Abstract
The use of synthetic messenger ribonucleic acid (mRNA) to express specific proteins is a highly promising therapeutic and vaccine approach that avoids many safety issues associated with viral or DNA-based systems. However, in order to optimize mRNA designs and delivery, technology advancements are required to study fundamental mechanisms of mRNA uptake and localization at the single-cell and tissue level. Here, we present a single RNA sensitive fluorescent labeling method which allows us to label and visualize synthetic mRNA without significantly affecting function. This approach enabled single cell characterization of mRNA uptake and release kinetics from endocytic compartments, the measurement of mRNA/protein correlations, and motivated the investigation of mRNA induced cellular stress, all important mechanisms influencing protein production. In addition, we demonstrated this approach can facilitate near-infrared imaging of mRNA localization in vivo and in ex-vivo tissue sections, which will facilitate mRNA trafficking studies in pre-clinical models. Overall, we demonstrate the ability to study fundamental mechanisms necessary to optimize delivery and therapeutic strategies, in order to design the next generation of novel mRNA therapeutics and vaccines.
INTRODUCTION
The use of exogenous, in vitro synthesized mRNA as an expression vector for therapeutic or antigenic proteins is highly promising. Expression of mRNA-encoded proteins is transient and more direct than DNA-based vectors, which requires intermediate steps such as nuclear localization and transcription. Additionally, mRNA vectors do not pose safety risks such as genomic integration, antibiotic resistance, or strong immunogenic responses due to a replicating vector (1).
Exogenous mRNA has been successfully utilized to generate proteins in both cell culture and in vivo (2,3). In order to obtain therapeutic levels of protein expression, strategies for improving in-vitro transcribed (IVT) mRNA, such as through the incorporation of modified nucleosides (4–8) and purification methods (9), have been the subject of intense study. Despite these improvements, mechanistic studies of mRNA delivery, protein production and innate immune activation at the single cell and single molecule level are needed. The primary reason why these studies have not been performed is the lack of approaches to measure cellular mRNA uptake without compromising translational potential, interactions with cellular proteins or altering the uptake pathway (10). Current studies are limited to direct-nucleotide labeling methods and the use of mathematical models to approximate mRNA–protein correlations (11,12). Lorenz et al. provided evidence that naked mRNA delivered in vitro enters cells via receptor-mediated endocytosis and predominantly remains in endosomes (13). Even though this mRNA was not functional, which is a significant drawback of direct labeling, this work highlighted the importance of tracking the subcellular location of delivered mRNA, and, in particular, the number of molecules that reach the cytosolic compartment, the cellular site of translation. In the siRNA community, the inability to measure cytosolic levels of siRNA has greatly limited the optimization of siRNA-based therapeutics, and only recently this barrier was overcome but using low throughput methods such as electron microscopy or single vesicle tracking (14,15); the approach presented here for mRNA, allows for a more rapid, quantitative assessment of cytoplasmic delivery.
Given the enormous potential for mRNA therapeutics and vaccines, we developed a strategy to address these limitations. Here, we present a general methodology for characterizing delivered mRNA at the level of single cells and single RNAs, in vitro and in vivo. We first constructed fluorescent imaging probes, multiply labeled tetravalent RNA imaging probes (MTRIPs), which bind, via nucleic-acid hybridization, to the 3΄ UTR region of synthetic mRNA (16–20). The 3΄ UTR of mRNA was used as a binding site in order to preclude any interference with ribosomal loading or progression, and thus prevent interference with translation. Due to the small size of the 3΄ UTR of most mRNAs, typically ∼100nt long, a limited number of probes can be bound to the mRNA thus multi-fluorophore probes are necessary to ensure adequate brightness. MTRIPs carry 8–10 fluorophores per probe and when 2–3 probes are bound to an mRNA, allow for 20–30 fluorophores per mRNA, making them easily visible with most fluorescent microscopes and in tissue. In previous publications, we successfully utilized MTRIPs to visualize endogenous mRNAs, viral genomic RNA, as well as plasmid-derived transcripts. Other approaches, such as molecular beacons, etc., only contain one fluorophore per probe, which given the short 3΄-UTR, would limit labeled-mRNA brightness and thus the detection of mRNAs. Labeling of mRNA with MTRIPs, in contrast to covalent incorporation of fluorophores, does not significantly affect the behavior or localization of target mRNA and does not illicit cellular immune responses (27,29–32). Finally, this method does not require large sequence incorporations into the mRNA itself or co-expression with reporters, such as with the MS2 aptamer and MS2-like systems (21).
In order to use MTRIPs to label IVT mRNA, we optimized binding conditions between the mRNA and MTRIP probes through an iterative process where temperature, salt concentration, incubation time and filtration method were varied. Heat was applied to remove secondary structure in the mRNA. Salt concentration allowed stabilization of the hybridization reaction over time. Finally, filtration was used to remove excess probe while minimizing loss of mRNA (see Materials and Methods). We labeled in vitro transcribed mRNA consistently with approximately two probes per mRNA and verified that they do not significantly affect translation.
We then applied this labeling strategy to perform a mechanistic characterization of mRNA delivery in cells. Key mechanisms of importance are noted in Figure 1A, including mRNA entry pathway, cytoplasmic release, translational efficiency (RNA/protein correlation), and PKR activation. Here, we demonstrate the ability of our tools to address these mechanisms. Fully addressing each mechanism across all conditions and cell types is beyond the scope of this work, but the measurements presented set the stage for future studies.
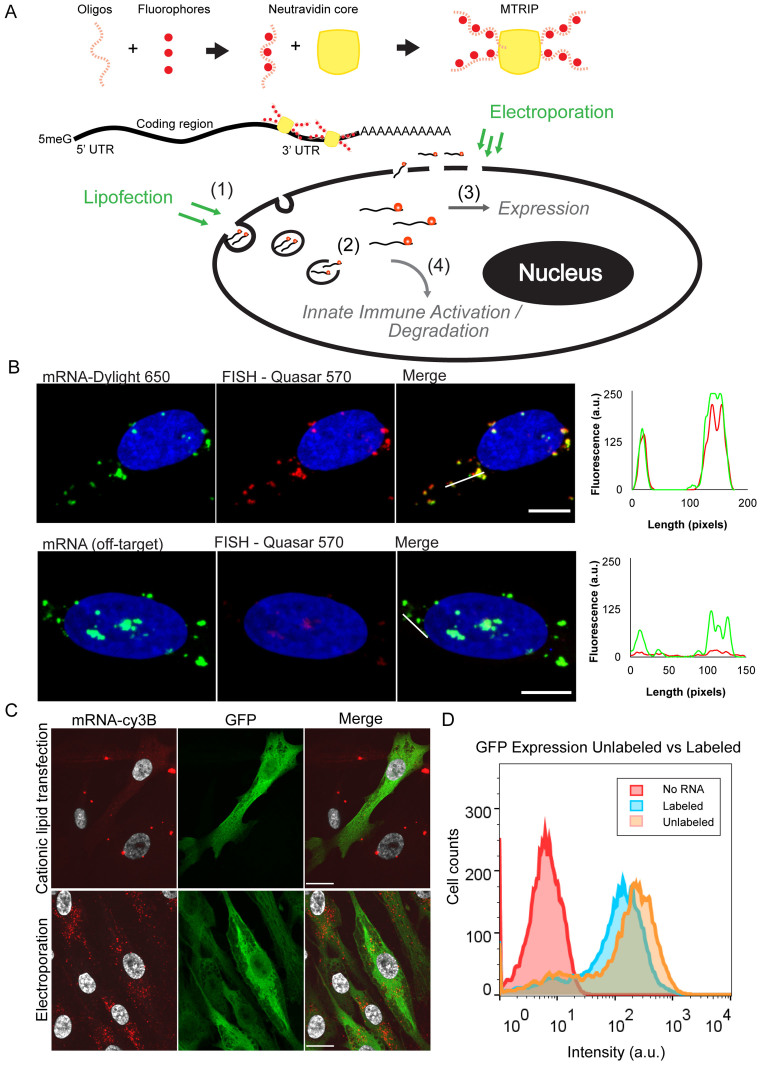
Figure 1.
mRNA labeling, validation, and transfection into cells using cationic lipids or electroporation. (A) Illustrative diagram of mRNA labeling and delivery. MTRIPS are composed of four biotinylated and fluorescently labeled oligonucleotides assembled on a Neutravidin core. They bind to IVT mRNA in the 3΄ UTR region. Fluorescently labeled mRNA is electroporated or lipofected into cells. Electroporated mRNA is immediately available in the cytosol, while lipofected mRNA must first travel through endosomes until released. Degradation and sequestration of mRNA reduces final expression of the target protein. Highlighted mechanisms of importance include (i) mRNA entry pathway, (ii) cytoplasmic release, (iii) translational efficiency (RNA/protein correlation) and (iv) innate immune (PKR) activation. (B) Labeled mRNA (green) delivered via lipofection colocalized with Stellaris FISH probe signal (red). Line profiles are indicated by white lines. No FISH signal was visible in cells transfected with an mRNA containing a different coding region. (scale bar = 10 μm). (C) Cationic lipid transfection and electroporation with labeled mRNA encoding EGFP resulted in similar levels of protein production (green) but distinct subcellular distributions of mRNA (red) at 2 h post-transfection. Upon electroporation, small mRNA granules were observed throughout the cell cytoplasm, while cationic lipid transfection resulted in few large mRNA granules (scale bar = 20 μm). (D) Flow cytometry of EGFP expression using labeled versus unlabeled EGFP mRNA showed only a slight reduction in protein expression.
