Abstract
Background
The Rh system is the most complex and polymorphic blood group system in humans with more than 460 alleles known for the RHD gene. The DAU cluster of RHD alleles is characterized by the single nucleotide change producing the p.Thr379Met amino acid substitution. It is called the DAU-0 allele and has been postulated to be the primordial allele, from which all other alleles of the DAU cluster have eventually evolved.
Study design and methods
For 2 novel DAU alleles, the nucleotide sequences of all 10 exons as well as adjacent intronic regions, including the 5’ and 3’ untranslated regions (UTR), were determined for the RHD and RHCE genes. A phylogenetic tree for all DAU alleles was established using the neighbor-joining method with Pan troglodytes as root. Standard hemagglutination and flow cytometry tests were performed.
Results
We characterized 2 DAU alleles, DAU-11 and DAU-5.1, closely related to DAU-3 and DAU-5 respectively. A phylogenetic analysis of the 18 known DAU alleles indicated point mutations and interallelic recombination contributing to diversification of the DAU cluster.
Conclusions
The DAU alleles encode a group of RhD protein variants, some forming partial D antigens known to permit anti-D in carriers; all are expected to cause anti-D alloimmunization in recipients of red cell transfusions. The DAU alleles evolved through genomic point mutations and recombination. These results suggest that the cluster of DAU alleles represent a clade, which is concordant with our previous postulate that they derived from the primordial DAU-0 allele.
Background
The D antigen, encoded by the RHD gene, is the second most immunogenic and clinically significant blood group antigen, next only to A and B antigens.1 Genetic rearrangements in the RHD gene, such as gene conversions and point mutations, led to a great variety of RHD alleles, more than 460 of which have been identified to date. They encode variants with either a normal or an altered RhD protein expression on the red blood cell (RBC) surface.2–6
In phylogenetic analysis, a group of sequences defined by a common DNA mutation are known as a “cluster”. We have applied the term “cluster” to describe, so far, 4 groups of phylogenetically related RHD alleles.7–10 A cluster of alleles may represent more specifically a clade, if they originated from a common ancestral allele, known as the primordial allele. 11,12 The DAU-0 has been the proposed primordial allele of the DAU cluster.7 It is characterized by the single nucleotide polymorphism (SNP) c.1136C>T (p.Thr379Met) in exon 8 of the RHD gene.7 Only 9 DAU alleles have formally been published between 20027 and 2009,10,13–15 with documented anti-D in carriers of DAU-37 and DAU-4,16 although several more DAU allele candidates have since accrued in online repositories.2,17,18
The DAU-3 partial D allele is defined by 2 non-synonymous mutations (p.Val279Met and p.Thr379Met) in the RHD exons 6 and 8 and comes in a haplotype with an RHce allele lacking further characterization.7,13,15 The DAU-3 allele has previously been associated with an anti-D immunization in a carrier.7 In 2012, the nucleotide sequence of a DAU-3 allele with the additional non-synonymous mutation p.Ala85Val has been deposited (GenBank accession number HE965768.1) in trans to the RHD*Ψ allele, but no serologic data were reported.
The DAU-5 partial D allele is defined by 3 non-synonymous mutations (p.Phe223Val, p.Glu233Gln and p.Thr379Met) in the RHD exons 5 and 8 and is associated with an RHce allele lacking further characterization.4,10,14,15 DAU-5 is reported to be a recombinant allele between the DAU-0 and DV type 1 alleles4,14 and has not been associated with anti-D alloimmunizations in carriers. In 2014, the nucleotide sequence of a DAU-5 allele with the additional synonymous mutation p.Ile374Ile has been deposited (GenBank accession number HG918112.1), but no serologic data were reported.
In the present study, we describe these 2 new DAU alleles that are closely related to DAU-3 and DAU-5. We also investigated the phylogenetic relationship among the 18 known DAU alleles and the distribution of their amino acid substitutions in the RhD protein. DAU-0 was confirmed to be the primordial allele for all of them.
Materials and Methods
Study subjects
EDTA-anticoagulated blood samples were obtained from the patients with written informed consent. The DNA was extracted using a BioRobot EZ1 workstation with EZ1 DNA blood kit (Qiagen, Valencia, CA).
Immunohematology
Hemagglutination tests were performed by standard tube and anti-IgG gel matrix testing with licensed reagents (Ortho, Raritan, NJ). Several monoclonal anti-D from RhD typing kits were used to establish the epitope patterns (D Screen; Diagast, Loos, France; and Advanced Partial RhD typing kit; Alba Bioscience, Edinburgh, UK). Additional monoclonal anti-D were BS226 and BS232 (both IgM, Seraclone anti-D (RH1); Bio-Rad, Dreieich, Germany), LDM1, RUM-1 and TH28 (all IgM, DiaClon ABO-Confirmation for Patients; Bio-Rad), P3x61 (IgM, Seraclone Anti-CDE (RH2, 1, 3); Bio-Rad) and TH28 (IgM) and MS26 (IgG) (microtiter plate, Galileo Neo; Immucor, Norcross, GA, USA). Antibody screening and identification were done with gel matrix (rabbit anti-IgG; Micro Typing Systems, Pompano Beach, FL, USA).
Flow cytometry
The D antigen density was estimated by flow cytometry (FACSCalibur; Becton Dickinson, Heidelberg, Germany) with 4 monoclonal anti-D as described previously19 (Birma D6 and BRAD 3; International Blood Group Reference Laboratory, Bristol, UK; and BS221 and H41; Bio-Rad, Dreieich, Germany). Cryopreserved RBCs with a D+C+E-c+e+ phenotype expressing 14,000 D antigens per RBC was used as reference, which had previously been calibrated by a published workshop standard.20
RHD molecular screening
Initial RHD genotyping was done with kits (BAGene Weak D-TYPE and Partial D-TYPE; BAG Health Care, Lich, Germany).
RHD and RHCE sequencing
The RHD and RHCE genes were sequenced at NIH21,22 or at Linz23 as previously described. The nucleotide sequences of all 10 exons as well as the adjacent intronic regions including the 5’ and 3’ untranslated regions (UTR) were determined for both genes. Zygosity testing for the RHD gene was done at NIH by restriction fragment length polymorphism (RFLP)24 and quantitative fluorescence polymerase chain reaction (QF-PCR)25, while in Linz a hybrid Rhesus Box assay was applied (RBC-Ready Gene ZygoFast; Inno-train Diagnostic, Kronberg, Germany).
RH Sequence analysis
Nucleotide sequences were aligned and compared with the RHD (NG_007494.1) and RHCE reference sequences (NG_009208.3). All variations are described according to current mutation nomenclature guidelines,26 ascribing the A of the first ATG translational initiation codon as nucleotide +1 in the mRNA coding region of RHD (NM_016124.4) and RHCE (NM_020485.4). Multiple sequence comparisons were carried out (MUSCLE, v3.8 with default settings).27
Database mining for DAU alleles
The human RhesusBase2 and NCBI GenBank28 genetic sequence databases were searched for RHD alleles fitting the definition of the DAU cluster.7 One allele (GenBank accession number EU557240) harboring a codon insertion (GTG) immediately following the start codon (ATG) in addition to p.Thr379Met was excluded from the study, as no 5’UTR or corroborating information could be obtained since its release in 2008.
