Abstract
Oligoethylene glycols are used as crowding agents in experiments that aim to understand the effects of intracellular environments on DNAs. Moreover, DNAs with covalently attached oligoethylene glycols are used as cargo carriers for drug delivery systems. To investigate how oligoethylene glycols interact with DNAs, we incorporated deoxythymidine modified with oligoethylene glycols of different lengths, such as tetraethylene glycol (TEG), into DNAs that form antiparallel G-quadruplex or hairpin structures such that the modified residues were incorporated into loop regions. Thermodynamic analysis showed that because of enthalpic differences, the modified G-quadruplexes were stable and the hairpin structures were slightly unstable relative to unmodified DNA. The stability of G-quadruplexes increased with increasing length of the ethylene oxides and the number of deoxythymidines modified with ethylene glycols in the G-quadruplex. Nuclear magnetic resonance analyses and molecular dynamics calculations suggest that TEG interacts with bases in the G-quartet and loop via CH–π and lone pair–π interactions, although it was previously assumed that oligoethylene glycols do not directly interact with DNAs. The results suggest that numerous cellular co-solutes likely affect DNA function through these CH–π and lone pair–π interactions.
INTRODUCTION
Biomolecules occupy a significant fraction of volume in living cells (up to 40%), resulting in a highly crowded intracellular environment (1–4). The thermodynamic stability of DNA structures is altered in a crowded environment. Oligoethylene glycol and polyethylene glycol (PEG) are used as co-solutes to mimic the physical properties inside cells. Oligoethylene glycol and PEG destabilize duplexes but stabilize non-canonical structures such as G-quadruplexes (3,5,6). Transcription, reverse transcription and translation are controlled by stable G-quadruplex formation by DNA or RNA (7–12). Therefore, methods for specifically stabilizing G-quadruplexes are relevant to therapeutic strategies involving these processes. To increase the stability of G-quadruplexes, certain G-quadruplex ligands, such as 5,10,15,20-tetrakis (N-methyl-4-pyridyl) porphyrin and N-methyl mesoporphyrin IX (NMN), have been developed, (7,13) although these ligands also stabilize duplex structures to some extent.
We recently demonstrated that tetraethylene glycol (TEG)-modified deoxythymidines stabilize G-quadruplexes with single G-tracts (14). The TEG-modified intermolecular G-quadruplexes are significantly stabilized relative to unmodified DNA and efficiently inhibit reverse transcription, (14) although it is unclear how PEG or oligoethylene glycol specifically stabilizes G-quadruplexes. Oligoethylene glycol and PEG covalently bind liposomes, nanoparticles, proteins and nucleic acids (15,16). These modifications facilitate liposome uptake into mammalian cells and enhance pharmacokinetics (17,18). Thus, it is important to understand how oligoethylene glycol and PEG affect DNA structures.
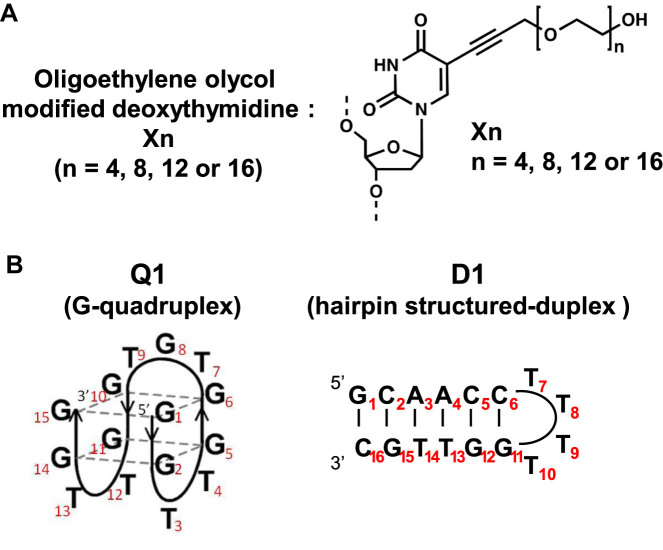
Here we evaluated the effect of oligoethylene glycol on DNA structure and structural thermodynamic stability. We synthesized deoxythymidine tethered to the oligoethylene glycols of TEG (molecular weight, 194; X4), octaethylene glycol (OEG, molecular weight, 370; X8), dodecaethylene glycol (DEG, molecular weight, 547; X12) and hexadecaethylene glycol (HEG, molecular weight, 723; X16) (Figure 1A). Pure, well-defined oligoethylene glycols were prepared using a recently published cost-effective method (19). These nucleotide derivatives were incorporated into the thrombin aptamer (Q1) as a model of a G-quadruplex and into a hairpin as a model of a duplex (D1) (Figure 1B). According to an NMR (nuclear magnetic resonance) study of the thrombin aptamer, the thymines at positions 4 (T4) and 13 (T13) stack on a G-quartet, although the T in position 7 (T7) does not (20,21). Thus, we replaced deoxythymidines in the loop regions of Q1 and D1 with X (Figure 1B and Supplementary Table S1). In our nomenclature, the type of modification is followed by the position number. For example, Q1-(X4)4 indicates the thrombin-binding aptamer Q1 with TEG-modified deoxythymidine (X4) at position 4. We report here the thermodynamic properties of structures adopted by DNAs modified with oligoethylene glycols and a comparison with those of unmodified DNAs.
Figure 1.
(A) Oligoethylene glycol-modified deoxythymine. (B) Antiparallel G-quadruplex (Q1) and antiparallel hairpin (D1) were used in the present study.
MATERIALS AND METHODS
Materials
The oligodeoxynucleotides used in this study, except for those modified with oligoethylene glycols, were purified using high-performance liquid chromatography by Japan Bio Services Co., LTD. Single-strand concentrations of DNA oligonucleotides were calculated by measuring their absorbance at 260 nm and 90°C using single-strand extinction coefficients calculated from the mononucleotide and dinucleotide data, according to the nearest-neighbor approximation model (22). Absorbance was measured using a Shimadzu 1700 spectrophotometer equipped with a thermoprogrammer. The amidite of oligoethylene glycol-modified deoxythymidine was synthesized according to a published procedure (See Supplementary Data) (14).
Thermodynamic analyses
Ultraviolet (UV) absorbance was measured using a Shimadzu 1700 spectrophotometer equipped with a temperature controller. Melting curves at 260 or 295 nm were measured in buffers containing 100 mM KCl, 10 mM K2HPO4 (pH 7.0 at 37°C) and 1 mM K2EDTA. Samples were heated at 0.5°C min−1 or 0.1°C min−1; melting curves of certain oligonucleotides differed at 0.1°C min−1 and 0.5°C min−1 (data not shown). Thermodynamic parameters (Table 1) were calculated using average values obtained from curve fitting at different DNA concentrations (2, 5, 10 and 20 μM) (23). Before measurements, DNA samples were heated to 90°C, cooled to 0°C at a rate of −0.5°C min−1 and incubated at 0°C for 30 min.