We started with entry pathway, followed by whole cell quantification of mRNA uptake, and the quantification of the fraction of mRNA released into the cytosolic compartment. As a model system, we used primary human skeletal muscle cells (HSkMC) in the myoblast stage of development, because IM injection is the most commonly used delivery method for vaccine applications and is relevant for therapeutic delivery due to its practical nature.
The entry pathway is a critical metric for assessing the mechanistic action of different formulations of cationic lipids, lipid nano-particles and other delivery vehicles. The mode of entry can determine downstream interactions with cellular machinery and thus modulate the efficiency of protein expression (22). Traditional methods of colocalization analysis are very useful but can be hindered by imaging limitations. In specific instances, mRNA and an endocytic marker were found to be adjacent with indications of partial encirclement but with little or no overlap. In order to clarify the relationship between the mRNA and pathways of endocytosis, we hypothesized that a proximity-based assay between delivered mRNA and specific endocytic markers would be a more accurate and easily quantifiable method for describing the entry pathway used by the mRNA. Therefore, we performed a proximity ligation assay (PLA), an ideal strategy for the quantitative assessment of the interactions of molecules in the cellular environment (23–27). RNA–protein PLA assays have been pioneered by our lab (16,28–31). This method allowed us to differentiate between clathrin-mediated endocytosis, caveolae-mediated endocytosis and clathrin/caveolin-independent pathways in a proximity dependent, and easily quantifiable fashion.
Next, we studied mRNA uptake and expression, which are necessary metrics for generating kinetic models for drug optimization and design (11,12) in the context of both unmodified and modified nucleotides. To date, studies on mRNA therapeutics or vaccines have focused on expression or uptake separately, but were not able to measure these factors simultaneously due to limitations in labeling technologies as described earlier. Using MTRIP-labeled mRNA encoding for a fluorescent reporter, we measured both total mRNA uptake and protein expression per cell using flow cytometry. We extended this measurement to include two delivery methods, cationic lipid complexes and electroporation, and both unmodified and modified mRNA, using 5-methyl-cytidine (5meC) and pseudouridine (Ψ), two commonly used and previously studied modifications. Cationic lipids have been in use for over two decades and are known to deliver nucleic acids into the cell via clathrin and caveolin-mediated endocytosis (32,33). In contrast, electroporation is known to deliver molecules directly to the cytoplasm, where they may interact with various cellular structures, such as microtubules or freely diffuse (34). Delivery via cationic lipids resulted in two populations of mRNA: mRNA trapped in the endocytic compartment, and mRNA released into the cytosol. This allowed us to relate protein production and mRNA localization directly for these two prominent delivery modalities and as a function of mRNA chemistry. Given cationic lipid delivery is complicated by endocytic trapping of mRNA, we next examined the contribution of cytosolic mRNA. The cytosolic fraction of the mRNA, following release from lipoplexes, represents, at any particular time, the fraction of delivered mRNA which has the potential to interact with translational complexes in order to produce protein (12).
In order to better assess the amount of translatable mRNA per cell, we specifically measured the amount of cytosolic mRNA by using MTRIP-labeled mRNA and performed intracellular antibody staining for the endocytic pathway, using multiple markers, in order to ensure maximum coverage of the entire system. We were able to measure the amount of cytosolic mRNA based on colocalization analysis via high resolution, high dynamic range imaging (15) combined with image processing tools. This approach would not be practical using single labeled probes as they would not be bright enough to detect cytoplasmic mRNA, due to the extremely high dynamic range of signals within the cells.
Cytosolic mRNA can also bind to innate immune sensors which detect exogenous nucleic acids, competing with the translational machinery (35,36). Thus, the copy number of mRNA in the cytosol is dependent on both translation initiation and activation of pattern recognition receptors, such as Protein Kinase R (PKR) (5,37). In order to better understand the contribution to protein production of translation versus innate immune activation, we again used an EGFP mRNA variant modified by the complete incorporation of 5-methyl-cytidine (5meC) and pseudouridine (Ψ), which is known to reduce immune activation (4). Flow cytometry analysis of HSkMCs following lipofection showed an increase of EGFP expression in cells lipofected with mRNA incorporating modified nucleotides, as well as higher transfection efficiencies. This increase was not observed in the case of electroporation. We hypothesized that this was the result of a stress response due to PKR. Interactions between double stranded sections of delivered mRNA and PKR trigger phosphorylation of the eukaryotic initiation translation factor 2α subunit (eIF2α). This decreases global translation by reducing formation of the ternary complex, stalling translation initiation and causing dissociation of ribosomes (8). A direct result of eIF2α phosphorylation is SG formation, which we used via intracellular antibody staining as an indicator for cellular stress upon transfection. We then verified that this was due to PKR using siRNA and a PKR-knockout cell line.
Finally, we verified that this labeling methodology allows for mRNA localization without precluding protein expression in vivo. To do so, we utilized the Fluobeam system, an in-vivo, near-IR fluorescence imaging system. It consists of a portable and adjustable fluorescence excitation and emission detection system, which allows for real-time intraoperative imaging during surgery. We imaged mRNA in living mice upon intra muscular injection (IM), during post-transfection surgery, and in ex-vivo tissue sections. We verified that protein production remained unhindered and that MTRIPs remain on the mRNA after intramuscular injection.
Overall, we demonstrate the ability of our labeled mRNA approach to make critical measurements necessary to optimize mRNA delivery and efficacy. Unfortunately, mRNA delivery, release and translation are complex, and optimization necessitates the ability to make multiple types of measurements, which we demonstrate here.
MATERIALS AND METHODS
IVT mRNA and multiply labeled tetravalent imaging probes (MTRIP) labeling
All IVT mRNAs were synthesized by Moderna Therapeutics (Boston, MA, USA) containing identical sequences and included 5΄ capping and polyadenylation. EGFP-encoding mRNAs either were synthesized without modified nucleosides or with total incorporation of 5meC and Pseudouridine. RNA was stored frozen in –80°C and subjected to minimal freeze-thaw cycles. MTRIPs were constructed as previously described (17,19). A detailed protocol for MTRIPs assembly and characterization was described in Santangelo et al. (38). Four oligos complementary to four adjacent sequences spanning the mouse alpha globin 3΄ UTR (NM_001083955.1) of the IVT mRNA were generated. Sequences were adjacent due to the small length of the UTR region. Probe sequences were as follows:
Biotin-T(C6-Amino)-TTTTT-T(C6-Amino)-G-C-A-A-G-C-C-C-C-G-C-A-G-A-A-G-G-T(C6-Amino)
Biotin-T(C6-Amino)-TTATT-T(C6-Amino)-A-G-A-G-A-A-G-A-A-G-G-G-C-A-T(C6-Amino)-G-G
Biotin-T(C6-Amino)-TTTT-T(C6-Amino)-A-C-C-A-A-G-A-G-G-T(C6-Amino)-A-C-A-G-G-T(C6-Amino)-G-C
Biotin-T(C6-Amino)-TTTTTT-C-T(C6-Amino)-A-C-U-C-A-G-G-C-T(C6-Amino)-U-U-A-U-T(C6-Amino)-C
Each sequence was analyzed via nucleotide BLAST to ensure minimal off-target binding. Sequences were purchased as 2΄-O-methyl RNA-DNA chimeric oligonucleotides 17–18 bases long with a short 5–7 poly(T) linker and 4 C6-amino-modified thymidines. The oligos included a 5΄ biotin modification and were purchased from Biosearch Technologies (Petaluma, CA, USA). The oligonucleotides were labeled with Cy3b-NHS ester (GE Healthcare) or Dylight 650/680-NHS esters (Pierce) using manufacturer protocols. MTRIPs were assembled by incubation with Neutravidin (Pierce) for 1 h at RT followed by filtration using 30 kD MWCO centrifugal filters (Millipore). mRNA was buffer exchanged into 1× PBS, heated to 70°C for 10 min and immediately placed on ice, combined with MTRIPs in a 1:1 mRNA:MTRIP ratio and then incubated overnight at 37°C. The next day, the labeled mRNA was filtered using a 200 kD MWCO ultrafiltration unit (Advantec MFS Inc.) and concentrated by 50 kD MWCO centrifugal filters (Millipore). Alternative filters tested during protocol optimization included 100 and 300 kD MWCO, but either did not filter unbound MTRIPs successfully or failed to successfully retain mRNA.
Microscopy
Stress granule (SG) imaging was performed using a Nikon Plan-Apo 40 × 0.95 NA air objective on a Nikon Eclipse TE2000 widefield microscope equipped with a Hamamatsu C9000-02 EM-CCD camera. All other samples, including tissue slides, were imaged using a Zeiss Plan-Apo 63 × 1.4 NA oil objective on an UltraVIEW Spinning Disk Confocal Microscope equipped with a Hamamatsu Flash 4.0v2 CMOS camera. The full dynamic range of the camera was necessary to capture intensities of large and small granules without undersampling or saturating images. All microscopes were controlled by the Volocity acquisition software (PerkinElmer).