Reference red cell genotyping
At the BloodCenter of Wisconsin (BCW), genomic DNA was extracted from patient samples and evaluated for SNPs, insertions and deletions associated with non-RhD antigens, including C, E, c and e, and the 7 Rh variant antigens, such as partial C, partial c, partial e, V, VS, hrB and hrS,29 or for partial D (BAGene Partial D-TYPE). To resolve 2 different variant RHD alleles in samples, called RHD compound heterozygotes, the coding sequence of the RHD gene was sequenced in full length30 with RHD-specific intron amplification primers. Results of all samples, sent between February 1, 2013 and February 15, 2016 by 20 outside institutions, were collated for reference red cell genotyping. Comparable data sets were established at the institutions in Linz and Springe.
Phylogenetic analysis
A possible phylogenetic tree for DAU alleles was developed, based on the RHD coding sequence and the presence of its associated RHCE allele. Each single nucleotide substitution was counted as one event. Clustering of the described DAU alleles was done manually. Sequences from chimpanzees (Pan troglodytes Rh-like protein IIR, GenBank accession number L37050.1)31 were used for external rooting, as previously described for RHD.7,9,10,32
Computational modeling of RhD protein and amino acid substitutions
The 3D structure for the RhD protein was modeled from the crystal structure of RhCG protein (Protein Data Bank accession code 3HD6)33 using SWISS-MODEL.34 Stereochemical quality and accuracy of the predicted RhD model was analyzed using Ramachandran plot analysis,35 ProSA28,29 and ProQ.36 The distances between the C-alpha atoms of the amino acids in the DAU alleles and a line traversing the central pore of the RhD protein were estimated.
Polymorphism Phenotyping algorithm (PolyPhen-2)37, Sorting Intolerant From Tolerant (SIFT)38, Protein Variation Effect Analyzer (PROVEAN)39 and Screening for Non-Acceptable Polymorphisms (SNAP2)40 were used to predict the functional impact of amino acid substitutions on RhD protein structure.
Nomenclature
New DAU alleles described in this study were named following the nomenclature in the human RhesusBase.2 DAU-0 to DAU-7 had been named previously.3,10,13–15 A new allele that differed by a non-synonymous substitution from any previously described DAU allele was denoted by a new number, such as DAU-8; whereas a new allele that differed by a synonymous substitution from a previously described allele, for example DAU-5, was designated as a subtype and denoted with a decimal, such as DAU-5.1. The numbers were in chronological order of deposition in any public database.
Results
We defined 2 novel DAU alleles in 3 patients (Table 1). A 32 year old African female patient in Linz carried the DAU-5.1 allele. A 50 year old African American male with bladder cancer at NIH and another 32 year old African pregnant patient in Salzburg carried the DAU-11 allele. The DAU-5.1 allele is probably occurring in one haplotype with the recently published, rare RHCE*ce48C, 105T allele41,42 (in cis on 1 chromosome) with the RHCE*ce reference allele in trans. The DAU-11 allele (NIH sample) could have either the RHCE*ce254G (RHCE*ceAG) or RHCE*733G allele in one haplotype (Table 1).
Table 1.
Molecular basis of DAU alleles described in this study
|
RHD Allele |
Nucleotide Substitution in RHD gene |
Effect on Protein sequence |
Exon involved |
Observed samples
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
RHD genotype
|
RHCE genotype (observed RHCE alleles) |
GenBank accession number |
||||||||
| Patients (n) |
Ethnicity | Phenotype | Zygosity |
RHD allele in trans |
||||||
| DAU-5.1 | 667T>G | F223V | 5 | 1 | African | D+C-E-c+e+ | Hemizygous | None | RHCE*ce48C, 105T RHCE*ce | HG918112.1 |
| 697G>C | E233Q | 5 | ||||||||
| 1122C>T | I374I | 8 | ||||||||
| 1136C>T | T379M | 8 | ||||||||
| DAU-11 | 254C>T | A85V | 2 | 1 | African American | D+C-E-c+e+ | Hemizygous | None | RHCE*ce254G RHCE*733G | KU248927.1 |
| 835G>A | V279M | 6 | ||||||||
| 1136C>T | T379M | 8 | ||||||||
| 1 | African | D+C-E-c+e+ | Compound heterozygous | RHD*Ψ | Unknown (DNA supply exhausted) | HE965768.1 | ||||
Immunohematology
All 3 patients were found in routine D antigen typing by discordant results with 2 different anti-D reagents (Table S1). Antibody screening and direct antiglobulin results were negative for all 3 samples. The D antigen density was approximately 6200 per RBC for DAU-5.1 and 2400 for DAU-11 (Table S2). The DAU-11 (NIH sample) reacted in variable strength with all 25 monoclonal anti-D tested (Table S3).
DAU alleles
We collated the 18 known DAU alleles characterized by harboring the c.1136C>T single nucleotide polymorphism encoding p.Thr379Met (Fig. 1 and Table S4). They differed by 1 or more additional missense (non-synonymous) or silent (synonymous) substitutions dispersed throughout the length of the RHD coding sequence (CDS).2 Almost all alleles were originally described in individuals with an African ethnic background,2 presented as D+C-E-c+e+ phenotypes (Table S5), and hence all occurred in a Dce haplotype. There were 18 nucleotide substitutions encoding 14 non-synonymous and 4 synonymous substitutions in the RHD CDS (Fig. 1). Most SNPs in the CDS had been listed in the dbSNP database, but 4 SNPs were novel (Table S4). Information about the variations in the non-coding regions (5’-UTR, introns and 3’-UTR) was lacking for many DAU alleles, while their association with distinct RHCE alleles has been shown for some of them (Table S6).
Figure 1. Known DAU alleles.
