Abstract
Spliceosome activity is tightly regulated to ensure adequate splicing in response to internal and external cues. It has been suggested that core components of the spliceosome, such as the snRNPs, would participate in the control of its activity. The experimental indications supporting this proposition, however, remain scarce, and the operating mechanisms poorly understood. Here, we present genetic and molecular evidence demonstrating that the LSM2–8 complex, the protein moiety of the U6 snRNP, regulates the spliceosome activity in Arabidopsis, and that this regulation is controlled by the environmental conditions. Our results show that the complex ensures the efficiency and accuracy of constitutive and alternative splicing of selected pre-mRNAs, depending on the conditions. Moreover, miss-splicing of most targeted pre-mRNAs leads to the generation of nonsense mediated decay signatures, indicating that the LSM2–8 complex also guarantees adequate levels of the corresponding functional transcripts. Interestingly, the selective role of the complex has relevant physiological implications since it is required for adequate plant adaptation to abiotic stresses. These findings unveil an unanticipated function for the LSM2–8 complex that represents a new layer of posttranscriptional regulation in response to external stimuli in eukaryotes.
INTRODUCTION
In eukaryotes, introns are removed from the precursor mRNAs (pre-mRNAs) in a process known as splicing. The splicing is catalyzed by a highly evolutionary conserved multimegadalton ribonucleoprotein (RNP) complex, the spliceosome, constituted by five small nuclear RNPs (snRNPs) and many non-snRNP proteins (1). The snRNPs are composed by small nuclear uridine-rich RNAs (U1, U2, U4, U5 and U6 snRNAs) and their corresponding interacting proteins (1), and can be classified in two groups depending on these proteins. One group comprises U1, U2, U4 and U5 snRNPs, whose snRNAs interact with the Sm multiprotein complex. The U6 snRNA, meanwhile, interacts with the related Sm-like (LSM) 2–8 heteroheptameric complex (2), and forms the other group. The splicing is a highly dynamic process in which the spliceosome undergoes many conformational and compositional changes to coordinate a sequential assembly of snRNPs and a variable array of non-snRNP proteins aimed to generate functional spliceosomal subcomplexes. These subcomplexes display specific roles, depending on the snRNPs they include. U1 and U2 snRNPs define those responsible for the recognition of the splicing sites (E and A subcomplexes), while subsequent substitution of U1 by U5 and U6 snRNPs ends up with the formation of the catalytic subcomplexes (B* and C) (1). Among all snRNPs, the U6 snRNP seems to have a critical function in the catalytic reaction that takes place in the splicing and, accordingly, the U6 is the snRNA with the highest evolutionary conservation (3).
In recent years, numerous studies have investigated the mechanisms controlling the spliceosome activity. Results revealed that the determination of pre-mRNA splicing patterns is subjected to a tight regulation involving several RNA sequence elements and non-spliceosomal protein regulators such as the serine/arginine (SR)-rich and heterogeneous nuclear RNPs (hnRNPs) proteins (4). The interaction of these regulators with the cis-elements largely determines the spliceosome activity. The different regulatory elements and proteins allow a variable assembling of introns and exons that, in turn, results in the formation of various mRNA transcript isoforms from the same pre-mRNA in one of the most versatile posttranscriptional regulatory mechanisms of gene expression named alternative splicing. This mechanism, prevalent in higher eukaryotes, significantly enhances transcriptome and proteome plasticity, and plays a relevant role in controlling the responses to external and internal stimuli (4–6). Interestingly, recent data suggest that, in addition to the non-spliceosomal proteins (i.e. SRs and hnRNPs), some core components of the spliceosome (i.e. snRNPs) might also be involved in modulating the alternative splicing. Thus, in human cells, low levels of SmB/B΄, an integral part of U1, U2, U4 and U5 snRNPs, originate a reduction in the number of alternative exons present in several transcripts encoding RNA-processing and RNA-binding factors (7). Moreover, the functional characterization of Arabidopsis mutants lsm4 and lsm5/sad1 revealed that the core components of the splicing machinery LSM4 and LSM5 proteins are required for the correct alternative splicing of several genes related to salt response and the circadian clock (8–10). Similarly, a decrease in the levels of human LSM2, 3, 4, 6 and 7 proteins, lead to alterations in the alternative splicing patterns of genes implicated in cell proliferation and/or apoptosis (11).
The LSMs are evolutionary conserved RNA-binding proteins, typically associated in heteroheptameric ring shaped complexes, with a function in mRNA metabolism. We reported that in Arabidopsis, as in yeast and animals, the eight canonical LSM proteins are organized in two heteroheptameric complexes, LSM1–7 and LSM2–8, specifically localized in the cytoplasm and nucleus, and defined by the subunits LSM1 and LSM8, respectively (12). The Arabidopsis LSM1–7 complex is involved in accurate mRNA turnover by promoting decapping and subsequent 5΄-3΄ degradation (12,13). The analysis of Arabidopsis null lsm8 mutants unveiled that LSM8 is essential for the assembly of the LSM nuclear complex and that this complex acts in pre-mRNA splicing through U6 snRNA stabilization, thus allowing the formation of the U6 snRNP (12). The capacity of the LSM2–8 complex to regulate the spliceosome activity, however, has not yet been established in any organism. As mentioned above, several studies indicate that some LSMs, including LSM2, 3, 4, 5, 6 and 7, may regulate alternative splicing in plants and animals (8–11,14). Still, it must be kept in mind that all these proteins, in addition to participating in the LSM2–8 complex, are also integral components of the LSM1–7 complex (12), which has been described to control the stability of transcripts encoding alternative splicing factors (15). Their role in alternative splicing regulation, therefore, might be more related to their participation in the decapping activation complex than in the U6 snRNP. Here, we present genetic and molecular evidence that the LSM2–8 complex, and consequently the U6 snRNP, regulates the spliceosome activity. Remarkably, our results show that the function of the complex in regulating the spliceosome activity is controlled by external signals. Indeed, genome-wide transcriptomic analyses of lsm8 plants demonstrate that the Arabidopsis LSM nuclear complex is essential for the correct constitutive and alternative splicing of selected pre-mRNAs, depending on the environmental situation, and to maintain adequate levels of the corresponding functional transcripts. The selective role of the complex has substantial physiological implications since it differentially modulates plant adaptation to abiotic stress conditions. Furthermore, our results also show that the LSM2–8 complex, and therefore the U6 snRNA, differentially accumulate in response to abiotic stresses, suggesting that the levels of U6 snRNP contribute to shape the spliceosome activity according to the external entourage. The uncovered function of LSM2–8 represents a new layer of posttranscriptional regulation in response to external stimuli in eukaryotes.
MATERIALS AND METHODS
Plant materials, growth conditions and tolerance assays
Arabidopsis thaliana Columbia (Col-0) ecotype was used in all experiments as wild-type (WT) plant. The lsm8-1 and lsm8-2 mutants and c-lsm8 plants were previously described (12). Plants were grown at 20°C in pots containing a mixture of organic substrate and vermiculite (3:1, v/v) or on Petri dishes containing  Murashige and Skoog medium, supplemented with 1% (w/v) sucrose and solidified with 0.9% (w/v) plant agar (GM medium), under long-day photoperiod (16 h of cool-white fluorescent light, photon flux of 90 μmol m−2 s−1). Low temperature treatments for gene expression and immunoblot analyses were performed by transferring 2-week-old plants to a growth chamber set to 4°C for different times under long-day photoperiods with a photon flux of 40 μmol m−2 s−1. Salt treatments for gene expression and immunoblot analyses were accomplished by transferring 2-week-old plants grown vertically in Petri dishes under standard conditions on nylon mesh to plates containing GM medium supplemented with 150 mM NaCl for different times. Tolerance to freezing temperatures was determined on 2-week-old plants grown on soil as previously described (16). Tolerance to salt stress was analyzed by transferring 7-day-old seedlings growing vertically in plates containing GM medium to new plates supplemented with 150 mM NaCl. The tolerance was quantified as the percentage of root length and fresh weight of the plants after one week of NaCl treatment with respect to non-stressed plants. In all cases, data reported are expressed as the mean and the standard error of the mean of six independent experiments with, at least, 40 plants each.
Murashige and Skoog medium, supplemented with 1% (w/v) sucrose and solidified with 0.9% (w/v) plant agar (GM medium), under long-day photoperiod (16 h of cool-white fluorescent light, photon flux of 90 μmol m−2 s−1). Low temperature treatments for gene expression and immunoblot analyses were performed by transferring 2-week-old plants to a growth chamber set to 4°C for different times under long-day photoperiods with a photon flux of 40 μmol m−2 s−1. Salt treatments for gene expression and immunoblot analyses were accomplished by transferring 2-week-old plants grown vertically in Petri dishes under standard conditions on nylon mesh to plates containing GM medium supplemented with 150 mM NaCl for different times. Tolerance to freezing temperatures was determined on 2-week-old plants grown on soil as previously described (16). Tolerance to salt stress was analyzed by transferring 7-day-old seedlings growing vertically in plates containing GM medium to new plates supplemented with 150 mM NaCl. The tolerance was quantified as the percentage of root length and fresh weight of the plants after one week of NaCl treatment with respect to non-stressed plants. In all cases, data reported are expressed as the mean and the standard error of the mean of six independent experiments with, at least, 40 plants each.
Microscopy analysis
Subcellular localization of LSM8-GFP fusion protein was accomplished by confocal microscopy in roots of 7-day-old c-lsm8 seedlings grown on GM medium under control conditions or exposed 48 h to 4°C or 10 h to 150 mM NaCl. Microscopy images were collected using a Confocal Laser Spectral microscope TCS SP5 (Leica Mycrosystem). The excitation line for imaging GFP fusions was 488 nm.
