Abstract
Purpose of review
Once and obscure disease, recent studies have transformed our understanding of angioimmunoblastic T-cell lymphoma (AITL). In this review we summarize new major advances in the genetics and biology of AITL.
Recent findings
Genome wide sequencing studies have dissected the repertoire of the genetic alterations driving AITL uncovering a highly recurrent Gly17Val somatic mutation in the small GTPase RHOA and major role for mutations in epigenetic regulators, such as TET2, DNMT3A and IDH2, and signaling factors (e.g. FYN and CD28). These findings support a multistep model of follicular T helper cell transformation in AITL and pinpoint novel candidates for the development of targeted therapies in this disease.
Summary
AITL originates from follicular T helper cells and is characterized by the presence of RHOA G17V mutation together with genetic alterations in TET2, DNMT3A and IDH2. Research efforts now focus on the elucidation of the specific roles and interplay of these genetic alterations in the pathogenesis of AITL.
Keywords: Angioimmunoblastic T cell lymphoma, follicular T helper cell, RHOA, TET2, DNMT3A
Introduction
Angioimmunoblastic T-cell lymphoma (AITL) is a clinically aggressive lymphoma derived from the malignant transformation of follicular T-helper (TFH) cells. AITL affects mostly elderly adults and accounts for 20% of peripheral T-cell lymphomas (1). Despite intensive therapy, 5 year overall survival rates in AITL are only about 30% (2), with little progress in the last two decades (3). However, recent studies have shed new light on the pathophysiology and genetics of this disease opening the field for the development of novel animal models and targeted therapies. Here we review the most recent advances on normal normal TFH cell development and the genetics of AITL.
Gene expression profiling insights in the classification and biology of AITL
Peripheral T-cell lymphomas are a highly heterogeneous and relatively poorly defined group of mature T-cell malignancies. Limitations in diagnosis and classification represent a clinical challenge that affects therapeutic options and clinical management. Systematic efforts to improve the classification of PTCLs using gene expression profiling has been instrumental in defining distinct molecular groups of PTCL and has provided new information on the pathobiology of these tumors (4–12).
Transcriptional characterization of PTCL tumor samples and their normal lymphocyte counterparts identified a unique transcriptional profile for AITL (7, 8). Importantly, this AITL-associated signature can be found in about 20% of tumors diagnosed as PTCL not otherwise specified (PTCL NOS) supporting that gene expression profiling provides a more accurate classification than histology for the diagnosis of PTCL (12–14). In addition, AITL gene expression programs reflect an important contribution of the tumor microenvironment and are enriched in B-cell and follicular dendritic cell (15). In this context, the AITL B-cell signature seems to be associated with a better outcome, while signatures related to immunosuppression could be linked to poor clinical outcome (13). Moreover, it is worth noting that AITL tumors show high levels of expression of VEGF, which may play a pathogenic role driving angiogenesis and stimulating lymphoma cell growth via autocrine or paracrine loops (16).
However, the most prominent finding of gene expression studies on the pathophysiology of AITL was the discovery of the cellular derivation of TFH cells, a subset of CD4+ T helper cells residing in the follicular centers, as the normal cellular counterpart of malignant AITL cells. This way, even though a relationship between AITL tumors cells and TFHs was first proposed based on the expression of the chemokine CXCL13 in AITLs samples (17, 18), this was ultimately and most clearly substantiated by gene expression studies, which established a close similarity between the expression signatures of TFH cells and AITL tumor biopsies (7, 8). In addition to CXCL13, TFH-characteristic genes highly expressed in AITL include the CD28-related inducible T-cell co-stimulator ICOS, CD154, CD40L and NFATC1 (7, 18).
TFH development and AITL
TFH cells were initially described as a separate CD4+ T-cell population characterized by high levels of expression of the chemokine receptor CXCR5 (19–21). Since then, TFH have emerged as a distinct subset of effector T helper cells with a characteristic gene expression signature and functionally separate from other known CD4 T-cell subsets (22–24). TFHs are required for germinal center formation and play important roles in germinal center B-cell differentiation and survival and in the development of long-lived plasma cells and memory T-cells (25).
TFH cell differentiation is initiated by the interaction of a naïve CD4+ T-lymphocyte with dendritic cells in a developing germinal center (26) (Figure 1). This interaction involves the activation of ICOS in the T-cell (27, 28), and the consequent activation of the PI3K pathway, which leads to expression of the BCL6 transcription factor, a critical regulator of TFH development (29–31). The master regulator role of BCL6 in TFH development is demonstrated by the failure of Bcl6−/− cells to differentiate into TFH cells in vivo (32, 33). Moreover, constitutive expression of Bcl6 enhances T cell differentiation towards the TFH lineage (32, 34) and transcriptional repression defective forms of BCL6 block TFH cells differentiation (35). Although the precise mechanisms operating downstream of Bcl6 are not fully clarified yet, this transcriptional repressor seems to participate in the restriction of alternative cell fates during TFH cell development via repression of critical factors implicated in Th1 (T-bet), Th2 (GATA3) and Th17 (RORγt) development (31, 32, 35, 36). Following ICOS activation and induction of BCL6 expression, activated T cells upregulate the expression of PD1 and CXCR5 becoming TFH precursors, which migrate to the border of the B-cell follicle to engage in secondary cell-cell interactions with antigen-specific B-cells (32, 37). Then, and as antigen stimulation builds up a germinal center reaction, these precursors complete maturation and acquire a definitive TFH phenotype characterized by expression of high levels of CXCR5, PD1, BCL6, MAF and SAP (37) (Figure 1). In addition to BCL6, TFH development depends on multiple other transcription factors including ASCL2, c-MAF, IRF4, and AP-1 (25, 32, 33, 35, 38–40). Moreover, in addition to ICOS engagement, activation of JAK-STAT signaling by IL6, IL21 and IL12 play important roles in TFH cell development (25, 41–47).
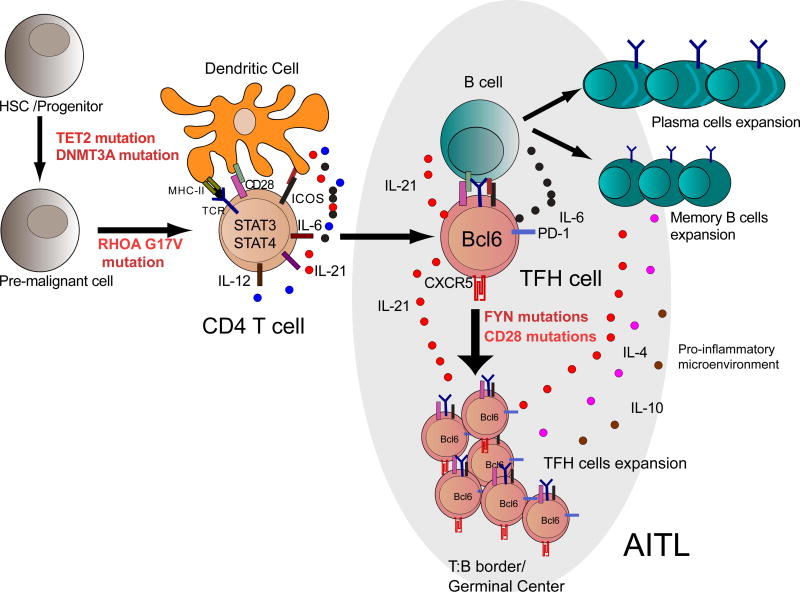
Figure 1. Normal development and malignant transformation of TFH cells.