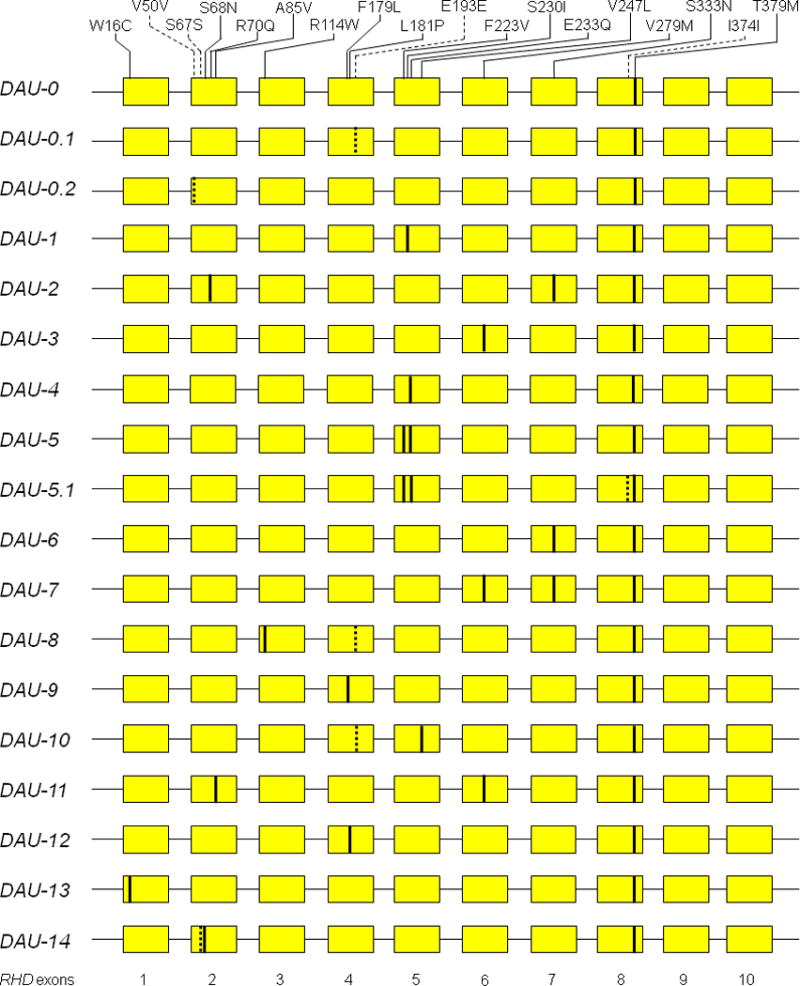
The mutations in RHD gene exons are shown for the 18 known DAU alleles. The RHD allele comprise 10 exons each (yellow boxes). Non-synonymous (solid lines) and synonymous nucleotide substitutions (dotted lines) depict differences to the RHD reference sequences (NM_016124.4).
Clinical patient samples
Within 4 years, Milwaukee has received requests for reference testing by red cell genotyping in 2257 patient samples (Table 2).41,43,44 Among the 379 samples tested for partial D, 155 (41%) were confirmed as partial D. Among those, 75 (48%) patients carried at least one DAU allele, of which 8 were shown to carry a DAU-4 or DAU-5 allele hemizygously. Springe analyzed 3147 patient referrals within 8 years and Linz 1271 within 4 years. The distribution of RHD alleles detected differed between the US and European centers. For 52 DAU samples from Milwaukee, the associated RHCE alleles were identified (Table S7), all being concordant with published associations (Table S6).
Table 2.
DAU alleles among samples tested by red cell genotyping at regional reference laboratories
| Patient samples (n) at reference laboratories
|
|||
|---|---|---|---|
| Red cell genotyping procedure and result | German Red Cross Springe |
BloodCenter of Wisconsin |
Austrian Red Cross Linz |
| Any procedure | |||
| Total, including weak D screening | 3147 * | 2257 | 1271 |
| Screening test for partial D | |||
| Partial D and normal D confirmed | 79 | 379 † | 532 |
| RHD*01 (normal) only | n.d. | 224 | 306 |
| Any partial D | 78 | 155 | 226 |
| Partial D test result | |||
| Partial D allele other than DAU | 56 | 80 | 219 |
| Any DAU allele | 22 | 75 | 7 |
| DAU test result | |||
| DAU allele not specified | 1 | 34 | 2 |
| DAU-0, 1, 2 or 3 allele hemizygous | n.a. | 28 | n.a. |
| DAU-0 allele hemizygous | 3 | n.a. | n.a. |
| DAU-2 allele hemizygous | 4 | n.a. | n.a. |
| DAU-4 or 5 allele hemizygous | n.a. | 8 | 3 |
| DAU-4 allele hemizygous | 2 | n.a. | n.a. |
| DAU-6 allele hemizygous | 1 | n.a. | 2 |
| DAU compound heterozygous | 11 ‡ | 5 ¶ | 0 |
| Time frame | 8 years | 4 years | 4 years |
includes 954 blood donor samples
Partial D analysis using kit (n = 370) or nucleotide sequencing (n = 9)
5 DAU-0/RHD*01; 1 DAU-0/RHD*Ψ; 1 DAU-0/DIIIa-CE(4–7)-D;40,42,43 1 DAU-1/RHD*Ψ; 1 DAU-3/RHD*01; 1 DAU-3/weak D type 4.2; and 1 DAU-3/ DIIIa-CE(4–7)-D40,42,43
2 DAU-0/DAU-5; 1 DAU/weak D type 4.2; 1 DAU-0/weak D type 41; and 1 DAU-5/RHD*01
n.d. – not determined, n.a. – not applicable
Phylogenetic analysis of DAU alleles
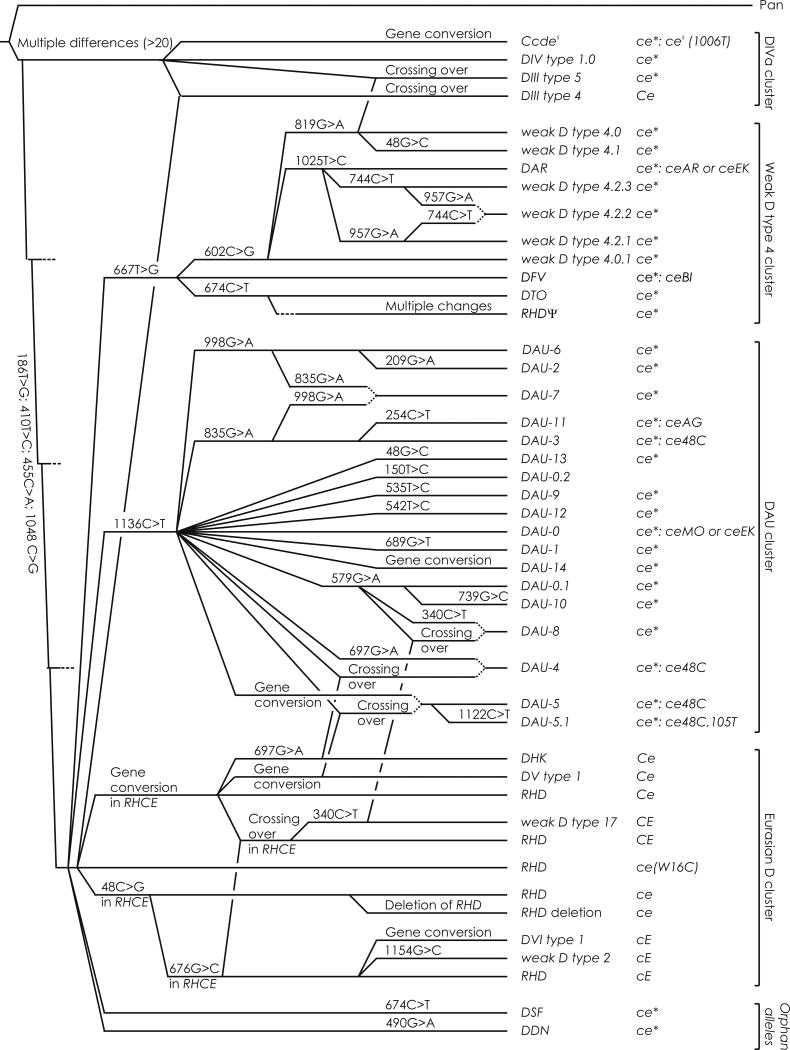
We parsed the new DAU alleles in our previously published phylogenic trees.7,10,32 The 8 DAU alleles DAU-0.1,-0.2, -1, -3, -6, -9, -12 and -13 could have originated by single synonymous or non-synonymous substitutions in the DAU-0 allele (Fig. 2). The 4 other DAU alleles DAU-2, -5.1, -10 and -11 could have originated through mutation in the previously established DAU alleles.7,10,13,14 The DAU-14 allele was likely a result of interlocus gene conversion between the DAU-0 allele and the exon 2 of an RHCE allele. The 4 remaining DAU alleles DAU-4, -5, -7 and -8 could have originated by single recombination events between 2 RHD alleles (Fig. 2).