Immunoblot analysis
Total protein extracts were obtained from c-lsm8 and WT plants grown under control conditions or exposed to 4°C or 150 mM NaCl for different times (see above). Proteins were separated electrophoretically in 15% SDS-polyacrylamide gels and blotted to Hybond-P PVDF membranes (Amersham) according to the manufacturer's protocol. A rabbit anti-GFP was used as primary antibody (Abcam; Cat. No. ab290), and a goat anti-rabbit IgG-HRP as secondary antibody (Santa Cruz Biotechnology; Cat. No. sc2030). Signals were detected with the ECL Western Blotting Detection Kit (Amersham), according to the manufacturer's protocol. In all cases, equal loading of samples was verified by staining the membranes with Coomassie Blue. All assays were performed in triplicate employing three independent biological samples.
Gene expression analysis and RNA sequencing experiments
For quantitative real-time PCR (qPCR) experiments, total RNA was extracted using TRIzol reagent (Life Technologies) and subsequently treated with DNase I (Roche). cDNA was synthesized with the iScript® cDNA Synthesis Kit, following the manufacturer's instructions (Bio-Rad), and used as a template for qPCR assays employing the SsoFast EvaGreen Supermix (Bio-Rad) in an iQ2 thermal cycler machine (Bio-Rad) with the specific primers listed in Supplementary Table S1. The relative expression values were calculated using the AT4G26410 gene as a reference (17). Fold change was calculated by means of the ΔΔCT method (18). All assays were performed in triplicate with three independent RNA samples.
For RNA sequencing (RNA-seq) experiments, total RNA was extracted with TRIzol reagent (Life Technologies) and purified with the RNeasy Plant Mini Kit (Qiagen). cDNA libraries were generated from three independent RNA preparations each. RNA quality determination, library preparation and subsequent sequencing in an Illumina HiSeq™ 2000 platform were performed by the staff of the Beijing Genome Institute. Approximately, 50 million 91 base pair (bp) paired-end reads per sample were generated and >90% reads were aligned to the TAIR10 Col-0 reference genome using SOAP2 (19) with default parameters. Gene expression levels were calculated using the RPKM (reads per kilobase per million reads) method (20). Differentially expressed genes (DEGs) were identified using the algorithm developed by Audic and Claverie (21) to obtain a P-value for each gene between any pair of samples. Then, a False Discovery Rate (FDR) analysis was performed to determine the threshold of P-values in multiple tests. We established a FDR ≤0.001 and a fold change ±2 as cut-offs for any given DEG.
Detection of differential alternative splicing events
To identify and quantify alternative splicing events, the reads from RNA-seq experiments were first mapped to the TAIR10 Col-0 reference genome using the TopHat software (22) with default parameters. Read alignments were then processed with the SAMtools software package (23) to generate a pileup file (coverage maps) for each sample. To detect alternative 5΄ splice site (A5΄SS), alternative 3΄ splice site (A3΄SS) and exon skipping (ES) events, we employed the SplAdder software (24) using the highest confidence level. This toolbox takes the RNA-Seq alignments generated by TopHat and the Arabidopsis annotation data (TAIR10) as inputs to yield a splicing graph representation of all annotated transcripts adding new exons and introns. To minimize the false positive rate, we only considered the events with at least five reads in any of the samples analyzed. In addition, for A5΄SS and A3΄SS events, we discarded reads with a length difference <6 bp between the detected alternative forms, and with a length difference respect to the corresponding annotated exons <4 bp. For ES events, we discarded events affecting exons <7 bp. To detect intron retention (IR) events, an in-house developed script was used. Introns were defined as regions that are flanked by two exons and do not overlap 100% with exons, or combinations of exons, annotated in TAIR10 reference genome. The number of reads mapping to these intron regions was calculated from the coverage maps generated with SAMtools. In these analyses, we considered as retained introns only those displaying a mean of more than five reads per base throughout their sequences in, at least, one of the samples. Intron retention levels were always calculated as a ratio between the expression levels of the introns and those of the corresponding genes. Introns detected as retained in WT plants were annotated as alternatively spliced. All introns not annotated as alternatively spliced in WT plants were classified as constitutively spliced. In all cases (A5΄SS, A3΄SS, ES and IR), events with fold change ±2 in lsm8-1 respect to WT plants and Q-value ≤ 0.05, calculated by Fisher's exact tests, were regarded as significant differential ones. Events exclusively observed in the lsm8-1 mutant, and with a mean of, at least, five reads per base throughout their sequences, were also considered significant and their assigned Q-value was zero.
Validation of intron retention events
Intron retention events were validated by qPCR using specific primers (Supplementary Table S1). qPCR reactions were always carried out in triplicate with three RNA samples different from those employed in the RNA-seq experiments. The expression of AT4G26410 was used as reference to normalize the data obtained (17).
Gene ontology enrichment analysis
Gene ontology (GO) categorization was performed with the ThaleMine data mining tool from Araport (www.araport.org). Significantly enriched GO terms (P-value ≤ 0.05) were established using a Holm-Bonferroni corrected hypergeometric test.
Sequence analysis
The size and GC content of introns and exons were calculated from the TAIR10 reference genome. The identification of mRNAs with nonsense-mediated decay (NMD) features generated by altered splicing events was based on the TAIR10 reference genome and the criteria previously described (25) (i.e. premature termination codon and a 3΄UTR longer than 350nt, 3΄UTR longer than 350nt, more than 55nt between the stop codon and a downstream intron, upstream open reading frame (uORF) longer than 35 amino acids, uORF overlapping with the start codon of the main ORF). Searching for sequence motifs in introns was carried out by scanning their whole sequence using the MEME suit (www.meme-suit.org). The nucleotide frequencies around the 5΄ and 3΄ splice sites (five nucleotides in each side of the exon/intron junctions) and in the branch site were determined and displayed as a sequence logo with the WebLogo application (http://weblogo.threeplusone.com/). The sequences of the exon/intron junctions were obtained from the TAIR10 reference genome. The sequence of the branch site for each intron was obtained from the database of plant splice sites and splicing signals ERISdb (26).
Statistical analysis
Data sets were statistically analyzed with Prism 6 software (GraphPad Software Inc., USA). Comparisons between two groups were made by using the one‐tailed t-test. Comparisons between multiple groups were made by one‐way or two-way ANOVA followed by Dunnett test depending whether one or two variables were considered, respectively.
Data availability
The complete genome-wide data from this publication were submitted to the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/) under accession number GSE87415. The full names of the genes mentioned in this article are included in Supplementary Table S2.
RESULTS
The LSM2–8 complex controls the efficiency and accuracy of constitutive and alternative pre-mRNA splicing in Arabidopsis
As mentioned above, we previously described that the Arabidopsis LSM2–8 complex is required for pre-mRNA splicing through U6 snRNA stabilization (12). To establish its possible function in regulating the efficiency and accuracy of constitutive and/or alternative pre-mRNA splicing, we performed a high-coverage RNA-seq analysis on 2-week-old WT and lsm8-1 plants grown in soil at 20°C. To identify and quantify alternative splicing events, the resulting reads (around 50 million, 91 bp paired-end per genotype) were mapped to the Arabidopsis genome (TAIR10 version) using the TopHat software (22), and read alignments were processed with SAMtools and SplAdder softwares (23,24). To identify IR events, in addition to the TopHat and SAMtools softwares, an in-house script was used. Introns were defined as regions flanked by two exons, as annotated in TAIR10, and having no overlap with them or with any other combination of exons. In addition, only introns displaying a mean of more than five reads per base throughout their sequence in, at least, one of the samples were considered as retained introns. Intron retention levels were always calculated as a ratio between the expression levels of the introns and those of the corresponding genes.