TFH cell differentiation is initiated by activation of CD4 naïve T cells by dendritic cells in presence of IL6, IL21 and IL12 leading to STAT3/STAT4 activation. Activation of ICOS induces the upregulation of BCL6 and CXCR5, allowing them to migrate to B cell follicles to induce germinal centers formation. Stimulation of TFH cells and antigen presentation by B cells leads to full development of TFH cells, whose mission is supporting B-cells and facilitating the generation of long-lived plasma cells and memory B cells. Malignant transformation of TFH leads to the development of AITL following a multistep tumor model where TET2 and/or DNMT3A mutations would be acquired first, followed by specification into the TFH lineage guided by expression of the RHOA G17V mutant and enhanced by hyper activation of the TCR signaling pathway. Deregulated expansion and/or function of TFH could induce the generation of cytokines (IL4, IL6, IL21 and IL10) which play a prominent role in the early stages of lymphoma progression and in setting the abundant inflammatory component of AITL tumor lesions.
Genomic analysis of AITL
Genomic profiling studies have started to dissect the repertoire of genetic alterations driving the pathogenesis of AITL and PTCL, NOS tumors. These studies have already uncovered a major role for mutations in the small GTPase RHOA and in epigenetic factor genes –including TET2, DNMT3A and IDH2– in the pathogenesis of these tumors and pointed to additional relevant pathways in these diseases.
The RHOA G17V mutation in AITL
The RHOA small GTPase protein regulates multiple biological processes, including cytoskeleton remodeling, cell adhesion, migration, proliferation and survival (48, 49). As other small GTPases, RHOA cycles between an active GTP-bound state and an inactive GDP-bound configuration. RHOA activation is catalyzed by RHO GEFs (Guanine Nucleotide Exchange Factors), which facilitate the incorporation of GTP. Conversely, RHOA GAPs (GTPase Activating Proteins), which stimulate the conversion of GTP to GDP to promote the transition of the RHOA protein to the inactive state (48, 49). Early studies using transgenics expressing dominant negative forms of RHOA or suppressing RhoA via expression of the exoenzyme C3 transferase, an ADP ribosyl transferase that selectively ribosylates and inactivates RhoA, RhoB and RhoC proteins, pointed to an essential role for RHOA in multiple T-cell functions including polarization, migration and signaling through the TCR (50–54). Moreover, RHOA inactivation causes a developmental blockade during thymocyte progression (55–57) and inactivation of RhoA in the thymus has been linked to development of T-cell lymphomas (58). More recently, the analysis of conditional RhoA knockout mice has demonstrated a role of RhoA in thymocyte proliferation and survival, beta-selection, positive selection, early single positive lineage commitment, and notably, mitochondrial function (59). Moreover, altered Rho GTPase activity has been linked with the development of autoimmunity (60), one of the hallmarks of AITL.
A central role of RHOA in the pathogenesis of AITL is supported by the identification of recurrent, highly prevalent heterozygous missense mutations in the RHOA gene in about 70% of AITLs (61–66). Among these, the RHOA G17V allele accounts for over 90% of RHOA mutations in AITL (61–63) (Figure 2). Biochemical analysis and cellular assays demonstrated that the RHOA G17V mutant does not bind GTP and functions as an inactive and dominant negative protein which interferes with the activity of wild type RHOA (61–63), most probably by sequestering and interfering with the activity of RHOA GEFs (61). The RHOA G17V mutation is highly characteristic of AITL. Thus, presence of RHOA G17V mutation in about 25% of PTCL NOS cases suggests that these tumors probably represent misdiagnosed AITLs (61). However, RHOA mutations have also been identified in Burkitt lymphoma, gastric carcinoma, and adult T-cell leukemia lymphoma samples (67–70).
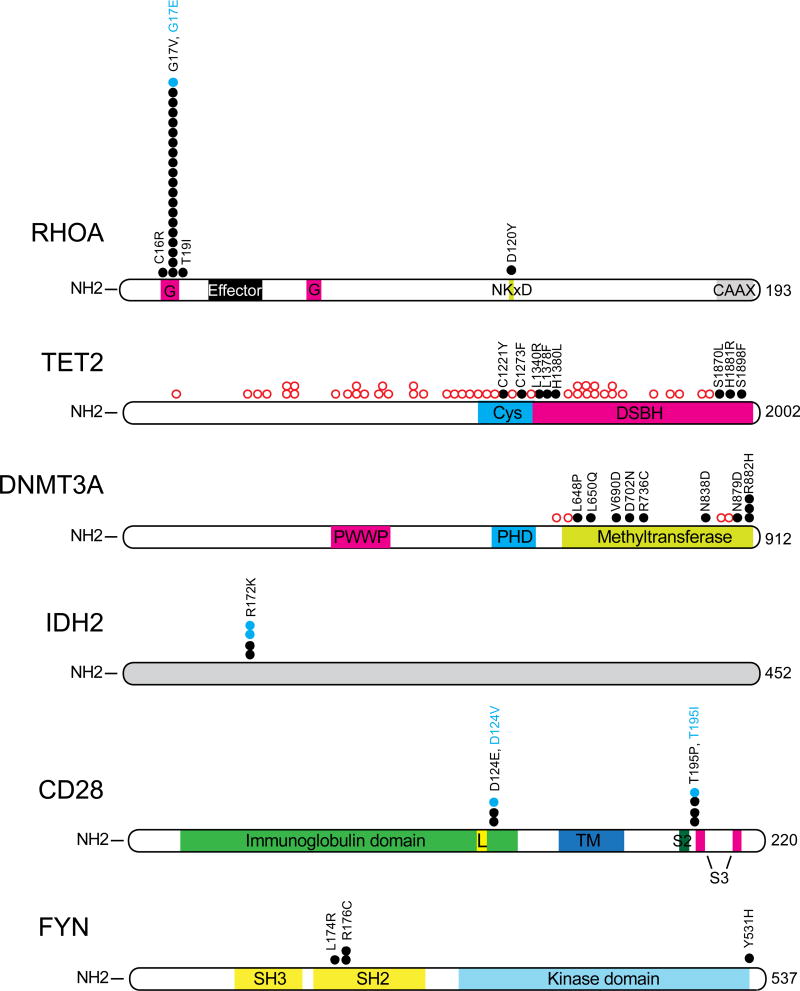
Figure 2. Recurrent mutations in AITL.
Schematic representation of the structure of the most frequently mutated proteins in AITL: RHOA, TET2, DNMT3A, IDH2, FYN and CD28. (Data adapted from Palomero et al, 2014 (for RHOA, TET2, DNMT3A, IDH2 and FYN) or Wang et al, 2015 (for CD28)).Black circles represent amino acid substitutions while open red circles indicate truncating mutations. G:GTP/GDP binding domain; Effector: effector interaction domain; NKXD: NKXD GTP-binding domain; CAAX: CAAX box prenylation domain; Cys: cysteine-rich domain; DSBH: double-stranded beta helix fold domain; PWWP: PWWP domain; PHD: plant homeodomain; Immunoglobulin domain: Ig variable region-like domain CD28 and CTLA4; TM, transmembrane domain. L: ligand interaction site; S2: SH2-binding motif; pink: S3: SH3-binding motifs; SH3: SH3 domain; SH2: SH2 domain.