Figure 2. Phylogeny of alleles in the DAU cluster.
A phylogenetic tree of the DAU cluster is shown for the 18 known alleles. For each evolutionary step, the event is indicated; the depicted distances of the alleles are arbitrary, as previously described for RHD7 The extended molecular phylogenetic analysis of RHD alleles delineated 4 clusters: the Eurasian D cluster with the consensus RHD (NM_016124.4) and 3 African clusters designated DIVa, DAU, and weak D type 4. 7–10 “Gene conversion” denotes a gene conversion in RHD using RHCE as template, if not mentioned otherwise. In this genealogy, DIII type 5 is assumed to be derived from the “basal” DIVa cluster rather than DIII type 4, because DIII type 4 is a rare allele caused by a recombination of an allele of the DIVa cluster with Eurasian RHD6 In this analysis, the RHCE allele polymorphisms were not considered, and the actual phylogeny may be even more complex. However, the typically associated RHCE allele is indicated for each RHD allele, if known (see Tables S5 and S6). ce* indicates a ce-like allele which may frequently be a variant.
Predicted effect of non-synonymous substitutions
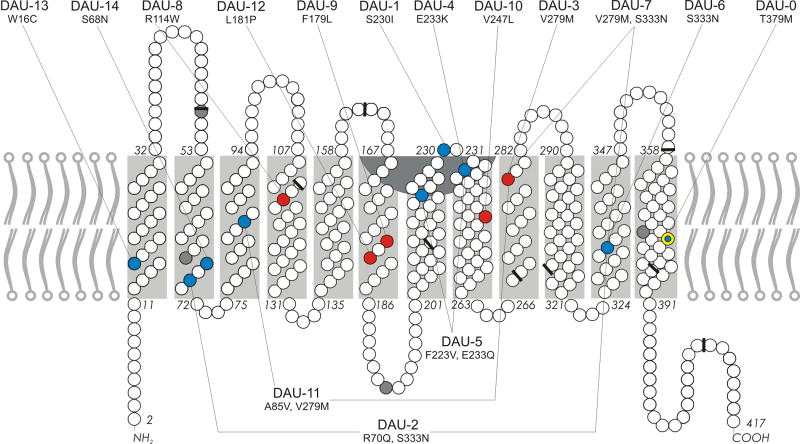
The PolyPhen-2, SIFT, PROVEAN, SNAP2 and INPS bioinformatic programs predicted deleterious structural changes induced by the non-synonymous p.Arg114Trp, p.Phe179Leu, p.Leu181Pro, p.Val247Leu and p.Val279Met substitutions (Table S8). The 14 non-synonymous substitutions were distributed along the whole length of the RHD CDS without any apparent clustering (Fig. 3).
Figure 3. Model of RhD protein in the red cell membrane.
The RhD protein consist of 417 amino acids (circles). The first amino acid is lacking from the mature protein in the membrane. The extracellular Rh vestibule (inverted black arc) is in part bordered by amino acids of loops 3 and 4.3,4 There are 9 exon boundaries in the RHD cDNA as reflected in the amino acid sequence (black bars).5 All known amino acid substitutions encoding DAU alleles are labeled (colored circles). The 4 synonymous SNPs cause no amino acid change (grey). The other SNPs are non-synonymous and cause amino acid changes that are predicted to affect the RhD protein structure (red) or to be neutral (blue). The p.Thr379Met amino acid change (yellow ring), defining the DAU cluster, is predicted to having no effect on the RhD protein structure (neutral).
Comparative homology modeling of the RhD protein
The template-based homology model of RhD protein was consistent with the model proposed on the basis of computational hydropathy plots45 (Fig. 4). The model comprised 408 amino acids from Lys4 to Pro411 and lacked 9 residues (3 in the N terminus and 6 in the C terminus). An analysis of the stereochemistry using RAMPAGE software showed all the main chain atoms falling within the generously allowed region of the Ramachandran plot35 with 394 residues in the most favored region (96.5%), 10 residues in the additionally allowed region (2.5%), and 4 residues in the outlier region (1%). The ProSA-web z-score of the model was −6.2 (Fig. S1), a value within the range of other experimentally determined protein structures of the same size.28,29 ProQ results predicted LGscore and MaxSub as 6.66 and 0.63, respectively, indicating a very good model.36
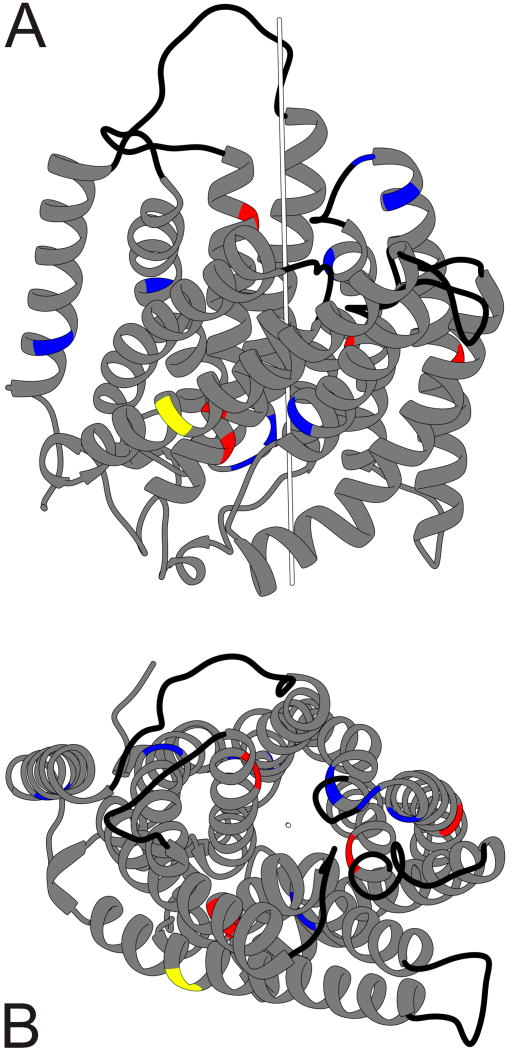
Figure 4. Molecular structure of RhD protein.