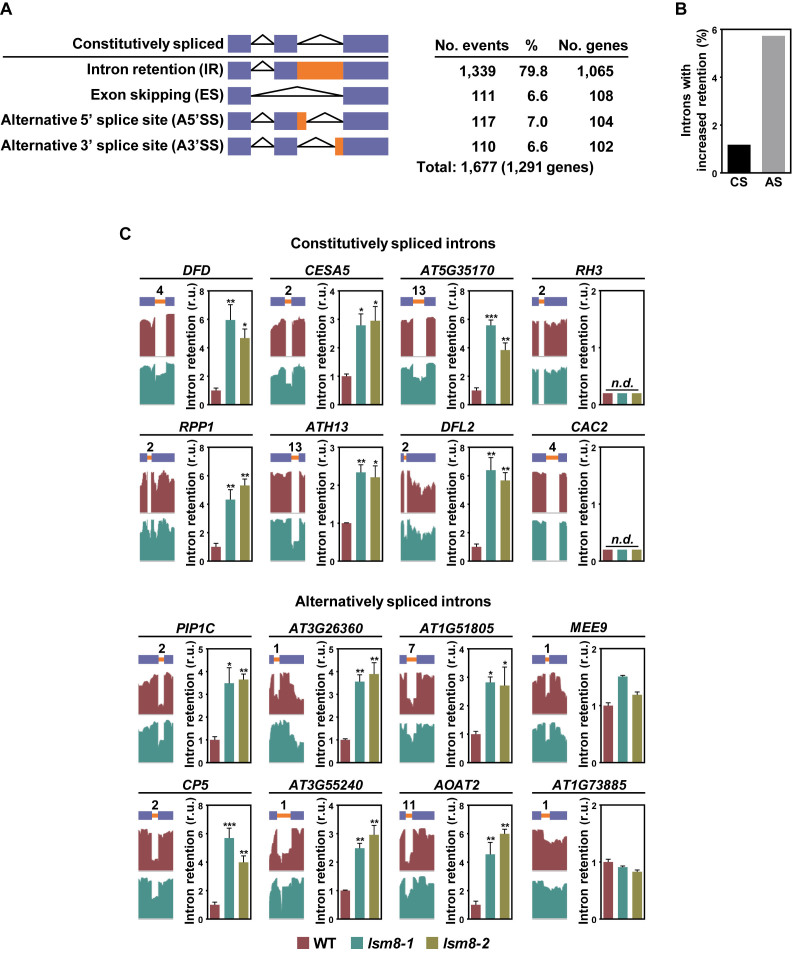
When comparing data obtained from lsm8-1 with those from WT plants, mutants showed alterations in 1677 splicing events corresponding to 1291 different genes (fold change ±2; Fisher's exact test, Q ≤ 0.05) (Supplementary Table S3). These alterations could be pooled into four main categories, namely IR (79.8%), ES (6.6%), A5΄SS (7.0%) and A3΄SS (6.6%) (Figure 1A). Intron retention was by far the most abundant category and, consequently, we mainly focused on this type of anomalies when proceeding with our study. Data revealed that the LSM2–8 complex controlled the adequate removal of 1339 introns from 1065 genes, 932 corresponding to constitutively and 407 to alternatively spliced introns (Figure 1A and Supplementary Table S3). These numbers represented around 1.2% and 5.7% of all constitutively (79 081) and alternatively (7099) spliced introns identified in WT plants under our standard conditions, which were similar to those reported earlier (10) (Figure 1B). Results from RNA-seq experiments were validated by analyzing, in independent RNA samples, the retention of a number of constitutively and alternatively spliced introns in lsm8-1 and lsm8-2 mutants by means of qPCR assays. Constitutively and alternatively spliced introns not affected by the lsm8-1 mutation were also validated. In all cases, the retention patterns were coincident with those inferred from RNA-seq experiments (Figure 1C), indicating a low false positive rate.
Figure 1.
The LSM2–8 complex controls constitutive and alternative splicing in Arabidopsis. (A) Quantification of altered splicing events (IR, ES, A5΄SS, A3΄SS) identified in lsm8-1 respect to WT plants under control conditions. Blue bars represent exons. Orange bars represent intron regions retained in some events. The total number of different genes affected by the ensemble of altered splicing events identified is shown in parentheses. (B) Percentage of constitutively (CS) and alternatively spliced (AS) introns with increased retention in lsm8-1 plants relative to the total number of the corresponding introns identified in WT plants. (C) Different IR events identified in lsm8-1 plants. For each event, the name of the gene containing the corresponding constitutively or alternatively spliced intron is indicated. A diagram of the pre-mRNA regions, including the retained introns (orange bars) with their relative positions in the representative gene model and the flanking exons (blue bars), together with the captures of the corresponding read coverage tracks obtained from the IGV software is shown in the left. The quantification of retained introns in WT, lsm8-1 and lsm8-2 plants by qPCR assays is displayed in the right. Bars indicate the standard error of the mean (n = 3). Asterisks (*) indicate significant differences (*P ≤ 0.01, **P ≤ 0.001, ***P ≤ 0.0001) between lsm8 mutants and WT plants, as determined by ANOVA-test. n.d. indicates non-detected events by qPCR.
Intron retention events frequently originate transcripts with NMD signatures that are poorly translated (25). These events, therefore, may influence the levels of functional transcripts and, thus, the corresponding proteins. Interestingly, the introns retained in lsm8-1 plants, constitutively and alternatively spliced, gave rise to 890 mRNAs (83.6% of all mRNAs with IR events) displaying one or more features of NMD targeted transcripts (Table 1). We concluded, therefore, that the LSM2–8 complex plays a critical role in controlling the efficiency and accuracy of both constitutive and alternative pre-mRNA splicing in Arabidopsis, this role being more significant on the latter, and the levels of functional transcripts.
Table 1. Introns retained in lsm8-1 mutants under control (20°C), cold (4°C) or high salt (NaCl) conditions that generate transcripts with NMD features.
| Environmental condition | Intron type | PTCa | 3΄UTR > 350ntb | SC→intron > 55ntc | 5΄uORFd | uORF overlape | Genesf (%) | Non-NMD genesg (%) |
|---|---|---|---|---|---|---|---|---|
| 20°C | Constitutive | 735 | 8 | 3 | 7 | 7 | 890 (83.6) | 175 (16.4) |
| Alternative | 313 | 7 | 4 | 3 | 2 | |||
| 4°C | Constitutive | 442 | 7 | 3 | 11 | 4 | 531 (81.7) | 119 (18.3) |
| Alternative | 161 | 2 | 0 | 2 | 0 | |||
| NaCl | Constitutive | 1442 | 20 | 18 | 56 | 16 | 1454 (85.4) | 249 (14.6) |
| Alternative | 294 | 10 | 3 | 7 | 6 |
aPremature termination codon (PTC) and a 3΄UTR longer than 350nt.
b3΄UTR longer than 350nt.
cMore than 55 nt between stop codon (SC) and a downstream intron.
dUpstream open reading frame (uORF) longer than 35 amino acids.
euORF overlapping with the start codon of the main ORF.
fTotal number of genes containing at least one retained intron causing NMD features.
gTotal number of genes containing retained introns that do not generate NMD features.
The Arabidopsis LSM2–8 complex controls the constitutive and alternative splicing of specific introns from selected pre-mRNAs depending on the environmental conditions
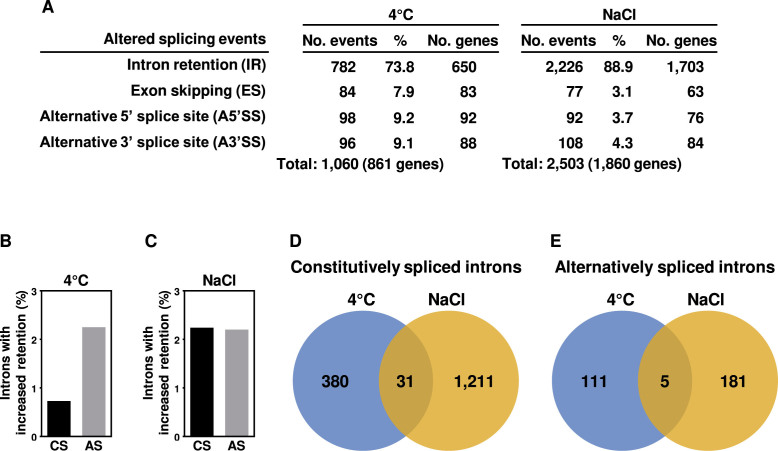
Recent findings from our laboratory uncovered that the Arabidopsis LSM1–7 cytoplasmic complex targets specific transcripts for decapping and subsequent degradation, depending on the environment (13). Then, we decided to explore the possibility that the LSM2–8 complex could also differentially control the constitutive and/or alternative splicing of specific introns, depending on the environmental conditions. With this aim, we first carried out a high-coverage RNA-seq analysis on 2-week-old WT and lsm8-1 plants exposed to low temperature (4°C, 24 h) or high salt (150 mM NaCl, 10 h). In the case of cold treatment, plants were grown in soil. For salt application, plants were grown on GM medium. After processing reads (∼50 million, 91 bp paired-end per genotype and condition) as described above, we identified 1060 and 2503 altered splicing events (fold change ±2; Fisher΄s exact test, Q ≤ 0.05) in lsm8-1 plants subjected to cold and salt stress, corresponding to 861 and 1860 different genes, respectively (Figure 2A and Supplementary Tables S4 and S5). Again, IR was the most represented event in both challenging conditions (73.8% under cold and 88.9% under high salt) (Figure 2A and Supplementary Tables S4 and S5). Mutant plants exposed to low temperature exhibited 782 introns from 650 genes with anomalous splicing, 575 corresponding to constitutively and 207 to alternatively spliced introns (Supplementary Table S4). These introns represented about 0.7% and 2.3% of all constitutively and alternatively spliced introns identified in cold-treated WT plants, respectively (Figure 2B). When subjected to high salt, lsm8-1 plants displayed alterations in the splicing of 2226 introns belonging to 1703 genes. On this occasion, 1845 corresponded to constitutively and 381 to alternatively spliced introns (Supplementary Table S5), which roughly represented 2.2% of all constitutively and alternatively spliced introns detected in salt-treated WT plants (Figure 2C). Hence, the LSM2–8 complex differentially regulates the efficiency and accuracy of both constitutive and alternative pre-mRNA splicing in Arabidopsis in response to cold and salt stress. Notably, while in the case of cold stress the function of the complex is more influential on alternative than on constitutively pre-mRNA splicing, under salt stress the complex acts in a similar way on both types of splicing.
Figure 2.
The LSM2–8 complex differentially controls constitutive and alternative splicing in Arabidopsis plants exposed to low temperature and high salt conditions. (A) Quantification of altered splicing events (IR, ES, A5΄SS, A3΄SS) identified in lsm8-1 respect to WT plants under cold (4°C) or high salt (NaCl) conditions. The total number of different genes affected by the ensemble of altered splicing events identified is shown in parentheses. (B and C) Percentage of constitutively (CS) and alternatively spliced (AS) introns with increased retention in lsm8-1 plants exposed to cold (4°C) (B) or salt stress (NaCl) (C) relative to the total number of the corresponding introns identified in WT plants subjected to the same conditions. (D and E) Venn diagrams showing the overlap between the constitutively (D) and alternatively (E) spliced introns specifically retained in lsm8-1 with respect to WT plants under cold (4°C) or high salt (NaCl) conditions. The number of specific and common genes identified is indicated.