Mutations in epigenetic regulators: TET2, DNMT3A and IDH2
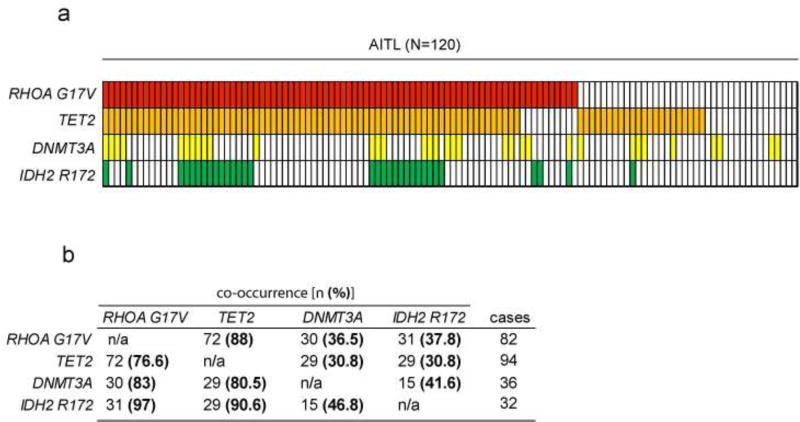
Mutations in TET2, DNMT3A and IDH2 are common in hematological malignancies. Originally described in myeloid malignancies, they were subsequently found in PTCL, being especially high in AITL and in a subgroup of PTCL, NOS with TFH features (71–73). Mutations in these epigenetic regulators are strongly associated with the expression of the RHOA G17V allele. In fact, analysis of 120 AITL samples from published data identifies mutations in at least one of these genes in 94% of RHOA G17V positive cases (Figure 3).
Figure 3. Co-occurrence of frequent mutations in AITL.
(a) Analysis of the mutational status of RHOA G17V, TET2, DNMT3A and IDH2 in a cohort of 120 AITL (information extracted from published data (61–64)). Each column represents a patient sample; each row represent mutations in each of the genes of interest. (b) Quantification of the co-occurrence of mutations in RHOA G17V and epigenetic regulators. In the column on the right are represented the cases mutated for RHOA G17V, TET2, DNMT3A and IDH2 R172; on the upper row the co-occurrence indicated by the number of cases on the left category that also carry the mutation in the genes indicated on the top (n) and the percentage of co-occurrence calculated from the total number of cases [(%) in bold].
The Ten-Eleven Translocation 2 (TET2) encodes a 2 oxoglutarate/Fe2+–dependent oxygenase that participates in the epigenetic control of gene expression through the oxidation of methylated cytosines and DNA demethylation by catalyzing the oxidation of DNA 5-methylcytosine to 5-hydroxymethylcytosine (74–76). TET2 was originally identified as a tumor suppressor in myeloid malignancies (77, 78); however, several later studies have also demonstrated a high frequency of loss of function mutations in TET2 in 70–80% of PTCLs (61, 62, 72, 79, 80), being particularly prevalent in AITL and PTCL, NOS (Figure 2). In PTCL, TET2 mutations are associated with advanced stage disease, adverse clinical parameters at presentation and shorter progression free survival (80).
The role of loss of function mutations in Tet2 has been investigated using mouse models, which showed that loss of Tet2 leads to higher frequency of hematopoietic stem cells, increased competitive repopulation abilities and biased differentiation towards the myeloid lineage (71, 81–87). Knockout mouse models of Tet2 are also associated with the development of hematopoietic malignancies, of both myeloid (71, 81, 82) and T-cell origin (88).
The DNA (cytosine-5) methyltransferase 3A (DNMT3A) gene encodes a methyltransferase involved in the epigenetic regulation of gene expression via methylation of cytosines in the DNA. Mutations in DNMT3A gene were originally described as highly recurrent in acute myeloid leukemia (AML) (89, 90), where they are associated with adverse survival (91). Analysis of DNMT3A in PTCL identified the presence of recurrent loss of function mutations in this gene in 10%–40% of AITL samples (61–64, 72) (Figure 2), frequently co-occurring with TET2 mutations and maybe even predating them in the process of malignant transformation (72) (Figure 3).
The Isocitrate Dehydrogenase 2 (IDH2) gene is mutated in about 30–40% of AITL cases (64, 73) (Figure 2). IDH2 encodes a metabolic mitochondrial enzyme physiologically involved in the oxidative decarboxylation of isocitrate to 2-oxoglutarate. Notably, the resulting mutant enzymes have a neomorphic enzymatic activity catalyzing the conversion of alpha ketoglutarate to 2 hydroxyglutarate (2-HG), an oncometabolite that antagonizes the activity of alpha ketoglutarate dependent dioxygenases, including the TET family enzymes, leading to impairment of DNA and histone demethylation and abnormal regulation of gene transcription (64, 92). While recurrent mutations in IDH2 can be frequently identified in other types of cancer, including AML, AITL is the only PTCL subgroup where IDH2 mutations are found, and remarkably, they occur exclusively in position R172 (R172K, R172S) (64, 93), which is associated to increased production of 2-HG compared to other IDH2 mutants alleles (94, 95). Of note, the presence of IDH2 mutations is associated with poor prognosis in a subset of AML patients (96), but there is no significant association of IDH2 mutations with survival in AITL (64). In contrast to AML, where IDH2 and TET2 mutations are mutually exclusive, AITLs frequently present co-occurring mutations in these epigenetic regulators (64) (Figure 3). Specifically, analysis of an AITL cohort (N=120) indicates than over 90% of the IDH2 mutated samples also harbor mutations in TET2 (Figure 3). Although methylation profiling reflected only a moderate effect of the double TET2/IDH2 versus the TET2 only mutant cases, gene expression profiling supports a cooperative effect of IDH2 and TET2 mutations on the regulation of the expression of TFH specific genes leading to a more polarized TFH signature that achieved by the presence of TET2 mutations alone (64).
Loss of function mutations in TET2 and DNMT3A seem to occur at an early stage of hematopoietic development, as mutations in those genes have been found in normal elderly individuals and as germline events in AML and PTCL patients (62, 71, 72, 97). Actually, the presence of somatic mutations in the blood of otherwise healthy individuals is associated with a higher risk of developing hematopoietic tumors (98, 99). Clonality and germline analysis in the context of AITL supports a transformation model in which TET2 and DNMT3A mutations constitute an initial or pre-malignant lesion in hematopoietic progenitors that could eventually lead to clonal expansion and malignant transformation both within the T-cell and myeloid lineages. In this context, the presence of mutations in IDH2 and RHOA, usually at a lower allelic frequency (62, 64) reflects a second hit that likely modulates lineage specification towards TFH cells orchestrating the cells into developing AITL.
Additional mutations in the TCR pathway
Gene expression profiling studies have proposed that AITL tumors may be driven by increased T-cell receptor signaling (12). Consistently, genomic studies have uncovered the presence of recurrent, albeit relatively rare, genetic alterations affecting the FYN and CD28 genes, two important elements in the TCR signaling cascade (61, 100).
The FYN tyrosine kinase is, together with LCK, the predominant SRC family kinase found in T lymphocytes and plays an important role in T-cell activation upon T-cell receptor stimulation (101). FYN mutations found in PTCL NOS and AITL cases specifically disrupt the intramolecular inhibitory interaction of the FYN SH2 domain with C-terminal phosphorylated FYN Tyr531 resulting in increased tyrosine kinase signaling (61) (Figure 2).
Recurrent mutations in CD28, a member of the immunoglobulin subfamily and the major co-stimulatory molecule for TCR-mediated activation, have been recently described in AITL (100, 102). PTCL-associated CD28 mutations affect the D124 and T195 residues (Figure 2) and result in increased signaling via increased ligand-receptor interaction and signal transduction (100). CD28-mutated AITL patients have inferior survival to non-mutated cases (100). Given the prominent role of CD28 signaling in normal TFH development, and the presence of activating mutations in CD28 in AITL, it is possible that CD28 directed therapies may be of relevance for the treatment of this disease.