Table 1. Thermodynamic parameters for G-quadruplex formationa.
| Sequence | ΔH° (kcal mol−1) | TΔS° (kcal mol−1) | ΔG°25 (kcal mol−1) | T m b (°C) |
|---|---|---|---|---|
| D1 | −51.2 ± 3.7 | −44.6 ± 3.4 | −6.6 ± 0.5 | 66.7 |
| D1-(X4)7 | −41.0 ± 3.3 | −35.9 ± 2.9 | −5.1 ± 0.4 | 66.0 |
| D1-(X4)10 | −43.7 ± 3.4 | −37.8 ± 2.7 | −5.9 ± 0.4 | 66.5 |
| Q1 | −42.6 ± 2.8 | −39.1 ± 2.0 | −3.5 ± 0.3 | 50.7 |
| Q1-(X4)4 | −55.4 ± 3.3 | −48.8 ± 2.9 | −6.6 ± 0.4 | 58.8 |
| Q1-(X4)7 | −41.7 ± 8.7 | −37.5 ± 1.6 | −4.2 ± 0.7 | 54.9 |
| Q1-(X4)13 | −49.7 ± 3.4 | −44.0 ± 2.7 | −5.7 ± 0.4 | 57.6 |
| Q1-(X4)4,13 | −57.2 ± 1.5 | −49.7 ± 5.4 | −7.5 ± 0.1 | 66.2 |
| Q1-(X4)4,7,13 | −57.9 ± 1.1 | −49.9 ± 3.6 | −8.0 ± 0.0 | 69.4 |
| Q1-(X8)4 | −48.7 ± 1.3 | −41.6 ± 1.2 | −7.1 ± 0.2 | 64.3 |
| Q1-(X8)7 | −44.3 ± 3.3 | −39.8 ± 2.9 | −4.4 ± 0.4 | 55.2 |
| Q1-(X12)4 | −49.6 ± 2.9 | −43.1 ± 8.1 | −6.5 ± 0.4 | 64.9 |
| Q1-(X12)7 | −42.8 ± 1.2 | −38.3 ± 1.2 | −4.5 ± 0.1 | 56.2 |
| Q1-(X16)4 | −49.1 ± 2.1 | −42.8 ± 1.8 | −6.3 ± 0.3 | 64.9 |
| Q1-(X16)7 | −45.1 ± 2.1 | −38.9 ± 1.9 | −4.6 ± 0.2 | 56.5 |
aExperiments were performed in a buffer containing 100 mM KCl, 10 mM K2HPO4 (pH 7.0) and 1 mM K2EDTA. Thermodynamic parameters were calculated from the average values obtained from curve fitting.
bMelting temperature was calculated at a strand concentration of 5 μM.
NMR spectroscopy
Oligonucleotides Q1-(X4)4 or Q1-(X4)7 were dissolved in 10 mM K2HPO4 buffer (pH 7.0) containing 100 mM KCl, 1 mM K2EDTA and 10% H2O. The final DNA concentration in 300 μl was ∼0.5 mM. DNA solutions were transferred to Shigemi NMR tubes. All spectra were acquired at 25°C using an Agilent VNMRS 600 MHz NMR spectrometer equipped with a cold probe. The fingerprint imino regions of 1D 1H NMR spectra were used to confirm G-quadruplex folding. To confirm G-quadruplex topology, 2D NOESY spectra were acquired using a mixing time of 200 ms. Double-pulsed field gradient spin echo water suppression was used with 1D and 2D pulse sequences.
Molecular dynamics (MD) simulations
The structure of the G-quadruplex formed by 5΄-GGTTGGTGTGGTTGG-3΄, determined using NMR, was acquired from the Protein Data Bank (PDB ID: 1C35) (24) and adopted as an initial structure of Q1. We added K+ ions around the phosphate groups of the DNA backbone, and two Cl− ions were added to neutralize the charge of the systems. The structures of Q1-(X4)4 and Q1-(X4)7 were constructed by replacing the methyl groups of fourth and seventh thymines with TEG, respectively. Similarly, the structures of Q1-(X8)4 and Q1-(Pro)4 used to investigate the effects of propynyl groups in X4 and X8 were prepared by replacing the methyl group of the fourth thymine with OEG or with a propynyl group. Subsequently, TIP3P water molecules were merged into the system, and MD simulations were performed using AMBER14 software (25). The force fields of X4 and X8 and the propynyl group were obtained using the GAFF module included in Amber Tools. The AMBERff99bsc0 force field was applied to the other nucleotides and ions (26). Optimization protocols and MD simulation were as follows: (i) optimization of water molecules and ions was performed in 1500 steps, (ii) the entire system was optimized in 1500 steps without constraints, (iii) the water molecules and ions in system were heated to 298 K for 100 ps with fixed conformations for TEG or OEG-modified Q1, (iv) solvated structures were generated at constant pressure (1 atm) and temperature (298 K) for 1 ns to equilibrate the system and (v) a 35-ns simulation was performed under identical conditions. During optimization and MD simulations, the cut-off distance for non-bonded interactions was set to 10 Å. For MD simulations, the SHAKE algorithm (27,28) was applied to all bond lengths and periodic boundary conditions were used. Supplementary Movies S1, 2 and 3 show the trajectories of Q1-(X4)4 for 12.5–13.5 ns, Q1-(X4)7 for 12.3–13.3 ns and Q1-(X8)4 for 12.8–13.8 ns. Guanine and thymine bases are green and dark cyan, respectively. K+ ions are shown as purple spheres. X4 and X8 are shown in ball-and-stick representations. The color scheme of the modified thymines is as follows: gray, carbon; red, oxygen; blue, nitrogen and white, hydrogen.
RESULTS AND DISCUSSION
Stability and structures of TEG-modified DNAs
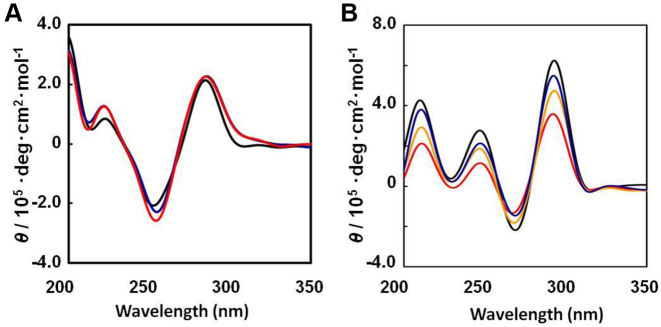
To evaluate the effect of TEG modification on duplex and G-quadruplex structures, the circular dichroism (CD) spectra of oligonucleotide D1, D1-(X4)7, D1-(X4)10, Q1, Q1-(X4)4, Q1-(X4)7 and Q1-(X4)13 were acquired using a solution containing 100 mM KCl, 10 mM K2HPO4 (pH 7.0) and 1 mM K2EDTA. B-form DNA exhibits two positive bands at ∼220 and 280 nm and one negative band at ∼248 nm. At 37°C, the CD spectra of D1, D1-(X4)7 and D1-(X4)10 were typical of B-form duplexes (Figure 2A) with two positive bands near 220 and 280 nm and a negative band near 250 nm. Moreover, the CD spectra of Q1, Q1-(X4)4, Q1-(X4)7 and Q1-(X4)13 exhibited the characteristic of antiparallel stranded G-quadruplexes (Figure 2B) with a positive band near 295 nm and a negative band near 265 nm. These results indicate that TEG modifications of oligonucleotides did not significantly alter the adopted structures.
Figure 2.
CD spectra for 20 μM (A) D1 (black), D1-(X4)7 (blue) and D1-(X4)10 (red), and (B) Q1 (black), Q1-(X4)4 (red), Q1-(X4)7 (orange) and Q1-(X4)13 (blue) at 4°C. The buffer was 100 mM KCl, 10 mM K2HPO4 (pH 7.0 at 25 °C) and 1 mM K2EDTA.
