity responses associated with slow progression of HIV infection. Immunology. 2013;138(2):116–123. doi: 10.1111/imm.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laoprasopwattana K, et al. Antibody-dependent cellular cytotoxicity mediated by plasma obtained before secondary dengue virus infections: potential involvement in early control of viral replication. J Infect Dis. 2007;195(8):1108–1116. doi: 10.1086/512860. [DOI] [PubMed] [Google Scholar]
- 43.Hung IF, et al. Convalescent plasma treatment reduced mortality in patients with severe pandemic influenza A (H1N1) 2009 virus infection. Clin Infect Dis. 2011;52(4):447–456. doi: 10.1093/cid/ciq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hung IF, et al. Hyperimmune IV immunoglobulin treatment: a multicenter double-blind randomized controlled trial for patients with severe 2009 influenza A(H1N1) infection. Chest. 2013;144(2):464–473. doi: 10.1378/chest.12-2907. [DOI] [PubMed] [Google Scholar]
- 45.Stevens NE, Hatjopolous A, Fraser CK, Alsharifi M, Diener KR, Hayball JD. Preserved antiviral adaptive immunity following polyclonal antibody immunotherapy for severe murine influenza infection. Sci Rep. 2016;6:29154. doi: 10.1038/srep29154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dugast AS, et al. Independent evolution of Fc- and Fab-mediated HIV-1-specific antiviral antibody activity following acute infection. Eur J Immunol. 2014;44(10):2925–2937. doi: 10.1002/eji.201344305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahmed MS, et al. Cross-reactive immunity against influenza viruses in children and adults following 2009 pandemic H1N1 infection. Antiviral Res. 2015;114:106–112. doi: 10.1016/j.antiviral.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 48.Mahallawi WH, et al. Infection with 2009 H1N1 influenza virus primes for immunological memory in human nose-associated lymphoid tissue, offering cross-reactive immunity to H1N1 and avian H5N1 viruses. J Virol. 2013;87(10):5331–5339. doi: 10.1128/JVI.03547-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roos A, et al. Protection against H5N1 by multiple immunizations with seasonal influenza vaccine in mice is correlated with H5 cross-reactive antibodies. Vaccine. 2015;33(14):1739–1747. doi: 10.1016/j.vaccine.2015.01.070. [DOI] [PubMed] [Google Scholar]
- 50.Henry Dunand CJ, et al. Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe. 2016;19(6):800–813. doi: 10.1016/j.chom.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seymour CW, et al. Assessment of clinical criteria for sepsis: for the third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315(8):762–774. doi: 10.1001/jama.2016.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Baalen CA, et al. ViroSpot microneutralization assay for antigenic characterization of human influenza viruses. Vaccine. 2017;35(1):46–52. doi: 10.1016/j.vaccine.2016.11.060. [DOI] [PubMed] [Google Scholar]
- 53.Cox JH. HIV-1-Specific Antibody-dependent cellular cytotoxicity (ADCC) Methods Mol Med. 1999;17:373–381. doi: 10.1385/0-89603-369-4:373. [DOI] [PubMed] [Google Scholar]
- 54.Gooneratne SL, Richard J, Lee WS, Finzi A, Kent SJ, Parsons MS. Slaying the Trojan horse: natural killer cells exhibit robust anti-HIV-1 antibody-dependent activation and cytolysis against allogeneic T cells. J Virol. 2015;89(1):97–109. doi: 10.1128/JVI.02461-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.