The promise for targeted therapies
Understanding the mechanisms relevant for TFH differentiation, proliferation or function offers a number of novel therapeutic opportunities for targeting malignant TFH cells (Figure 4). In this perspective, BCL6 arises as one the most promising targets, since within T cells; BCL6 expression is restricted to TFH subset and is necessary for TFH survival (103). Thus, blocking BCL6 is an attractive targeted therapy for AITL. Indeed, small molecule or peptomimetic BCL6 inhibitors have been developed as targeted therapies in diffuse large cell lymphoma (104–106) and breast cancer (106).
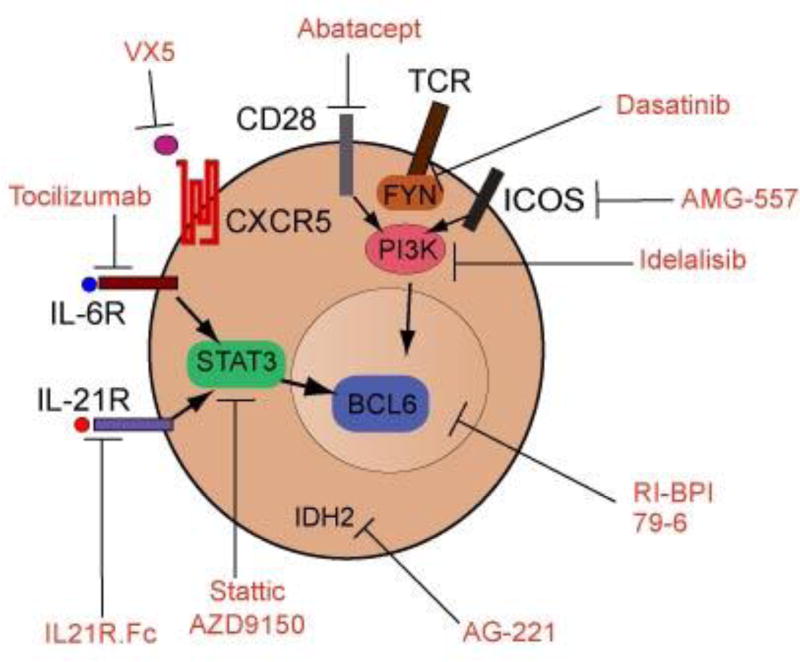
Figure 4. Targeted therapies in AITL.
Schematic diagram depicting pharmacologic agents that target relevant proteins in AITL and TFH cells that could be used for personalized therapies against AITL lymphomas.
Given the close relationship between TFH cells and AITL tumor lymphocytes, potentially same therapies evaluated for autoimmune disorders where TFH cells are involved might be relevant for AITL. Thus, Abatacept, a recombinant antibody which blocks both CTLA4 and CD28 signaling, suppresses TFH generation in an experimental model of autoimmunity (107), suggesting a potential therapeutic use in AITL. However, recent studies have shown the prominent role of ICOS in the development and maintenance of TFH cells (27–29, 37), indicating that ICOS could be a more specific target for therapy in AITL. A phase I trial of ICOS blockade in systemic lupus erythematosus demonstrates the feasibility of administering this antibody to human subjects (108). As ICOS function is critically dependent on the activity of phosphoinositide 3-kinase (PI3K), small molecule inhibitors of this kinase offers an alternative approach to abrogate ICOS signaling. Pharmacological inhibition of the PI3K/AKT signaling pathway has already shown promise as an effective therapy in experimental models of autoimmunity and is currently being evaluated in clinical trials in CLL and follicular lymphoma (109). Both CD28 and ICOS function as co-stimulators of TCR, consequently, the inhibition of FYN, a key molecule in TCR signaling could also be evaluated in AITL. In this sense, PTCL-associated mutant active FYN proteins can be effectively inhibited with dasatinib, a multikinase inhibitor with activity against ABL1 and SRK kinases supporting a potential role for this inhibitor as targeted therapy (61).
IL6 and IL21 are key cytokines required for TFH induction and differentiation through a STAT3 dependent mechanism, therefore, inhibition of those signals may abrogate TFH differentiation and survival. Tocilizimab, an anti-IL6R antibody reduces circulating TFH cell numbers and IL21 production in patients with rheumatoid arthritis (110) and IL-21R blockade arrested the disease progression and mortality in a mouse model of SLE (111). An alternative may be the use of STAT3 specific inhibitors. Thus, Stattic, a specific STAT3 small molecule inhibitor has been shown to induce apoptosis in Sézary syndrome cells (112) and AZD9150, an antisense oligonucleotide inhibitor of STAT3 has been reported to have effect in vivo in lymphoma models (113).
CXCL13 determines TFH homing by engaging with the cell surface receptor CXCR5, higly expressed in AITL tumor cells. Anti-CXCL13 treatment diminishes the number of germinal centers in immunized mice and demonstrates efficacy in models of rheumatoid arthritis and multiple sclerosis (114). Currently pre-clinical studies have been initiated with anti-CXCL13 antibodies (VX5).
Finally, both IDH2 and TET2 mutations can lead to promoter hypermethylation, suggesting a potential role for hypomethylating agents as possible therapy for AITL. In this regard, the use of hypomethylating agents has already been reported to induce responses in TET2 mutated myelodysplastic syndromes (115), and a recent case report showed efficacy of 5-AZT in an AITL patient whose tumor carried a TET2 mutation (116). Additionally, AG-221, a small molecule inhibitor targeting mutant IDH2, has shown promising results in AML (117), and its use as a targeted therapy in AITL is being currently tested in a clinical trial (118). In the case of AITL, the frequent co-occurrence of TET2 mutations in IDH2 mutated cases might support the combination of hypomethylating agents with IDH2 inhibitors to increase efficacy for targeting AITL (64).
Conclusion
AITLs constitute a group of poor prognosis lymphoma derived from T-cell follicular helper cells, a subset of T-cells normally present in germinal centers with a helper function to germinal center B-cells. Gene expression profiling has improved the diagnosis and classification of PTCLs, demonstrating the association of AITL, as well as a subgroup of AITL-like PTCL NOS cases, with a TFH-like gene expression signature. Genomic analyses have identified multiple genes frequently mutated in AITL, including mutations epigenetic regulators TET2, DNMT3A and IDH2; the small GTPase RHOA gene, and components of the TCR pathway, including CD28 and FYN. The process of malignant transformation leading to AITL fits a multistep model with pre-malignant mutations in TET2 or DNMT3A occurring as initiating events followed by the acquisition of the RHOA G17V or IDH2 R172 mutation (Figure 1). Significant efforts are underway to develop cell and animal models for AITL. These models will provide powerful tools for the study of the pathogenesis of AITL and the identification and development of molecularly targeted therapies to treat this disease.
Key points.
Mutations in the small GTPase RHOA gene, specifically the dominant negative G17V allele, are present in over 70% of the patients with AITL and almost 20% of PTCL, NOS cases.
Loss of function mutation in epigenetic regulators TET2 and DNMT3A are a frequent event in the pathogenesis of AITL
IDH2 R172 is the only mutated allele found in AITL, is generally associated with TET2 mutations and define a specific subgroup within this disease
Mutations in elements of the TCR pathway –CD28 and FYN- lead to increased TCR signaling in AITL
The current model for AITL development suggest the existence of a pre-malignant lesion in the TET2 or DNMT3A epigenetic regulators, followed by a secondary mutation in RHOA G17V or IDH2 R172 that results in the malignant transformation of mature T-cells with a TFH phenotype.