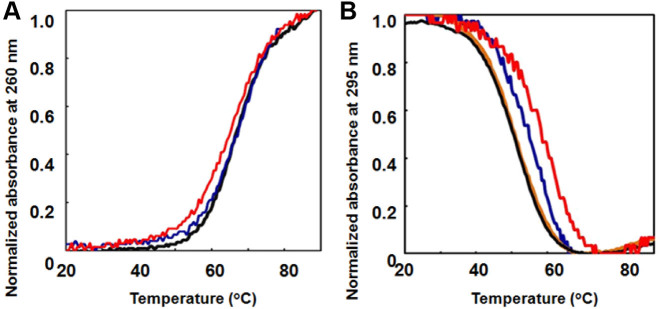
To determine how the stabilities of DNA structures were affected by TEG modification, the midpoints of the melting curves (Tms) of 5 μM DNA were determined in a buffer containing 100 mM KCl, 10 mM K2HPO4 (pH 7.0) and 1 mM K2EDTA. The Tm values of 5 μM D1, D1-(X4)7 and D1-(X4)10 were 66.7, 66.0 and 66.5°C, respectively (Figure 3A and Table 1). The Tm values of 5 μM Q1, Q1-(X4)4, Q1-(X4)7 and Q1-(X4)13 were 50.7, 58.8, 54.9 and 57.6°C, respectively (Figure 3B and Table 1). The results show that the incorporation of TEG-modified deoxythymidine significantly stabilized the G-quadruplex but slightly destabilized the duplex in a position-specific manner. Moreover, we synthesized Q1 analogues with TEG tethered to the phosphate group of the 5΄ terminus (TEG-pQ1) (Supplementary Figure S1) and Q1 with a 5΄ phosphate group (pQ1). TEG-pQ1 was destabilized relative to pQ1 indicating that the negative charge of the phosphate group at the 5΄ end the G-quadruplex decreased stability. We also compared the TEG modification effect on the G-quadruplex stability. The Tm values for TEG-pQ1 and pQ1 were 44.7 and 47.5°C, respectively, indicating that the position of TEG modification was critical for influencing DNA stability.
Figure 3.
Typical UV melting curves of (A) D1 (black), D1-(X4)7 (blue) and D1-(X4)10 (red) and (B) Q1 (black), Q1-(X4)4 (red), Q1-(X4)7 (orange) and Q1-(X4)13 (blue). The buffer was 100 mM KCl, 10 mM K2HPO4 (pH 7.0 at 25°C) and 1 mM K2EDTA. Strand concentrations were 5 μM.
To confirm the effect on different types of G-quadruplexes, we evaluated this modification in the context of an intramolecular mixed-type G-quadruplex according to the sequence of the human telomere (Q2) (Supplementary Figure S2a) and an intermolecular antiparallel G-quadruplex (Q3) (Supplementary Figure S2b). We evaluated a second TEG-modified hairpin (D2) (Supplementary Figure S2c) to investigate how the TEG-modified deoxythymidine influenced duplex stability with different nucleotides in the loop. As we expected, the modification stabilized the G-quadruplexes but destabilized the duplex compared with the unmodified DNAs (Supplementary Figure S3). Thus, the effects of TEG modifications of thymidines were independent of sequence but were dependent on the duplex or G-quadruplex.
We determined the thermodynamic parameters of the structures formed by the duplexes and G-quadruplexes (Table 1). According to the CD spectra of unmodified and modified DNAs as a function of temperature (Supplementary Figure S4), we estimated thermodynamic parameters using the two-state model. The free energy change during the formation of DNA structures (ΔG°25) for D1, D1-(X4)7 and D1-(X4)10 at 25°C were −6.6, −5.1 and −5.9 kcal mol−1, respectively (Table 1), indicating that TEG did not stabilize the hairpin. The stability difference of D1-(X4)7 and D1-(X4)10 may be due to base stacking of T7 and T10 with closing base pairs in the stem region of the hairpin. In contrast, the ΔG°25 values of G-quadruplexes formed by Q1, Q1-(X4)4, Q1-(X4)7 and Q1-(X4)13 were −3.5, −6.6, −4.2 and −5.7 kcal mol−1, respectively. Although Q1-(X4)7 was stabilized slightly compared with Q1, the differences in ΔH° and TΔS° values between Q1 and Q1-(X4)7 were within the experimental error range. Significantly increased stabilization of Q1-(X4)4 and Q1-(X4)13 was achieved when the TEG-modified deoxythymidines were in a position in which stacking was observed in the NMR study (20,21). In each case, the higher stability of TEG-modified G-quadruplexes compared with unmodified Q1 was imparted by a favorable enthalpic contribution (Table 1), indicating that TEG interacted with bases in the G-quadruplexes. Hence, we focused on the modification at position 4 and 7 of Q1 because the effect of the oligoethlene glycols for position 4 [Q1-(X4)4] and 7 [Q1-(X4)7] on the G-quadruplexes were drastically change.
The propynyl group stacks with bases in duplexes and triplexes (29,30). Therefore, we designed propynyl-modified deoxythymidine (Pro) (Supplementary Figure S5a) and incorporated this residue into duplex- and G-quadruplex-forming oligonucleotides. UV melting curves (Supplementary Figure S5b) demonstrate that propynyl modification slightly destabilized the duplex when incorporated at position 7 (D1-(Pro)7(Supplementary Table S2). In contrast, propynyl modification stabilized the G-quadruplex in a position-dependent manner imparted by a favorable enthalpic contribution (Q1-(Pro)4 and Q1-(Pro)7 (Supplementary Table S2). Stabilization imparted by a favorable enthalpic contribution was observed for TEG modification (Table 1), and the stabilization was larger for TEG modification than for propynyl modification.
TEG comprises hydrocarbons, ether groups and a hydroxyl group that interact via CH–π interactions, lone pair–π interactions and hydrogen bonds with the G-quadruplex. Therefore, we hypothesized that TEG in the loop of the G-quadruplex adopts a conformation suitable for interactions between TEG and the G-quadruplex. To confirm that TEG interacted with the G-quadruplex, the stabilities of Q1 and Q1-(X4)4 were measured in the presence of the G-quadruplex-stabilizing ligand NMN, which stacks on a G-quartet via π–π interactions (31) and was expected to compete with tethered TEG. Upon addition of NMN, the ΔG°25 value of Q1 decreased by 0.6 kcal mol−1 (Supplementary Figure S6). The ΔG°25 values of Q1-(X4)4, with or without NMN, were similar (Supplementary Figure S6), indicating that TEG stabilized Q1. This suggests that TEG in Q1-(X4)4 stacks on a G-quartet and that the interactions between the G-quartet and TEG are stronger compared with the π-π interaction between the G-quartet and NMN.
Effect of the number of hydroxyl groups and length of ethylene oxide on the stability of the G-quadruplex
We estimated the effect of the number of hydroxyl groups and length of the ethylene oxides in the G-quadruplex on its stability. The ΔG°25 values of Q1-(X4)4,13 and Q1-(X4)4,7,13 were −7.5 and −8.0 kcal mol−1, respectively (Table 1), indicating that increasing the number of positions modified by TEG groups increased with the stability of G-quadruplexes.
We investigated how longer ethylene oxides influenced the stability of the G-quadruplex. For this purpose, we synthesized G-quadruplexes with longer oligoethylene oxides (X8, X12 and X16) at positions T4 and T7 (Figure 1 and Supplementary Table S1). We confirmed that all modifications did not affect the overall architectures of the antiparallel G-quadruplexes (Supplementary Figure S7). The ΔG°25 values of Q1-(X8)4 and Q1-(X8)7 were −7.1 and −4.4 kcal mol−1, respectively (Table 1), indicating their higher stability compared with the structures with TEG modification at the same positions. Stabilization by OEG was imparted by a favorable enthalpic contribution (Table 1), suggesting that the hydrocarbons and ether groups in the ethylene oxides interacted with the G-quartet via CH-π and lone pair-π interactions. The ΔG°25 values of Q1-(X12)4 and Q1-(X16)4 were −6.5 and −6.3 kcal mol−1, respectively (Table 1). This indicates that the stabilization effect by oligoethylene glycol at T4 was saturated by the oligoethylene glycols. Because the oligoethylene glycol located in the narrow space in the two lateral loops of the G-quadruplex when the oligoethylene glycol is modified at T4, longer oligoethylene glycols may prevent favorable interactions. In contrast, the ΔG°25 values of Q1-(X12)7 and Q1-(X16)7 were −4.5 and −4.6 kcal mol−1, respectively (Table 1), indicating that the oligoethylene glycol modified at T7 may prevent favorable interactions, because the thymine base at position 7 flipped out.