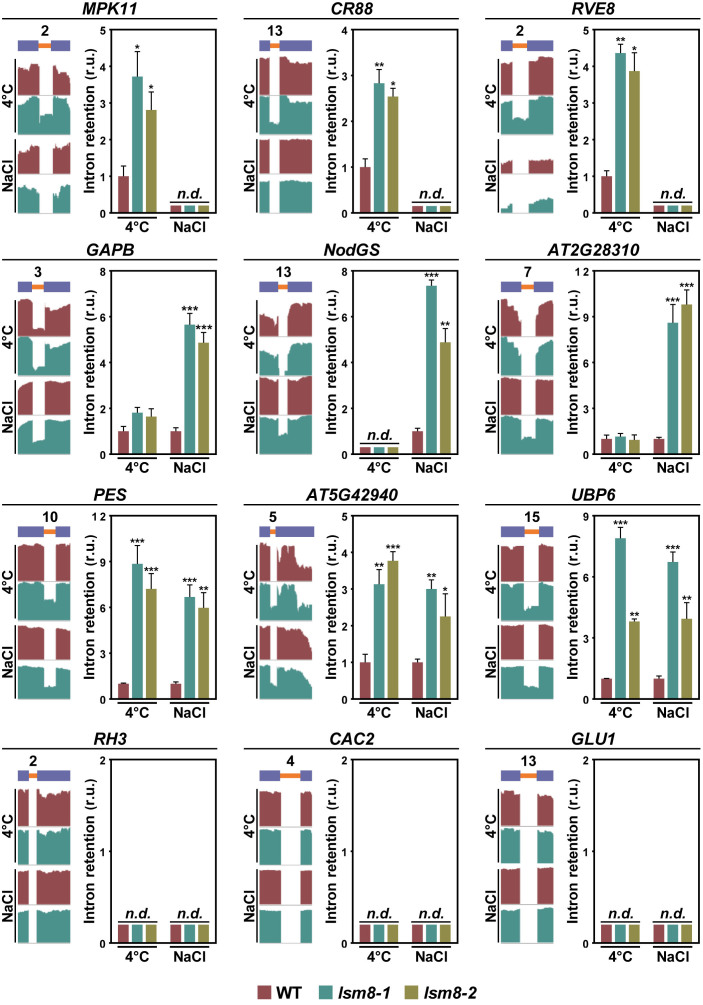
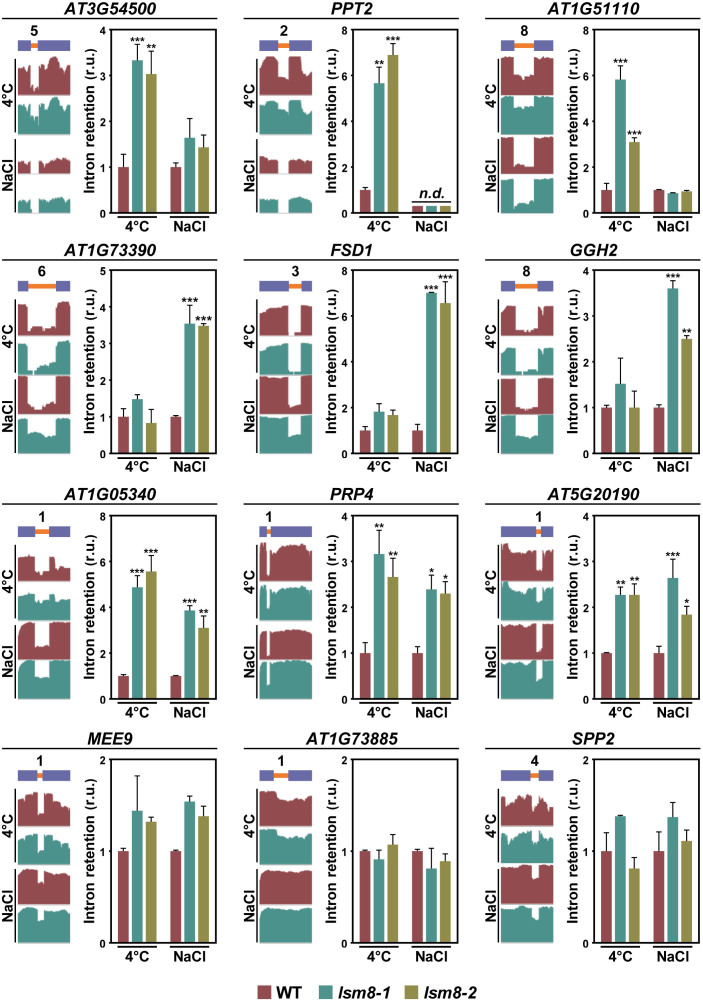
The different impact of the lsm8-1 mutation on constitutive and alternative splicing of pre-mRNAs in response to low temperature and salinity strongly suggested that, indeed, according to our assumption, the Arabidopsis LSM nuclear complex could control the splicing of specific introns from selected pre-mRNAs, depending on the environmental circumstances. To confirm this possibility, we compared the introns targeted by the complex under cold (Supplementary Table S4) or high salt (Supplementary Table S5) but not under any of the control conditions [soil (Supplementary Table S3) and GM medium (Supplementary Table S6)]. Remarkably, 380 (92.5%) and 1211 (97.5%) constitutively spliced introns resulted to be specifically regulated by LSM2–8 in response to low temperature and high salt, respectively (Figure 2D and Supplementary Table S7). In the case of alternatively spliced introns, 111 (95.7%) and 181 (97.3%) were specifically controlled by the complex under cold and salt stress, respectively (Figure 2E and Supplementary Table S8). Only 31 constitutively (Figure 2D) and 5 alternatively (Figure 2E) spliced introns were regulated by LSM2–8 under both abiotic stresses (Supplementary Table S9). qPCR assays of several specific and nonspecific IR events in independent RNA samples from WT, lsm8-1 and lsm8-2 plants exposed to low temperature or high salt were performed to validate the data from RNA-seq analyses. The events assayed corresponded to constitutively (Figure 3) and alternatively (Figure 4) spliced introns, affected and not affected by the lsm8-1 mutation. In all cases, the retention patterns were coincident with those inferred from RNA-seq experiments, indicating that the rate of false positives was very low.
Figure 3.
The Arabidopsis LSM2–8 complex controls the splicing of specific constitutively spliced introns in response to different environmental conditions. Different constitutively spliced introns specifically retained in lsm8 plants under cold (top line), high salt (second line) or both conditions (third line). The bottom line shows constitutively spliced introns non-affected in lsm8 plants under any condition. The name of the gene containing the corresponding constitutively spliced intron is indicated in each case. A diagram of the pre-mRNA regions, including the retained introns (orange bars) with their relative positions in the representative gene model and the flanking exons (blue bars), together with the captures of the corresponding read coverage tracks obtained from the IGV software is shown in the left. The quantification of retained introns in WT, lsm8-1 and lsm8-2 plants exposed to low temperature (4°C) or salt stress (NaCl) by qPCR assays is displayed in the right. Bars indicate the standard error of the mean (n = 3). Asterisks (*) indicate significant differences (*P ≤ 0.01, **P ≤ 0.001, ***P ≤ 0.0001) between lsm8 mutants and WT plants, as determined by ANOVA-test. n.d. indicates non-detected events by qPCR.
Figure 4.
The Arabidopsis LSM2–8 complex controls the splicing of specific alternatively spliced introns in response to different environmental conditions. Different alternatively spliced introns specifically retained in lsm8 plants under cold (top line), high salt (second line) or both conditions (third line). The bottom line shows alternatively spliced introns non-affected in lsm8 plants under any condition. The name of the gene containing the corresponding alternatively spliced intron is indicated in each case. A diagram of the pre-mRNA regions, including the retained introns (orange bars) with their relative positions in the representative gene model and the flanking exons (blue bars), together with the captures of the corresponding read coverage tracks obtained from the IGV software is shown in the left. The quantification of retained introns in WT, lsm8-1 and lsm8-2 plants exposed to low temperature (4°C) or salt stress (NaCl) by qPCR assays is displayed in the right. Bars indicate the standard error of the mean (n = 3). Asterisks (*) indicate significant differences (*P ≤ 0.01, **P ≤ 0.001, ***P ≤ 0.0001) between lsm8 mutants and WT plants, as determined by ANOVA-test. n.d. indicates non-detected events by qPCR.
A detailed sequence analysis of all introns, constitutively and alternatively spliced, retained in lsm8-1 under cold or salt stress revealed that, as in control conditions, most of them led to pre-mRNAs with characteristics of NMD targets. Particularly, 81.7% and 85.4% of the pre-mRNAs containing IR events in lsm8-1 plants exposed to 4°C or NaCl, respectively, displayed one or more NMD features (Table 1). Moreover, as expected, most introns retained in lsm8-1 specifically under these stress conditions also led to pre-mRNAs with NMD signatures (79.8% in response to low temperature and 83.9% to high salt) (Table 2), strongly suggesting a function for the LSM nuclear complex in maintaining the levels of selected functional transcripts when plants are exposed to challenging situations. On the whole, all results described above provided genetic and molecular evidence that the Arabidopsis LSM2–8 complex controls the correct splicing of selected pre-mRNAs in response to different environmental conditions and, more important, that this differential control is carried out by targeting specific constitutively and alternatively spliced introns, depending on the condition to which plants are exposed. Furthermore, our data indicated that the complex is essential to guarantee appropriate levels of functional transcripts, and in all likelihood proteins, corresponding to the genes containing the targeted introns.