Acknowledgments
We would like to thank Drs. Adolfo Ferrando and Laura Belver for their suggestions and critical comments on the manuscript.
Financial support and sponsorship
This work was supported by a grant from the National Cancer Institute 1R01CA197945-01 (TP).
Footnotes
Conflicts of interest
There are no conflicts of interest.
References and recommended reading
Papers of particular interest, published within the past two years (2014–2015), have been highlighted as:
* of special interest
** of outstanding interest
- 1.de Leval L, Gisselbrecht C, Gaulard P. Advances in the understanding and management of angioimmunoblastic T-cell lymphoma. British journal of haematology. 2010;148(5):673–89. doi: 10.1111/j.1365-2141.2009.08003.x. [DOI] [PubMed] [Google Scholar]
- 2.Federico M, Rudiger T, Bellei M, Nathwani BN, Luminari S, Coiffier B, et al. Clinicopathologic characteristics of angioimmunoblastic T-cell lymphoma: analysis of the international peripheral T-cell lymphoma project. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31(2):240–46. doi: 10.1200/JCO.2011.37.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3*.Xu B, Liu P. No survival improvement for patients with angioimmunoblastic T-cell lymphoma over the past two decades: a population-based study of 1207 cases. PloS one. 2014;9(3):e92585. doi: 10.1371/journal.pone.0092585. Outcome study on a large panel of AITL patients diagnosed from 1973 to 2010 showing no improvement in survival in the past two decades. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez-Delgado B, Cuadros M, Honrado E, Ruiz de la Parte A, Roncador G, Alves J, et al. Differential expression of NF-kappaB pathway genes among peripheral T-cell lymphomas. Leukemia. 2005;19(12):2254–63. doi: 10.1038/sj.leu.2403960. [DOI] [PubMed] [Google Scholar]
- 5.Ballester B, Ramuz O, Gisselbrecht C, Doucet G, Loi L, Loriod B, et al. Gene expression profiling identifies molecular subgroups among nodal peripheral T-cell lymphomas. Oncogene. 2006;25(10):1560–70. doi: 10.1038/sj.onc.1209178. [DOI] [PubMed] [Google Scholar]
- 6.Martinez-Delgado B. Peripheral T-cell lymphoma gene expression profiles. Hematological oncology. 2006;24(3):113–9. doi: 10.1002/hon.781. [DOI] [PubMed] [Google Scholar]
- 7.de Leval L, Rickman DS, Thielen C, Reynies A, Huang YL, Delsol G, et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood. 2007;109(11):4952–63. doi: 10.1182/blood-2006-10-055145. [DOI] [PubMed] [Google Scholar]
- 8.Piccaluga PP, Agostinelli C, Califano A, Carbone A, Fantoni L, Ferrari S, et al. Gene expression analysis of angioimmunoblastic lymphoma indicates derivation from T follicular helper cells and vascular endothelial growth factor deregulation. Cancer research. 2007;67(22):10703–10. doi: 10.1158/0008-5472.CAN-07-1708. [DOI] [PubMed] [Google Scholar]
- 9.Piccaluga PP, Agostinelli C, Califano A, Rossi M, Basso K, Zupo S, et al. Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. The Journal of clinical investigation. 2007;117(3):823–34. doi: 10.1172/JCI26833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuadros M, Dave SS, Jaffe ES, Honrado E, Milne R, Alves J, et al. Identification of a proliferation signature related to survival in nodal peripheral T-cell lymphomas. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007;25(22):3321–9. doi: 10.1200/JCO.2006.09.4474. [DOI] [PubMed] [Google Scholar]
- 11.Iqbal J, Weisenburger DD, Greiner TC, Vose JM, McKeithan T, Kucuk C, et al. Molecular signatures to improve diagnosis in peripheral T-cell lymphoma and prognostication in angioimmunoblastic T-cell lymphoma. Blood. 2010;115(5):1026–36. doi: 10.1182/blood-2009-06-227579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12**.Iqbal J, Wright G, Wang C, Rosenwald A, Gascoyne RD, Weisenburger DD, et al. Gene expression signatures delineate biological and prognostic subgroups in peripheral T-cell lymphoma. Blood. 2014;123(19):2915–23. doi: 10.1182/blood-2013-11-536359. Analysis of gene-expression profiles in a large cohort of PTCL patients identifies molecular classifiers highly specific for different PTCL subgroups, and identifies microenvironment associated signatures in AITL that are predictive of survival in an independent cohort. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13*.Iqbal J, Wilcox R, Naushad H, Rohr J, Heavican TB, Wang C, et al. Genomic signatures in T-cell lymphoma: How can these improve precision in diagnosis and inform prognosis? Blood reviews. 2015 doi: 10.1016/j.blre.2015.08.003. Review on recent advances in the molecular diagnosis and prognosis of different PTCL subgroups using genome-wide techniques (gene expression profiling and mutational analysis) with emphasis on the identification of molecular targets for precision therapy. [DOI] [PubMed] [Google Scholar]
- 14.Piccaluga PP, Navari M, Etebari M, Pileri S. Molecular Genetics of Peripheral T-cell Lymphomas. eLS [Internet] 2015 Dec; [Google Scholar]
- 15*.Gaulard P, de Leval L. The microenvironment in T-cell lymphomas: emerging themes. Seminars in cancer biology. 2014;24:49–60. doi: 10.1016/j.semcancer.2013.11.004. Role of the microenvironment in PTCL, describing AITL as a paradigm for crosstalk between tumor cells and the microenvironment. [DOI] [PubMed] [Google Scholar]
- 16*.Piccaluga PP, Tabanelli V, Pileri SA. Molecular genetics of peripheral T-cell lymphomas. International journal of hematology. 2014;99(3):219–26. doi: 10.1007/s12185-014-1522-1. Review on the use of gene expression profiling and how it has diagnosis in PTCL and identified signaling pathways that have provided the rationale for the introduction of novel drugs for the treatment of PTCLs. [DOI] [PubMed] [Google Scholar]
- 17.Dupuis J, Boye K, Martin N, Copie-Bergman C, Plonquet A, Fabiani B, et al. Expression of CXCL13 by neoplastic cells in angioimmunoblastic T-cell lymphoma (AITL): a new diagnostic marker providing evidence that AITL derives from follicular helper T cells. The American journal of surgical pathology. 2006;30(4):490–4. doi: 10.1097/00000478-200604000-00009. [DOI] [PubMed] [Google Scholar]
- 18.Grogg KL, Attygalle AD, Macon WR, Remstein ED, Kurtin PJ, Dogan A. Expression of CXCL13, a chemokine highly upregulated in germinal center T-helper cells, distinguishes angioimmunoblastic T-cell lymphoma from peripheral T-cell lymphoma, unspecified. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2006;19(8):1101–7. doi: 10.1038/modpathol.3800625. [DOI] [PubMed] [Google Scholar]
- 19.Schaerli P, Willimann K, Lang AB, Lipp M, Loetscher P, Moser B. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. The Journal of experimental medicine. 2000;192(11):1553–62. doi: 10.1084/jem.192.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim CH, Rott LS, Clark-Lewis I, Campbell DJ, Wu L, Butcher EC. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. The Journal of experimental medicine. 2001;193(12):1373–81. doi: 10.1084/jem.193.12.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, et al. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. The Journal of experimental medicine. 2000;192(11):1545–52. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS, et al. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. Journal of immunology. 2004;173(1):68–78. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- 23*.Ueno H, Banchereau J, Vinuesa CG. Pathophysiology of T follicular helper cells in humans and mice. Nature immunology. 2015;16(2):142–52. doi: 10.1038/ni.3054. Review summarizing the biology of TFH cells in mice and humans and how they participate in immune responses in various human diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vinuesa CG, Cook MC, Angelucci C, Athanasopoulos V, Rui L, Hill KM, et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435(7041):452–8. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- 25**.Vinuesa CG, Linterman MA, Yu D, MacLennan IC. Follicular Helper T Cells. Annual review of immunology. 2016 doi: 10.1146/annurev-immunol-041015-055605. Most recent review on function, differentiation, migration and plasticity of TFH cells and their role in cancer and immunity. [DOI] [PubMed] [Google Scholar]
