Abstract
RNase P is primarily responsible for the 5΄ maturation of transfer RNAs (tRNAs) in all domains of life. Archaeal RNase P is a ribonucleoprotein made up of one catalytic RNA and five protein cofactors including L7Ae, which is known to bind the kink-turn (K-turn), an RNA structural element that causes axial bending. However, the number and location of K-turns in archaeal RNase P RNAs (RPRs) are unclear. As part of an integrated approach, we used native mass spectrometry to assess the number of L7Ae copies that bound the RPR and site-specific hydroxyl radical-mediated footprinting to localize the K-turns. Mutagenesis of each of the putative K-turns singly or in combination decreased the number of bound L7Ae copies, and either eliminated or changed the L7Ae footprint on the mutant RPRs. In addition, our results support an unprecedented ‘double K-turn’ module in type A and type M archaeal RPR variants.
INTRODUCTION
RNase P catalyzes removal of the 5΄ leader in all precursor tRNAs (pre-tRNAs) transcribed as either part of a polycistronic transcript or separately with a 5΄ extension (1–5). Although a proteinaceous form has been described in several eukaryotes (6), the ribonucleoprotein (RNP) form is found in all domains of life (1–5). In all organisms where it has been examined, the RNP form has one catalytic RNase P RNA (RPR) with conserved core structural elements, and a varying complement of RNase P proteins (RPPs): one in bacteria, up to five in archaea, and as many as ten in eukaryotes (1–5,7–9). The large RNase P complex from eukaryotes has been intractable to assembly and biochemical studies in vitro. In contrast, the simpler RNP complex from several archaea has been successfully reconstituted in vitro, providing valuable insights into protein-aided RNA catalysis in this multi-RPP holoenzyme (9–16). Archaeal RNase P has thus served as an experimental proxy for the eukaryotic enzyme because all archaeal RPPs are orthologous to their cousins in eukaryotic RNase P (1,17–19).
Euryarchaeal RNase P has mainly been classified into two groups based on the RPR secondary structure (20) [see (21,22) for crenarchaeal variants]. Type A RPRs resemble the ancestral (or type A) bacterial RPRs (Supplementary Figure S1) and can similarly process pre-tRNAs without protein cofactors, though not as efficiently (8,9,11,12,16). Type M RPRs–so called due to their presence mainly in Methanococcales–are active only when aided by RPPs or when a pre-tRNA is tethered in cis (9,13).
All five archaeal RPPs (POP5, RPP30, RPP21, RPP29 and L7Ae) were identified by sequence homology to eukaryotic RPPs (17,23) and confirmed experimentally by their co-elution with partially purified native archaeal RNase P and by their ability to boost RPR catalysis in vitro (13,14,17,23). Four of these RPPs function as pairs, with POP5•RPP30 enhancing the cleavage rate and RPP21•RPP29 improving substrate binding (9,11,16). While the in vitro reconstituted complex with RPR and these four RPPs is active, addition of L7Ae raises the reaction temperature optimum and kcat/KM close to those observed with the native enzyme (13,23).
L7Ae binds a well-studied and widespread RNA structural module called a kink-turn (K-turn) (24–26). This module is characterized by a 3-nucleotide (nt) bulge flanked by a 5΄ canonical (C) helix and a 3΄ non-canonical (NC) helix that is capped with tandem G•A and A•G trans Hoogsteen/sugar edge pairs (Figure 1, inset). An RNA duplex harboring a K-turn can exist in a conformational equilibrium between a tightly kinked structure with an included angle of ∼50° between the two helices and an extended helix, with a bias toward the latter (27). The kinked structure is accompanied by a twist and is stabilized by Mg2+ (in some K-turns) (28), tertiary interactions, and protein binding (24–27,29–31). While it is clear that L7Ae is part of type A and type M archaeal RNase P (13,23), the number and localization of its binding sites on these RPRs need further investigation.
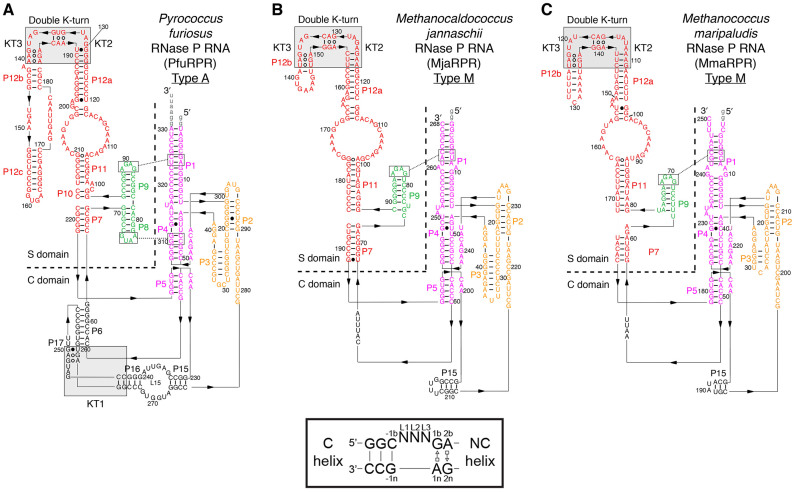
Figure 1.
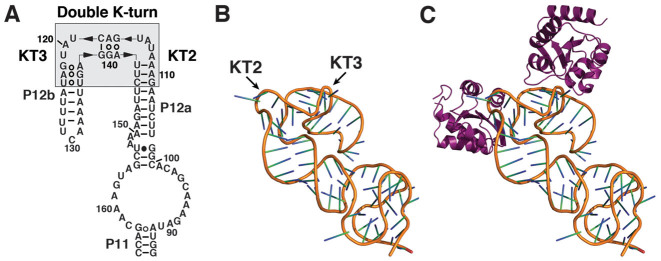
Putative K-turns in type A and type M archaeal RPRs. Putative K-turns are indicated by gray boxes in the secondary structures of (A) PfuRPR, (B) MjaRPR and (C) MmaRPR. Each RPR has two functional domains, the catalytic (C) and the substrate specificity (S) domain, which are demarcated here by a dashed line. Inset shows a standard K-turn with a canonical (C) and a non-canonical (NC) helix. We termed the K-turn in P16-17 of PfuRPR KT1, and the two in P12 of all RPRs KT2 and KT3. Note that KT2 and KT3 overlap, forming a double K-turn motif. The K-turn mutants were created by mutating the As in the G•A pairs of the specified K-turn(s) to Cs to replace the G•A with the Watson–Crick GC pairs (see Figure 4B F for the PfuRPR K-turn mutants).