Quantification of cytosolic mRNA
mRNA quantification was performed using Volocity software in images obtained on the spinning disk confocal microscope described above and a 63× objective. Briefly, thresholds were set to detect the dimmest mRNA granules (Cy3b), which were near the detectable limit of the camera (∼500/65536). All mRNA were identified as objects and sorted based on size (greater than or less than 1 μm3) and overlap with endocytic markers. Large granules either overlapped with endocytic markers or were found to be outside the cell, which was verified by visual inspection. Such mRNAs were discarded from subsequent analysis. Smaller granules (<1 μm3) were considered to be cytosolic if they did not overlap endosomal markers. For each cell, the sum of cytosolic mRNA and GFP expression was recorded and plotted using Sigmaplot. At least 30 cells were used per condition. In detail, in Volocity software, the ‘Find Objects’ function was used to automatically select RNA granules using intensity set above background levels. The objects found by the ‘find objects’ tool, for a given intensity setting, were verified by visual inspection (see Figure 4). A manual threshold was applied to sort objects into populations (‘Filter Population’ function) based on size (∼1 μm3). ‘Find Objects’ was applied again to generate an object population representing endosomal markers (CD63/EEA1/LAMP1) with manual intensity threshold set above background values. All populations were clipped and compartmentalized to ROIs which were manually drawn around individual cells. Populations were subdivided using the ‘Exclude touching’ and ‘Exclude non-touching’ functions between RNA granules of every size and endosome objects. Large and small RNA granules touching endosome objects were considered ‘trapped’ RNA granules. Large granules in contact with endosome objects, which were extremely rare, were individually inspected and removed from analysis as all appeared to be located above the cell. Small RNA granules not touching endosome objects were considered ‘free’ mRNA granules. The sum of all free mRNA per cell was calculated using the ‘Analysis’ tab in Volocity. Protein expression was calculated in Volocity for each ROI as well. At least 30 cells were counted in this manner per condition per timepoint.
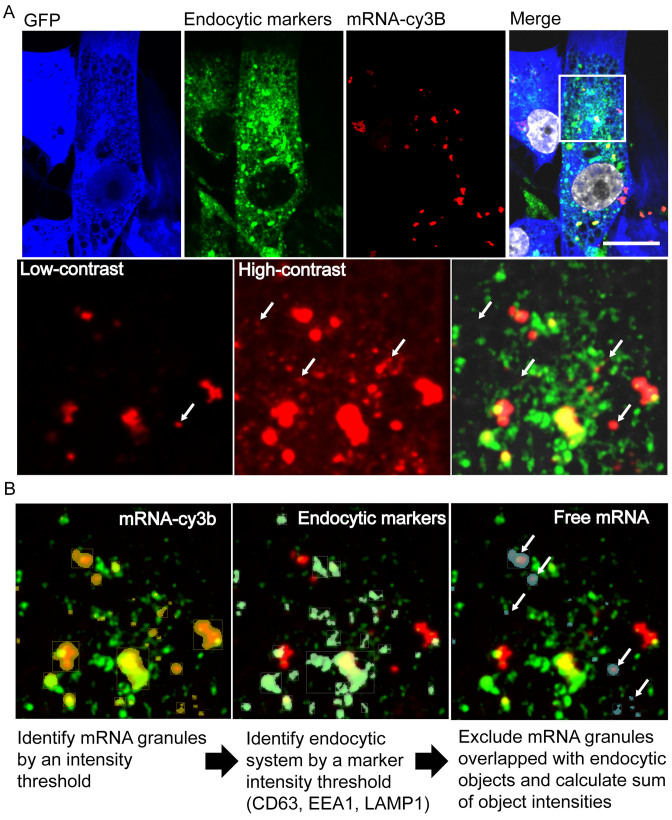
Figure 4.
Imaging and analysis of transfected cells fixed and stained with endocytic markers EEA1, CD63 and LAMP1 allows quantification and correlation of cytosolic mRNA and EGFP synthesis. (A) Representative images of 5meC + PseudoU modified EGFP mRNA lipofected into HSkMCs and fixed at 24 h post-transfection. Due to high dynamic range, the contrast enhancement necessary to visualize small mRNAs resulted in large mRNA granules to appear larger. White arrows indicate mRNA granules (red) that did not colocalize with endocytic markers (green), indicating cytosolic mRNA. (scale bar = 20 μm). (B) Representative images describing the protocol for the identification of mRNA granules and endocytic vesicles based on voxel intensity in each channel using Volocity software. mRNA granules not overlapping with endocytic objects were considered free, cytosolic mRNA (white arrows). Here, the number of identified objects shown was simplified for clarity.
Colocalization between pre-labeled mRNAs and Stellaris FISH
100 ng of unmodified mRNA was pre-labeled with Dylight-650 labeled MTRIPs, as previously described. After filtration to remove the unbound MTRIPS, the mRNA was transfected to HeLa cells using Lipofectamine 2000 according to manufacturer's instructions. The cells were fixed 5 h post-transfection in 4% PFA and permeabilized in 70% ethanol at 4°C overnight. The following day we performed FISH using Stellaris FISH RNA probes labeled with Quasar 570 (Biosearch) designed against the codon optimized coding region. FISH was performed according to the Stellaris protocol. Briefly, cells were washed 5 min in wash buffer (10% formamide, 2% SSC). Cells were then incubated at 37°C in the presence of 200 nM 570-labeled Stellaris probes in hybridization buffer (10% formamide, 2% SSC, 10% dextran sulfate, .5% tRNA, 0.5% ssDNA, 0.2% bovine serum albumin—BSA) in a humid chamber. After 4 h, cells were washed in wash buffer for 30 min at 37°C, nuclei were stained with DAPI and coverslips were mounted on glass slides using Prolong gold (Life Technologies). Mander's coefficients were measured using Costes threshold calculation in >15 cells using Volocity software. Controls consisted of FISH on vehicle only-transfected cells, cells transfected with an mRNA containing a different coding region (Chemokine ligand 3), and DNA oligos with the same sequence as the mRNA 3΄ UTR region.
For experiments performed in tissue sections, 10 μg of unmodified mRNA was pre-labeled with Cy3b labeled MTRIPs. After filtration to remove the unbound MTRIPS, the mRNA was complexed with a PEI polymer (Viromer Red, Lipocalyx) and injected intramuscularly in a Ringer's Lactate (RiLa) buffer into the anterior tibialis muscle of BALB/C mice. Controls consisted of an intramuscular sham injection of RiLa buffer. The mice were sacrificed and the muscle harvested 2 h post-injection. The harvested tissue was fixed in 4% PFA overnight at 4°C, soaked in PBS containing 30% sucrose overnight at 4°C, and then embedded in optimal cutting temperature solution. The tissue blocks were then snap frozen in isopentane and dry ice. Finally, the frozen tissue blocks were cut into ∼10 um thick sections with a cryostat and put onto slides to be used for FISH. The tissue was fixed and permeabilized in 50:50 methanol:acetone at -20°C for 10 min. Stellaris FISH probes labeled with Quasar 670 (Biosearch) were designed matching the probes used for the in vitro experiment. FISH was then performed according to the Stellaris protocol. Briefly, after methanol:acetone fixation, the tissue was equilibrated in wash buffer (10% formamide, 2% SSC). Tissue was then incubated at 37°C in the presence of 20 nM of 670-labeled Stellaris probes in hybridization buffer (10% formamide, 2% SSC, 10% dextran sulfate, 0.5% tRNA, 0.5% ssDNA, 0.2% BSA) in a humid chamber overnight. The next morning, tissue was washed in wash buffer for 30 min at 37°C. Finally, the nuclei were stained with DAPI and coverslips were mounted on top the tissue using Prolong gold (Life Technologies).
Proximity ligation assays
Protein/mRNA PLA has been previously described (28,29) and a detailed protocol can be found in Zurla et al. (39). Briefly, neutravidin was tagged with a V5 epitope through Solulink conjugation technology. A maleimide hynic linker (Solulink) was conjugated to the V5 tag, while an S-4FB linker (Solulink) was conjugated to the neutravidin, following manufacturer's instructions. After conjugation, the two reagents were mixed with the Turbolink catalyst (Solulink) to covalently bind the V5 tag to neutravidin. MTRIPs were then assembled as previously described using V5 labeled neutravidin (Na-V5), mRNA was labeled as above, and then used for transfection. Two hours post-transfection, cells were fixed with 1% paraformaldehyde and permeabilized with 0.2% Triton X in 1× PBS. Cells were then blocked for nonspecific interactions for 30 min at 37°C with PLA blocking buffer (0.1% gelatin, 2% donkey serum and 1% BSA in 1× PBS). Primary antibodies consisted of rabbit anti-V5 (Abcam), mouse anti-V5 (abcam), mouse anti-caveolin-1 (Sigma), mouse anti-clathrin light chain (Sigma) and rabbit anti-ARF6 (Pierce). Primary antibodies were then delivered (1:1000 V5 Ab and 1:250 ARF6 Ab, 1:500 clathrin light chain Ab or 1:10 000 caveolin Ab in PLA primary diluent (1% BSA, 1% donkey serum and 0.2% Triton-X in 1× PBS) for 30 min at 37°C. Cells were washed in wash buffer A (Sigma) at RT for 10 min. Secondary antibodies for rabbit and mouse antibodies (Sigma) were delivered at a concentration indicated by the manufacturer in PLA secondary diluent (0.05% tween 20 in 1× PBS) for 30 min at 37°C. This was followed by another 10 min wash in wash buffer A (Sigma) at RT. PLA ligation and rolling circle amplification were performed as specified by the manufacturer. Finally, cells were washed and mounted on a slide with DAPI mounting medium (Sigma). PLA interactions were quantified using a 63× (oil) objective on a spinning disk confocal microscope. Thiry cells were measured per experimental condition and analyzed by Volocity software.