- 26.Goenka R, Barnett LG, Silver JS, O’Neill PJ, Hunter CA, Cancro MP, et al. Cutting edge: dendritic cell-restricted antigen presentation initiates the follicular helper T cell program but cannot complete ultimate effector differentiation. Journal of immunology. 2011;187(3):1091–5. doi: 10.4049/jimmunol.1100853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stone EL, Pepper M, Katayama CD, Kerdiles YM, Lai CY, Emslie E, et al. ICOS coreceptor signaling inactivates the transcription factor FOXO1 to promote Tfh cell differentiation. Immunity. 2015;42(2):239–51. doi: 10.1016/j.immuni.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weber JP, Fuhrmann F, Feist RK, Lahmann A, Al Baz MS, Gentz LJ, et al. ICOS maintains the T follicular helper cell phenotype by down-regulating Kruppel-like factor 2. The Journal of experimental medicine. 2015;212(2):217–33. doi: 10.1084/jem.20141432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leavenworth JW, Verbinnen B, Yin J, Huang H, Cantor H. A p85alpha-osteopontin axis couples the receptor ICOS to sustained Bcl-6 expression by follicular helper and regulatory T cells. Nature immunology. 2015;16(1):96–106. doi: 10.1038/ni.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30**.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41(4):529–42. doi: 10.1016/j.immuni.2014.10.004. This is a very complete review on TFH cell differentiation and role of TFH cells as cell of origin of AITL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Nurieva RI, Dong C. Transcriptional regulation of follicular T-helper (Tfh) cells. Immunological reviews. 2013;252(1):139–45. doi: 10.1111/imr.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325(5943):1001–5. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu D, Batten M, Mackay CR, King C. Lineage specification and heterogeneity of T follicular helper cells. Current opinion in immunology. 2009;21(6):619–25. doi: 10.1016/j.coi.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 34.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325(5943):1006–10. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Hatzi K, Nance JP, Kroenke MA, Bothwell M, Haddad EK, Melnick A, et al. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. The Journal of experimental medicine. 2015;212(4):539–53. doi: 10.1084/jem.20141380. A in depth analysis on the direct targets of BCL6 in TFH cells and their role in the differentiation towards the TFH lineage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi H, Chen X, Chu C, Liu D, Ma W, Wang Y, et al. Tfh cell differentiation and their function in promoting B-cell responses. Advances in experimental medicine and biology. 2014;841:153–80. doi: 10.1007/978-94-017-9487-9_6. [DOI] [PubMed] [Google Scholar]
- 37.Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, et al. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. 2011;34(6):932–46. doi: 10.1016/j.immuni.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F, et al. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 2014;507(7493):513–8. doi: 10.1038/nature12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, et al. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. Journal of immunology. 2012;188(8):3734–44. doi: 10.4049/jimmunol.1103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwon H, Thierry-Mieg D, Thierry-Mieg J, Kim HP, Oh J, Tunyaplin C, et al. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 2009;31(6):941–52. doi: 10.1016/j.immuni.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29(1):138–49. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao W, Lin JX, Wang L, Li P, Leonard WJ. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nature immunology. 2011;12(6):551–9. doi: 10.1038/ni.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi YS, Yang JA, Yusuf I, Johnston RJ, Greenbaum J, Peters B, et al. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. Journal of immunology. 2013;190(8):4014–26. doi: 10.4049/jimmunol.1202963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suto A, Kashiwakuma D, Kagami S, Hirose K, Watanabe N, Yokote K, et al. Development and characterization of IL-21-producing CD4+ T cells. The Journal of experimental medicine. 2008;205(6):1369–79. doi: 10.1084/jem.20072057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma CS, Suryani S, Avery DT, Chan A, Nanan R, Santner-Nanan B, et al. Early commitment of naive human CD4(+) T cells to the T follicular helper (T(FH)) cell lineage is induced by IL-12. Immunology and cell biology. 2009;87(8):590–600. doi: 10.1038/icb.2009.64. [DOI] [PubMed] [Google Scholar]
- 46.Schmitt N, Bustamante J, Bourdery L, Bentebibel SE, Boisson-Dupuis S, Hamlin F, et al. IL-12 receptor beta1 deficiency alters in vivo T follicular helper cell response in humans. Blood. 2013;121(17):3375–85. doi: 10.1182/blood-2012-08-448902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmitt N, Morita R, Bourdery L, Bentebibel SE, Zurawski SM, Banchereau J, et al. Human dendritic cells induce the differentiation of interleukin-21-producing T follicular helper-like cells through interleukin-12. Immunity. 2009;31(1):158–69. doi: 10.1016/j.immuni.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annual review of cell and developmental biology. 2005;21:247–69. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 49.Boulter E, Estrach S, Garcia-Mata R, Feral CC. Off the beaten paths: alternative and crosstalk regulation of Rho GTPases. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012;26(2):469–79. doi: 10.1096/fj.11-192252. [DOI] [PubMed] [Google Scholar]
- 50.del Pozo MA, Vicente-Manzanares M, Tejedor R, Serrador JM, Sanchez-Madrid F. Rho GTPases control migration and polarization of adhesion molecules and cytoskeletal ERM components in T lymphocytes. European journal of immunology. 1999;29(11):3609–20. doi: 10.1002/(SICI)1521-4141(199911)29:11<3609::AID-IMMU3609>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 51.Heasman SJ, Carlin LM, Cox S, Ng T, Ridley AJ. Coordinated RhoA signaling at the leading edge and uropod is required for T cell transendothelial migration. The Journal of cell biology. 2010;190(4):553–63. doi: 10.1083/jcb.201002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heasman SJ, Ridley AJ. Multiple roles for RhoA during T cell transendothelial migration. Small GTPases. 2010;1(3):174–9. doi: 10.4161/sgtp.1.3.14724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mou F, Praskova M, Xia F, Van Buren D, Hock H, Avruch J, et al. The Mst1 and Mst2 kinases control activation of rho family GTPases and thymic egress of mature thymocytes. The Journal of experimental medicine. 2012;209(4):741–59. doi: 10.1084/jem.20111692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corre I, Gomez M, Vielkind S, Cantrell DA. Analysis of thymocyte development reveals that the GTPase RhoA is a positive regulator of T cell receptor responses in vivo. The Journal of experimental medicine. 2001;194(7):903–14. doi: 10.1084/jem.194.7.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henning SW, Galandrini R, Hall A, Cantrell DA. The GTPase Rho has a critical regulatory role in thymus development. The EMBO journal. 1997;16(9):2397–407. doi: 10.1093/emboj/16.9.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galandrini R, Henning SW, Cantrell DA. Different functions of the GTPase Rho in prothymocytes and late pre-T cells. Immunity. 1997;7(1):163–74. doi: 10.1016/s1074-7613(00)80519-1. [DOI] [PubMed] [Google Scholar]