Table 2. Introns retained in lsm8-1 mutants specifically under cold (4°C) or high salt (NaCl) conditions that generate transcripts with NMD features.
| Environmental condition | Intron type | PTCa | 3΄UTR > 350ntb | SC→intron > 55ntc | 5΄uORFd | uORF overlape | Genesf (%) | Non-NMD genesg (%) |
|---|---|---|---|---|---|---|---|---|
| 4°C | Constitutive | 283 | 4 | 2 | 8 | 2 | 344 (79.8) | 87 (20.2) |
| Alternative | 85 | 2 | 0 | 0 | 0 | |||
| NaCl | Constitutive | 955 | 12 | 11 | 18 | 6 | 1009 (83.9) | 194 (16.1) |
| Alternative | 145 | 4 | 1 | 3 | 2 |
aPremature termination codon (PTC) and a 3΄UTR longer than 350nt.
b3΄UTR longer than 350nt.
cMore than 55 nt between stop codon (SC) and a downstream intron.
dUpstream open reading frame (uORF) longer than 35 amino acids.
euORF overlapping with the start codon of the main ORF.
fTotal number of genes containing at least one retained intron causing NMD features.
gTotal number of genes containing retained introns that do not generate NMD features.
The Arabidopsis LSM2–8 complex differentially regulates plant tolerance to abiotic stresses by controlling the constitutive and alternative splicing of specific introns from selected abiotic stress-related pre-mRNAs
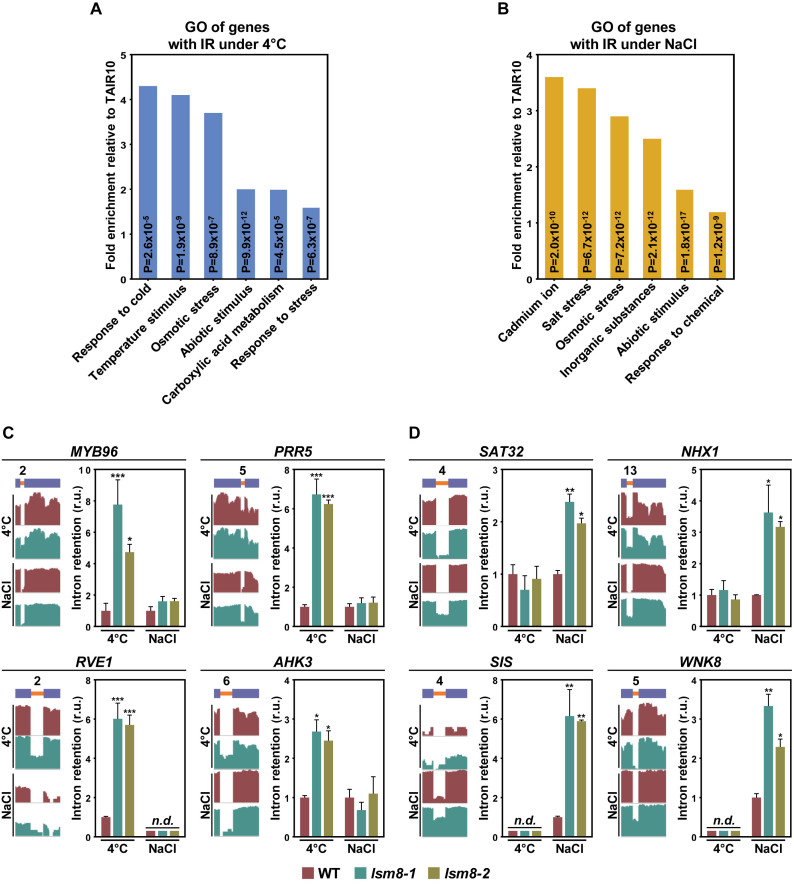
Interestingly, gene ontology (GO) analysis revealed that a significant number of the genes identified in our RNA-seq experiments that contained introns specifically targeted by the LSM2–8 complex under cold or high salt conditions were involved in abiotic stress responses. Out of the first six enriched GO categories, ‘response to cold’ (GO: 0009409) was the one with a highest value of fold enrichment (fold change = 4.3; P = 2.6 × 10−5) among the genes with introns targeted at 4°C (Figure 5A and Supplementary Table S10). Among the genes with introns targeted in response to salt stress, ‘response to salt stress’ (GO: 0009651) was the second GO category with higher fold enrichment (fold change = 3.4; P = 6.7 × 10−12) (Figure 5B and Supplementary Table S11). It is worth noting, furthermore, that some of them had even been reported to play a role in Arabidopsis tolerance to freezing and/or salt stress. These RNA-seq results were validated by analyzing the retention of several constitutively and alternatively spliced introns, whose adequate removal was specifically controlled by the LSM nuclear complex under low temperature or high salt conditions, in independent RNA samples from WT, lsm8-1 and lsm8-2 plants exposed 24 h to 4°C or 10 h to 150 mM NaCl through qPCR assays. Thus, we investigated introns belonging to genes described to regulate freezing tolerance, such as the transcriptional regulators MYB96, PRR5 and RVE1 or the cytokine receptor AHK3 (27–30), or salt tolerance, including the interferon-related developmental regulator SAT32, the Na+/H+ antiporter NHX1, the E3 ubiquitin ligase SIS or the protein kinase WNK8 (31–34), in Arabidopsis. Figures 5C and D display that, in all cases, the retention patterns observed were as those deduced from RNA-seq experiments.
Figure 5.
The LSM2–8 complex controls the splicing of specific introns from genes encoding key regulators of Arabidopsis tolerance to freezing temperatures and high salt conditions. (A and B) First six gene ontology (GO) terms enriched in genes containing constitutively and alternatively spliced introns specifically retained in lsm8-1 plants exposed to low temperature (A) or high salt conditions (B). Bars represent the fold enrichment of genes with retained introns in lsm8-1 plants relative to total number of annotated genes in the Arabidopsis genome (TAIR10) mapped to each term. The P value of each GO term is indicated inside its corresponding bar. (C and D) Different constitutively and alternatively spliced introns specifically retained in lsm8 plants under cold or high salt conditions that belong to genes involved in Arabidopsis tolerance to freezing temperatures (C) or high salinity (D). The name of the gene containing the corresponding intron is indicated in each case. A diagram of the pre-mRNA regions, including the retained introns (orange bars) with their relative positions in the representative gene model and the flanking exons (blue bars), together with the captures of the corresponding read coverage tracks obtained from the IGV software is shown in the left. The quantification of retained introns in WT, lsm8-1 and lsm8-2 plants exposed to low temperature (4°C) or salt stress (NaCl) by qPCR assays is displayed in the right. Bars indicate the standard error of the mean (n = 3). Asterisks (*) indicate significant differences (*P ≤ 0.01, **P ≤ 0.001, ***P ≤ 0.0001) between lsm8 mutants and WT plants, as determined by ANOVA-test. n.d. indicates non-detected events by qPCR.
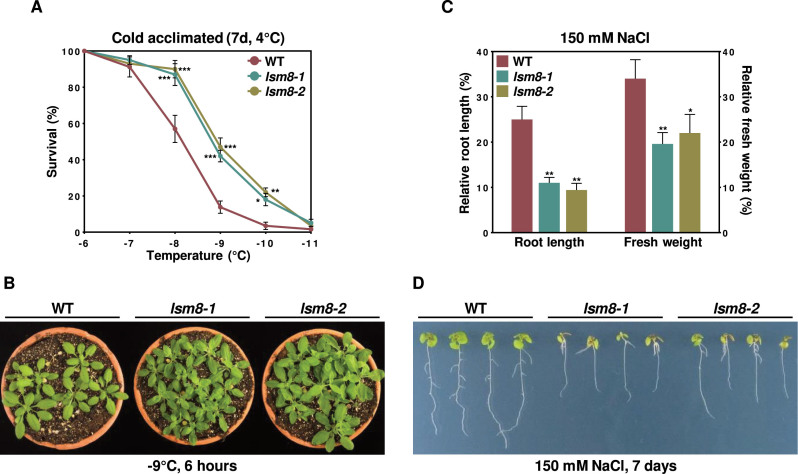
Considering that 79.8% and 83.9% of the introns specifically retained in lsm8-1 plants under cold or salt stress, respectively, led to the generation of pre-mRNA isoforms with features of NMD targets (Table 2), the data reported above indicated that the LSM2–8 complex could be implicated in Arabidopsis tolerance to freezing and high salt. Freezing tolerance was analyzed in non-acclimated and cold acclimated (4°C, 7 days) 2-week-old WT, lsm8-1 and lsm8-2 plants exposed 6 h to various freezing temperatures. Non-acclimated mutants presented a similar capacity to tolerate freezing as the WT, the LT50 (temperature that causes 50% lethality) values being in both cases around –6.2°C (Supplementary Figure S1). In contrast, the freezing tolerance of cold acclimated lsm8-1 and lsm8-2 plants was significantly higher than that of WT. In this case, the calculated LT50 values were –8.2°C for WT plants and –9.0°C for lsm8 mutants (Figure 6A and B). Salt tolerance was assayed in 7-day-old WT, lsm8-1 and lsm8-2 seedlings grown one additional week on plates containing 150 mM NaCl. Mutants displayed shorter main roots and lower fresh weights than WT seedlings (Figure 6C and D), manifesting their higher sensitivity to salt stress. All these findings suggested that, in fact, the LSM2–8 complex would differentially regulate Arabidopsis tolerance to abiotic stresses by ensuring the adequate levels of functional transcripts from selected genes having critical functions in tolerance to such adverse environments through the accurate splicing of specific introns contained in those genes.