L7Ae binds P12 and P16–17 in type A archaeal RPRs (23,32), but it binds only P12 in type M RPRs (13) as P16–17 is missing in these RPRs (Figure 1). Our sequence alignment of 10 different type M RPRs highlighted a conserved region in P12 (13) that could fold into a structure resembling two overlapping K-turns, which we now term a “double K-turn”; site-directed mutagenesis of each of these K-turns in Methanococcus maripaludis (Mma; type M) RPR showed that L7Ae binding at one of the sites is required for cleavage rate enhancement (13). Recently, we used site-specific hydroxyl radical (OH•)-mediated footprinting to show that Pyrococcus furiosus (Pfu; type A) RPR is bound by L7Ae within P12a-b as well as P16–17, where a K-turn motif could be identified (Figure 1A) (32). As observed in MmaRPR, the P12 region of PfuRPR footprinted by L7Ae could also be folded into a double K-turn, a module that has not been recognized thus far despite an appreciation of the prevalence of single K-turns. A recent report based on L7Ae pull-down analysis using Pyrococcus horikoshii (Pho; type A) RPR lacking P12, P15–17, or both also concluded that there are three L7Ae binding sites in PhoRPR (33), though only the sequence of the predicted K-turn in P16–17 of PhoRPR (34) agrees with ours in PfuRPR (32). However, the Pho RNase P experiments were conducted with either large deletions within PhoRPR or RNA fragments modified from the native context in PhoRPR, both of which are likely to promote structural alterations. Therefore, there is a clear need to better characterize these predicted K-turns in native archaeal RPRs, particularly those forming the double K-turn.
Here, we leveraged two recent advances (32,35) to validate the number and location of all K-turns in a type A and a type M archaeal RPR, especially the double K-turn in P12 that is common to both (Figure 1). First, we sought to establish the maximal copy number of L7Ae bound to the RPR (wild-type or K-turn mutants), as it is a direct measure of the number of K-turns. To this end, we used native mass spectrometry (MS), which we recently demonstrated (35) as an effective method for studying RNPs, including those assembled in Mg2+. Second, to correlate the MS data with occupancy of specific K-turns, we used the directed OH•-probing method (32) to map PfuL7Ae binding to each K-turn in PfuRPR and its K-turn mutants. Our results both confirmed the predicted K-turns and validated the double K-turn as a new RNA architectural element.
MATERIALS AND METHODS
Mutagenesis, in vitro transcription and refolding of RPRs
Transcription of Pfu, Methanocaldococcus jannaschii (Mja) and Mma RPRs has been previously described (9,11,13,16). RPRs were refolded as described earlier for MS (35), footprinting (32) and pre-tRNA processing (11,16) experiments. See Supplementary Information for additional details.
Purification of recombinant Pfu, Mja and Mma L7Ae
To obtain archaeal L7Ae variants, we followed established purification schemes (13,32,36) with some critical changes to ensure that final preparations were free of nucleic acids (see Supplementary Information for details). Note that PfuL7Ae used for MS is PfuL7Ae-C71V, a Cys-less version that showed a higher activity than the wild type (32). PfuL7Ae-C71V was used as the template from which single Cys-substituted mutant derivatives were generated for the footprinting studies (32).
Native MS analysis
All MS experiments were performed using an Exactive™ Plus Extended Mass Range (EMR) Orbitrap™ mass spectrometer (Thermo Scientific). Pfu, Mja and Mma L7Ae were dialyzed in 10 mM NH4OAc for 12–16 h before use. All RNP complexes were reconstituted in 800 mM NH4OAc plus 1 mM Mg(OAc)2 (unless otherwise specified) and incubated for 10 min at 55°C (for Pfu and Mja) or 37°C (for Mma) immediately before analysis. Each sample was infused directly into the EMR mass spectrometer using nano-electrospray ionization by first being placed in a borosilicate glass capillary (Kimble Chase), individually pulled in-house using a P-97 micropipette puller (Sutter Instrument Company). A platinum wire was then inserted into the sample solution, and a voltage of 0.8–1.7 kV was applied for ionization. The instrument settings were as follows: capillary temperature, 275°C; source DC offset, 60 V; injection flatapole DC, 8 V; inter flatapole DC, 3 V; bent flatapole DC, –5 V; transfer multipole DC tune offset, –10 V; C-trap entrance lens tune offset, 5 V; trapping gas pressure setting, 8; and resolving power, 8750 [full width at half maximum (FWHM) at 200 m/z)]. Voltages for source-induced dissociation and higher-energy collisional dissociation (HCD) were set to 20 and 90 V, respectively.
PfuL7Ae footprinting experiments
OH•-mediated footprinting experiments with assemblies of a PfuL7Ae mutant derivative plus either a wild-type or mutant PfuRPR and subsequent reverse transcription reactions were performed largely as described previously (32), except that sequencing ladders were generated using 1 mM dNTPs. Sequences of the primers used for reverse transcription are provided in Supplementary Table S2. Reverse transcription products (cDNAs of footprinted RNAs and their respective DNA sequencing ladders) were separated on a pre-warmed 8% (w/v) polyacrylamide/7 M urea gel (SequaGel; National Diagnostics); the gel was then fixed, dried, scanned with a Typhoon phosphorimager (GE Healthcare), and analyzed using ImageQuant (GE Healthcare).
RESULTS
Approach
We chose PfuRPR as a representative of the type A archaeal RPR and Mja and Mma RPRs of the type M. Figure 1 depicts the putative K-turns in each RPR: KT1 in P16–17 is only present in PfuRPR, while KT2 and KT3 are found in P12 of all three RPRs. KT1 is localized to the catalytic (C) domain while KT2 and KT3 are in the substrate specificity (S) domain; the archaeal RPRs, like their bacterial counterparts, utilize the C domain to cleave the pre-tRNA and the S domain to recognize the elbow (D/T loops) of the pre-tRNA (9,16,37–41).
KT2 and KT3 together comprise a double K-turn motif because they are not merely two K-turns in tandem—the NC helix of KT2 also serves as the C helix of KT3. Additionally, this helix deviates from the standard K-turn in having only three base pairs; in PfuRPR, a U•A pair at positions 2b,2n substitutes for the typical A•G pair (b and n refer to the bulged and non-bulged strand, respectively; Figure 1, inset). Existence of this double K-turn is supported by sequence conservation at nearly identical positions in euryarchaeal RPRs from Thermococcales, Methanococcales and Methanobacteriales (Figure 1) (13,32).
Site-directed mutagenesis was used to mutate each K-turn individually (e.g. PfuRPR-mKT2, where KT2 was mutated) and in combination (e.g. PfuRPR-mKT23, where both KT2 and KT3 were mutated). In each K-turn, the 1n and 2b nucleotides were both changed to a C; the 2n nucleotide of PfuRPR-mKT2 was also mutated (A → G). Thus, the two central, sheared G•A pairs (or G•A and U•A pairs in KT2 of PfuRPR) of each K-turn were altered to two Watson–Crick GC pairs to weaken/disrupt binding by L7Ae, which makes side-chain and backbone contacts to the NC helix.