- 57.Cleverley S, Henning S, Cantrell D. Inhibition of Rho at different stages of thymocyte development gives different perspectives on Rho function. Current biology : CB. 1999;9(12):657–60. doi: 10.1016/s0960-9822(99)80289-9. [DOI] [PubMed] [Google Scholar]
- 58.Cleverley SC, Costello PS, Henning SW, Cantrell DA. Loss of Rho function in the thymus is accompanied by the development of thymic lymphoma. Oncogene. 2000;19(1):13–20. doi: 10.1038/sj.onc.1203259. [DOI] [PubMed] [Google Scholar]
- 59**.Zhang S, Konstantinidis DG, Yang JQ, Mizukawa B, Kalim K, Lang RA, et al. Gene targeting RhoA reveals its essential role in coordinating mitochondrial function and thymocyte development. Journal of immunology. 2014;193(12):5973–82. doi: 10.4049/jimmunol.1400839. This article describes the critical role of RHOA in regulating thymocyte development by coordinating multiple developmental events using a conditional mouse model bearing T cell specific deletion of RhoA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pernis AB. Rho GTPase-mediated pathways in mature CD4+ T cells. Autoimmunity reviews. 2009;8(3):199–203. doi: 10.1016/j.autrev.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 61**.Palomero T, Couronne L, Khiabanian H, Kim MY, Ambesi-Impiombato A, Perez-Garcia A, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nature genetics. 2014;46(2):166–70. doi: 10.1038/ng.2873. This study was one of the two initial reports on the presence of mutations in RHOA G17V in PTCL, their role as dominant negatives and the co-occurrence with mutations in TET2 epigenetic regulator. Additionally, identifies the presence of mutation in the FYN kinase and its potential as therapeutic target due to sensitivity to the kinase inhibitor dasatinib. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62**.Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nature genetics. 2014;46(2):171–5. doi: 10.1038/ng.2872. This study was one of the two initial reports on the presence of mutations in RHOA G17V in PTCL, their role as dominant negatives and the co-occurrence with mutations in TET2 epigenetic regulator. [DOI] [PubMed] [Google Scholar]
- 63**.Yoo HY, Sung MK, Lee SH, Kim S, Lee H, Park S, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nature genetics. 2014;46(4):371–5. doi: 10.1038/ng.2916. This study reports on the presence of mutations in RHOA G17V in AITL, their role as dominant negatives. [DOI] [PubMed] [Google Scholar]
- 64**.Wang C, McKeithan TW, Gong Q, Zhang W, Bouska A, Rosenwald A, et al. IDH2R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. Blood. 2015;126(15):1741–52. doi: 10.1182/blood-2015-05-644591. This study analyzed the presence of mutations in IDH2, showing they are restricted to the R172 residue and frequently co-occur with TET2 mutations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manso R, Sanchez-Beato M, Monsalvo S, Gomez S, Cereceda L, Llamas P, et al. The RHOA G17V gene mutation occurs frequently in peripheral T-cell lymphoma and is associated with a characteristic molecular signature. Blood. 2014;123(18):2893–4. doi: 10.1182/blood-2014-02-555946. [DOI] [PubMed] [Google Scholar]
- 66.Ondrejka SL, Grzywacz B, Bodo J, Makishima H, Polprasert C, Said JW, et al. Angioimmunoblastic T-cell Lymphomas With the RHOA p.Gly17Val Mutation Have Classic Clinical and Pathologic Features. The American journal of surgical pathology. 2015 doi: 10.1097/PAS.0000000000000555. [DOI] [PubMed] [Google Scholar]
- 67.Rohde M, Richter J, Schlesner M, Betts MJ, Claviez A, Bonn BR, et al. Recurrent RHOA mutations in pediatric Burkitt lymphoma treated according to the NHL-BFM protocols. Genes, chromosomes & cancer. 2014;53(11):911–6. doi: 10.1002/gcc.22202. [DOI] [PubMed] [Google Scholar]
- 68.Kakiuchi M, Nishizawa T, Ueda H, Gotoh K, Tanaka A, Hayashi A, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nature genetics. 2014;46(6):583–7. doi: 10.1038/ng.2984. [DOI] [PubMed] [Google Scholar]
- 69.Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nature genetics. 2014;46(6):573–82. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 70.Nagata Y, Kontani K, Enami T, Kataoka K, Ishii R, Totoki Y, et al. Variegated RHOA mutations in adult T-cell leukemia/lymphoma. Blood. 2015 doi: 10.1182/blood-2015-06-644948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer cell. 2011;20(1):25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 72.Couronne L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. The New England journal of medicine. 2012;366(1):95–6. doi: 10.1056/NEJMc1111708. [DOI] [PubMed] [Google Scholar]
- 73.Cairns RA, Iqbal J, Lemonnier F, Kucuk C, de Leval L, Jais JP, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012;119(8):1901–3. doi: 10.1182/blood-2011-11-391748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–33. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cimmino L, Abdel-Wahab O, Levine RL, Aifantis I. TET family proteins and their role in stem cell differentiation and transformation. Cell stem cell. 2011;9(3):193–204. doi: 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pronier E, Delhommeau F. Role of TET2 mutations in myeloproliferative neoplasms. Current hematologic malignancy reports. 2012;7(1):57–64. doi: 10.1007/s11899-011-0108-8. [DOI] [PubMed] [Google Scholar]
- 78.Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114(1):144–7. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vose JM. Update on T-cell lymphoma. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2008;(19 Suppl 4):74–6. doi: 10.1093/annonc/mdn203. [DOI] [PubMed] [Google Scholar]
- 80.Lemonnier F, Couronne L, Parrens M, Jais JP, Travert M, Lamant L, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120(7):1466–9. doi: 10.1182/blood-2012-02-408542. [DOI] [PubMed] [Google Scholar]
- 81.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer cell. 2011;20(1):11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118(17):4509–18. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(35):14566–71. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ko M, Rao A. TET2: epigenetic safeguard for HSC. Blood. 2011;118(17):4501–3. doi: 10.1182/blood-2011-08-373357. [DOI] [PubMed] [Google Scholar]
- 85.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell stem cell. 2011;8(2):200–13. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kunimoto H, Fukuchi Y, Sakurai M, Sadahira K, Ikeda Y, Okamoto S, et al. Tet2 disruption leads to enhanced self-renewal and altered differentiation of fetal liver hematopoietic stem cells. Scientific reports. 2012;2:273. doi: 10.1038/srep00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shide K, Kameda T, Shimoda H, Yamaji T, Abe H, Kamiunten A, et al. TET2 is essential for survival and hematopoietic stem cell homeostasis. Leukemia. 2012;26(10):2216–23. doi: 10.1038/leu.2012.94. [DOI] [PubMed] [Google Scholar]
- 88*.Muto H, Sakata-Yanagimoto M, Nagae G, Shiozawa Y, Miyake Y, Yoshida K, et al. Reduced TET2 function leads to T-cell lymphoma with follicular helper T-cell-like features in mice. Blood cancer journal. 2014;4:e264. doi: 10.1038/bcj.2014.83. This article demonstrates the major role of TET2 in the development of T-cell lymphoma with Tfh-like features using Tet2 knockdown mice. The Tet2 knockdown mice develop T-cell lymphoma with long latency that presents TFH features and epigenetic changes associated to the downregulation of Tet2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. The New England journal of medicine. 2010;363(25):2424–33. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shah MY, Licht JD. DNMT3A mutations in acute myeloid leukemia. Nature genetics. 2011;43(4):289–90. doi: 10.1038/ng0411-289. [DOI] [PubMed] [Google Scholar]