Cell culture and transfection
HeLa cells and Mouse Embryonic Fibroblasts were obtained from ATCC and maintained in DMEM or EMEM (Lonza) and supplemented with 10% FBS (Hyclone), 100 U/ml penicillin and 100 μg/ml streptomycin (Life Technologies). Primary human skeletal muscle cells (Promocell) were grown in HSkMC growth media (Promocell) supplemented without FBS and only with penicillin and streptomycin as above. Cells were plated the day before an experiment in preparation for lipofectamine transfection. Lipofectamine was combined with labeled mRNA in Optimem (Life Technologies) using manufacturer protocols. Lipofectamine solutions were replaced by fresh media 5 h post-transfection. For electroporation, cells were placed into suspension using trypsin which was subsequently deactivated by the addition of media containing serum. Cells were counted, pelleted, and re-suspended at a final concentration of 5 million cells/ml in electroporation buffer R (Life Technologies) containing 200 ng/10 ul of mRNA. All electroporations were performed using the Neon Transfection System (Life Technologies) using a 10 μl reaction size per well in a 24-well plate and following manufacturer protocols. Pulse voltage was set to 1100 V with a pulse width of 30 ms and a pulse number of 2. Media was replaced after 5 h. Knockdown experiments for PKR were performed by Neon electroporation with anti-PKR siRNA (Smartpool, Dharmacon) 48 h prior to mRNA transfection and knockdown efficiency was quantified to be >80% using PCR.
Immunofluorescence
Cells were fixed in 4% PFA for 10 min, blocked in 25% BSA and immunostained as previously described (47) using appropriate antibodies. Endosomal route markers used were caveolin, clathrin light chain and ARF6 (Santa Cruz Biotechnology). General endocytic markers used for evaluating cytosolic mRNA were CD63 (mouse anti-CD63, Developmental Studies Hybridoma Bank- DSHB), EEA1 (mouse anti-EEA1, BD Biosciences) and LAMP1 (mouse anti-LAMP1, DSHB). Stress granule markers included G3BP (mouse anti-G3BP, BD Biosciences) and TIAR (goat anti-TIAR, Santa Cruz). EGFP was stained using a rabbit anti-GFP polyclonal antibody (Life Technologies). Secondary antibodies were purchased pre-conjugated to either Alexa Fluor 488 (Life Technologies), Cy3 (Jackson Immuno) or Alexa Fluor 647 (Life Technologies). Cells were finally stained with DAPI for 5 min and mounted on glass slides with Prolong gold. Tissue immunofluorescence staining was performed following 4% PFA fixation, paraffinization and antigen retrieval with standard protocols using antibodies as above or anti-cd11b (Abcam) and anti-Vimentin (Santa Cruz Biotechnology). Tissues were imaged with a 40× objective on the Ultraview Spinning Disk microscope using stitching algorithms in Volocity.
Flow cytometry
Cells were prepared for flow cytometry using warm Versene-EDTA (Lonza) for 5 min for detachment followed by 10 min fixation in 4% paraformaldehyde at 4°C, multiple washing steps and resuspension using FACS buffer (Dulbecco's phosphate buffered saline–Ca2+–Mg2+ supplemented with 1% FBS and 5 mM EDTA). Dylight-650 labeled mRNA was used for flow cytometry experiments, performed using a BD FACS-Canto II flow cytometer and analyzed using FlowJo software. Experiments were performed in duplicate with >5000 cells per condition.
Gel shift to demonstrate MTRIPS binding to mRNA
100 ng of unmodified EGFP mRNA was pre-labeled with Dylight-650 using the above protocol. After filtration to remove unbound MTRIPS, the labeled mRNA was loaded on a 2% agarose gel and run with constant voltage (50 V for 2 h). As a control, we ran naked mRNA (incubated over night at 37°C without MTRIPS) or labeled mRNA boiled at 95°C for 10 min.
Size exclusion chromatography to assess degree of labeling
In order to assess labeling efficiency, mRNA and MTRIPs labeled with Dylight-650 or the same amount of MTRIPs without mRNA was analyzed by size exclusion chromatography using an SEC-4000 Yarra column (Phenomenex). Samples were flowed using 1× phosphate buffer and analyzed by fluorescence reading in real-time on a Shimadzu Prominence HPLC system.
Mouse experiments
Mice were injected in the anterior tibialis muscle using 10 μg of Dylight680 or Cy3b-labeled mRNA in 40 μl of Ringer's Lactate (RiLa) or with RiLa alone. For in vivo EGFP production, 2 ul of Viromer Red (Lipocalyx) delivery vehicle was mixed with mRNA. Imaging during surgery was performed using the Fluobeam 700 Near-IR Imaging System (Fluoptics). The anterior tibialis muscle was removed 16 h post-delivery, fixed in 4% PFA and cryopreserved in optimum cutting temperature solution. Staining was performed on slices and imaged as described above. All animal handling and experiments were performed in accordance with protocols approved by the IACUC at Georgia Institute of Technology.
Statistics
Significance in proximity ligation assays was determined by running a one-way ANOVA on the data, using the Kruskal–Wallis test and Dunn's multiple comparisons test. Correlation statistics (Pearson, two-tailed P values), and Mann–Whitney t-tests were calculated in Graphpad Prism software. All other data was tested with two-way ANOVA at a = 0.05 followed by Holm–Sidak method for multiple comparison testing. All measurements were analyzed with Microsoft Excel, Graphpad Prism, and/or Sigmaplot 13.
RESULTS
Labeling of exogenous mRNA with MTRIPs
In order to use MTRIPs (Figure 1A) to label IVT mRNA, we designed four MTRIPs complementary to the 3΄ untranslated region (UTR) of our in vitro transcribed EGFP mRNA, a 93 bp sequence derived from the mouse alpha globin 3΄ UTR. We optimized binding conditions through an iterative process where temperature, salt concentration, incubation time and filtration method were varied. We first verified binding of probes to the target mRNA via agarose gel, where labeled mRNA exhibited reduced migration (Supplementary Figure S1) with respect to naked mRNA or denatured mRNA. Labeling efficiency was tested using exclusion chromatography, which resulted in a degree of labeling of approximately two MTRIPs per mRNA, based on the difference in fluorescence signal between MTRIP-labeled mRNA and the same amount of MTRIPs alone prior to filtering (Supplementary Figure S2). We measured the effects of variations in incubation time and temperature, and determined that optimal binding occurs through a short, 10 min heat induced denaturation of mRNA to remove secondary structures, followed by addition of MTRIPs and an incubation overnight at 37°C. We also examined the effect of different buffers, including PBS and SSC at multiple concentrations, and found that PBS provided an adequate salt concentration to stabilize binding of MTRIPs to mRNA. Finally, filtration was necessary to remove unbound MTRIPs. We found that a 200 kD molecular weight cutoff filters provided the highest yield of mRNA while removing unbound MTRIPs.
To demonstrate that MTRIPS remained bound to mRNA upon delivery, HeLa cells were lipofected with Dylight 650-labeled mRNA. Cells were subsequently hybridized post-fixation with Quasar 570 Stellaris RNA FISH probes specific for the coding region (40). MTRIPs exhibited a high degree of colocalization (Costes overlap coefficient of ∼0.6, Manders overlap coefficients m1 and m2 were ∼0.6 and ∼1 respectively) with FISH probes compared to controls using mRNA with an identical UTR but a different coding region corresponding to Chemokine Ligand 3 (Figure 1B, Supplementary Figure S3).
Electroporation and transfection via Lipofectamine 2000 (L2K) of HSkMCs with labeled EGFP mRNA yielded highly distinct distributions of intracellular mRNA. Electroporation resulted in a large number of lower intensity, dispersed mRNA granules while lipofection resulted in a few bright, large granules with a lower number of smaller granules dispersed through the cell (Figure 1C).
A strong benefit of using MTRIPs bound to the 3΄ UTR region is that they do not significantly inhibit the translation of endogenuous mRNA (16,20). To demonstrate that the labeling of mRNA does not affect translation, we measured differences of protein production following electroporation of HSkMCs cells with labeled and unlabeled mRNA using flow cytometry. Labeled mRNA showed a similar protein expression distribution as unlabeled mRNA. The small decrease in protein production is likely attributable to RNA loss during the purification step following the labeling reaction (Figure 1D).
Entry pathway and subcellular localization of L2K transfected and electroporated mRNA
We then investigated the entry pathway of labeled mRNA into the cell. Endocytosis of mRNA can be detected via co-staining with antibodies against clathrin-light chain, caveolin-1 and ARF6 (41) and subsequent colocalization analysis. These markers were used to differentiate clathrin-mediated endocytosis, caveolae-mediated endocytosis and clathrin/caveolin-independent pathways, respectively. We initially used 3D images of single cells to determine colocalization between labeled mRNA and relevant pathway markers (Supplementary Figure S4). Based on line profiles (Supplementary Figure S4), mRNA delivered by lipofectamine colocalized with both clathrin and caveolin, but less frequently with ARF6. This was consistent with published transmembrane routes of cationic liposomal delivery (32). In order to confirm and better quantify these results, we verified entry pathway using a RNA–protein proximity ligation assay.