To assess the number of K-turns in archaeal RPRs, we used native MS to accurately measure the number of copies of L7Ae that bind these RPRs. Weak signal intensity and poor resolution caused by ionization suppression (42,43) and peak broadening (44,45), respectively, have historically posed obstacles for the study of RNPs assembled in Mg2+. We have overcome some of these barriers by using native MS with collision-induced cleaning and surface-induced dissociation coupled with ion mobility-MS (35). Due to the anticipated heterogeneity associated with different RPR:L7Ae stoichiometries, we utilized an EMR Orbitrap mass spectrometer that affords high sensitivity and spectral resolution. The high transmission efficiency of even large macromolecular complexes with this instrument enabled the use of only 50 nM RPR, a concentration 100-fold lower than what we had employed previously (35) and one that minimizes the possibility of non-specific binding.
To validate the number of copies of L7Ae bound to the wild-type RPR, we examined L7Ae stoichiometry with a panel of mutant RPRs. Moreover, to correlate binding of L7Ae to the multiple sites, the occupancy of L7Ae at each site in the wild-type and mutant PfuRPRs was mapped using OH•-mediated footprinting (32).
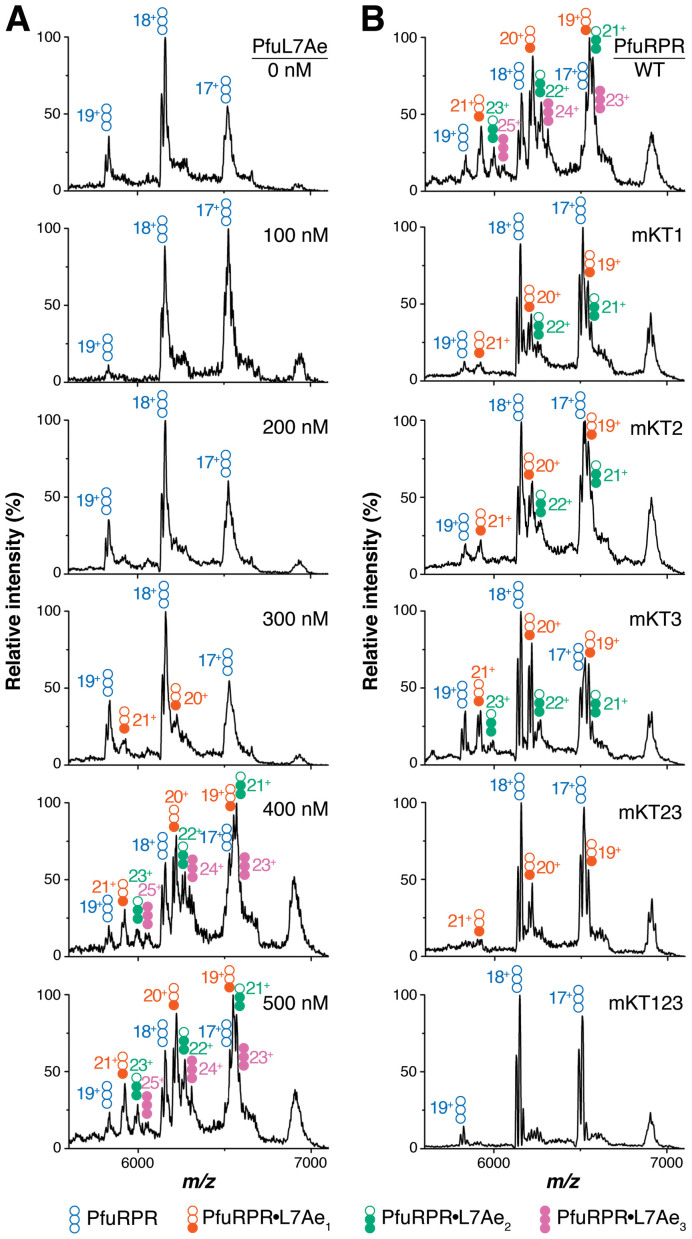
Three PfuL7Ae copies are bound to PfuRPR (type A)
Because conditions for preservation of macromolecular complexes in the gas phase may differ from those in solution, we began by examining two experimental conditions. First, the optimal Mg2+ concentration was determined by assessing RNP assembly with 50 nM PfuRPR and 500 nM PfuL7Ae in 800 mM NH4OAc and 1, 2, 3, 4, 5 or 6 mM Mg(OAc)2. The broad peaks observed in 4–6 mM Mg(OAc)2 complicated mass assignment (data not shown). However, because the complexes observed in 1–3 mM were similar to those in 4–6 mM Mg(OAc)2, we decided to perform all subsequent MS experiments in 1 mM Mg(OAc)2 to facilitate data analysis. We also used simulated Gaussian peaks corresponding to different charge states of the RPR, RPR•L7Ae1, RPR•L7Ae2, and RPR•L7Ae3 to assess the fit to the species present in the mixture (Supplementary Figure S2). Second, complex formation was evaluated using a PfuL7Ae titration to determine the optimal L7Ae concentration in 1 mM Mg(OAc)2 (Figure 2A). Peaks corresponding to only PfuRPR were observed between 0 and 200 nM PfuL7Ae. At 300 nM PfuL7Ae, the RPR•L7Ae1 complex was evident. At higher PfuL7Ae concentrations, RPR•L7Ae2 and RPR•L7Ae3 were observed. Although the peaks resolved better with increasing L7Ae concentration as high as 1 μM (data not shown), we chose 500 nM PfuL7Ae (Figure 2A) for subsequent MS experiments to minimize non-specific interactions between PfuRPR and PfuL7Ae. Additionally, these conditions mirrored those of our previous site-specific OH•-mediated footprinting of PfuL7Ae on PfuRPR (32).
Figure 2.
Pfu RPR•L7Ae (type A) complexes detected by native MS. (A) Titration of 50 nM PfuRPR with PfuL7Ae (0–500 nM). (B) Complex formation with 50 nM of the indicated PfuRPR and 500 nM PfuL7Ae. Loss of specific PfuL7Ae binding was observed when the two sheared G•A pairs (or G•A and U•A pairs in the case of KT2) in the indicated K-turn(s) were mutated to Watson–Crick GC pairs. The different PfuRPR•L7Ae complexes are color-coded as noted. The charge states of each complex are indicated.
At 500 nM PfuL7Ae (and even up to 1 μM), the largest complex observed by native MS was RPR•L7Ae3. This observation is consistent with our prediction of three K-turns in PfuRPR, an inference that we further corroborated with a panel of K-turn mutants. When the K-turn mutants of PfuRPR were each used in place of the wild-type counterpart, L7Ae occupancy decreased by the number of mutated K-turns (Figure 2B): two with PfuRPR-mKT1, -mKT2, and -mKT3; one with PfuRPR-mKT23; and none with PfuRPR-mKT123. We were unable to determine the KD values from the MS data because of overlapping peaks in some cases and rapid dissociation in others. While these issues do not affect inferences about L7Ae stoichiometry, they serve to highlight limitations in the use of native MS for studies of RNPs.