- 91.Thiede C. Mutant DNMT3A: teaming up to transform. Blood. 2012;119(24):5615–7. doi: 10.1182/blood-2012-04-423905. [DOI] [PubMed] [Google Scholar]
- 92.Parker SJ, Metallo CM. Metabolic consequences of oncogenic IDH mutations. Pharmacology & therapeutics. 2015;152:54–62. doi: 10.1016/j.pharmthera.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dang L, Jin S, Su SM. IDH mutations in glioma and acute myeloid leukemia. Trends in molecular medicine. 2010;16(9):387–97. doi: 10.1016/j.molmed.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 94.Lu C, Venneti S, Akalin A, Fang F, Ward PS, Dematteo RG, et al. Induction of sarcomas by mutant IDH2. Genes & development. 2013;27(18):1986–98. doi: 10.1101/gad.226753.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ward PS, Lu C, Cross JR, Abdel-Wahab O, Levine RL, Schwartz GK, et al. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. The Journal of biological chemistry. 2013;288(6):3804–15. doi: 10.1074/jbc.M112.435495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Kronke J, Bullinger L, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28(22):3636–43. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 97*.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nature medicine. 2014;20(12):1472–8. doi: 10.1038/nm.3733. This article describes the frequent existence of mutations in peripheral blood of healthy individuals. The frequency of damaging mutations increases with age and may represent premalignant events that cause clonal hematopoietic expansion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. The New England journal of medicine. 2014;371(26):2488–98. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. The New England journal of medicine. 2014;371(26):2477–87. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100**.Rohr J, Guo S, Huo J, Bouska A, Lachel C, Li Y, et al. Recurrent activating mutations of CD28 in peripheral T-cell lymphomas. Leukemia. 2015 doi: 10.1038/leu.2015.357. This study is the first article characterizing the T195 and D124 mutations in CD28 in AITL, describing their role enhancing signaling downstream the T-cell receptor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Palacios EH, Weiss A. Function of the Src-family kinases, Lck and Fyn, in T-cell development and activation. Oncogene. 2004;23(48):7990–8000. doi: 10.1038/sj.onc.1208074. [DOI] [PubMed] [Google Scholar]
- 102**.Lee SH, Kim JS, Kim J, Kim SJ, Kim WS, Lee S, et al. A highly recurrent novel missense mutation in CD28 among angioimmunoblastic T-cell lymphoma patients. Haematologica. 2015;100(12):e505–7. doi: 10.3324/haematol.2015.133074. This study describes mutations in CD28 in position T195 in AITL and their function on the signaling downstream of the TCR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hollister K, Kusam S, Wu H, Clegg N, Mondal A, Sawant DV, et al. Insights into the role of Bcl6 in follicular Th cells using a new conditional mutant mouse model. Journal of immunology. 2013;191(7):3705–11. doi: 10.4049/jimmunol.1300378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huang C, Hatzi K, Melnick A. Lineage-specific functions of Bcl-6 in immunity and inflammation are mediated by distinct biochemical mechanisms. Nature immunology. 2013;14(4):380–8. doi: 10.1038/ni.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cerchietti L, Melnick A. Targeting BCL6 in diffuse large B-cell lymphoma: what does this mean for the future treatment? Expert review of hematology. 2013;6(4):343–5. doi: 10.1586/17474086.2013.826928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dupont T, Yang SN, Patel J, Hatzi K, Malik A, Tam W, et al. Selective targeting of BCL6 induces oncogene addiction switching to BCL2 in B-cell lymphoma. Oncotarget. 2016;7(3):3520–32. doi: 10.18632/oncotarget.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Platt AM, Gibson VB, Patakas A, Benson RA, Nadler SG, Brewer JM, et al. Abatacept limits breach of self-tolerance in a murine model of arthritis via effects on the generation of T follicular helper cells. Journal of immunology. 2010;185(3):1558–67. doi: 10.4049/jimmunol.1001311. [DOI] [PubMed] [Google Scholar]
- 108.Ahearne MJ, Allchin RL, Fox CP, Wagner SD. Follicular helper T-cells: expanding roles in T-cell lymphoma and targets for treatment. British journal of haematology. 2014;166(3):326–35. doi: 10.1111/bjh.12941. [DOI] [PubMed] [Google Scholar]
- 109.Yang Q, Modi P, Newcomb T, Queva C, Gandhi V. Idelalisib: First-in-Class PI3K Delta Inhibitor for the Treatment of Chronic Lymphocytic Leukemia, Small Lymphocytic Leukemia, and Follicular Lymphoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(7):1537–42. doi: 10.1158/1078-0432.CCR-14-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chavele KM, Merry E, Ehrenstein MR. Cutting edge: circulating plasmablasts induce the differentiation of human T follicular helper cells via IL-6 production. Journal of immunology. 2015;194(6):2482–5. doi: 10.4049/jimmunol.1401190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang M, Yu G, Chan B, Pearson JT, Rathanaswami P, Delaney J, et al. Interleukin-21 receptor blockade inhibits secondary humoral responses and halts the progression of preestablished disease in the (NZB x NZW)F1 systemic lupus erythematosus model. Arthritis & rheumatology. 2015;67(10):2723–31. doi: 10.1002/art.39233. [DOI] [PubMed] [Google Scholar]
- 112.van der Fits L, Out-Luiting JJ, Tensen CP, Zoutman WH, Vermeer MH. Exploring the IL-21-STAT3 axis as therapeutic target for Sezary syndrome. The Journal of investigative dermatology. 2014;134(10):2639–47. doi: 10.1038/jid.2014.199. [DOI] [PubMed] [Google Scholar]
- 113.Hong D, Kurzrock R, Kim Y, Woessner R, Younes A, Nemunaitis J, et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Science translational medicine. 2015;7(314):314ra185. doi: 10.1126/scitranslmed.aac5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Klimatcheva E, Pandina T, Reilly C, Torno S, Bussler H, Scrivens M, et al. CXCL13 antibody for the treatment of autoimmune disorders. BMC immunology. 2015;16:6. doi: 10.1186/s12865-015-0068-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bejar R, Lord A, Stevenson K, Bar-Natan M, Perez-Ladaga A, Zaneveld J, et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood. 2014;124(17):2705–12. doi: 10.1182/blood-2014-06-582809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116*.Cheminant M, Bruneau J, Kosmider O, Lefrere F, Delarue R, Gaulard P, et al. Efficacy of 5-azacytidine in a TET2 mutated angioimmunoblastic T cell lymphoma. British journal of haematology. 2015;168(6):913–6. doi: 10.1111/bjh.13170. Case report indicating sensitivity to hypomethylating agents in an AITL patient with a TET2 loss of function mutation. [DOI] [PubMed] [Google Scholar]
- 117.DiNardo CEMS, Altman JK, Collins R, DeAngelo DJ, Fathi AT, et al. AG-221, an oral, selective, first-in-class, potent inhibitor of the IDH2 mutant enzyme, induced durable responses in a phase 1 study of IDH2 mutation-positive advanced hematologic malignancies Haematol Eur Hematol Assoc Annu Meet. 2015:569. [Google Scholar]
- 118.Stein EM. IDH2 inhibition in AML: Finally progress? Best practice & research Clinical haematology. 2015;28(2–3):112–5. doi: 10.1016/j.beha.2015.10.016. [DOI] [PubMed] [Google Scholar]