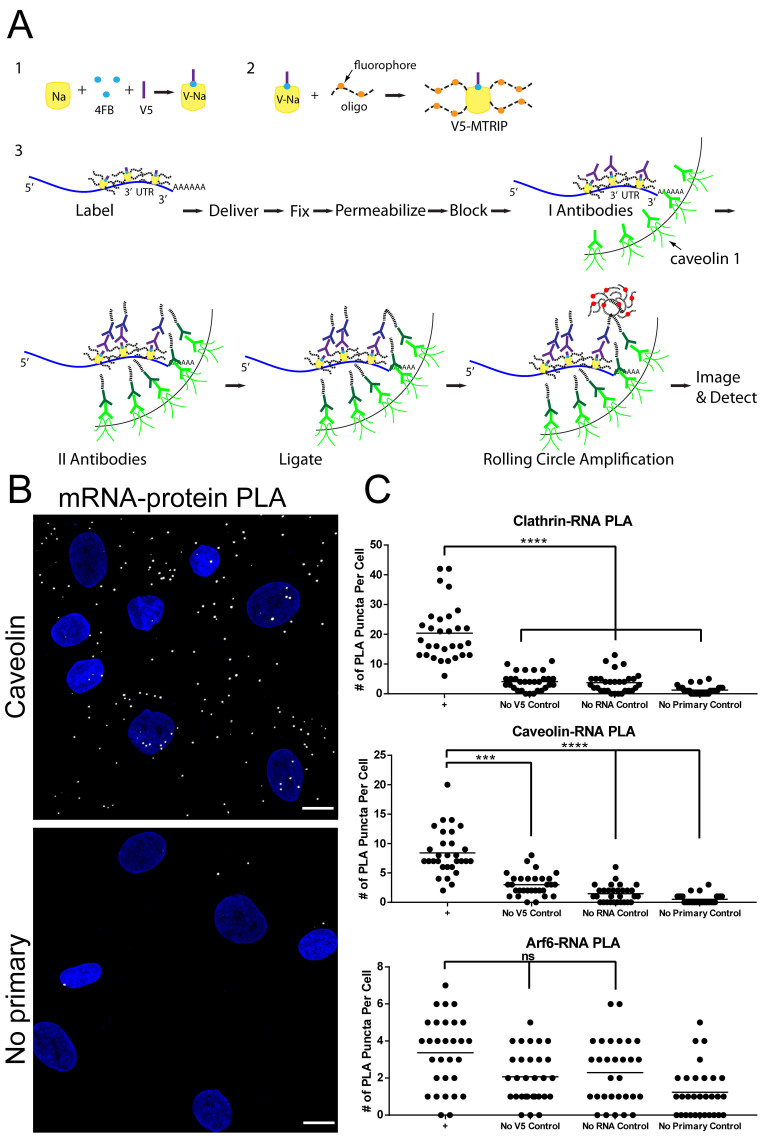
The overall PLA process is summarized in Figure 2A. Briefly, mRNA was labeled with V5-peptide-tagged MTRIPs (the peptide is covalently attached to neutravidin) and delivered to HSkMCs by lipofection. Cells were fixed 2 h post-transfection. Primary antibodies specific for the V5 tag and different endocytic pathway markers, clathrin light chain, caveolin-1 or ARF6, were then added, followed by PLA proximity probes, oligonucleotide-labeled secondary antibody compatible for PLA. If the mRNA and the protein of interest are close to each other (<40 nm), the oligonucleotides are ligated by the addition of a ligase. Rolling circle amplification then creates a DNA ‘ball’, which is visualized by fluorescence in situ hybridization. This results in very bright (>30 fluorophores), diffraction-limited fluorescent puncta which are easily detected and quantified via microscopy (23). PLA puncta (Figure 2B) due to interactions between mRNA and endosomal markers were quantified via ‘puncta’ detection and counting, and compared to controls including mock transfection and no primary antibody. In the case of clathrin-light chain and caveolin-1, statistically significant differences from controls indicated that lipofectamine-delivered mRNA interacted with clathrin or caveolin-1, but not with ARF6 (Figure 2C). Significantly, this result confirmed observations obtained by colocalization and overlap coefficient measurements.
Figure 2.
Analysis of lipofection-mediated mRNA uptake pathway via PLA. (A) Overall schematic of PLA experiments to quantify interactions between mRNA and endocytic markers. A V5 tag is covalently bound to the MTRIPs prior to mRNA labeling. Following mRNA delivery by lipofectamine, primary antibodies target the V5 tag and either clathrin light chain, caveolin-1, or ARF6. PLA proximity probes recognize the primary antibodies and provide the substrate for ligation and rolling circle amplification. mRNA–protein interactions result in PLA puncta of homogeneous size and brightness. Standard controls include experiments performed without primary antibodies, without mRNA or using MTRIPS without the V5 tag. (B) PLA between V5-tagged MTRIPs and caveolin-1 in HSkMCs two hours post-lipofection resulted in significantly higher numbers of PLA puncta when compared to no primary control. (scale bar = 10 μm). (C) The number of PLA puncta per cell was quantified for 30 cells per condition for each endocytic marker. Significant proximity with endocytic markers and mRNA was detected for clathrin and caveolin but not ARF6 when compared to controls. (****P < 0.0001, ***P = 0.0005).
Whole cell RNA uptake and protein expression measured via flow cytometry
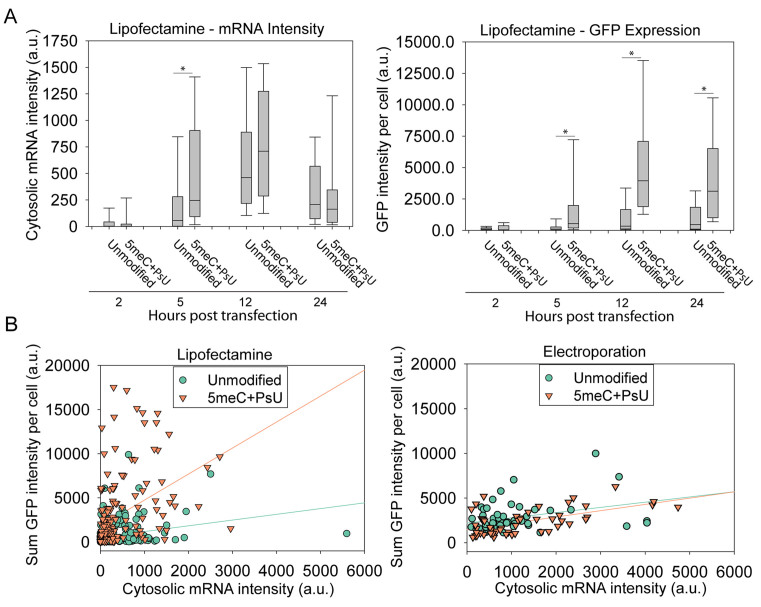
In order to compare efficiencies of delivery methods, electroporation and lipofection, we used flow cytometry, which yields an integration of mRNA signal and protein-reporter fluorescence on a per cell basis. We electroporated HSkMCs with increasing amounts of labeled EGFP mRNA and observed that between 0 and 1000 ng of delivered mRNA, the mean mRNA intensity per cell scaled in a linear fashion (Supplementary Figure S5). In order to maintain similar protein expression between delivery methods, as well as to avoid possible cellular stress response to unfolded protein, in subsequent experiments we delivered 200 ng per 100 000 cells. In order to measure EGFP production and quantify, at the same time, the mean total mRNA delivered per cell, we transfected HSkMCs with 200 ng of labeled mRNA and analyzed cells at 24 h post-transfection via flow cytometry. Cells were categorized into EGFP ‘expressing’ and ‘non-expressing’ populations, using a threshold of six standard deviations above the intensity of mock transfected cells. From this experiment, we found that upon electroporation, up to 90% of cells expressed GFP, whereas only 40% of lipofected cells expressed GFP at 24 h post-transfection (Supplementary Figure S6). Importantly, the mean mRNA intensity within expressing cells was 10 times higher upon L2K transfection than electroporation (Figure 3A). As previously described in Figure 1C, it is clear that much of the mRNA delivered via lipofection localizes in cytoplasmic vesicles, possibly due to compartmentalization in the endosomal system (32,33). Such mRNA likely does not contribute to translation (12). Additionally, we observed that cell-to-cell heterogeneity in protein expression was much higher with L2K.
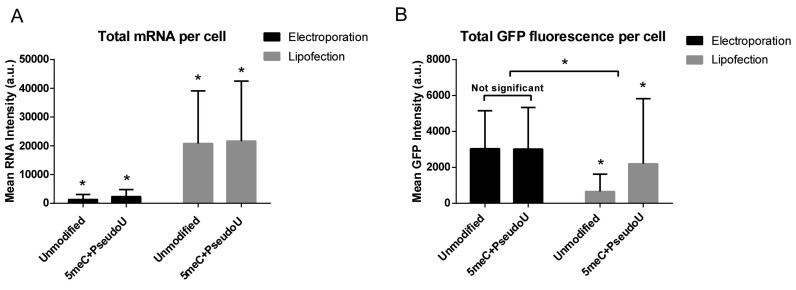
Figure 3.
Flow cytometry analysis of HSkMCs following electroporation or lipofection with labeled 5meC + Pseudouridine or unmodified EGFP mRNA showed differences in mRNA uptake and protein expression. (A) The mean total labeled mRNA intensity per cell was evaluated at 24 h post-transfection. Lipofection resulted in ∼10 times higher mRNA per cell than electroporation. All comparisons are statistically significant based on two way ANOVA (P < 0.0001) (bars = S.D. of population). (B) The mean total EGFP intensity per cell was measured at 24 hours post-transfection. A significant drop in intensity was observed upon lipofection of unmodified mRNA. Also, protein expression upon lipofection did not correlate with the amount of mRNA per cell. Statistical comparisons performed by two-way ANOVA with a = 0.05, P < 0.0001 for all conditions except as indicated. Experiment was performed in duplicate (bars = S.D. of population).
We repeated the experiment using an EGFP mRNA variant modified by the complete incorporation of 5-methyl-cytidine (5meC) and pseudouridine (Ψ), which is known to reduce immune activation (4). We determined that the mean mRNA intensity within expressing cells was similar to the one measured for unmodified mRNA. The mean total protein expression per cell was similar between unmodified and modified mRNA upon electroporation, but a significant increase in protein production was observed upon lipofection using modified mRNA (Figure 3B).