Figure 6.
The LSM2-8 complex differentially regulates abiotic stress tolerance inArabidopsis. (A) Freezing tolerance of cold acclimated 2-week-old WT, lsm8-1 and lsm8-2 plants exposed 6 h to the indicated freezing temperatures. Freezing tolerance was estimated as the percentage of plants surviving each specific temperature after 7 d of recovery under control conditions. (B) Representative cold acclimated plants 7 d after being exposed to -9°C for 6 h. (C) Salt tolerance of 7-day-old WT, lsm8-1 and lsm8-2 seedlings. Salt tolerance was calculated as the relative root length and fresh weight of seedlings exposed 7 d to 150 mM NaCl respect to seedlings grown under control conditions. (D) Representative seedlings exposed to 150 mM NaCl for 7 d. In all graphs, bars indicate the standard error of the mean (n=6). Asterisks (*) indicate significant differences (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001) between lsm8-1 or lsm8-2 and WT plants or seedlings, as determined by ANOVA test.
The Arabidopsis LSM2–8 complex differentially accumulates in response to abiotic stresses resulting in distinctive levels of U6 snRNA
Results reported above indicated that the Arabidopsis LSM2–8 complex is involved in controlling plant responses to abiotic stresses. It was conceivable, therefore, that the levels of the complex could themselves be regulated by those challenging situations in order to function adequately. We first assessed this possibility by investigating the expression patterns of LSM8, the gene encoding the subunit that defines and determines the LSM nuclear complex (12), in 2-week-old WT plants exposed to 4°C or 150 mM NaCl. qPCR assays showed that the expression of LSM8 was positively and specifically regulated by low temperature, as described for the other Arabidopsis LSM genes (13). The levels of LSM8 transcripts increased under cold conditions, reaching a peak of induction after 48 h of treatment, but not by salt stress (Figure 7A). Then, we monitored whether the LSM8 protein accumulated in parallel with transcripts in response to abiotic stresses. Western-blot experiments using lsm8-1 mutants complemented with a genomic LSM8-GFP fusion driven by the LSM8 promoter (c-lsm8) (12) showed that, concomitantly with the transcript increase, the levels of LSM8-GFP protein progressively accumulated with the cold exposure. Salt stress, however, did not alter the levels of LSM8 protein (Figure 7B). Previous data revealed that LSM8 localizes diffusely into the nuclei of Arabidopsis cells at 20°C (12). Hence, we determined whether the subcellular localization of the LSM2–8 complex could be altered under cold or salt stresses. Confocal microscopy analyses pointed out that, consistent with the western-blot experiments, the levels of green fluorescence only increased in root tip cells from c-lsm8 seedlings exposed 48 h to 4°C. More important, they evidenced that LSM8 remained with a diffused distribution into the nuclei regardless of the stress conditions (Figure 7C). Together, these results demonstrated that the Arabidopsis LSM nuclear complex differentially accumulates in response to abiotic stresses.
Figure 7.
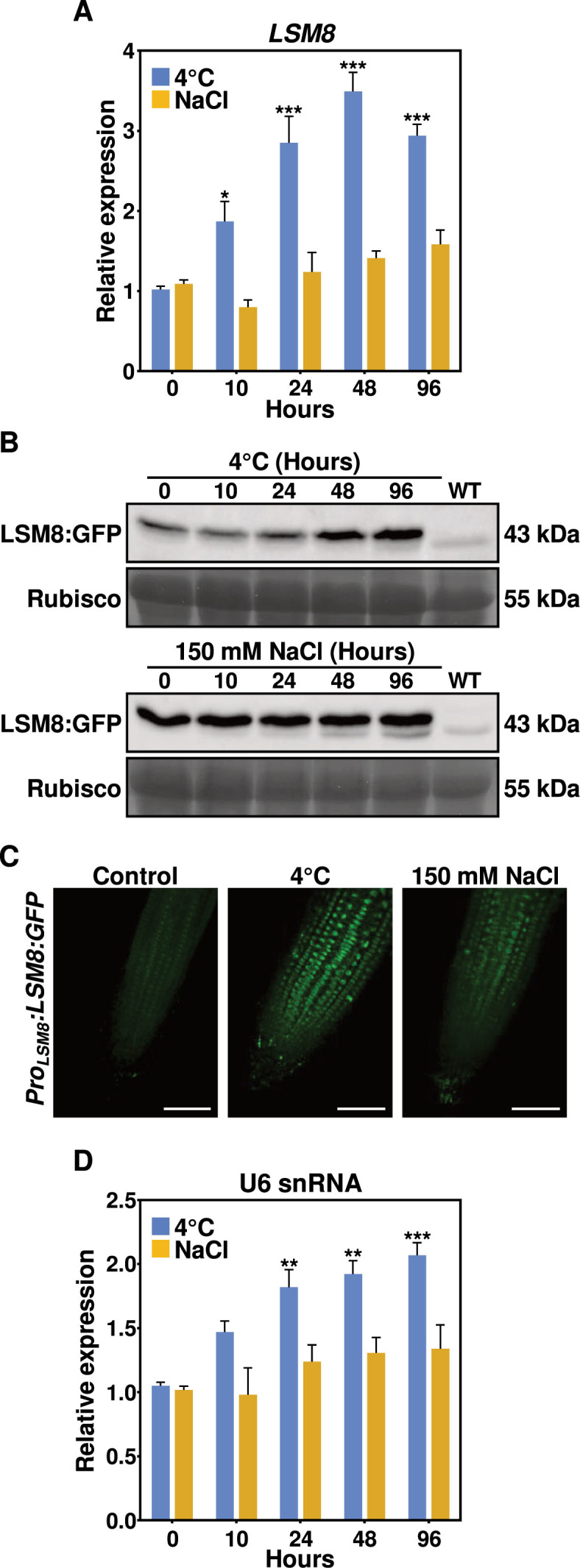
Arabidopsis LSM8 protein and U6 snRNA specifically accumulate in response to low temperature. (A) Expression of LSM8 in 2-week-old WT plants exposed for the indicated hours to 4°C or 150 mM NaCl. Levels, determined by qPCR, are represented as relative to their corresponding control values (0 h). (B) Immunoblots showing levels of LSM8-GFP protein in 2-week-old transgenic Arabidopsis plants exposed to 4°C (left) or 150 mM NaCl (right) for the indicated hours. A lane with proteins from WT plants was added to the immunoblots as a negative control. The large subunit of Rubisco, visualized by Coomassie staining, was used as a loading control. (C) Subcellular localization of LSM8-GFP in root tip cells from 7-day-old transgenic Arabidopsis seedlings grown under control conditions (control), or exposed 48 h to 4°C or 10 h to 150 mM NaCl. Bars = 75 μm. (D) Expression of U6 snRNA in 2-week-old WT plants exposed for the indicated hours to 4°C or 150 mM NaCl. Levels, determined by qPCR, are represented as relative to their corresponding values at 0 h. In all graphs, error bars indicate the standard error of the mean (n = 3). Asterisks (*) indicate significant differences (*P ≤ 0.01, **P ≤ 0.001, ***P ≤ 0.0001) between stressed and control plants, as determined by ANOVA test.
Since we had reported that the LSM2–8 complex of Arabidopsis is essential for stabilizing and maintaining the correct levels of U6 snRNA (12), we considered that, consequently with the LSM8 accumulation described above, the levels of U6 snRNA should also increase under cold conditions but not in the presence of high salt. This assumption was evaluated by determining the amounts of U6 snRNA present in 2-week-old WT plants subjected for different times at 4°C or 150 mM NaCl by means of qPCR assays. Figure 7D shows that, as expected, low temperature induced a significant (fold change ≥ 2) increase of U6 snRNA. Paralleling the accumulation of the LSM2–8 complex, the levels of U6 snRNA did not significantly augment by salt stress (Figure 7D). Intriguingly, a downregulation of U6 snRNA levels under high salt conditions has been reported (35). The discrepancy between these results and our results must be due, in all likelihood, to the important differences in the experimental conditions (i.e. plant growing conditions, NaCl concentration, and time and mode of treatment) used in both works. All in all, we concluded that the LSM nuclear complex of Arabidopsis differentially accumulates in response to abiotic stresses, which results in distinctive levels of U6 snRNA depending on the stress condition.
DISCUSSION
Understanding the molecular mechanisms involved in regulating the spliceosome activity is essential for deciphering the post-transcriptional control of the genome. Here, we demonstrate that, in Arabidopsis, the LSM2–8 complex, a core component of the spliceosome, is a regulator of the activity of this macromolecular machinery, and that this regulation is controlled by the environmental conditions. We show that it ensures the adequate constitutive and alternative splicing of selected pre-mRNAs under distinct environments, depending on the conditions, as well as the levels of the corresponding functional transcripts. Interestingly, the selective role of the complex has a significant physiological impact since it differentially modulates plant adaptation to abiotic stresses. Furthermore, we also show that the LSM2–8 complex, and consequently the U6 snRNA, differentially accumulate in response to abiotic stresses, suggesting that the concentration of U6 snRNP contributes to determine the spliceosome activity according to the external surroundings.