To independently validate the binding of two copies of L7Ae to the double K-turn motif, we performed gel-shift assays using a 43-nt PfuRPR fragment that includes part of P12a, the double K-turn, and P12b, terminating in a non-native GAAA tetraloop (Supplementary Figure S3). We observed two shifted bands upon titration of increasing amounts of PfuL7Ae, indicative of two L7Ae binding sites (Supplementary Figure S3). When KT2 or KT3 was mutated in this 43-nt P12 RNA, the slower migrating P12•PfuL7Ae2 complex disappeared, leaving the P12•PfuL7Ae1 complex. These results parallel those observed with MS using the full-length PfuRPRs (Figure 2B).
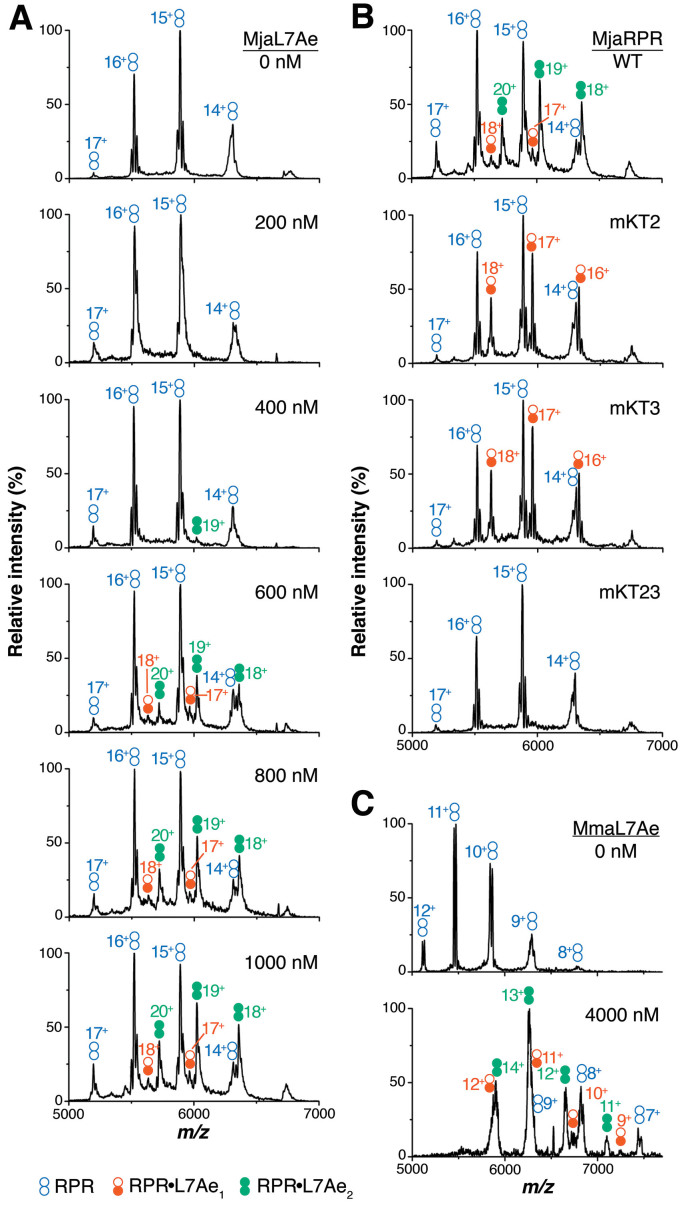
Two L7Ae copies are bound to type M RPRs
Since type M archaeal RPRs also appear to have the double K-turn motif (13), we next investigated L7Ae binding in two type M representatives. In 1 mM Mg(OAc)2, 50 nM MjaRPR was first titrated against increasing MjaL7Ae concentrations from 0 to 1000 nM (Figure 3A). Here, reliable complex formation of both RPR•L7Ae1 and RPR•L7Ae2 first appeared at 600 nM MjaL7Ae, and the largest complex found even at 2 μM MjaL7Ae was RPR•L7Ae2 (data not shown). The maximal L7Ae occupancy of 2 matches the two predicted K-turns in type M RPRs like MjaRPR. Due to the potential for non-specific L7Ae binding, we chose to examine the K-turn mutants of MjaRPR using 1 μM MjaL7Ae. While the maximal occupancy of MjaL7Ae on MjaRPR was 2, it decreased to 1 in each of the single K-turn mutants MjaRPR-mKT2 and -mKT3, and to 0 in the double mutant MjaRPR-mKT23 (Figure 3B). These findings showed that our prediction of two K-turns was correct, and that they are part of the double K-turn motif.
Figure 3.
Type M RPR•L7Ae complexes detected by native MS. (A) Titration of 50 nM MjaRPR with MjaL7Ae (0–1000 nM). (B) Complex formation with 50 nM of the indicated MjaRPR and 1000 nM MjaL7Ae. Loss of specific MjaL7Ae binding was observed when the two sheared G•A pairs in the indicated K-turn(s) were mutated to Watson–Crick GC pairs. (C) Mma RPR•L7Ae complex formation with 50 nM wild-type MmaRPR and 0 or 4000 nM MmaL7Ae. Both RPR•L7Ae1 and RPR•L7Ae2 complexes were observed. The different RPR•L7Ae complexes are color-coded as noted. The charge states of each complex are indicated.
Although we had previously conducted functional tests of MmaRPR K-turn mutants (13), we sought to verify the number of L7Ae copies bound to the wild-type MmaRPR. Indeed, MmaRPR also displayed an RPR•L7Ae2 stoichiometry (Figure 3C), mirroring MjaRPR.
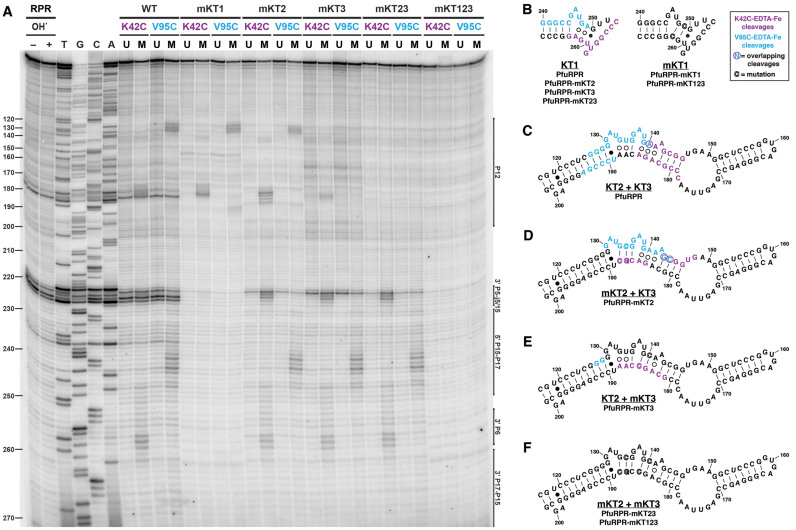
PfuL7Ae footprints on each of the three putative K-turns in PfuRPR (type A)
To examine the specific interaction of PfuL7Ae with its cognate RPR, we turned to a site-specific OH•-mediated method that we had successfully used previously to localize the footprints of PfuL7Ae on PfuRPR (32). Briefly, this approach entails covalently attaching an EDTA-Fe moiety to PfuL7Ae at an engineered single-Cys residue positioned at or proximal to the RNA-binding surface, thus converting the protein into a nuclease that can cleave a bound RNA but only at sites within 10 Å of the Fe.