The side view of a homology model of the RhD protein is depicted as it is situated in the RBC membrane; the top faces the RBC surface and bottom the RBC inside (A). The view to the RhD protein from the inside of an RBC is depicted as it is embedded in the RBC membrane (B). Panel A is rotated 90° relative to panel B. Most amino acid substitutions (red and blue) occur in the transmembraneous helices (grey ribbons) rather than the extracellular loops (black lines). The position of the central pore is indicated by the white rod (
 ) and the open circle (○). The 14 non-synonymous amino acid substitutions are predicted to either affect (red) or not affect the RhD protein structure (blue). The p.Thr379Met amino acid change is predicted to having no effect on the RhD protein structure (yellow).
) and the open circle (○). The 14 non-synonymous amino acid substitutions are predicted to either affect (red) or not affect the RhD protein structure (blue). The p.Thr379Met amino acid change is predicted to having no effect on the RhD protein structure (yellow).
The central pore of the modeled RhD protein coincided with the crystal structure of the RhCG protein model (Fig. 4).33 We estimated the distance between the C-alpha atom of each amino acid and the central pore of the RhD protein (Fig. 4): there was no statistically significant difference between the 14 amino acids in DAU variants (12.4 Å ± 5.8 Å; mean ± SD) and the remaining 394 amino acids (14.8 Å ± 5.8 Å; p>0.05, Mann-Whitney U-test, 2 sided).
SNPs at CpG sites
Nucleotide substitutions are known to occur frequently at CpG sites, which are defined by a cytosine followed by a guanine in the linear nucleotide sequence along its 5’ → 3’ direction. We found 38 CpG sites in the 1254 nucleotides of the RHD CDS. Among the 18 mutated positions in DAU alleles, including the primordial DAU-0 allele (Table S4), 4 sites represented C>T transition in CpG sites (Fig. S2). The mutations at the CpG sites in the DAU alleles were overrepresented (4 of 38 compared to 18 of 1254; p<0.01, Fisher’s exact test, 2 sided). The CpG site mutation at position 201 was excluded from this calculation because it likely resulted from a gene conversion event with RHCE that also comprised the position 203.
Discussion
The 14 non-synonymous DAU mutations were found to be dispersed over the entire RhD protein with no evidence of clustering at specific sites. All non-synonymous DAU mutations occurred inside the red cell membrane (Fig. 3) with the only exception of the previously described DAU-1 (p.Ser230Ile).7 The recurrent p.Thr379Met mutation in exon 8 (Fig. 1) may represent a possibly neutral amino acid substitution that became originally fixed in an isolated African population.
The serology of DAU phenotypes (Table S1 and S2) exemplified the potential relevance of testing the D antigen in the clinical routine with 2 different anti-D monoclonals,46 not mandatory in the US, but widely applied in Europe for 2 decades. Current serologic routine procedures detected DAU variants (Table 2 and Table S1). The choice of the right monoclonal anti-D reagents will obviously determine that the clinically relevant D variants47 are preferentially recognized and forwarded to red cell genotyping (Table 2). The distribution of RHD alleles detected differed much between the US and European centers, which can be explained by differences in the populations, the routine serologic screening procedures and the approaches to the molecular work-up, which are yet to be standardized. A more detailed immunohematologic workup is still possible for many DAU variants (Table S5), which are primarily needed to determine the clinical relevance of distinct alleles. These in vivo data may also elucidate the molecular mechanisms integrating proteins into cell membranes.
Various RHD alleles are associated with frequent anti-D alloimmunization, especially in chronically transfused patients such as patients with hemoglobinopathies.14,48 Knowledge of RH alleles, their phylogeny and prevalence will aid in identifying the clinically relevant RHD alleles occurring in patient samples by high throughput technologies, such as next generation sequencing (NGS).49 Haplotypes can refer to the specific combination of alleles at different locations on a single chromosome.50 At a given RH gene locus, the 1 RHD and 1 RHCE allele represent 1 haplotype. Distinct RHD alleles have been documented to accompany distinct RHCE alleles, each combination thus constituting a unique haplotype. We sequenced the RHCE gene in many different DAU samples (Table S6 and S7)51–55 and identified the most probable RHCE allele associated with a given DAU allele as a haplotype (Fig. 2).
The DAU-5.1 allele harbored the p.Ile374Ile substitution in combination with the 3 previously described DAU-5 mutations (p.Phe223Val, p.Glu233Gln and p.Thr379Met), while the DAU-11 allele harbored the p.Ala85Val substitution in combination with the 2 previously described DAU-3 mutations (p.Val279Met and p.Thr379Met). The 2 new DAU-5.1 and DAU-11 phenotypes were both found to express a lower D antigen density than their parent DAU phenotypes, with 6236 and 2483 D antigens per RBC respectively. The D antigen densities for these DAU-0, DAU-3 and DAU-5 phenotypes have been reported to be 15,285,7 10,8797 and 10,131 D antigens per RBC (Table S2 and Table S5).
The p.Ala85Val amino acid substitution observed in DAU-11 is predicted to reside in the transmembrane region of the RhD protein.5 Alanine, hydrophobic like valine but smaller, is a much better helix-forming residue.56 Because position 85 resides in the middle of the 3rd helix (Fig. 3), the disruptive effect by Valine on the helix structure was predicted to be stronger and this perturbation of the helix may hamper lodging of the RhD protein in the RBC membrane.57 Because it is in direct contact with the lipid bilayer, the substitution may also affect the tertiary interactions and stabilization of the RhD protein (Fig. 4).58 A different nucleotide substitution (c.254C>G; GenBank accession number HE613970.1) at the same codon position causing an p.Ala85Gly substitution expressed even less D antigens with 618 D antigens per RBC.58 Glycine, the smallest amino acid but hydrophilic, may more strongly disrupt the RhD folding, lipid membrane integration or interaction with other proteins of Rhesus complex.58
The potential impact of non-synonymous nucleotide substitutions on protein expression has recently been well illustrated in vitro for the Dombrock blood group system.59 The p.Ile374Ile synonymous nucleotide substitution observed in DAU-5.1 is an excellent in vivo example that synonymous substitutions are also neither random nor neutral. The much reduced D antigen density of DAU-5.1 as compared to DAU-5.0 can be explained on the basis of well-documented molecular effects, such as changing mRNA splicing,60 mRNA folding,61 codon usage bias,62 and RNA-RNA interactions, all influencing gene function.63 According to the codon usage database,64 ATC coding for Isoleucine (I - Ile) in the normal RhD protein is used in humans 1.3-fold more frequently than the ATT coding for Ile in DAU-5.1. In the reference RHD CDS, the ATC codon is utilized 15 times and the alternate ATT codon used 7 times, a 2.1-fold difference. Translation efficiency and protein folding can be disturbed by this codon bias mechanism.