Cytosolic mRNA correlates with protein production
The flow cytometry data described in Figure 3 indicated an order of magnitude increase in total transfected RNA per cell upon L2K transfection compared to electroporation. However, levels of protein production were similar, indicating that L2K delivered mRNA trapped in the endosomal compartments does not contribute to translation. We hypothesized that cytosolic RNA and not total RNA contributed to protein production. In order to discriminate and quantify the cytosolic and endocytic mRNA populations, we lipofected HSkMCs with modified and unmodified labeled EGFP mRNA, and co-stained with endocytic markers at 2, 5, 12, 24 and 48 h post-transfection (Figure 4A, Supplementary Figure S7). We used multiple endocytic markers including Early endosome antigen 1 (EEA1), CD63 and Lysosomal-associated membrane protein 1 (LAMP1), which provide broad coverage of the endosomal compartments (42,43). Within single cells, we identified labeled mRNA granules, determined their volume and colocalization with endocytic markers (Figure 4B). Large mRNA granules (∼>1 μm3) colocalized with endocytic markers or were typically outside the cell, and therefore were excluded from further analysis. Smaller RNA granules were classified based on their colocalization with endocytic markers. Small granules which did not colocalize with endocytic markers were classified as ‘free’ cytosolic molecules (see Materials and Methods section). The quantification of protein expression and cytosolic mRNA per cell over time is shown in Figure 5A. It is important to note that detected RNA granules varied in intensity; this intensity difference reflects the diversity in the number of RNAs per granule.
Figure 5.
Cytosolic mRNA intensity and protein levels indicated differences in release rate as well as differential expression for given amounts of mRNA. (A) Cytosolic mRNA and GFP intensity per cell at different time points post-lipofection. Data were collected from 30 cells per sample (box = median, upper and lower quartile, and min/max). (B) Cytosolic mRNA and protein expression correlation per cell based on microscopy imaging combining all timepoints. Lines are provided as a qualitative measure to show differences in distribution. The distributions between modified and unmodified mRNA upon lipofection were significantly different (P < 0.0001) but not electroporation distributions (P = 0.6677). All comparisons between mRNA and GFP in each distribution were statistically correlated (Lipofection: unmodified P = 0.0002 | Modified correlated P < 0.0001 and Electroporation: unmodified P = 0.0195 | Modified, P < 0.0001).
By quantifying the sum of intensity of cytosolic mRNA granules at multiple timepoints, we were able to examine the release kinetics of mRNA. We observed a significant increase of free modified mRNA 5 h post-transfection upon lipofection (Figure 5A), suggesting a faster release rate into the cytosol from the endosomal compartment. These are the first experiments, to our knowledge, that permitted the quantification of mRNA cytosolic release upon lipofection.
The amount of free RNA and GFP expression, on a per-cell basis, can serve as a measure of mRNA transfection efficiency. A plot of the sum of GFP intensity versus free RNA for electroporation and lipofection (Figure 5B) yielded approximately three times more cytosolic RNA in electroporated cells than in lipofected cells (see Supplementary Table S1 for mean, SEM and statistical comparisons). Consistent with the flow cytometry data described in Figure 3, we observed that similar levels of mRNA yielded different levels of GFP expression. This result was particularly evident comparing the efficiency of lipofection of unmodified versus modified mRNA. During lipofection, the use of 5meC+Ψ modified mRNA resulted in a significantly higher protein–RNA ratio. However, no significant difference was observed using electroporation (see Supplementary Table S1). Statistical analysis of correlations between free mRNA and protein expression showed that they were statistically correlated (Lipofection: unmodified P = 0.0002 | modified P < 0.0001 and electroporation: unmodified P = 0.0195 | modified P < 0.0001), though cell-to-cell heterogeneity was still evident. Additionally, levels of cytosolic mRNA were higher upon electroporation but did not result in correspondingly high levels of protein expression (Figure 5B, Supplementary Table S1).
Stress granule (SG) formation negatively correlates with protein production
Differences in protein expression for given amounts of cytosolic mRNA, especially between modified and unmodified mRNA upon lipofection, suggested that protein production may be affected by innate immune activation (44). In order to further understand how changes in mRNA chemistry, as well as delivery method, impacted protein production, we used the formation of stress granules as a marker for innate immune activation.
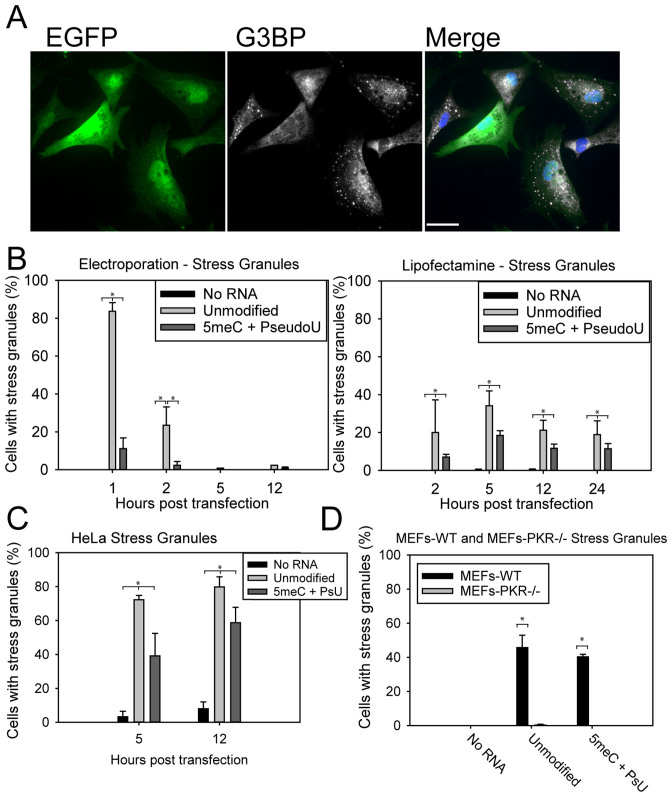
We transfected HSKMCs with unlabeled modified and unmodified mRNA and stained for SG markers Ras GAP-binding protein 1 (G3BP) and T-cell-restricted intracellular antigen 1 related protein (TIAR), in order to characterize the kinetics of SG formation over time (Figure 6A). SG formation upon electroporation was readily observable with unmodified mRNA (>80% of cells) 1-h post-transfection, and quickly diminished to become undetectable 5 h post-transfection (Figure 6B). Transfection with lipofectamine resulted in ∼20% of cells containing SGs, which diminished only slightly over 24 h. When HeLa cells were transfected with lipofectamine and mRNA, higher levels of SGs were observed (>70%), indicating that SG formation is cell-type dependent (Figure 6C). Transfection with modified mRNA significantly reduced, but did not completely eliminate, SGs. Mock transfections did not result in any significant SG formation.
Figure 6.
PKR-dependent stress granule formation correlates with decreased protein production and is cell type-dependent. (A) HSkMCs were fixed 1 h post-electroporation with EGFP mRNA and stained for stress granules (G3BP, white). Cells with stress granules produced less GFP (green) (scale bar = 20 μm). (B) Kinetics measurements in HSkMCs show a brief but strong increase in the number of cells with SGs 1 h after electroporation, which rapidly diminishes. The extended SG response upon lipofectamine transfection is still detectable 24 h post-transfection. 5meC + PseudoU reduced but did not completely eliminate SGs. (C) SG formation was found to be cell type-dependent. HeLa cells more readily formed SGs upon lipofection regardless of mRNA modification. (D) SGs formed readily 5 h post-lipofection in MEF cells but not in MEF PKR–/– cells, indicating that mRNA sensing by PKR resulted in SG formation. MEF cells 5 h post-lipofection produced extensive SGs regardless of mRNA modification. MEF PKR-/- cells produced no detectable SGs (bars in B, C, and D = S.D. of triplicate measurements of 100 cells each).
In order to test PKR-dependence of SG formation, we delivered PKR–siRNA into HSKMCs, which were then electroporated with unmodified mRNA. One hour post transfection, only 8% of cells formed SGs, versus >95% treated with control siRNA (data not shown). Then, mRNA transfection by lipofectamine was repeated in mouse embryonic fibroblasts (MEF) and MEF PKR–/– cells (Figure 6D). Modified mRNA slightly reduced the number of MEF WT cells containing SGs from 45% to 40%; as expected, MEF PKR–/– transfected cells formed no visible SGs. These cells expressed higher levels of GFP overall, with no differences between modified and unmodified mRNA (Supplementary Figure S8). This strongly suggested a cell type dependence on stress granule formation, and provided further evidence that stress granule formation occurs in a PKR-dependent manner.