This approach was employed again here to compare the footprints of PfuL7Ae on the K-turn mutants to those on the wild-type PfuRPR. In this study, we used two PfuL7Ae derivatives, one with a modification at K42C and the other at V95C (32). Based on crystal structures of L7Ae bound to a K-turn in several RNAs (29–31), K42C–EDTA-Fe was expected to produce cleavages in the non-bulged strand of the NC helix and V95C–EDTA-Fe in the bulged strand, especially around the bulge (32).
As observed in our previous report (32), PfuL7Ae footprinted in the predicted K-turns in P16-17 (KT1) and P12 (KT2 and KT3) of wild-type PfuRPR (Figure 4). At the putative KT1 (Figure 4A and B), K42C–EDTA-Fe footprinted at positions 253–263 in the non-bulged strand, including U3n to A1n; V95C–EDTA-Fe cleaved at positions 240–249, which include the three bulged nucleotides, G1b, and A2b. In the vicinity of KT2 and KT3 (Figure 4A and C; see Supplementary Figure S4 for a close-up), the cleavages produced by each L7Ae derivative were found on both strands, perhaps indicative of L7Ae binding to KT2 and KT3, which have an intervening twist. Cleavages by K42C–EDTA-Fe were at positions 140–146 and 178–185, which include the entire NC helix of KT3. Those produced by V95C–EDTA-Fe were at positions 125–140, which encompass the KT2 and KT3 bulges, and 189–194, which are in the non-bulged strand of the C helix of KT2.
Figure 4.
Mapping the footprint of PfuL7Ae on the three K-turns in PfuRPR. (A) Attachment of EDTA-Fe to two single Cys-substituted derivatives of PfuL7Ae, K42C and V95C, led to directed cleavages of proximal RNA sites upon binding to wild-type (WT) PfuRPR or a K-turn mutant. RNA cleavage products were reverse transcribed using 5΄-radiolabeled PfuRPRj15/2-R and resolved on a sequencing gel. U, unmodified; M, EDTA-Fe–modified. (B–F) Summary of PfuL7Ae–EDTA-Fe cleavages proximal to the three K-turns of PfuRPR. As indicated in the legend, specific and overlapping cleavages are color-coded and circled, respectively, and mutated nucleotides are outlined. (B) KT1 with specific cleavages in PfuRPR, PfuRPR-mKT2, -mKT3, and -mKT123 that were lost in PfuRPR-mKT1 and -mKT123. (C–F) The double K-turn comprising KT2 and KT3. The range of cleavages in the WT PfuRPR (C) was altered in PfuRPR-mKT2 (D) or decreased in PfuRPR-mKT3 (E). (F) All cleavages were completely lost when both KT2 and KT3 were mutated.
Next, we performed footprinting of PfuL7Ae on the PfuRPR K-turn mutants. Changing the sheared G•A to Watson–Crick GC pairs was expected to abolish L7Ae binding. Indeed, neither K42C–EDTA-Fe nor V95C–EDTA-Fe produced any footprint on the mutated KT1 in either PfuRPR-mKT1 or -mKT123 (Figure 4A and B). Meanwhile, the footprinting pattern in the P12 region of PfuRPR-mKT1 was the same as the one in the wild-type PfuRPR (Figure 4 and Supplementary Figure S4).
The pattern of cleavages at the mutated double K-turn, however, was more complex. Compared to the footprints in wild-type PfuRPR, the footprint of K42C–EDTA-Fe in PfuRPR-mKT2 (Figure 4A and D; Supplementary Figure S4) shifted by a few nucleotides in the bulged strand (positions 143–148) and in the non-bulged strand (positions 184–188); while the footprint of V95C–EDTA-Fe in the bulged strand (positions 130–144) also shifted, there was no footprint in the non-bulged strand (Figure 4A and Supplementary Figure S4). In contrast to the footprints in wild-type PfuRPR and PfuRPR-mKT2, the footprint in PfuRPR-mKT3 (Figure 4A and E; Supplementary Figure S4) for each PfuL7Ae derivative appeared in only one strand: K42C–EDTA-Fe led to cleavages at positions 181–188 and V95C–EDTA-Fe at positions 127–128.
When both KT2 and KT3 were mutated together, as in PfuRPR-mKT23 and -mKT123, the footprint of PfuL7Ae in P12 disappeared altogether, as expected (Figure 4A and F; Supplementary Figure S4). Meanwhile, both K42C- and V95C–EDTA-Fe yielded the wild-type pattern of footprints in the P16-17 region of PfuRPR-mKT2, -mKT3, and -mKT23.
Finally, when KT1, KT2, and KT3 were all mutated (PfuRPR-mKT123), PfuL7Ae did not footprint anywhere, as expected (Figure 4A and Supplementary Figure S4).
Mg2+ dependence of Pfu RNase P
Although an active Pfu RNase P complex can be reconstituted with the RPR + 4 RPPs (POP5, RPP30, RPP21 and RPP29), it requires 20–30 mM Mg2+ for optimal catalysis (9,11,16). Since these Mg2+ concentrations are not physiological (cellular [Mg2+] < 2 mM) (46), we postulated that L7Ae could readily bind the K-turns and restructure the RPR at low [Mg2+] to enable subsequent binding of the other four RPPs. Indeed, analysis of Pfu RNase P activity revealed that addition of PfuL7Ae to RPR + 4 RPPs at 55°C decreases KMg by 3-fold to ∼4 mM (Supplementary Figure S5).
DISCUSSION
Number and location of K-turns in type A and type M archaeal RPRs
Our predictions about the number and location of K-turns in PfuRPR (type A; three K-turns) and MjaRPR (type M; two K-turns) have been validated by our results, which show that individual and combined mutation of these K-turns proportionately decreased L7Ae stoichiometry and eliminated/altered the corresponding L7Ae footprint (Figures 2–4). Our results also support the possibility of a new double K-turn RNA module in the P12 helix (Figure 5), which is conserved in type A and type M RPRs from Thermococcales, Methanococcales and Methanobacteriales, and may reflect its possible functional importance to the resident RNase P.