We used the RhCG protein (Protein Data Bank accession code 3HD6) as template for our homology modeling of the RhD protein (Fig. 4). RhCG is the protein with known crystal structure, most homologous to RhD. However, due to low sequence similarity between RhD and RhCG proteins (33.1%), refinement in the accuracy of RhD protein modeling will be possible.65 The 14 non-synonymous mutated positions were not found to be clustered around the central pore of the RhD protein (Fig. 4) and thus may not directly affect the yet unknown function of the central pore. The 5 bioinformatic programs predicted deleterious effects for 5 out of the 14 amino acid substitutions (Table S8); their damaging effect may involve destabilizing the RhD protein, its integration in the RBC membrane or its interaction with other proteins in the Rh complex.
It has previously been proposed that ancestral African populations were structured.66 Hence mutations arising in isolated populations were prevented from recombining with one another, and differentiated haplotypes emerged with very little recombination between lineages.67 Later, local selective pressures might have favored the spread of different alleles and haplotypes in the populations of distinct geographic areas, such as Eurasia and Africa. The primordial allele of DAU cluster, DAU-0, may have originated in such an isolated population, probably as a premeiotic mutation, where it became fixed.68 Premeiotic mutations pass through meiosis and recombination; during these events, the ancestral DAU-0 allele was joined to different RHCE alleles while accumulating additional nucleotide substitutions, forming a variety of new, often more than 1, RH haplotypes (Fig. 2).
Our analysis prominently indicates the role of interallelic recombination in the evolution of DAU alleles, a conclusion based on the observation of 5 shared substitutions between at least 10 different DAU alleles. The present study supports the previous postulate7 that the 18 known DAU alleles evolved through random mutation in the primordial DAU-0 allele or through recombination among DAU and other RHD alleles.
Supplementary Material
Acknowledgments
We thank Phillip Cruz for advice and help in modeling the RhD protein; Kurt Ralph Wollenberg for discussing terms related to phylogenetic computer algorithms and description of phylogenetic trees; Barbara Becker for kindly providing anti-D clones BS221 and H41; Harvey Gordon Klein for critical review of the manuscript; Rebecca Perry Coward for serological analysis; Sharon Dolores Adams for sample coordination; the staff of HLA laboratory for DNA extraction; and Elizabeth Jane Furlong for English edits.
This work was supported by the Intramural Research Program (project ID Z99 CL999999) of the NIH Clinical Center.
Footnotes
Conflict of interest disclosure: The authors declared having no competing financial interest relevant to this article.
Statement of Disclaimer: The views expressed do not necessarily represent the view of the National Institutes of Health, the Department of Health and Human Services, or the U.S. Federal Government.
Author contributions: KS, HP, SLS and GAD performed experimental parts of the study and HP and CGa analyzed the 2 Austrian samples; CGr tested the Salzburg sample serologically; FFW collated DAU alleles and compiled the phylogeny; GAD compiled BCW data; WAF designed the study and experiments; all authors discussed the data; KS and WAF analyzed the allele and protein data and wrote the manuscript.
Web Resource
dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/)
ISBT website (http://www.isbtweb.org/working-parties/red-cell-immunogenetics-and-blood-group-terminology/) accessed on January 26, 2016
Codon usage database (http://www.kazusa.or.jp/codon/)
Multiple sequence alignment (http://www.ebi.ac.uk/Tools/msa/muscle/)
ProQ - Protein quality prediction (http://www.sbc.su.se/~bjornw/ProQ/ProQ.cgi)
ProSA-web (https://prosa.services.came.sbg.ac.at/prosa.php)
RAMPAGE (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php)
SWISS-MODEL (http://swissmodel.expasy.org/)
The human RhesusBase version 2.0 (http://www.rhesusbase.info/) accessed on January 12, 2016
The human RhesusBase, DAU cluster page (http://www.rhesusbase.info/U_RHDDAUcluster.htm) accesses on January 01, 2016
References
- 1.Wagner FF, Flegel WA. Review: the molecular basis of the Rh blood group phenotypes. Immunohematology. 2004;20:23–36. [PMC free article] [PubMed] [Google Scholar]
- 2.Wagner FF, Flegel WA. The Rhesus Site. Transfus Med Hemother. 2014;41:357–63. doi: 10.1159/000366176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conroy MJ, Bullough PA, Merrick M, Avent ND. Modelling the human rhesus proteins: implications for structure and function. Br J Haematol. 2005;131:543–51. doi: 10.1111/j.1365-2141.2005.05786.x. [DOI] [PubMed] [Google Scholar]
- 4.Flegel WA, von Zabern I, Doescher A, Wagner FF, Vytiskova J, Pisacka M. DCS-1, DCS-2, and DFV share amino acid substitutions at the extracellular RhD protein vestibule. Transfusion. 2008;48:25–33. doi: 10.1111/j.1537-2995.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 5.Flegel WA. Molecular genetics and clinical applications for RH. Transfus Apher Sci. 2011;44:81–91. doi: 10.1016/j.transci.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner FF, Moulds JM, Flegel WA. Genetic mechanisms of Rhesus box variation. Transfusion. 2005;45:338–44. doi: 10.1111/j.1537-2995.2005.04339.x. [DOI] [PubMed] [Google Scholar]
- 7.Wagner FF, Ladewig B, Angert KS, Heymann GA, Eicher NI, Flegel WA. The DAU allele cluster of the RHD gene. Blood. 2002;100:306–11. doi: 10.1182/blood-2002-01-0320. [DOI] [PubMed] [Google Scholar]
- 8.Flegel WA, Wagner FF. Molecular genetics of RH. Vox Sang. 2000;78(Suppl 2):109–15. [PubMed] [Google Scholar]
- 9.Grootkerk-Tax MG, van Wintershoven JD, Ligthart PC, van Rhenen DJ, van der Schoot CE, Maaskant-van Wijk PA. RHD(T201R, F223V) cluster analysis in five different ethnic groups and serologic characterization of a new Ethiopian variant DARE, the DIII type 6, and the RHD(F223V) Transfusion. 2006;46:606–15. doi: 10.1111/j.1537-2995.2006.00759.x. [DOI] [PubMed] [Google Scholar]
- 10.Chen Q, Flegel WA. Random survey for RHD alleles among D+ European persons. Transfusion. 2005;45:1183–91. doi: 10.1111/j.1537-2995.2005.00181.x. [DOI] [PubMed] [Google Scholar]