Validation of the mRNA labeling protocol for tissue studies in mice
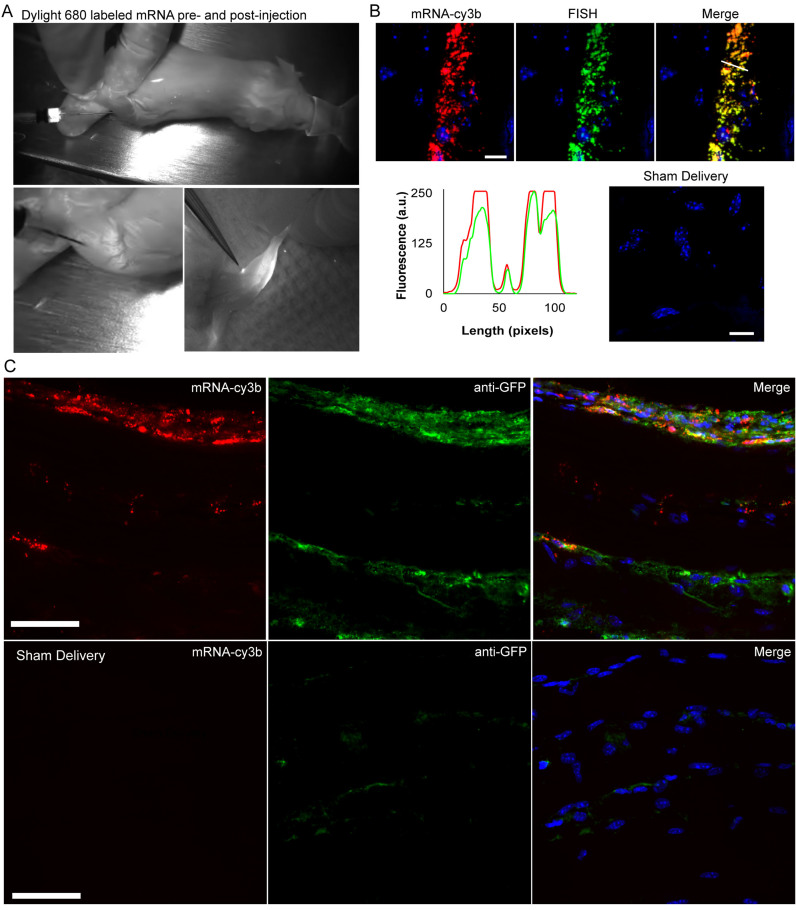
We next examined the applicability of IVT mRNA labeling for in vivo experiments in an IM injection mouse model. We utilized MTRIPs labeled with Dylight 680 to visualize the distribution of mRNA upon IM injection in live mice using the Fluobeam 700 live fluorescent imaging system. IM injection of mRNA into the anterior tibialis muscle of mice showed immediate saturation of muscle tissue with mRNA at the injection site, which remained 16 h post-injection along with fluorescence visible in neighboring lymph nodes (Figure 7A). Muscle tissue was then extracted, fixed and sectioned to visualize mRNA distributions in the muscle. We verified that MTRIPs remained bound to mRNA upon injection using FISH, as previously performed in vitro, on muscle sections fixed 2 h post-injection. We found that the FISH signal highly colocalized with MTRIP signal in tissue sections (Figure 7B).
Figure 7.
Visualization of labeled mRNA during IM injection and in extracted tissue sections. (A) Dylight-680 labeled mRNA (white) was imaged during surgery prior to and immediately following IM injection into the anterior tibialis of a live mouse using the Fluobeam near-IR fluorescence imaging system. The third panel obtained 16 h post-injection shows localization of mRNA to a lymph node. (B) Labeled mRNA (red) colocalized with Stellaris FISH probe signal (green), as indicated by the intensity line profile (white line). No FISH signal was visible in mock transfected tissue (scale bar = 10 μm). (C) Cy3b-labeled mRNA (red) and antibody staining for GFP expression (green) imaged in anterior tibialis muscle sections removed from a mouse 16 h post-IM injection. Sham delivery (without mRNA) shows comparatively low levels of background signal in the GFP channel due to non-specific binding of antibodies.
In order to correlate localization of mRNA with protein expression at the tissue level, we combined Cy3b-labeled EGFP mRNA with a PEI-based delivery vehicle and injected it into the anterior tibialis muscle. Staining for EGFP showed that the majority of protein expression was localized in the perinuclear region of muscle cells and in cells present in the interstitial spaces (Figure 7C), where most of the mRNA was found.
Last, we examined Cy3b-labeled mRNA localization in relation with markers for cell structure and cell type. Following injection of Cy3b-labeled mRNA, we stained fixed sections of muscle with antibodies against CD11b, a marker for leukocytes, as well as for vimentin, an intermediate filament marker, which enabled the visualization of striated muscle (Supplementary Figure S9). Skeletal muscle cells are the intended target during IM injection, while uptake by leukocytes is critical for vaccine applications. Labeled RNA was visibly located in the perinuclear region of muscle cells, and also colocalized with CD11b-marked leukocytes. Red blood cells were visible and indicative of blood vessels, though they did not contain labeled mRNA. Overall, these experiments serve as a proof of concept that RNA uptake and expression can be assessed for specific cell populations and localization within tissues.
DISCUSSION
A serious concern for the future of mRNA therapeutics is the efficiency of protein production in a given organ for a single dose. Delivery vehicles must be designed and optimized to ensure efficient delivery of mRNA to target cells and their cytosol, as well as to encourage translation. Likewise, the design of the mRNA sequences along with delivery formulations must be carefully considered. It is important to modulate the negative immune effects of mRNA, based on pathogen sensing mechanisms within various cell types. All three of these factors require new tools and methodologies to study mRNA uptake and kinetics of protein expression.
We have developed a labeling strategy for exogenous mRNA, which allows single RNA-sensitive detection (∼2 probes and 16–20 fluors per mRNA), without significant reductions in the translational potential of therapeutic molecules. Probes were hybridized to in vitro transcribed mRNA in controlled conditions, and were purified and characterized using syringe filters and size exclusion chromatography. Encoding secondary proteins for visualization or co-expression with reporter molecules is not required. These probes can potentially be designed against any 3΄ untranslated region. UTR regions can also be extended with specific binding sequences for MTRIP binding, without affecting the coding region or reading frame of the mRNA. The resulting labeled mRNAs are functional in both cells in culture and after in vivo delivery in animal models. Labeling of therapeutic RNA allowed fast and accurate screening of RNA formulations through compatibility with high-throughput tools such as flow cytometers and plate readers. Combining MTRIPs with a proximity ligation assay provided a novel method for defining uptake pathway by discrete quantification of interactions between mRNA and specific endocytic markers. For in vivo studies, this methodology enabled the imaging of RNA localization in tissue slices co-stained with markers for cell type or target proteins. Future mechanistic studies will be performed in order to evaluate the effectiveness of different formulations.
Translation of mRNA occurs in the cytosol, and thus delivery vehicles have the two-fold requirement of first reaching the target cells within tissues, and then either escaping from an endocytic compartment or entering the cytosol directly. Our results indicated that while both lipofectamine-mediated delivery and electroporation of mRNA resulted in protein expression, the amount of protein expression was based upon the amount of mRNA that reached the cytosol as well as a complex interplay between mRNA translation and innate immune activation. Lipofectamine delivery resulted in large amounts of mRNA trapped in intracellular vesicles. Measures of protein–RNA correlation indicated a higher efficiency of protein production per cytosolic mRNA using lipofection. This implied that mRNA were released from the endosome slowly over time, and can more readily interact with the translational machinery. In addition, L2K delivered mRNA may be protected from degradation during the mRNA assembly process with proteins (forming a messenger ribonucleoprotein (mRNP)). Therefore, controlling the kinetics of mRNA release may be critical to the optimization of protein production with cationic lipids. In contrast, electroporation removed the endosomal escape aspect entirely and resulted in a large amount of mRNA entering the cytosol directly.
It is also important to note that MTRIPs remain bound to the mRNA in vivo. This allows subsequent studies for biodistribution and mRNA trafficking on a whole-animal level as well as cell-type specific uptake and expression. MTRIPs can be labelled with different fluorophores depending on the imaging application. They can also be labeled with radionuclides, such as 64Cu, which would enable the use of position emission tomography detection of the mRNA in the deep tissue of live animals. This can be a critical aspect of testing mRNA therapeutics prior to translation into humans.
The presence of stress granules is indicative of PKR activation and translational repression via eIF2α phosphorylation, and is linked to abrogating apoptosis (45,46). Not only does SG formation negatively correlate with the expression of the therapeutic protein of interest, but they also influence host gene expression regulation, an undesirable side-effect for most therapies. Incorporation of modified nucleotides into mRNA substantially reduced this effect, but in a cell-type and delivery-dependent manner. Decreased SG levels in HSKMCs compared to HeLa and MEF cells might be due to lower overall PKR levels in human skeletal muscle. Electroporation resulted in a strong but short-lasting stress response, and thus SGs had a partial effect on the RNA–protein correlation. The longer-lasting, SG response observed using lipofectamine is likely due to slow release of mRNA from endosomes over time, and, thus, continuous activation of PKR and eIF2α phosphorylation. It is also possible that ‘priming’ of the stress response occurs through activation of toll-like receptors (TLRs) in the endosomal compartment, which does not happen during electroporation (34,47). Results with MEF PKR–/– cells indicate that the benefits of mRNA modification primarily stem from reduction in the stress response. Overall, SG formation should be avoided in order not to perturb cell state and produce the maximum amount of desired protein.
The biggest challenge to the development of mRNA therapeutics is efficiency. Reduction of eIF2α phosphorylation and efficient delivery of mRNA and subsequent release into the cytosolic compartment are instrumental to overcoming this barrier. mRNP formation kinetics may also limit protein production but this will be addressed in future work using the MTRIP-PLA approach. Overall, this methodology allows for the measurements of critical metrics to not only compare prospective mRNAs and their formulations, but also in probing the biological mechanisms limiting expression. Overcoming barriers of efficiency will enable new mRNA-based therapeutics which are safer and more effective.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the Georgia Tech administration for supporting the UltraView spinning disk microscope, as well as the support of core facilities at the Parker H. Petit Institute for Bioengineering and Bioscience. We also are thankful for support from the Emory Children's Pediatric Research Center Flow Cytometry Core. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Contributions: J.L.K. and P.J.S. conceived of the project, designed experiments, and wrote the manuscript. C.Z. performed FISH experiments and gels for probe verification. S.B. and K.H.L. performed mouse experiments and assisted with tissue staining. D.V. assisted in live cell experiments and statistical analysis. E.L.B. performed experiments involving FISH and proximity ligation assay. All other experiments were performed by J.L.K., K.M. and B.C.L in consultation with P.J.S.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Defense Advanced Research Projects Agency ADEPT PROTECT program; National Science Foundation Graduate Research Fellowship Program [DGE-1650044]. Funding for open access charge: DARPA ADEPT PROTECT program.
Conflict of interest statement. None declared.