Figure 5.
A preliminary model of MmaRPR P12 bound by two L7Ae copies. (A) Secondary structure of the P11–12 helix from MmaRPR. (B) A model of the structure in (A), with two kinks in the double K-turn module, was built and refined with Assemble (55) using available crystallographic structures of K-turns as templates (29–31). Note that the two-kink conformations at the double K-turn module enable the long P12 helix to fold back. (C) Docking of two copies of MjaL7Ae (PDB: 1XBI) on the RNA model illustrates approximate positioning and lack of steric constraints for double occupancy.
The double K-turn is a new RNA structural element
While KT1 is a stand-alone K-turn, KT2 and KT3 are interconnected, with the NC helix of KT2 also serving as the C helix of KT3. Given that K-turns are known to create a twist in the NC helix in addition to the eponymous kink, extensive cleavages by each L7Ae–EDTA-Fe derivative in both strands of P12 in PfuRPR suggest a compact and tight RPR fold facilitated by the binding of two L7Ae copies, one each at KT2 and KT3 (Figure 5C). Although the results from MS, gel-shift and footprinting studies indicate that the three K-turns operate independently of each other in their ability to bind L7Ae, there are also clearly some local structural changes when either KT2 or KT3 is mutated in the double K-turn motif (Figure 4 and Supplementary Figure S4).
When the sheared G•A pairs in the NC helix of KT2 were altered to GC pairs in PfuRPR-mKT2 and MjaRPR-mKT2, not only did mKT2 lose the ability to bind L7Ae (Figures 2B and 3B), but KT3 was expected to have a more typical C helix and thus an increased affinity for L7Ae. This premise is supported by two observations. First, the L7Ae footprint in PfuRPR-mKT2 did not differ much from the one in the wild-type PfuRPR, particularly in the bulged strand (Figure 4 and Supplementary Figure S4). Second, a similar KT2 mutation in MmaRPR left its catalytic activity intact (if not moderately higher) when it was reconstituted with all RPPs, including L7Ae (13). Conversely, mutating the G•A pairs in KT3 disabled mKT3 binding to L7Ae (Figures 2B and 3B) but should have no effect on KT2. While KT2 in PfuRPR-mKT3 can still bind L7Ae, it is likely to bind only weakly due to the absence of stabilizing interactions between L7Ae and the distal positions of the NC helix (e.g. 4n and 5n) (24). Indeed, gel-shift assays confirmed a large decrease in L7Ae binding to PfuP12-mKT3 (Supplementary Figure S3), and as expected there was a diminished L7Ae footprint in PfuRPR-mKT3 (Figure 4 and Supplementary Figure S4). The dramatic changes in the cleavage pattern in PfuRPR-mKT3 also suggest an altered local structure that could have eliminated any gain in activity mediated by L7Ae binding at KT2, as was observed with a similar KT3 mutation in Mma RNase P (13).
Implications for assembly and function of RNase P based on L7Ae copy number
K-turns, which enable axial bending and flexibility in RNA duplexes, are stabilized by Mg2+ ions, tertiary interactions, and/or binding by proteins such as L7Ae (24–26). Some (but not all) K-turns display Mg2+-induced folding, an attribute dictated by the identity of the 3b•3n pair (28). In PfuRPR, these positions are G•U, G•C, and A•A in KT1, KT2 and KT3, respectively; in MjaRPR, they are C•G and U•U in KT2 and KT3, respectively. Based on sequence rules for Mg2+-induced folding of K-turns (28), the PfuRPR and MjaRPR K-turns (barring U•U, potentially) are predicted to require either L7Ae binding, distal tertiary interactions mediated by other RPPs, or both to stabilize the kinked conformation. Findings from this study help advance testable postulates that are predicated on the idea that K-turns in archaeal RPRs are unlikely to adopt a stable kinked conformation with Mg2+ alone and therefore require L7Ae binding to facilitate the function of other RPPs at near-cellular Mg2+ concentrations (Supplementary Figure S5).
KT1 in the C domain
In type A archaeal RPRs, KT1 is flanked at its 5΄ end by L15, known to bind the pre-tRNA 3΄-CCA motif by Watson–Crick base pairing (41,47,48), and at its 3΄ end by the P6 pseudoknot. Interestingly, many type A bacterial RPRs contain a kink, termed a pK-turn (Supplementary Figure S1A), created by the juxtaposition of P16 and P17 around an asymmetric internal loop (38,49). This pK-turn was first reported in the crystal structure of the Thermotoga maritima (Tma) RNase P holoenzyme bound to a tRNA (38,49). The pK-turn can be superimposed on a K-turn with an RMSD of 1.5 Å (49), and these motifs can be functionally exchanged (50) even though they share none of the base pairing and sequence requirements. Hence, both bacterial and archaeal type A RPRs have a kink between P16 and P17, suggesting that this structural module is important for function, likely for improved substrate recognition mediated by the proximal L15 and for overall folding through formation of the P6 pseudoknot. Moreover, at low Mg2+ concentrations, we expect L7Ae to promote a tighter interaction between the RPR and POP5•RPP30, which is known to bind the RPR C domain and enhance the rate of cleavage by 60-fold (11,16,40).
The double K-turn in the S domain
In both type A and type M RPRs, a long-range tertiary interaction may be facilitated by the double K-turn module in P12. In a number of type A RPRs like PfuRPR, P12 terminates with a GNRA tetraloop, whose docking to a distal receptor would allow this long helix to fold back. A preliminary model shows that two copies of L7Ae can indeed bind P12 without any steric constraint, and facilitate a fold-back due to the double K-turn (Figure 5).
At low Mg2+ concentrations, L7Ae could bind the double K-turn to engender RPR remodeling and a tighter assembly with RPP21•RPP29, which is known to bind the S domain (including P12) and increase the enzyme's apparent substrate affinity (KS) by 16-fold (11). This postulate is supported by our RPP29–EDTA-Fe footprinting experiments which show that the terminal L12 tetraloop likely docks near the P7–10 cruciform in the RPR and that L7Ae increases the extent of OH•-mediated cleavages at low Mg2+ concentrations (Lai, Lai & Gopalan, unpublished data). Payoffs from this remodeling likely include improved positioning of the two universally conserved, interleaving T-loop motifs [one each in J11/12 and J12/11 (5,38,51)] that bind the pre-tRNA elbow, as described for the corresponding region in bacterial RPR (38).