- 11.Tan P-N, Steinbach M, Kumar V. Introduction to Data Mining. 1. Addison-Wesley Longman Publishing Co., Inc.; 2005. pp. 487–568. [Google Scholar]
- 12.Huson DH, Rupp R, Scornavacca C. Phylogenetic Networks: Concepts, Algorithms and Applications. Cambridge University Press; 2011. p. 127. [Google Scholar]
- 13.Wagner FF, Moulds JM, Tounkara A, Kouriba B, Flegel WA. RHD allele distribution in Africans of Mali. BMC Genet. 2003;4:14. doi: 10.1186/1471-2156-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denomme GA, Wagner FF, Fernandes BJ, Li W, Flegel WA. Partial D, weak D types, and novel RHD alleles among 33,864 multiethnic patients: implications for anti-D alloimmunization and prevention. Transfusion. 2005;45:1554–60. doi: 10.1111/j.1537-2995.2005.00586.x. [DOI] [PubMed] [Google Scholar]
- 15.Touinssi M, Chapel-Fernandes S, Granier T, Bokilo A, Bailly P, Chiaroni J. Molecular analysis of inactive and active RHD alleles in native Congolese cohorts. Transfusion. 2009;49:1353–60. doi: 10.1111/j.1537-2995.2009.02161.x. [DOI] [PubMed] [Google Scholar]
- 16.Ipe TS, Wilkes JJ, Hartung HD, Westhoff CM, Chou ST, Friedman DF. Severe hemolytic transfusion reaction due to anti-D in a D+ patient with sickle cell disease. J Pediatr Hematol Oncol. 2015;37:e135–7. doi: 10.1097/MPH.0000000000000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fichou Y, Le Marechal C, Bryckaert L, Guerry C, Benech C, Dupont I, Jamet D, Ferec C, Chen JM. Variant screening of the RHD gene in a large cohort of subjects with D phenotype ambiguity: report of 17 novel rare alleles. Transfusion. 2012;52:759–64. doi: 10.1111/j.1537-2995.2011.03350.x. [DOI] [PubMed] [Google Scholar]
- 18.Garcia F, Rodriguez MA, Goldman M, Azcarate MN, Rodriguez MI, Muniz-Diaz E, Puente F, Alshatti H, Haimila K, Molano A, Garaizar A, Ochoa-Garay G. New RHD variant alleles. Transfusion. 2015;55:427–9. doi: 10.1111/trf.12828. [DOI] [PubMed] [Google Scholar]
- 19.Polin H, Danzer M, Gaszner W, Broda D, St-Louis M, Proll J, Hofer K, Gabriel C. Identification of RHD alleles with the potential of anti-D immunization among seemingly D- blood donors in Upper Austria. Transfusion. 2009;49:676–81. doi: 10.1111/j.1537-2995.2008.02046.x. [DOI] [PubMed] [Google Scholar]
- 20.Flegel WA, Curin-Serbec V, Delamaire M, Donvito B, Ikeda H, Jorgensen J, Kumpel B, Le Pennec PY, Pisacka M, Tani Y, Uchikawa M, Wendel S, Wagner FF. Section 1B: Rh flow cytometry. Coordinator's report. Rhesus index and antigen density: an analysis of the reproducibility of flow cytometric determination. Transfus Clin Biol. 2002;9:33–42. doi: 10.1016/s1246-7820(01)00213-0. [DOI] [PubMed] [Google Scholar]
- 21.Wagner FF, Gassner C, Müller TH, Schönitzer D, Schunter F, Flegel WA. Molecular Basis of Weak D Phenotypes: Presented at the 25th Congress of the International Society of Blood Transfusion held in Oslo on June 29, 1998 and published in abstract form in Vox Sang 74:55, 1998 (suppl) Blood. 1999;93:385–93. [PubMed] [Google Scholar]
- 22.Fasano RM, Monaco A, Meier ER, Pary P, Lee-Stroka AH, Otridge J, Klein HG, Marincola FM, Kamani NR, Luban NL, Stroncek D, Flegel WA. RH genotyping in a sickle cell disease patient contributing to hematopoietic stem cell transplantation donor selection and management. Blood. 2010;116:2836–8. doi: 10.1182/blood-2010-04-279372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legler TJ, Maas JH, Kohler M, Wagner T, Daniels GL, Perco P, Panzer S. RHD sequencing: a new tool for decision making on transfusion therapy and provision of Rh prophylaxis. Transfus Med. 2001;11:383–8. doi: 10.1046/j.1365-3148.2001.00327.x. [DOI] [PubMed] [Google Scholar]
- 24.Wagner FF, Flegel WA. RHD gene deletion occurred in the Rhesus box. Blood. 2000;95:3662–8. [PubMed] [Google Scholar]
- 25.Pirelli KJ, Pietz BC, Johnson ST, Pinder HL, Bellissimo DB. Molecular determination of RHD zygosity: predicting risk of hemolytic disease of the fetus and newborn related to anti-D. Prenat Diagn. 2010;30:1207–12. doi: 10.1002/pd.2652. [DOI] [PubMed] [Google Scholar]
- 26.Wildeman M, van Ophuizen E, den Dunnen JT, Taschner PE. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum Mutat. 2008;29:6–13. doi: 10.1002/humu.20654. [DOI] [PubMed] [Google Scholar]
- 27.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013;41:D36–42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flegel WA, Gottschall JL, Denomme GA. Integration of red cell genotyping into the blood supply chain: a population-based study. Lancet Haematol. 2015;2:e282–e8. doi: 10.1016/S2352-3026(15)00090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yassai MB, Annen K, Bensing KM, Denomme GA. RHCE*cE94G encodes variable expression of c (RH4) Transfusion. 2015;55:2519–20. doi: 10.1111/trf.13164. [DOI] [PubMed] [Google Scholar]
- 31.Salvignol I, Blancher A, Calvas P, Clayton J, Socha WW, Colin Y, Ruffie J. Molecular genetics of chimpanzee Rh-related genes: their relationship with the R-C-E-F blood group system, the chimpanzee counterpart of the human rhesus system. Biochem Genet. 1994;32:201–21. doi: 10.1007/BF00554623. [DOI] [PubMed] [Google Scholar]
- 32.Flegel WA, von Zabern I, Doescher A, Wagner FF, Strathmann KP, Geisen C, Palfi M, Pisacka M, Poole J, Polin H, Gabriel C, Avent ND. D variants at the RhD vestibule in the weak D type 4 and Eurasian D clusters. Transfusion. 2009;49:1059–69. doi: 10.1111/j.1537-2995.2009.02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gruswitz F, Chaudhary S, Ho JD, Schlessinger A, Pezeshki B, Ho C-M, Sali A, Westhoff CM, Stroud RM. Function of human Rh based on structure of RhCG at 2.1 Å. Proc Natl Acad Sci U S A. 2010;107:9638–43. doi: 10.1073/pnas.1003587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 35.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–50. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 36.Cristobal S, Zemla A, Fischer D, Rychlewski L, Elofsson A. A study of quality measures for protein threading models. BMC Bioinformatics. 2001;2:5. doi: 10.1186/1471-2105-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–74. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hecht M, Bromberg Y, Rost B. Better prediction of functional effects for sequence variants. BMC Genomics. 2015;16(Suppl 8):S1. doi: 10.1186/1471-2164-16-S8-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reid ME, Halter Hipsky C, Hue-Roye K, Hoppe C. Genomic analyses of RH alleles to improve transfusion therapy in patients with sickle cell disease. Blood Cells Mol Dis. 2014;52:195–202. doi: 10.1016/j.bcmd.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fichou Y, Le Maréchal C, Scotet V, Jamet D, Férec C. Insights into RHCE Molecular Analysis in Samples with Partial D Variants: the Experience of Western France. Transfus Med Hemother. 2015;42:372–7. doi: 10.1159/000382086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moulds JM, Noumsi GT, Billingsley KL. A comparison of methods for the detection of the r’(s) haplotype. Transfusion. 2015;55:1418–22. doi: 10.1111/trf.12956. [DOI] [PubMed] [Google Scholar]