REFERENCES
- 1. Schlake T., Thess A., Fotin-Mleczek M., Kallen K.-J.. Developing mRNA-vaccine technologies. RNA Biol. 2012; 9:1319–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nair S.K., Boczkowski D., Morse M., Cumming R.I., Lyerly H.K., Gilboa E.. Induction of primary carcinoembryonic antigen (CEA)-specific cytotoxic T lymphocytes in vitro using human dendritic cells transfected with RNA. Nat. Biotech. 1998; 16:364–369. [DOI] [PubMed] [Google Scholar]
- 3. Weissman D., Ni H., Scales D., Dude A., Capodici J., McGibney K., Abdool A., Isaacs S.N., Cannon G., Karikó K.. HIV Gag mRNA transfection of dendritic cells (DC) delivers encoded antigen to MHC class I and II molecules, causes DC maturation, and induces a potent human in vitro primary immune response. J. Immunol. 2000; 165:4710–4717. [DOI] [PubMed] [Google Scholar]
- 4. Kormann M.S.D., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A. et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotech. 2011; 29:154–157. [DOI] [PubMed] [Google Scholar]
- 5. Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Karikó K.. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010; 38:5884–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nallagatla S.R., Bevilacqua P.C.. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008; 14:1201–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anderson B.R., Muramatsu H., Jha B.K., Silverman R.H., Weissman D., Karikó K.. Nucleoside modifications in RNA limit activation of 2΄-5΄-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011; 39:9329–9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anderson P., Kedersha N.. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 2008; 33:141–150. [DOI] [PubMed] [Google Scholar]
- 9. Karikó K., Muramatsu H., Ludwig J., Weissman D.. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011; 39:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weil T.T., Parton R.M., Davis I.. Making the message clear: visualizing mRNA localization. Trends Cell Biol. 2010; 20:380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ligon T.S., Leonhardt C., Rädler J.O.. Multi-level kinetic model of mRNA Delivery via transfection of lipoplexes. PLoS ONE. 2014; 9:e107148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leonhardt C., Schwake G., Stögbauer T.R., Rappl S., Kuhr J.-T., Ligon T.S., Rädler J.O.. Single-cell mRNA transfection studies: Delivery, kinetics and statistics by numbers. Nanomed.: Nanotechno. Biol. Med. 2014; 10:679–688. [DOI] [PubMed] [Google Scholar]
- 13. Lorenz C., Fotin-Mleczek M., Roth G., Becker C., Dam T.C., Verdurmen W.P.R., Brock R., Probst J., Schlake T.. Protein expression from exogenous mRNA: Uptake by receptor-mediated endocytosis and trafficking via the lysosomal pathway. RNA Biol. 2011; 8:627–636. [DOI] [PubMed] [Google Scholar]
- 14. Sahay G., Querbes W., Alabi C., Eltoukhy A., Sarkar S., Zurenko C., Karagiannis E., Love K., Chen D., Zoncu R. et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotech. 2013; 31:653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wittrup A., Ai A., Liu X., Hamar P., Trifonova R., Charisse K., Manoharan M., Kirchhausen T., Lieberman J.. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotech. 2015; 33:870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alonas E., Lifland A.W., Gudheti M., Vanover D., Jung J., Zurla C., Kirschman J., Fiore V.F., Douglas A., Barker T.H. et al. Combining single RNA sensitive probes with subdiffraction-limited and live-cell imaging enables the characterization of virus dynamics in cells. ACS Nano. 2014; 8:302–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lifland A.W., Zurla C., Santangelo P.J.. Single molecule sensitive multivalent polyethylene glycol probes for RNA imaging. Bioconj. Chem. 2010; 21:483–488. [DOI] [PubMed] [Google Scholar]
- 18. Lifland A.W., Zurla C., Yu J., Santangelo P.J.. Dynamics of native β-actin mRNA transport in the cytoplasm. Traffic (Copenhagen, Denmark). 2011; 12:1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Santangelo P.J., Lifland A.W., Curt P., Sasaki Y., Bassell G.J., Lindquist M.E., Crowe J.E.. Single molecule-sensitive probes for imaging RNA in live cells. Nat. Methods. 2009; 6:347–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zurla C., Lifland A.W., Santangelo P.J.. Characterizing mRNA interactions with RNA granules during translation initiation inhibition. PLoS ONE. 2011; 6:e19727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buxbaum A.R., Haimovich G., Singer R.H.. In the right place at the right time: visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell. Biol. 2015; 16:95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rejman J., Bragonzi A., Conese M.. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol. Ther. 2005; 12:468–474. [DOI] [PubMed] [Google Scholar]
- 23. Soderberg O., Gullberg M., Jarvius M., Ridderstrale K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G. et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006; 3:995–1000. [DOI] [PubMed] [Google Scholar]
- 24. Koos B., Andersson L., Clausson C.M., Grannas K., Klaesson A., Cane G., Soderberg O.. Analysis of protein interactions in situ by proximity ligation assays. Curr. Top. Microbiol. Immunol. 2014; 377:111–126. [DOI] [PubMed] [Google Scholar]
- 25. Leuchowius K.J., Weibrecht I., Soderberg O.. In situ proximity ligation assay for microscopy and flow cytometry. Curr. Protoc. Cytom. 2011; doi:10.1002/0471142956.cy0936s56. [DOI] [PubMed] [Google Scholar]
- 26. Soderberg O., Leuchowius K.J., Gullberg M., Jarvius M., Weibrecht I., Larsson L.G., Landegren U.. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods. 2008; 45:227–232. [DOI] [PubMed] [Google Scholar]
- 27. Weibrecht I., Lundin E., Kiflemariam S., Mignardi M., Grundberg I., Larsson C., Koos B., Nilsson M., Soderberg O.. In situ detection of individual mRNA molecules and protein complexes or post-translational modifications using padlock probes combined with the in situ proximity ligation assay. Nat. Protoc. 2013; 8:355–372. [DOI] [PubMed] [Google Scholar]
- 28. Jung J., Lifland A.W., Alonas E.J., Zurla C., Santangelo P.J.. Characterization of mRNA-cytoskeleton interactions in situ using FMTRIP and proximity ligation. PLoS ONE. 2013; 8:e74598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jung J., Lifland A.W., Zurla C., Alonas E.J., Santangelo P.J.. Quantifying RNA–protein interactions in situ using modified-MTRIPs and proximity ligation. Nucleic Acids Res. 2013; 41:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lifland A.W., Jung J., Alonas E., Zurla C., Crowe J.E., Santangelo P.J.. Human respiratory syncytial virus nucleoprotein and inclusion bodies antagonize the innate immune response mediated by MDA5 and MAVS. J. Virol. 2012; 86:8245–8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wigington C.P., Jung J., Rye E.A., Belauret S.L., Philpot A.M., Feng Y., Santangelo P.J., Corbett A.H.. Post-transcriptional regulation of programmed cell death 4 (PDCD4) mRNA by the RNA-binding proteins human antigen R (HuR) and T-cell intracellular antigen 1 (TIA1). J. Biol. Chem. 2015; 290:3468–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cui S., Wang B., Zhao Y., Chen H., Ding H., Zhi D., Zhang S.. Transmembrane routes of cationic liposome-mediated gene delivery using human throat epidermis cancer cells. Biotechnol. Lett. 2014; 36:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wasungu L., Hoekstra D.. Cationic lipids, lipoplexes and intracellular delivery of genes. J. Controlled Rel. 2006; 116:255–264. [DOI] [PubMed] [Google Scholar]
- 34. Vaughan E.E., Dean D.A.. Intracellular trafficking of plasmids during transfection is mediated by microtubules. Mol. Ther. 2006; 13:422–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heinicke L.A., Nallagatla S.R., Hull C.M., Bevilacqua P.C.. RNA helical imperfections regulate activation of the protein kinase PKR: effects of bulge position, size, and geometry. RNA. 2011; 17:957–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Toroney R., Bevilacqua P.C.. PKR and the ribosome compete for mRNA. Nat. Chem. Biol. 2009; 5:873–874. [DOI] [PubMed] [Google Scholar]
- 37. Warren L., Manos P.D., Ahfeldt T., Loh Y.-H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A. et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010; 7:618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Santangelo P.J., Alonas E., Jung J., Lifland A.W., Zurla C.. Probes for intracellular RNA imaging in live cells. Methods Enzymol. 2012; 505:383–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zurla C., Jung J., Blanchard E.L., Santangelo P.J.. A novel method to quantify RNA–protein interactions in situ using FMTRIP and proximity ligation. Methods Mol. Biol. 2017; 1468:155–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Raj A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., Tyagi S.. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008; 5:877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mayor S., Pagano R.E.. Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell. Biol. 2007; 8:603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bampton E.T., Goemans C.G., Niranjan D., Mizushima N., Tolkovsky A.M.. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy. 2005; 1:23–36. [DOI] [PubMed] [Google Scholar]
- 43. Das S., Pellett P.E.. Spatial relationships between markers for secretory and endosomal machinery in human cytomegalovirus-infected cells versus those in uninfected cells. J. Virol. 2011; 85:5864–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Loomis K.H., Kirschman J.L., Bhosle S., Bellamkonda R.V., Santangelo P.J.. Strategies for modulating innate immune activation and protein production of in vitro transcribed mRNAs. J. Mater. Chem. B. 2016; 4:1619–1632. [DOI] [PubMed] [Google Scholar]
- 45. Buchan J.R., Kolaitis R.-M., Taylor J.P., Parker R.. Eukaryotic stress granules are cleared by granulophagy and Cdc48/VCP function. Cell. 2013; 153:1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Buchan J.R., Parker R.. Eukaryotic stress granules: the ins and out of translation. Mol. Cell. 2009; 36:932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mehier-Humbert S., Guy R.H.. Physical methods for gene transfer: improving the kinetics of gene delivery into cells. Adv. Drug Del. Rev. 2005; 57:733–753. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.