The findings reported in this work uncover a prominent role of the LSM2–8 complex in ensuring correct pre-mRNA splicing depending on the environmental conditions. Indeed, we show that under distinct environments it mediates the processing of specific constitutively and alternatively spliced introns from selected pre-mRNAs. The impact of the complex on these two types of splicing, moreover, seems to be also influenced by the external circumstances. Thus, while it mainly controls alternatively spliced introns under control and cold situations, in the presence of high salt conditions it has a similar impact on constitutively and alternatively spliced introns. The function of the LSM nuclear complex, however, is not restricted to ensure adequate intron splicing in response to abiotic stresses. It also prevents the occurrence of specific ES, A5΄SS and A3΄SS splicing events in selected pre-mRNAs, depending on the stress situations (Supplementary Figure S2 and Supplementary Table S12). Perez-Santangelo et al. (10) proposed that Arabidopsis LSM4 and LSM5 participate in the control of pre-mRNA splicing, with a predominant effect on the control of alternatively spliced introns. In addition, they described that each protein selectively regulates the splicing of a particular group of introns, but whether this selectively could be modulated by external stimuli was not determined (10). More recently, human LSM2, 3, 4, 6 and 7 have also been related with splicing site selection (11). The simultaneous participation of all these LSM proteins in the LSM1–7 and the LSM2–8 complexes, however, hamper to discriminate the real relevance of these complexes in the regulation of pre-mRNA splicing. As mentioned above, we previously demonstrated that LSM8 is the subunit that defines and determines the LSM2–8 complex of Arabidopsis, and is essential for the proper establishment of the U6 snRNP (12). Therefore, the results presented here definitively establish that the LSM nuclear complex, and consequently the U6 snRNP, actively determine the splicing patterns. This critical function, furthermore, is differentially modulated by the environmental conditions, which represents a new regulatory mechanism of the spliceosome activity specificity in eukaryotes.
The main question that arises from the results discussed above is which are the molecular determinants underlying the specific function of the LSM nuclear complex under different environments. It is plausible that the selected introns targeted by the complex belong to genes only or highly transcribed under a particular external stimulus. This, however, is not very likely because most genes containing selected introns targeted by the LSM2–8 complex are not differentially transcribed under cold or high salt conditions (Supplementary Figure S3 and Supplementary Tables S13 and S14). We also evaluated the possibility that the specific introns could include particular sequence motifs. Neither the scanning of introns, using the MEME-suit to detect sequence motif enrichments, nor the analysis of the frequencies of nucleotide sequences around their 5΄ and 3΄ splice sites or in their branch site, using the WebLogo application, revealed significant differences (Supplementary Figure S4). Furthermore, there were also no significant differences with the splice and branch site consensus sequences described for Arabidopsis (26) in any case (Supplementary Figure S4). In a recent work, Mahrez and colleagues (36) have demonstrated that the Arabidopsis splicing factor BRR2a mediates the excision of a particular subset of introns to control flowering time. They propose that BRR2a would select its targets based on the size and GC content of the introns. This could also be the case of the LSM2–8 complex, since we have observed significant differences in GC content and/or size between some subsets of introns specifically targeted by the complex. Constitutively spliced introns specifically targeted by LSM2–8 in response to cold show a higher GC content (33.2% versus 32.6%; P = 0.007) and shorter sequence length (128.6 versus 142.4 bp; P = 0.04) than those targeted under high salt conditions (Supplementary Figure S5A). Furthermore, the adjacent 5΄exons to the introns targeted at 4°C are significantly shorter than those adjacent to the introns targeted in response to NaCl (176.0 versus 209.1 bp; P = 0.042) (Supplementary Figure S5A). Finally, alternatively spliced introns specifically targeted by the LSM2–8 complex under cold stress are significantly shorter than those targeted under salt stress (144.2 versus 212.6 bp; P = 0.0002) (Supplementary Figure S5B). It has also been proposed that modulation of the relative concentration of core components of the spliceosome can function as a physiological mechanism for specific splicing regulation (10,11). Our findings demonstrate that the LSM2–8 complex and the U6 snRNA, and consequently the U6 snRNP, differentially accumulate in response to distinct environmental situations, which could account for the functional specificity of the spliceosome under such circumstances. Nevertheless, the fact that under control and high salt conditions the levels of the complex and U6 snRNA are very similar weakens this possibility. As proposed for the increase of U1 snRNA levels that takes place when Arabidopsis plants are subjected to 10°C (37), the accumulation of the LSM nuclear complex and the U6 snRNA at 4°C would be necessary to counteract the detrimental effect of low temperatures in the activity of the spliceosome. Finally, studies in humans and plants have shown that the dimethylation of LSM4 by the methyltransferase PRMT5 is essential for spliceosome assembly and proper pre-mRNA splicing (8,38). The possibility that posttranslational modifications of the LSM2–8 complex could contribute to its specificity remains to be determined.
Interestingly, the data reported here reveal that, under cold and high salt conditions, several introns targeted by the LSM nuclear complex of Arabidopsis are located in genes encoding important intermediates of plant tolerance to these abiotic stresses. Miss-splicing of more than 80% of these introns ends up in the generation of pre-mRNA isoforms with characteristics of NMD targets, which have been reported to be barely translated (25). Hence, our results indicate that the LSM2–8 complex guarantees adequate levels of functional transcripts and, consequently, proteins of the corresponding genes. Among the introns whose correct splicing is ensured by LSM2–8 in response to low temperature, several belong to genes coding for proteins, such as PRR5, RVE1 or AHK3, that have been described to be negative regulators of freezing tolerance (28–30). Under salt stress, LSM2–8 controls the accurate splicing of introns that are part of genes encoding proteins known to be positive regulators of Arabidopsis tolerance to high salt, including SAT32, NHX1 or SIS (31–33). In addition, when analyzing the few ES, A5΄SS and A3΄SS events controlled by the LSM nuclear complex in response to 4°C or 150 mM NaCl (Figure 2A and Supplementary Tables S4 and S5), some of them were also found to be located in genes encoding proteins active in freezing tolerance (the ankyrin repeat protein ACD6 and the transcription factors CCA1 and VOZ1) or salt tolerance (the WWE domain-containing protein RCD1) (39–42) (see validation of RNA-seq results of these events by qPCR assays in Supplementary Figure S6). The function of the Arabidopsis LSM2–8 complex, therefore, and consequently that of U6 snRNP, is required for the adequate splicing of selected, specific and non-specific, pre-mRNAs from proteins involved in abiotic stress tolerance, depending on the stress to which plants are confronted. Furthermore, this function would be important to shape stress-specific translatome profiles. As expected from these results, we demonstrate that the complex plays an essential role in plant adaptation to challenging environments. Indeed, it negatively regulates the capacity of Arabidopsis to cold acclimate but acts as a positive regulator of Arabidopsis tolerance to high salt. Arabidopsis lsm4 and lsm5/sad1 mutants have also been described to show enhanced sensitivity to salt stress as a consequence of altered pre-mRNA splicing (8,9). The comparison between genes with anomalous spliced introns detected in lsm8-1 (1703) with those in lsm5/sad1 (908), revealed that 291 are common (Supplementary Figure S7), suggesting that most probably they correspond to bona fide targets of the LSM nuclear complex. The absence of a broader overlap must reflect, in all likelihood, the different experimental conditions used in both analyses, as well as the fact mentioned before that LSM5, in addition of being part of the LSM2–8 complex, also participates in the LSM1–7 complex (12). In spite of this, some of the common genes encode important positive regulators of Arabidopsis tolerance to high salt (i.e., SAT32, SIS, RCD1, the transcription factors SZF1 and WRKY33, and the membrane protein RCI2A) (32,33, 42–45), suggesting that this set of genes could be enriched in important players of plant response to salt stress. Other components of the spliceosome such as the yeast BRR1 protein and the SKIP, PRP31 and GEMIN2 proteins from Arabidopsis, all of them being non-snRNP proteins, have been implicated in regulating the tolerance to adverse environmental conditions by modulating pre-mRNA splicing (37,46–49). In no case, however, the specificity of their functions in response to abiotic stresses has been studied.
In conclusion, the results presented in this work uncover an unanticipated functional capacity of a core component of the spliceosome, the LSM2–8 complex, to ensure the efficiency and accuracy of constitutive and alternative splicing of selected pre-mRNAs, depending on the environmental conditions. This function represents a new layer of posttranscriptional regulation in response to external stimuli in eukaryotes that seems to be essential for their correct adaptation. Understanding the molecular mechanisms that control the function of the LSM nuclear complex depending on the environmental conditions constitutes an interesting challenge for future studies.
ACCESSION NUMBERS
The complete genome-wide data from this publication were submitted to the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/) under accession number GSE87415.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr José Antonio Jarillo and Dr Roberto Solano for discussions and comments on the manuscript.
Footnotes
Present addresses:
Tamara Hernández-Verdeja, Umeå Plant Science Centre, Department of Plant Physiology, Umeå University, SE-901 87 Umeå, Sweden.
Carlos Perea-Resa, Department of Molecular Biology, Massachusetts General Hospital, Boston, MA 02114, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
MINECO [BIO2013-47788-R] and AEI/FEDER, UE [BIO2016-79187-R] to J.S. Funding for open access charge: MINECO [BIO2016-79187-R].
Conflict of interest statement. None declared.