These results indicate that the number of hydroxyl groups and the length of ethylene oxides are important for significant stabilization of the G-quadruplex by oligoethylene glycol modification.
NMR analysis for identifying the effect of TEG modification on G-quadruplex structures
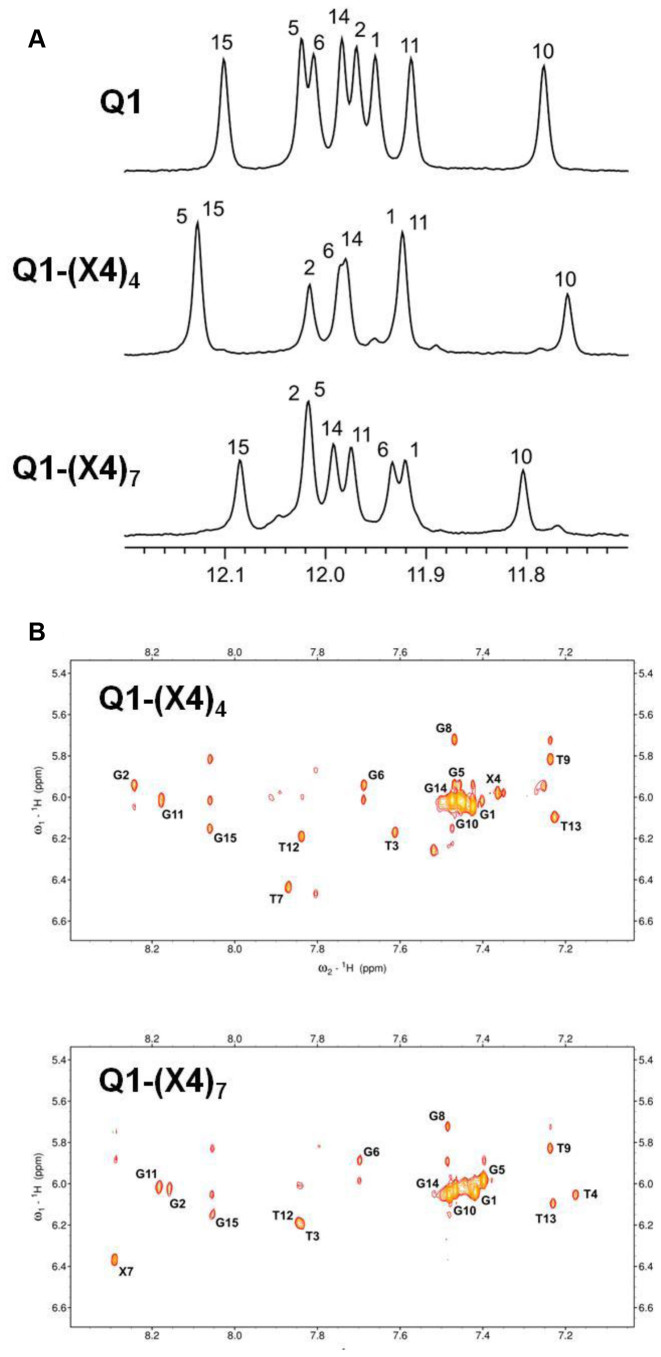
To analyze the interactions between TEG and DNA, NMR spectra of certain modified oligonucleotides were evaluated. The 1H NMR spectra of oligonucleotides Q1-(X4)4 and Q1-(X4)7 exhibited eight sharp imino resonances in the region from 11.7 to 12.2 ppm (Figure 4A), which indicates the formation of a single G-quadruplex species. 2D NOESY experiments were used to assign the relevant resonances of Q1-(X4)4 and Q1-(X4)7 (Figure 4B). NOE connectivity reveals that the topologies as well as syn/anti guanine conformations and hydrogen bond directionalities within G-quartets of Q1-(X4)4 or Q1-(X4)7 G-quadruplexes are the same as those in non-modified Q1 (21). In general, aromatic and anomeric chemical shifts in the spectra of Q1-(X4)4 and Q1-(X4)7 differed only slightly from the pattern of Q1. Only T3, T4 and T7 aromatic H6/H8 resonances in Q1-(X4)4 and Q1-(X4)7 shifted by ≥0.2 ppm compared with Q1 spectrum (Figure 4B). Further, anomeric H1΄ resonance shifts of Q1-(X4)4 and Q1-(X4)7 were within 0.1 ppm compared with the Q1 spectrum.
Figure 4.
(A) Imino regions of 1D 1H NMR spectra of Q1, Q1-(X4)4 and Q1-(X4)7 (in the presence of 100 mM KCl) with assignments. (B) Aromatic-anomeric regions of 2D NOESY spectra of Q1-(X4)4 and Q1-(X4)7. Intranucleotide H6/H8-H1΄ cross-peaks are labeled.
The imino peaks of modified oligonucleotides exhibited larger perturbations (Figure 4A). The imino pattern of Q1-(X4)4 with the TEG group on T4, which was near the G-quartet with two edge-type loops, exhibited a noticeable downfield shift of G5 imino resonances compared with the spectrum of unmodified Q1. Although modification of T4 in Q1-(X4)4 might have interfered with T4:T13 base pairing, no significant change in the G14 imino chemical shift was observed. The largest imino resonance shifts in Q1-(X4)7, compared with the spectrum of Q1, were downfield shifts of G10 and G11. It appears that the positioning of the TEG group exerts a larger influence on the diagonal G-tract than on the adjacent G6. We performed 2D NOESY NMR experiments to determine the spatial organization of the TEG groups with respect to the DNA. However, no NOE cross-peaks were observed between TEG protons and aromatic, amino or imino protons in Q1-(X4)4 or Q1-(X4)7. These analyses indicate that the TEG groups adopted multiple conformations.
Computational simulations to determine the effect of TEG modification on G-quadruplex structures
To understand how TEG interacts with G-quadruplexes, we performed 35-ns MD calculations of Q1, Q1-(X4)4 and Q1-(X4)7. To obtain the equilibrated structures from MD trajectories, we analyzed the temporal changes in root mean square deviations (RMSDs) of the sugar-phosphate backbones (Supplementary Figure S8). We acquired 24 000 snapshots (11–35 ns) to understand TEG-induced stabilization. Supplementary Movies S1 and 2 show the results of MD calculations for Q1-(X4)4 and Q1-(X4)7, respectively. In the simulations indicated by the NMR analyses, TEG was dynamic; however, the ethylene oxides in the TEG group of Q1-(X4)4 were located close to T13 during most of the simulation because of CH–π and Lone pair–π interactions (Supplementary Movie S1). Moreover, in Q1-(X4)7, the formation of a hydrogen bond was observed, and the hydroxyl group of TEG in Q1-(X4)7 formed a hydrogen bond with the phosphate moiety at G6 (Supplementary Movie S2). These interactions with TEG may therefore stabilize G-quadruplexes.
Snapshots taken during MD simulations were classified into 10 groups (clusters), depending on the RMSD values for all TEG atoms, using the hierarchical agglomerative clustering method (Supplementary Figures S9 and 10). NMR analysis shows that the TEG groups adopted multiple conformations; however, our simulations suggest that key TEG conformations stabilize the G-quadruplex. For Q1-(X4)4, 33.4% of snapshots grouped into cluster 1 (Figure 5A and Supplementary Figure S9a). In these snapshots, TEG bridged the two lateral loops (Figure 5A). Moreover, TEG adopted the bridged conformation in ∼59.1% of snapshots (clusters 1, 4, 5, 7 and 8) (Figure 5A and Supplementary Figure S9b). Further, the positions of the TEG groups in Q1-(X4)4 in MD simulations suggest that hydrogen bonds formed between the hydroxyl group of TEG and phosphate oxygens of the G-quadruplexes. In contrast, in the simulation of Q1-(X4)7, 35.3% of snapshots grouped into cluster 1 (Figure 5B and Supplementary Figure S10a) and there was no similar conformation among other clusters of Q1-(X4)7 (Supplementary Figure S10b).