- 44.Westhoff CM, Vege S, Hipsky CH, Horn T, Hue-Roye K, Keller J, Velliquette R, Lomas-Francis C, Chou ST, Reid ME. RHCE*ceAG (254C>G, Ala85Gly) is prevalent in blacks, encodes a partial ce-phenotype, and is associated with discordant RHD zygosity. Transfusion. 2015;55:2624–32. doi: 10.1111/trf.13225. [DOI] [PubMed] [Google Scholar]
- 45.Chérif-Zahar B, Bloy C, Le Van Kim C, Blanchard D, Bailly P, Hermand P, Salmon C, Cartron JP, Colin Y. Molecular cloning and protein structure of a human blood group Rh polypeptide. Proc Natl Acad Sci U S A. 1990;87:6243–7. doi: 10.1073/pnas.87.16.6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner FF, Kasulke D, Kerowgan M, Flegel WA. Frequencies of the blood groups ABO, Rhesus, D category VI, Kell, and of clinically relevant high-frequency antigens in south-western Germany. Infusionsther Transfusionsmed. 1995;22:285–90. doi: 10.1159/000223144. [DOI] [PubMed] [Google Scholar]
- 47.von Zabern I, Wagner FF, Moulds JM, Moulds JJ, Flegel WA. D category IV: a group of clinically relevant and phylogenetically diverse partial D. Transfusion. 2013;53:2960–73. doi: 10.1111/trf.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rujirojindakul P, Flegel WA. Applying molecular immunohaematology to regularly transfused thalassaemic patients in Thailand. Blood Transfus. 2014;12:28–35. doi: 10.2450/2013.0058-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johansen Taber KA, Dickinson BD, Wilson M. The promise and challenges of next-generation genome sequencing for clinical care. JAMA Intern Med. 2014;174:275–80. doi: 10.1001/jamainternmed.2013.12048. [DOI] [PubMed] [Google Scholar]
- 50.Ong RT-H, Liu X, Poh W-T, Sim X, Chia K-S, Teo Y-Y. A method for identifying haplotypes carrying the causative allele in positive natural selection and genome-wide association studies. Bioinformatics. 2011;27:822–8. doi: 10.1093/bioinformatics/btr007. [DOI] [PubMed] [Google Scholar]
- 51.Westhoff C, Vege S, Horn T, Hue-Roye K, Hipsky CH, Lomas-Francis C, Reid ME. RHCE*ceMO is frequently in cis to RHD*DAU0 and encodes a hr(S)-, hr(B)-, RH:-61 phenotype in Blacks; Clinical Significance. Transfusion. 2013;53:2983–9. doi: 10.1111/trf.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moser I, Vrignaud C, Roussel M, Mendonca PF, Fraga C, Pecquet F, Rodrigues M, Peyrard T. RHCE*ce48C,662G: a Novel RHCE Allele with RHD*DAU0 in cis in a Proband from the Azores Islands. Transfusion. 2015;55:153A. [Google Scholar]
- 53.Westhoff CM, Vege S, Hipsky CH, Horn T, Hue-Roye K, Keller J, Velliquette R, Lomas-Francis C, Chou ST, Reid ME. RHCE*ceAG (254C>G, Ala85Gly) is prevalent in blacks, encodes a partial ce-phenotype, and is associated with discordant RHD zygosity. Transfusion. 2015;55:2624–32. doi: 10.1111/trf.13225. [DOI] [PubMed] [Google Scholar]
- 54.Chou ST, Jackson T, Vege S, Smith-Whitley K, Friedman DF, Westhoff CM. High prevalence of red blood cell alloimmunization in sickle cell disease despite transfusion from Rh-matched minority donors. Blood. 2013;122:1062–71. doi: 10.1182/blood-2013-03-490623. [DOI] [PubMed] [Google Scholar]
- 55.Keller JA, Horn T, Chiappa C, Melland C, Vietz C, Castilho L, Keller MA. RHCE variant allele: RHCE*ce254G,733G. Immunohematology. 2014;30:121–2. [PubMed] [Google Scholar]
- 56.Padmanabhan S, Marqusee S, Ridgeway T, Laue TM, Baldwin RL. Relative helix-forming tendencies of nonpolar amino acids. Nature. 1990;344:268–70. doi: 10.1038/344268a0. [DOI] [PubMed] [Google Scholar]
- 57.Chakrabartty A, Schellman JA, Baldwin RL. Large differences in the helix propensities of alanine and glycine. Nature. 1991;351:586–8. doi: 10.1038/351586a0. [DOI] [PubMed] [Google Scholar]
- 58.Silvy M, Chapel-Fernandes S, Callebaut I, Beley S, Durousseau C, Simon S, Lauroua P, Dubosc-Marchenay N, Babault C, Mouchet C, Ferrera V, Chiaroni J, Bailly P. Characterization of novel RHD alleles: relationship between phenotype, genotype, and trimeric architecture. Transfusion. 2012;52:2020–9. doi: 10.1111/j.1537-2995.2011.03544.x. [DOI] [PubMed] [Google Scholar]
- 59.de Coulgeans CD, Silvy M, Halverson G, Chiaroni J, Bailly P, Chapel-Fernandes S. Synonymous nucleotide polymorphisms influence Dombrock blood group protein expression in K562 cells. Br J Haematol. 2014;164:131–41. doi: 10.1111/bjh.12597. [DOI] [PubMed] [Google Scholar]
- 60.Mueller WF, Larsen LS, Garibaldi A, Hatfield GW, Hertel KJ. The silent sway of splicing by synonymous substitutions. J Biol Chem. 2015;290:27700–11. doi: 10.1074/jbc.M115.684035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Behura SK, Severson DW. Codon usage bias: causative factors, quantification methods and genome-wide patterns: with emphasis on insect genomes. Biol Rev Camb Philos Soc. 2013;88:49–61. doi: 10.1111/j.1469-185X.2012.00242.x. [DOI] [PubMed] [Google Scholar]
- 63.Matveeva OV, Shabalina SA. Intermolecular mRNA-rRNA hybridization and the distribution of potential interaction regions in murine 18S rRNA. Nucleic Acids Res. 1993;21:1007–11. doi: 10.1093/nar/21.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 66.Garrigan D, Hammer MF. Reconstructing human origins in the genomic era. Nat Rev Genet. 2006;7:669–80. doi: 10.1038/nrg1941. [DOI] [PubMed] [Google Scholar]
- 67.Wall JD. Detecting Ancient Admixture in Humans Using Sequence Polymorphism Data. Genetics. 2000;154:1271–9. doi: 10.1093/genetics/154.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woodruff RC, Thompson JN. Have premeiotic clusters of mutation been overlooked in evolutionary theory? J evol Biol. 1992;5:457–64. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.