REFERENCES
- 1. Matera A.G., Wang Z.. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014; 15:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tharun S. Roles of eukaryotic Lsm proteins in the regulation of mRNA function. Int. Rev. Cell Mol. Biol. 2009; 272:149–189. [DOI] [PubMed] [Google Scholar]
- 3. Brow D.A., Guthrie C.. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988; 334:213–218. [DOI] [PubMed] [Google Scholar]
- 4. Lee Y., Rio D.C.. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 2015; 84:291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Staiger D., Brown J.W.S.. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell. 2013; 25:3640–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fu X.-D., Ares M.. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014; 15:689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saltzman A.L., Pan Q., Blencowe B.J.. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev. 2011; 25:373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang Z., Zhang S., Zhang Y.Y., Wang X., Li D., Li Q.Q., Yue M., Li Q.Q., Zhang Y.Y., Xu Y. et al. Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell. 2011; 23:396–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cui P., Zhang S., Ding F., Ali S., Xiong L.. Dynamic regulation of genome-wide pre-mRNA splicing and stress tolerance by the Sm-like protein LSm5 in Arabidopsis. Genome Biol. 2014; 15:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perez-Santángelo S., Mancini E., Francey L.J., Schlaen R.G., Chernomoretz A., Hogenesch J.B., Yanovsky M.J.. Role for LSM genes in the regulation of circadian rhythms. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:15166–15171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Papasaikas P., Tejedor J.R., Vigevani L., Valcárcel J.. Functional splicing network reveals extensive regulatory potential of the core spliceosomal machinery. Mol. Cell. 2015; 57:7–22. [DOI] [PubMed] [Google Scholar]
- 12. Perea-Resa C., Hernández-Verdeja T., López-Cobollo R., del Mar Castellano M., Salinas J.. LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. Plant Cell. 2012; 24:4930–4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Perea-Resa C., Carrasco-López C., Catalá R., Turečková V., Novak O., Zhang W., Sieburth L., Jiménez-Gómez J.M., Salinas J.. The LSM1–7 complex differentially regulates Arabidopsis tolerance to abiotic stress conditions by promoting selective mRNA decapping. Plant Cell. 2016; 28:505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xiong L., Gong Z., Rock C.D., Subramanian S., Guo Y., Xu W., Galbraith D., Zhu J.-K.. Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell. 2001; 1:771–781. [DOI] [PubMed] [Google Scholar]
- 15. Golisz A., Sikorski P.J., Kruszka K., Kufel J.. Arabidopsis thaliana LSM proteins function in mRNA splicing and degradation. Nucleic Acids Res. 2013; 41:6232–6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Catala R., Santos E., Alonso J.M., Ecker J.R., Martinez-Zapater J.M., Salinas J.. Mutations in the Ca2+/H+ transporter CAX1 increase CBF/DREB1 expression and the cold-acclimation response in Arabidopsis. Plant Cell. 2003; 15:2940–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.-R.. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005; 139:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 19. Li R., Yu C., Li Y., Lam T.-W., Yiu S.-M., Kristiansen K., Wang J.. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009; 25:1966–1967. [DOI] [PubMed] [Google Scholar]
- 20. Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B.. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008; 5:621–628. [DOI] [PubMed] [Google Scholar]
- 21. Audic S., Claverie J.M.. The significance of digital gene expression profiles. Genome Res. 1997; 7:986–995. [DOI] [PubMed] [Google Scholar]
- 22. Trapnell C., Pachter L., Salzberg S.L.. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The sequence alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kahles A., Ong C.S., Zhong Y., Rätsch G.. SplAdder: Identification, quantification and testing of alternative splicing events from RNA-Seq data. Bioinformatics. 2016; 32:1840–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu H., Tian C., Yu Y., Jiao Y.. Transcriptome survey of the contribution of alternative splicing to proteome diversity in Arabidopsis thaliana. Mol. Plant. 2016; 9:749–752. [DOI] [PubMed] [Google Scholar]
- 26. Szczesniak M.W., Kabza M., Pokrzywa R., Gudys A., Makalowska I.. ERISdb: a database of plant splice sites and splicing signals. Plant Cell Physiol. 2013; 54:e10. [DOI] [PubMed] [Google Scholar]
- 27. Lee H.G., Seo P.J.. The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF-COR pathway in Arabidopsis. Plant J. 2015; 82:962–977. [DOI] [PubMed] [Google Scholar]
- 28. Jeon J., Kim N.Y., Kim S., Kang N.Y., Novák O., Ku S.-J., Cho C., Lee D.J., Lee E.-J., Strnad M. et al. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J. Biol. Chem. 2010; 285:23371–23386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guan Q., Wu J., Zhang Y., Jiang C., Liu R., Chai C., Zhu J.. A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis. Plant Cell. 2013; 25:342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meissner M., Orsini E., Ruschhaupt M., Melchinger A.E., Hincha D.K., Heyer A.G.. Mapping quantitative trait loci for freezing tolerance in a recombinant inbred line population of Arabidopsis thaliana accessions Tenela and C24 reveals REVEILLE1 as negative regulator of cold acclimation. Plant. Cell Environ. 2013; 36:1256–1267. [DOI] [PubMed] [Google Scholar]
- 31. Apse M.P., Aharon G.S., Snedden W.A., Blumwald E.. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999; 285:1256–1258. [DOI] [PubMed] [Google Scholar]
- 32. Park M.-Y., Chung M.-S., Koh H.-S., Lee D.J., Ahn S.-J., Kim C.S.. Isolation and functional characterization of the Arabidopsis salt-tolerance 32 (AtSAT32) gene associated with salt tolerance and ABA signaling. Physiol. Plant. 2009; 135:426–435. [DOI] [PubMed] [Google Scholar]
- 33. Luhua S., Hegie A., Suzuki N., Shulaev E., Luo X., Cenariu D., Ma V., Kao S., Lim J., Gunay M.B. et al. Linking genes of unknown function with abiotic stress responses by high-throughput phenotype screening. Physiol. Plant. 2013; 148:322–333. [DOI] [PubMed] [Google Scholar]
- 34. Zhang B., Liu K., Zheng Y., Wang Y., Wang J., Liao H.. Disruption of AtWNK8 enhances tolerance of Arabidopsis to salt and osmotic stresses via modulating proline content and activities of catalase and peroxidase. Int. J. Mol. Sci. 2013; 14:7032–7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ding F., Cui P., Wang Z., Zhang S., Ali S., Xiong L.. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genomics. 2014; 15:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mahrez W., Shin J., Muñoz-Viana R., Figueiredo D.D., Trejo-Arellano M.S., Exner V., Siretskiy A., Gruissem W., Köhler C., Hennig L.. BRR2a affects flowering time via FLC splicing. PLoS Genet. 2016; 12:e1005924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schlaen R.G., Mancini E., Sanchez S.E., Perez-Santángelo S., Rugnone M.L., Simpson C.G., Brown J.W.S., Zhang X., Chernomoretz A., Yanovsky M.J.. The spliceosome assembly factor GEMIN2 attenuates the effects of temperature on alternative splicing and circadian rhythms. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:9382–9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brahms H., Meheus L., de Brabandere V., Fischer U., Lührmann R.. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B’ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 2001; 7:1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miura K., Ohta M.. SIZ1, a small ubiquitin-related modifier ligase, controls cold signaling through regulation of salicylic acid accumulation. J. Plant Physiol. 2010; 167:555–560. [DOI] [PubMed] [Google Scholar]
- 40. Seo P.J., Park M.-J., Lim M.-H., Kim S.-G., Lee M., Baldwin I.T., Park C.-M.. A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell. 2012; 24:2427–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nakai Y., Nakahira Y., Sumida H., Takebayashi K., Nagasawa Y., Yamasaki K., Akiyama M., Ohme-Takagi M., Fujiwara S., Shiina T. et al. Vascular plant one-zinc-finger protein 1/2 transcription factors regulate abiotic and biotic stress responses in Arabidopsis. Plant J. 2013; 73:761–775. [DOI] [PubMed] [Google Scholar]
- 42. Katiyar-Agarwal S., Zhu J.J., Kim K., Agarwal M., Fu X., Huang A., Zhu J.J.. The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:18816–18821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun J., Jiang H., Xu Y., Li H., Wu X., Xie Q., Li C.. The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 2007; 48:1148–1158. [DOI] [PubMed] [Google Scholar]
- 44. Jiang Y., Deyholos M.K.. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009; 69:91–105. [DOI] [PubMed] [Google Scholar]
- 45. Mitsuya S., Taniguchi M., Miyake H., Takabe T.. Disruption of RCI2A leads to over-accumulation of Na+ and increased salt sensitivity in Arabidopsis thaliana plants. Planta. 2005; 222:1001–1009. [DOI] [PubMed] [Google Scholar]
- 46. Noble S.M., Guthrie C.. Transcriptional pulse-chase analysis reveals a role for a novel snRNP-associated protein in the manufacture of spliceosomal snRNPs. EMBO J. 1996; 15:4368–4379. [PMC free article] [PubMed] [Google Scholar]
- 47. Noble S.M., Guthrie C.. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics. 1996; 143:67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Feng J., Li J., Gao Z., Lu Y., Yu J., Zheng Q., Yan S., Zhang W., He H., Ma L. et al. SKIP confers osmotic tolerance during salt stress by controlling alternative gene splicing in Arabidopsis. Mol. Plant. 2015; 8:1038–1052. [DOI] [PubMed] [Google Scholar]
- 49. Du J., Zhang S., Huang H.-W., Cai T., Li L., Chen S., He X.. The splicing factor PRP31 is involved in transcriptional gene silencing and stress response in Arabidopsis. Mol. Plant. 2015; 8:1053–1068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete genome-wide data from this publication were submitted to the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/) under accession number GSE87415. The full names of the genes mentioned in this article are included in Supplementary Table S2.