Figure 5.
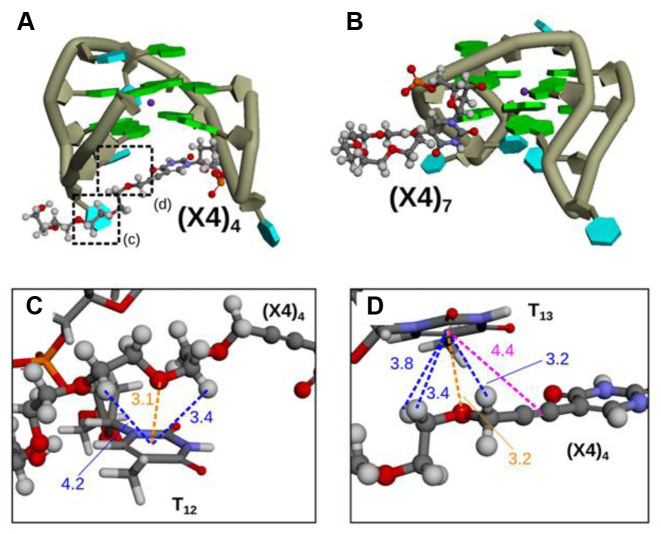
Representative conformations observed in (A) Q1-(X4)4 (cluster 1 in Supplementary Figure S9a) and (B) Q1-(X4)7 (cluster 1 in Supplementary Figure S10a). The backbone is shown in dark green, and guanine and thymine bases are shown as green and cyan blocks, respectively. K+ is represented by purple spheres. The X4 atoms are shown in ball-and-stick representation with orange for phosphorus, red for oxygen, blue for nitrogen, gray for carbon and white for hydrogen. (C and D) Enlarged view in Figure 5A. Blue, orange and pink dashed lines are CH–π interactions, lone pair–π interactions and π–π interaction, respectively. The values show distance (Å) between interacted regions.
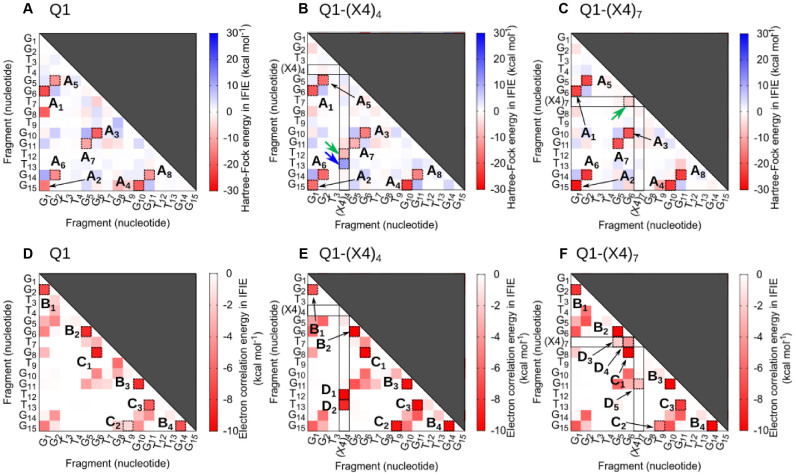
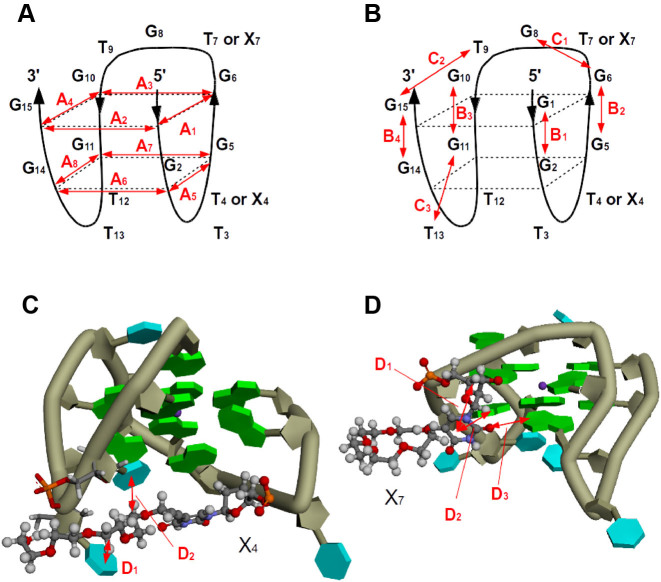
We calculated the electronic states of Q1, Q1-(X4)4 and Q1-(X4)7 using the fragment molecular orbital (FMO) method (32) at the MP2/6-31G(d) level. The nucleotides were divided into fragments (Supplementary Figure S11), and inter-fragment interaction energies (IFIEs) were calculated (Figure 6). First, we analyzed IFIEs for all fragment pairs for contributions from Hartree–Fock energies (EHF), which mainly reflect electrostatic interactions associated with the formation of hydrogen bonds and dipole–dipole interactions. Moreover, electron correlation energies (Ecorr), which represent the dispersion forces imparted by π–π stacking, CH–π interactions and lone pair–π interactions were estimated. The schematics of G-quadruplex structures and attractive interactions between DNA and TEG from Figure 6 are shown in Figure 7A and B (labels A1–A8, B1–B4 and C1–C3 in Figure 6) and in Figure 7C and D (labels D1–D3 in Figure 6).
Figure 6.
Interaction energies calculated from analysis of Hartree–Fock energies (kcal mol−1) for (A) Q1, (B) Q1-(X4)4 and (C) Q1-(X4)7, and electron correlation energies (kcal mol−1) between bases for (D) Q1, (E) Q1-(X4)4 and (F) Q1-(X4)7. The fragment number corresponds to the position of the nucleotide. Red and blue squares indicate attractive and repulsive interactions shown on scale on the right, respectively. Dashed squares labeled A and B indicate interactions within a G-quartet and between G-quartets, respectively (Figure 7A and B). Dashed squares labeled C indicate interactions of loop bases with G-quartets (Figure 7B). Dashed squares labeled D indicate specific interaction with TEG modified thymine (Figure 7C and D).
Figure 7.
Attractive interactions in the G-quadruplex for (A) Hartree–Fock energies in Figures 6A C, (B) electron correlation energy in Figures 6D-F and (C-D) electron correlation energy of TEG regions in Q1-(X4)4 (Figure 6E) and Q1-(X4)7 (Figure 6F). Double-headed arrows correspond to the dashed squares in Figure 6.
There were regions of low EHF (deep red points labeled A1 to A8) (Figure 6A–C). The interactions were mediated by hydrogen bond formation in G-quartets, and there was no critical difference among Q1, Q1-(X4)4 and Q1-(X4)7 (A1–A8 in Figure 6A–C). However, there were specific interactions with TEG (Figure 6B and C, green and blue arrows). Snapshot analysis showed that hydrogen bond formation between the hydroxyl group of TEG and a phosphate group in the G-quadruplex (Figure 6B and Supplementary Figure S12a and b; green lines) and the repulsion between T12 and X4 in thymine oxygens (Figure 6B, blue arrow; Supplementary Figure S12a and c) contributed to favorable and unfavorable interactions, respectively. Moreover, hydrogen bond formation between the thymines in X7 and G5 was observed (Figure 6C and Supplementary Figure S13a and b; green line). Therefore, hydrogen bonds likely play important roles in stabilizing G-quadruplexes through TEG modification.