A key inference from the P12 fold-back in PfuRPR (and possibly all type A RPRs) is the unexpected tertiary structure similarity between bacterial and archaeal RPRs. The overall stability of the bacterial RPR is mediated in part by tertiary interactions in the S domain between the P8–9 helical stack and tetraloops L14 and L18 (Supplementary Figure S1A); those structural elements thus act as a molecular strut that brings together distal helices (38). While archaeal RPRs lack P14 and L14 (Figure 1), their double K-turn in P12 could enable a similar local architecture to the bacterial cousin, enabling the tetraloop L12 to fulfill the role of the bacterial L14 (Supplementary Figure S1), a premise supported by the proximity of L12 to P8-9 in our RPP29–EDTA-Fe footprinting experiments (Lai, Lai & Gopalan, unpublished data). These findings exemplify the idea that different peripheral structural elements may be used to forge the same active-site core in large RNAs (52). Additional support for this notion is expected from studies of other RPRs including eukaryotic variants that lack the double K-turn in P12 (53).
Summary
Our findings should motivate the search for other RNAs with the double K-turn or similar modules that result in the unique fold-back topology. The latter expectation of structurally-equivalent swaps has a precedent in the interchange of K-turns and pK-turns in riboswitches and bacterial RPRs, respectively, from different phylogenetic groups (49,50,54). Such studies may provide a better framework for uncovering why evolution may have opted for a double K-turn motif over two tandem K-turns. Finally, the integrated approach of using high-resolution native MS and directed footprinting should find broad utility as a powerful experimental strategy for establishing the stoichiometry and placement of protein subunits in large RNPs, a prerequisite for understanding their assembly and function.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Drs. Mark Foster and Brandon Crowe (OSU) for helpful discussions on some of the structural aspects described in this study, and Dr. Kotaro Nakanishi (OSU) for advice on modeling. We thank Dr. Stuart Maxwell (North Carolina State University) for generously sharing a His6-tagged Mja L7Ae overexpression construct, and Dr. Mark Foster for use of the HPLC system in his laboratory. We also appreciate the efforts of Cecilia Go (OSU), who performed some of the initial gel-shift analyses with PfuRPR and L7Ae.
Footnotes
Present address: Wen-Yi Chen, Nexcelom Bioscience, San Diego, CA 92121, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Behrman Research Fund [to V.G.]; National Science Foundation [MCB-0843543 to V.G.]; National Institutes of Health [R01 GM113658 to V.W.; R01 GM120582 to V.G., V.W., Mark Foster, Julius Lucks and Michael Poirier]. Funding for open access charge: NIH [R01 GM120582].
Conflict of interest statement. E.W. is an Editor of Nucleic Acids Research.
REFERENCES
- 1. Lai L.B., Vioque A., Kirsebom L.A., Gopalan V.. Unexpected diversity of RNase P, an ancient tRNA processing enzyme: challenges and prospects. FEBS Lett. 2010; 584:287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jarrous N., Gopalan V.. Archaeal/eukaryal RNase P: subunits, functions and RNA diversification. Nucleic Acids Res. 2010; 38:7885–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu F., Altman S.. Ribonuclease P. 2010; NY: Springer. [Google Scholar]
- 4. Esakova O., Krasilnikov A.S.. Of proteins and RNA: the RNase P/MRP family. RNA. 2010; 16:1725–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Evans D., Marquez S.M., Pace N.R.. RNase P: interface of the RNA and protein worlds. Trends Biochem. Sci. 2006; 31:333–341. [DOI] [PubMed] [Google Scholar]
- 6. Lechner M., Rossmanith W., Hartmann R.K., Tholken C., Gutmann B., Giege P., Gobert A.. Distribution of ribonucleoprotein and protein-only RNase P in eukarya. Mol. Biol. Evol. 2015; 32:3186–3193. [DOI] [PubMed] [Google Scholar]
- 7. Kikovska E., Svard S.G., Kirsebom L.A.. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:2062–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pannucci J.A., Haas E.S., Hall T.A., Harris J.K., Brown J.W.. RNase P RNAs from some Archaea are catalytically active. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:7803–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pulukkunat D.K., Gopalan V.. Studies on Methanocaldococcus jannaschii RNase P reveal insights into the roles of RNA and protein cofactors in RNase P catalysis. Nucleic Acids Res. 2008; 36:4172–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boomershine W.P., McElroy C.A., Tsai H.-Y., Wilson R.C., Gopalan V., Foster M.P.. Structure of Mth11/Mth Rpp29, an essential protein subunit of archaeal and eukaryotic RNase P. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:15398–15403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen W.Y., Pulukkunat D.K., Cho I.M., Tsai H.Y., Gopalan V.. Dissecting functional cooperation among protein subunits in archaeal RNase P, a catalytic ribonucleoprotein complex. Nucleic Acids Res. 2010; 38:8316–8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen W.Y., Xu Y., Cho I.M., Oruganti S.V., Foster M.P., Gopalan V.. Cooperative RNP assembly: complementary rescue of structural defects by protein and RNA subunits of archaeal RNase P. J. Mol. Biol. 2011; 411:368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cho I.M., Lai L.B., Susanti D., Mukhopadhyay B., Gopalan V.. Ribosomal protein L7Ae is a subunit of archaeal RNase P. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:14573–14578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kouzuma Y., Mizoguchi M., Takagi H., Fukuhara H., Tsukamoto M., Numata T., Kimura M.. Reconstitution of archaeal ribonuclease P from RNA and four protein components. Biochem. Biophys. Res. Commun. 2003; 306:666–673. [DOI] [PubMed] [Google Scholar]
- 15. Chen W.Y., Singh D., Lai L.B., Stiffler M.A., Lai H.D., Foster M.P., Gopalan V.. Fidelity of tRNA 5΄-maturation: a possible basis for the functional dependence of archaeal and eukaryal RNase P on multiple protein cofactors. Nucleic Acids Res. 2012; 40:4666–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tsai H.Y., Pulukkunat D.K., Woznick W.K., Gopalan V.. Functional reconstitution and characterization of Pyrococcus furiosus RNase P. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:16147–16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hall T.A., Brown J.W.. Archaeal RNase P has multiple protein subunits homologous to eukaryotic nuclear RNase P proteins. RNA. 2002; 8:296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lai L.B., Cho I.-M., Chen W.-Y., Gopalan V.. Liu F, Altman S. Ribonuclease P. 2010; NY: Springer Verlag; 153–172. [Google Scholar]