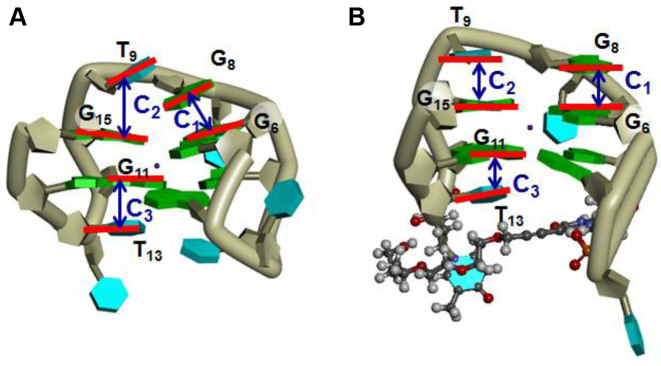
In contrast, the Ecorr values associated with the stacking interactions in the G-quartets (B1–B4 in Figure 6D–F) for Q1, Q1-(X4)4 and Q1-(X4)7 showed similar trends. The Ecorr values labeled C1, C2 and C3 for Q1-(X4)4 were clearly lower compared with those for Q1 and Q1-(X4)7 (Figure 6D–F). For example, the Ecorr values labeled C1, C2 and C3 for Q1 were −9.0, −1.6 and −6.6 kcal mol−1, and those for Q1-(X4)4 were −15.5, −9.3 and −10.5 kcal mol−1. The red bars in snapshots for Q1 and Q1-(X4)4 (Figure 8) show the orientations of bases in G-quartets and bases in loops, which stack on the G-quartets. The guanine bases in the G-quartets in Q1-(X4)4 were parallel, whereas those in Q1 were not. Moreover, loop bases G8 and T9 in Q1-(X4)4 were parallel to G15 and G6 (Figure 8B), respectively, indicating strong stacking interactions (C1 and C2) (Figure 6E). T13 was pushed up by the TEG group in Q1-(X4)4; therefore, the distance between G11 and T13 in Q1-(X4)4 was shorter than that in Q1 (C3 in Figure 6E), suggesting that the stacking interactions were enhanced by TEG modification.
Figure 8.
The structures of (A) Q1 and (B) Q1-(X4)4. (A) Snapshot at 35 ns. (B) Representative conformation of cluster 1 (Figure 5A). Dark-green tubes and green and cyan blocks indicate backbone, guanine and thymine, respectively. Orange, red, gray, blue and white balls indicate phosphorus, oxygen, carbon, nitrogen and hydrogen atoms of X4, respectively. Red lines show the orientations of bases in G-quadruplexes. Blue arrows indicate interactions between the loop bases and G-quartets shown in Figures 6 and 7.
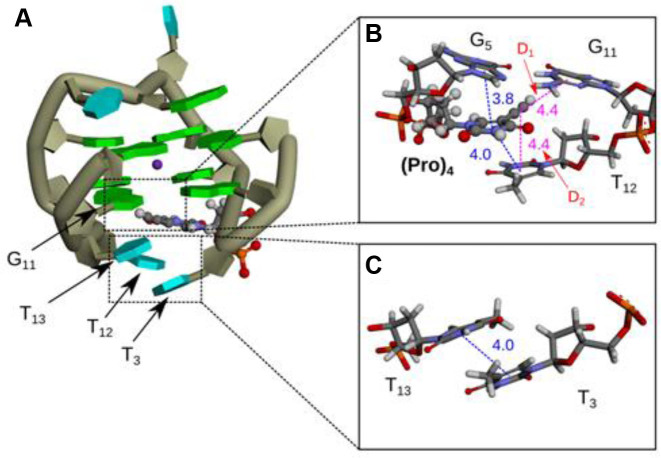
Moreover, interactions imparted by TEG-modified deoxythymidine induced lower Ecorr values (Figure 5, D1–D4 in Figure 6D and E). T-shaped π–π and dipole-induced dipole interactions were observed between TEG and G5 or G11 in Q1-(X4)7, which correspond to D3 and D4 (Supplementary Figure S13c). Figure 5C and D show enlarged views of the interactions between TEG and T12 or T13in Q1-(X4)4, which correspond to D1 and D2 (Figure 6E). The hydrocarbons in TEG group in Q1-(X4)4 formed CH–π and lone pair–π interactions with T12 or T13 (Figure 5C and D). The CH–π and lone pair–π interactions had low Ecorr values, reflected by the stabilizing interactions of D1 and D2. Moreover, the interaction of D1 for Q1-(X4)4 involved π-π interactions between the propynyl group of TEG and the base of T12.
Importance of oligoethylene glycols for stabilizing G-quadruplexes
The propynyl group stabilizes DNA structures via stacking interactions. Therefore, we performed MD calculations for Q1-(Pro)4 (Figure 9; Supplementary Figures S14 and 15). As shown in Figure 9, Supplementary Figure S15a and b, the hydrogen bonds in G-quartets (A1–A8) and stacking interactions between G-quartets (B1–B4) followed the same trends that were observed for Q1, Q1-(X4)4 and Q1-(X4)7 (Figure 6). However, there were differences among interactions between the bases around G-quartets and guanine bases of the G-quartet (C1–C3). (Pro)4 interacted with G11 (D1, −4.1 kcal mol−1) and T12 (D2, −7.5 kcal mol−1) (Supplementary Figure S15b). The propynyl group in Q1-(Pro)4 was located near G5 and G11, and the thymine in (Pro)4 stacked on G5 (Supplementary Figure S15a and b), suggesting π–π interactions between the π electrons of the base of G11 and triple bond in (Pro)4. A similar interaction has been reported in benzene and acetylene clusters (33). Therefore, these stacking interactions involving (Pro)4 may stabilize the G-quadruplex. However, the propynyl group in Q1-(X4)4 was not located near G5 (Figure 5A), suggesting that the position of the propynyl group in Q1-(X4)4 differed from that in Q1-(Pro)4 (Figures 5A and 9A). Therefore, the stacking interactions between the propynyl group and bases in Q1-(X4)4 were weaker than the interactions between TEG and bases. Our results indicate that the interactions of G-quadruplexes with TEG should be more favorable compared with those with the propynyl group.
Figure 9.
(A) Representative structure of Q1-(Pro)4 at 31 ns and (B and C) enlarged views around (Pro). Green and cyan blocks and the dark-green tube represent guanine, thymine and the backbone of the G-quadruplex, respectively. Orange, red, blue, gray and white sticks or balls illustrate phosphorus, oxygen, nitrogen, carbon and hydrogen atoms, respectively. Blue and pink dashed lines indicate π–π stacking interactions and π–π interactions, respectively. The values show the distances in Å between interacting regions.
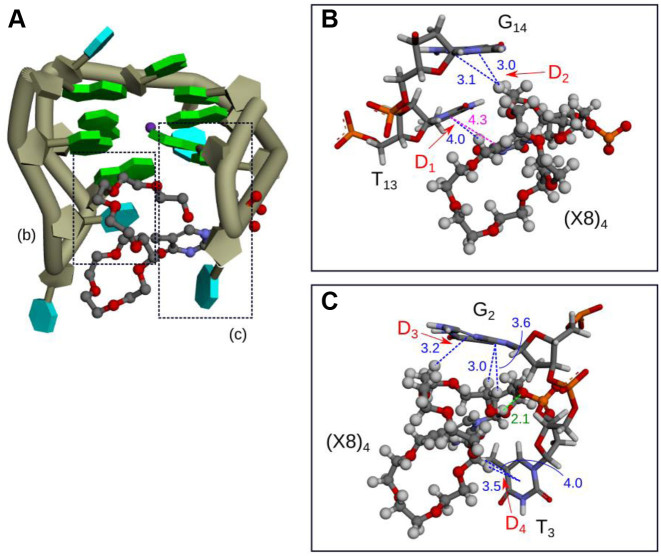
Moreover, we analyzed Q1-(X8)4 structures. Supplementary Movie S3 shows the MD trajectory of Q1-(X8)4 in equilibrium. The OEG group of X8 was flexible as was X4 in Q1-(X4)4 and Q1-(X4)7. We clustered snapshots of Q1-(X8)4 according to the OEG conformations (Supplementary Figure S16). The ethylene oxide chain of OEG rotated around the G-quartet and fit between two loops in the G-quadruplex (Supplementary Figure S16). Figure 10 shows the typical OEG conformations of Q1-(X8)4. The ethylene oxides in OEG were located close to bases in the loop of the G-quadruplex at distances indicative of CH–π and lone pair–π interactions. Snapshots of cluster 1 of Q1-(X8)4 reveal that hydrogen bonds in G-quartets (A1–A9) (Supplementary Figure S17a), stacking interactions between G-quartets (B1–B4) (Supplementary Figure S17b) and stacking interactions between the bases around the G-quartet were similar to those in Q1-(X4)4 cluster 1. These results indicate that the longer chain pushed up the G-quartets. Further, specific interactions of D1–D4 were observed in Q1-(X8)4 (Supplementary Figure S17b). Although the interaction of D1 (between T13 and X8) was the same as that observed in Q1-(X4)4, the interactions of D2, D3 and D4 were specific to the longer oligoethylene glycol-modified deoxythymine (Supplementary Figure S17).
Figure 10.
(A) Representative structure of cluster 1 for Q1-(X8)4 and (B and C) enlarged views around X8. Green and cyan blocks and the dark-green tube indicate guanine, thymine and the backbone of Q1, respectively. Orange, red, blue, gray and white sticks or spheres illustrate phosphorus, oxygen, nitrogen, carbon and hydrogen atoms, respectively. Green, blue and violet dashed lines are hydrogen bonds, π–π stacking interactions, and π–π interactions, respectively. The values show the distances in Å between interacting regions.
These results show that OEG induced stabilization of G-quadruplexes compared with TEG because of its numerous donor sites for CH–π and lone pair–π interactions and the donor and acceptor sites. However, the ΔG°25 values of Q1-(X8)4, Q1-(X12)4 and Q1-(X16)4 were −7.1, −6.5 and −6.3 kcal mol−1, respectively (Table 1). The longer oligoethylene glycols DEG and HEG comprise numerous hydrocarbon moieties that form CH–π interactions. The conformations of OEG observed in MD simulations, therefore, reveal the mechanism of G-quadruplex stabilization.
Biological significance of G-quadruplex stabilization by oligoethylene glycols
Synthetic saccharides with hydrocarbons stacked onto the ends of DNA duplexes via CH–π interactions stabilize the duplexes, (34) suggesting that CH–π interactions of TEG-modified deoxythymidine affect duplex stability. Our MD and FMO calculations show the importance of TEG positions for stabilizing DNA structures. TEG-modified deoxythymidine specifically stabilized the G-quadruplex but not the G-duplex, because the TEG positions in the duplex may be unsuitable for the formation of CH–π interactions. Electrostatic interactions, hydrogen bonding and stacking interactions are the best understood non-covalent interactions between DNA and proteins, metabolites and drugs (35). CH–π interactions exert multiple effects, and these interactions are critical to the association of organic small molecules, (36) self-assembly of super molecules, (37) interactions between drugs and proteins (38) and associations between sugars and proteins (39). However, the impact of these interactions on DNA structures is not considered significant. Numerous co-solutes with hydrocarbons are present in cells, such as polysaccharides and metabolites. The structure and stability of DNA in a solution with co-solutes, such as oligoethylene glycols, PEGs and polysaccharides, were investigated to understand the effect of intercellular co-solutes on DNA function in cells. For example, evidence indicates that co-solutes change DNA function through alterations in physical properties of the solution rather than through specific interactions with DNA (3). However, co-solutes may directly interact with DNA (40,41). We reported that co-solutes alter DNA structure and stability through enthalpic contributions, (3) suggesting the possibility of direct interaction of co-solutes with DNA.
Our present results suggest that the oligoethylene glycol specifically interacts with DNA via CH–π and lone pair–π interactions. Further, oligoethylene glycols covalently linked to DNA interacted with G-quadruplexes and increased the stability of these DNA structures compared with free oligoethylene glycols in solution, which was imparted by entropic contributions. Our analyses of these modified DNAs suggest that oligoethylene glycols may interact in a structure-specific manner with DNA. Therefore, our present results will facilitate understanding PEG–DNA interactions as well as co-solute–DNA interactions such as CH–π interactions between DNA and co-solutes that regulate the function of DNA in cells. A significant finding of the present study demonstrates that PEG changed the stability of DNAs with structural specificity. Our results are therefore applicable to the design and control of DNA structures within the cell to regulate biological reactions via the formation of non-canonical structures.
Supplementary Material
ACKNOWLEDGEMENTS
Financial support by JSPS and Slovenian Research Agency under JSPS Bilateral Joint Research Program to the two P.I.s is also gratefully acknowledged. The computations were performed using Research Center for Computational Science, Okazaki, Japan. We thank Mrs Y. Kametani and Ms M. Ito for collecting the UV melting data and Dr Rainy Chowdhury for supporting the synthesis of modified deoxythymidine. Special thanks to Dr M. Trajkovski for the NMR measurement of Q1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-Aid for Scientific Research, Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan (in part); MEXT-Supported Program for the Strategic Research Foundation at Private Universities (2014-2019), Japan; JSPS and Slovenian Research Agency under JSPS Bilateral Joint Research Program; Hirao Taro; Okazaki Kazuo Foundation of KONAN GAKUEN for Advanced Scientific Research; the Chubei Itoh Foundation; Foundation for the promotion of ion engineering. The open access publication charge for this paper has been waived by Oxford University Press—NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Zimmerman S.B., Minton A.P.. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 1993; 22:27–65. [DOI] [PubMed] [Google Scholar]
- 2. Hall D., Minton A.P.. Macromolecular crowding: qualitative and semiquantitative successes, quantitative challenges. Biochim. Biophys. Acta. 2003; 1649:127–139. [DOI] [PubMed] [Google Scholar]
- 3. Nakano S., Miyoshi D., Sugimoto N.. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem. Rev. 2014; 114:2733–2758. [DOI] [PubMed] [Google Scholar]
- 4. Tateishi-Karimata H., Nakano S., Sugimoto N.. Quantitative analyses of nucleic Acid stability under the molecular crowding condition induced by cosolutes. Curr. Protoc. Nucleic Acid Chem. 2013; doi:10.1002/0471142700.nc0719s53. [DOI] [PubMed] [Google Scholar]
- 5. Goobes R., Kahana N., Cohen O., Minsky A.. Metabolic buffering exerted by macromolecular crowding on DNA-DNA interactions: origin and physiological significance. Biochemistry. 2003; 42:2431–2440. [DOI] [PubMed] [Google Scholar]
- 6. Spink C.H., Chaires J.B.. Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry. 1999; 38:496–508. [DOI] [PubMed] [Google Scholar]