- 19. Samanta M.P., Lai S.M., Daniels C.J., Gopalan V.. Sequence analysis and comparative sudy of the protein subunits of archaeal RNase P. Biomolecules. 2016; 6:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harris J.K., Haas E.S., Williams D., Frank D.N., Brown J.W.. New insight into RNase P RNA structure from comparative analysis of the archaeal RNA. RNA. 2001; 7:220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chan P.P., Brown J.W., Lowe T.M.. Modeling the thermoproteaceae RNase P RNA. RNA Biol. 2012; 9:1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lai L.B., Chan P.P., Cozen A.E., Bernick D.L., Brown J.W., Gopalan V., Lowe T.M.. Discovery of a minimal form of RNase P in Pyrobaculum. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:22493–22498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fukuhara H., Kifusa M., Watanabe M., Terada A., Honda T., Numata T., Kakuta Y., Kimura M.. A fifth protein subunit Ph1496p elevates the optimum temperature for the ribonuclease P activity from Pyrococcus horikoshii OT3. Biochem. Biophys. Res. Commun. 2006; 343:956–964. [DOI] [PubMed] [Google Scholar]
- 24. Huang L., Lilley D.M.J.. The molecular recognition of kink-turn structure by the L7Ae class of proteins. RNA. 2013; 19:1703–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lilley D.M. The K-turn motif in riboswitches and other RNA species. Biochim. Biophys. Acta. 2014; 1839:995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lilley D.M.J. The structure and folding of kink turns in RNA. Wiley Interdiscipl. Rev.: RNA. 2012; 3:797–805. [DOI] [PubMed] [Google Scholar]
- 27. Goody T.A., Melcher S.E., Norman D.G., Lilley D.M.. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA. 2004; 10:254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McPhee S.A., Huang L., Lilley D.M.. A critical base pair in k-turns that confers folding characteristics and correlates with biological function. Nat. Commun. 2014; 5:5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hamma T., Ferré-D’Amaré A.R.. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 Å resolution. Structure. 2004; 12:893–903. [DOI] [PubMed] [Google Scholar]
- 30. Moore T., Zhang Y., Fenley M.O., Li H.. Molecular basis of box C/D RNA-protein interactions: cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 2004; 12:807–818. [DOI] [PubMed] [Google Scholar]
- 31. Xue S., Wang R., Yang F., Terns R.M., Terns M.P., Zhang X., Maxwell E.S., Li H.. Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle. Mol. Cell. 2010; 39:939–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lai S.M., Lai L.B., Foster M.P., Gopalan V.. The L7Ae protein binds to two kink-turns in the Pyrococcus furiosus RNase P RNA. Nucleic Acids Res. 2014; 42:13328–13338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oshima K., Kakiuchi Y., Tanaka Y., Ueda T., Nakashima T., Kimura M., Yao M.. Structural basis for recognition of a kink-turn motif by an archaeal homologue of human RNase P protein Rpp38. Biochem. Biophys. Res. Commun. 2016; 474:541–546. [DOI] [PubMed] [Google Scholar]
- 34. Kimura M., Kakuta Y.. Satyanarayana T, Johri BN, Prakash A. Microorganisms in Sustainable Agriculture and Biotechnology. 2012; Springer Science+Business Media B.V; 487–508. [Google Scholar]
- 35. Ma X., Lai L.B., Lai S.M., Tanimoto A., Foster M.P., Wysocki V.H., Gopalan V.. Uncovering the stoichiometry of Pyrococcus furiosus RNase P, a multi-subunit catalytic ribonucleoprotein complex, by surface-induced dissociation and ion mobility mass spectrometry. Angew. Chem. Int. Ed. Engl. 2014; 53:11483–11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gagnon K., Zhang X., Maxwell E.S.. In vitro reconstitution and affinity purification of catalytically active archaeal box C/D sRNP complexes. Methods Enzymol. 2007; 425:263–282. [DOI] [PubMed] [Google Scholar]
- 37. Pan T. Higher order folding and domain analysis of the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry. 1995; 34:902–909. [DOI] [PubMed] [Google Scholar]
- 38. Reiter N.J., Osterman A., Torres-Larios A., Swinger K.K., Pan T., Mondragon A.. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010; 468:784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Torres-Larios A., Swinger K.K., Krasilnikov A.S., Pan T., Mondragón A.. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005; 437:584–587. [DOI] [PubMed] [Google Scholar]
- 40. Xu Y., Amero C.D., Pulukkunat D.K., Gopalan V., Foster M.P.. Solution structure of an archaeal RNase P binary protein complex: formation of the 30-kDa complex between Pyrococcus furiosus RPP21 and RPP29 is accompanied by coupled protein folding and highlights critical features for protein-protein and protein-RNA interactions. J. Mol. Biol. 2009; 393:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sinapah S., Wu S., Chen Y., Pettersson B.M.F., Gopalan V., Kirsebom L.A.. Cleavage of model substrates by archaeal RNase P: role of protein cofactors in cleavage-site selection. Nucleic Acids Res. 2011; 39:1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Annesley T.M. Ion suppression in mass spectrometry. Clin. Chem. 2003; 49:1041–1044. [DOI] [PubMed] [Google Scholar]
- 43. King R., Bonfiglio R., Fernandez-Metzler C., Miller-Stein C., Olah T.. Mechanistic investigation of ionization suppression in electrospray ionization. J. Am. Soc. Spectrom. 2000; 11:942–950. [DOI] [PubMed] [Google Scholar]
- 44. Crain P.F., McCloskey J.A.. Applications of mass spectrometry to the characterization of oligonucleotides and nucleic acids. Curr. Opin. Biotechnol. 1998; 9:25–34. [DOI] [PubMed] [Google Scholar]
- 45. Pan J., Xu K., Yang X., Choy W.-Y., Konermann L.. Solution-phase chelators for suppressing nonspecific protein−metal interactions in electrospray mass spectrometry. Anal. Chem. 2009; 81:5008–5015. [DOI] [PubMed] [Google Scholar]
- 46. Cowan J.A. Cowan JA. The Biological Chemistry of Magnesium. 1995; NY: VCH Publishers, Inc; 1–23. [Google Scholar]
- 47. Kirsebom L.A., Svärd S.G.. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBo J. 1994; 13:4870–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Terada A., Yoshida T., Kimura M.. Identification of nucleotide residues essential for RNase P activity from the hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biosci. Biotechnol. Biochem. 2007; 71:1940–1945. [DOI] [PubMed] [Google Scholar]
- 49. Meyer M., Westhof E., Mesquida B.. A structural module in RNase P expands the variety of RNA kinks. RNA Biol. 2011; 9:254–260. [DOI] [PubMed] [Google Scholar]
- 50. Daldrop P., Masquida B., Lilley D.M.J.. The functional exchangeability of pk- and k-turns in RNA structure. RNA Biol. 2013; 10:445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gopalan V. Uniformity amid diversity in RNase P. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:2031–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Reiter N.J., Chan C.W., Mondragon A.. Emerging structural themes in large RNA molecules. Curr. Opin. Struct. Biol. 2011; 21:319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Piccinelli P., Rosenblad M.A., Samuelsson T.. Identification and analysis of ribonuclease P and MRP RNA in a broad range of eukaryotes. Nucleic Acids Res. 2005; 33:4485–4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lescoute A., Leontis N.B., Massire C., Westhof E.. Recurrent structural RNA motifs, isostericity matrices and sequence alignments. Nucleic Acids Res. 2005; 33:2395–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jossinet F., Ludwig T.E., Westhof E.. Assemble: an interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels. Bioinformatics. 2010; 26:2057–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.