- 7. Mergny J.L., Helene C.. G-quadruplex DNA: a target for drug design. Nat. Med. 1998; 4:1366–1367. [DOI] [PubMed] [Google Scholar]
- 8. Tateishi-Karimata H., Isono N., Sugimoto N.. New insights into transcription fidelity: thermal stability of non-canonical structures in template DNA regulates transcriptional arrest, pause, and slippage. PLoS One. 2014; 9:e90580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Endoh T., Kawasaki Y., Sugimoto N.. Suppression of gene expression by G-quadruplexes in open reading frames depends on G-quadruplex stability. Angew. Chem. Int. Ed. Engl. 2013; 52:5522–5526. [DOI] [PubMed] [Google Scholar]
- 10. Mitchell T., Ramos-Montoya A., Di Antonio M., Murat P., Ohnmacht S., Micco M., Jurmeister S., Fryer L., Balasubramanian S., Neidle S. et al. Downregulation of androgen receptor transcription by promoter g-quadruplex stabilization as a potential alternative treatment for castrate-resistant prostate cancer. Biochemistry. 2013; 52:1429–1436. [DOI] [PubMed] [Google Scholar]
- 11. Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H.. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang J.Y., Zheng K.W., Xiao S., Hao Y.H., Tan Z.. Mechanism and manipulation of DNA:RNA hybrid G-quadruplex formation in transcription of G-rich DNA. J. Am. Chem. Soc. 2014; 136:1381–1390. [DOI] [PubMed] [Google Scholar]
- 13. Rodriguez R., Pantos G.D., Goncalves D.P., Sanders J.K., Balasubramanian S.. Ligand-driven G-quadruplex conformational switching by using an unusual mode of interaction. Angew. Chem. Int. Ed. Engl. 2007; 46:5405–5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tateishi-Karimata H., Muraoka T., Kinbara K., Sugimoto N.. G-quadruplexes with tetraethylene glycol-modified deoxythymidines are resistant to nucleases and inhibit HIV-1 reverse transcriptase. Chembiochem. 2016; 17:1399–1402. [DOI] [PubMed] [Google Scholar]
- 15. Lachelt U., Wagner E.. Nucleic Acid Therapeutics Using Polyplexes: a Journey of 50 Years (and Beyond). Chem. Rev. 2015; 115:11043–11078. [DOI] [PubMed] [Google Scholar]
- 16. Turecek P.L., Bossard M.J., Schoetens F., Ivens I.A.. PEGylation of biopharmaceuticals: a review of chemistry and nonclinical safety information of approved drugs. J. Pharm. Sci. 2016; 105:460–475. [DOI] [PubMed] [Google Scholar]
- 17. Rameez S., Palmer A.F.. Simple method for preparing poly(ethylene glycol)-surface-conjugated liposome-encapsulated hemoglobins: physicochemical properties, long-term storage stability, and their reactions with O2, CO, and NO. Langmuir. 2011; 27:8829–8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Litzinger D.C., Buiting A.M., van Rooijen N., Huang L.. Effect of liposome size on the circulation time and intraorgan distribution of amphipathic poly(ethylene glycol)-containing liposomes. Biochim. Biophys. Acta. 1994; 1190:99–107. [DOI] [PubMed] [Google Scholar]
- 19. Wawro A.M., Muraoka T., Kinbara K.. Chromatography-free synthesis of monodisperse oligo(ethylene glycol) mono-p-toluenesulfonates and quantitative analysis of oligomer purity. Polym. Chem. 2016; 7:2389–2394. [Google Scholar]
- 20. Macaya R.F., Schultze P., Smith F.W., Roe J.A., Feigon J.. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:3745–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trajkovski M., Sket P., Plavec J.. Cation localization and movement within DNA thrombin binding aptamer in solution. Org. Biomol. Chem. 2009; 7:4677–4684. [DOI] [PubMed] [Google Scholar]
- 22. Richards E.G., In Fasman G.D.. Handbook of Biochemistry and molecular Biology: Nucleic Acids. 1975; 1, 3rd edn, Cleveland: CRC Press. [Google Scholar]
- 23. Tateishi-Karimta H., Sugimoto N.. Control of stability and structure of nucleic acids using cosolutes. Methods. 2014; 67:151–158. [DOI] [PubMed] [Google Scholar]
- 24. Marathias V.M., Bolton P.H.. Structures of the potassium-saturated, 2:1, and intermediate, 1:1, forms of a quadruplex DNA. Nucleic Acids Res. 2000; 28:1969–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Case D.A., Berryman J.T., Betz R.M., Cerutti D.S., Cheatham T.E. III, Darden T.A., Duke R.E., Giese T.J., Gohlke H., Goetz A.W. et al. AMBER14. 2015; San Francisco: University of California. [Google Scholar]
- 26. Pérez A., Marchán I., Svozil D., Sponer J., Cheatham T.E., Laughton C.A., Orozco M.. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007; 92:3817–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ryckaert J.-P., Ciccotti G., Berendsen H.J.C.. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1997; 23:327–341. [Google Scholar]
- 28. Miyamoto S., Kollman P.A.. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992; 13:952–962. [Google Scholar]
- 29. Cassidy R.A., Kondo N.S., Miller P.S.. Triplex formation by psoralen-conjugated chimeric oligonucleoside methylphosphonates. Biochemistry. 2000; 39:8683–8691. [DOI] [PubMed] [Google Scholar]
- 30. Barnes T.W. 3rd, Turner D.H.. Long-range cooperativity in molecular recognition of RNA by oligodeoxynucleotides with multiple C5-(1-propynyl) pyrimidines. J. Am. Chem. Soc. 2001; 123:4107–4118. [DOI] [PubMed] [Google Scholar]
- 31. Nicoludis J.M., Miller S.T., Jeffrey P.D., Barrett S.P., Rablen P.R., Lawton T.J., Yatsunyk L.A.. Optimized end-stacking provides specificity of N-methyl mesoporphyrin IX for human telomeric G-quadruplex DNA. J. Am. Chem. Soc. 2012; 134:20446–20456. [DOI] [PubMed] [Google Scholar]
- 32. Tanaka S., Mochizuki Y., Komeiji Y., Okiyama Y., Fukuzawa K.. Electron-correlated fragment-molecular-orbital calculations for biomolecular and nano systems. Phys. Chem. Chem. Phys. 2014; 16:10310–10344. [DOI] [PubMed] [Google Scholar]
- 33. Majumder M., Mishra B.K., Sathyamurthy N.. CH...π and π...π interaction in benzene–acetylene clusters. Chem. Phys. Lett. 2013; 557:59–65. [Google Scholar]
- 34. Lucas R., Gomez-Pinto I., Avino A., Reina J.J., Eritja R., Gonzalez C., Morales J.C.. Highly polar carbohydrates stack onto DNA duplexes via CH/pi interactions. J. Am. Chem. Soc. 2011; 133:1909–1916. [DOI] [PubMed] [Google Scholar]
- 35. Saenger W. Principles of Nucleic Acid Structure. 1984; NY: Springer-Verlag. [Google Scholar]
- 36. Obenchain D.A., Bills B.J., Christenholz C.L., Elmuti L.F., Peebles R.A., Peebles S.A., Neill J.L., Steber A.L.. C-H...pi interactions in the CHBrF2...HCCH weakly bound dimer. J. Phys. Chem. A. 2011; 115:12228–122234. [DOI] [PubMed] [Google Scholar]
- 37. Chun Y., Singh N.J., Hwang I.C., Lee J.W., Yu S.U., Kim K.S.. Calix[n]imidazolium as a new class of positively charged homo-calix compounds. Nat. Commun. 2013; 4:1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kondo J., Koganei M., Kasahara T.. Crystal structure and specific binding mode of sisomicin to the bacterial ribosomal decoding site. ACS Med. Chem. Lett. 2012; 3:741–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hudson K.L., Bartlett G.J., Diehl R.C., Agirre J., Gallagher T., Kiessling L.L., Woolfson D.N.. Carbohydrate-aromatic interactions in proteins. J. Am. Chem. Soc. 2015; 137:15152–15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Buscaglia R., Miller M.C., Dean W.L., Gray R.D., Lane A.N., Trent J.O., Chaires J.B.. Polyethylene glycol binding alters human telomere G-quadruplex structure by conformational selection. Nucleic Acids Res. 2013; 41:7934–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nakano M., Tateishi-Karimata H., Tanaka S., Tama F., Miyashita O., Nakano S., Sugimoto N.. Thermodynamic properties of water molecules in the presence of cosolute depend on DNA structure: a study using grid inhomogeneous solvation theory. Nucleic Acids Res. 2015; 43:10114–10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.