Abstract
eIF5A is an essential protein involved in protein synthesis, cell proliferation and animal development. High eIF5A expression is observed in many tumor types and has been linked to cancer metastasis. Recent studies have shown that eIF5A facilitates the translation elongation of stretches of consecutive prolines. Activated eIF5A binds to the empty E-site of stalled ribosomes, where it is thought to interact with the peptidyl-tRNA situated at the P-site. Here, we report a genome-wide analysis of ribosome stalling in Saccharomyces cerevisiae eIF5A depleted cells using 5Pseq. We confirm that, in the absence of eIF5A, ribosomes stall at proline stretches, and extend previous studies by identifying eIF5A-dependent ribosome pauses at termination and at >200 tripeptide motifs. We show that presence of proline, glycine and charged amino acids at the peptidyl transferase center and at the beginning of the peptide exit tunnel arrest ribosomes in eIF5A-depleted cells. Lack of eIF5A also renders ribosome accumulation at the stop codons. Our data indicate specific protein functional groups under the control of eIF5A, including ER-coupled translation and GTPases in yeast and cytoskeleton organization, collagen metabolism and cell differentiation in humans. Our results support a broad mRNA-specific role of eIF5A in translation and identify the conserved motifs that affect translation elongation from yeast to humans.
INTRODUCTION
Translation is the process by which ribosomes read genetic information (mRNAs) to produce new proteins. As all fundamental processes in life, translation is highly regulated from initiation to elongation and termination. Translation elongation does not occur uniformly along messenger RNA (mRNA) and is modulated by different factors, e.g. the chemical characteristics of the acceptor and of the donor amino acid can affect peptide bond formation and slow down elongation. In other cases, the nascent peptide can interact with the ribosomal exit tunnel and can also stall translation. Major effects in ribosome pausing have been described specifically for proline and positively charged amino acids. Given its unique side chain cyclized onto backbone nitrogen, proline acts as a bad acceptor and donor for peptide bond formation, and results in stalled ribosomes (1–6). In some cases, positively charged amino acids can stall ribosomes by interacting with the peptide exit tunnel (4,6–8). Besides single amino acids, the combination of specific residues can also block protein synthesis. This is the case of the polyproline motifs formed by three consecutive prolines or more. Recent evidence suggests that polyproline motifs induce ribosome pauses that require translation elongation factor eIF5A in eukaryotes (or its homologue EF-P in prokaryotes) to resume protein synthesis (9–12).
eIF5A is a translation elongation factor and an essential protein in all eukaryotes. However, its effect in translation has a direct impact only on 30–75% of global translation (13,14). eIF5A is the only known protein to follow a post-translational modification called hypusination. This modification is rare, but essential, and the enzymes that synthesize the hypusine on an eIF5A lysine residue are conserved through evolution (reviewed in (15,16)). Research interest in eIF5A is currently growing because this protein is highly expressed during tumorigenesis in different cancer types (e.g. pancreatic, hepatic, colon, lung and ovarian) and a high expression of specific eIF5A isoforms has been linked to cancer metastasis (reviewed in (17–19)). In vitro experiments initially showed that eIF5A is needed for the formation of the first peptide bond and was, therefore, classified as a translation initiation factor (20,21). Later experiments have shown accumulation of ribosomes blocked during translation elongation in cells that lack functional eIF5A, which indicates its role as a non-essential factor for translation elongation (22,23). Biochemical and crystallography studies have revealed that hypusinated eIF5A can bind ribosomes (24,25), and that a clear structural similarity exists between eukaryotic eIF5A and bacterial EF-P, which resembles the structure of a tRNA (reviewed in (15,16)). Current data suggest that eIF5A could bind stalled ribosomes with a free E-site, and then eIF5A and its hypusine moiety would force a productive positioning of the CCA-end of the P-tRNA to facilitate the transfer of the nascent chain from P-tRNA to A-tRNA (26). Moreover, studies with EF-P have shown that the elongation factor contributes to the formation of a peptide bond with proline in an entropic way by providing a better orientation of Pro-tRNA for the catalytic reaction at the peptidyl transfer center (27). Therefore, these results explain how eIF5A promotes the elongation of paused ribosomes at polyproline motifs.
eIF5A depletion results in pleiotropic phenotypes, implicating eIF5A in multiple cellular processes; e.g., cell cycle progression (28), actin dynamics (29), cotranslational translocation of proteins into the endoplasmic reticulum (ER) (30), apoptosis, cell migration and metastasis (17–19). Except for polarized growth during yeast mating, where we previously showed that hypusinated eIF5A is required for the translation of the polyproline motifs of formin Bni1 (12), there is no direct connection between the translation role of eIF5A and its physiological phenotypes. In bacteria, EF-P has proven important for the translation of not only PPP motifs, but also of distinct diprolyl motifs (e.g., DPP, PPD, APP, PPG, PPW) (31–34), where the effect on translation is modulated by the specific amino acids that surround the proline-rich motif (34,35). Recent proteomic studies conducted in eukaryotic cells that lack functional eIF5A have identified proteins with altered levels. However, not all PPP-containing proteins are down-regulated and many regulated proteins do not contain polyproline motifs (36–38). Although proteomic approaches are sensitive to not only protein synthesis, but also to protein degradation, lack of a good correlation between regulated proteins and polyproline motifs renders the determination of the ribosome pausing provoked by eIF5A depletion in eukaryotes on a high-precision and genomic scale necessary.
Here, we report a genome-wide analysis of ribosome protected regions in S. cerevisiae eIF5A-depleted cells using 5Pseq (39). This method allows ribosome stalls to be studied by sequencing 5΄ phosphorylated mRNA co-translational degradation intermediates (40). This approach offers an in vivo snapshot or ribosome footprints, and does not require the use of translation inhibitors or in vitro mRNA digestion (41). Our 5PSeq data indicate that presence of proline, glycine or charged amino acids at specific positions of the peptidyl transferase center and at the beginning of the peptide exit tunnel stalls translation in the absence of eIF5A. We confirmed the role of eIF5A in facilitating the translation of polyproline sequences at different yeast proteins at a single nucleotide resolution. We also extend previous studies by showing how eIF5A-depletion stalls translation at more than 200 tripeptides (3% of all possible motifs), many of which with no proline. Lack of eIF5A also renders ribosome accumulation at the stop codons. Our data also point to specific gene functional groups under the control of eIF5A in yeast and humans.
MATERIALS AND METHODS
Growth conditions and sample preparation
Saccharomyces cerevisiae haploid strains wild-type BY4741 (MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0) and temperature-sensitive eIF5A mutants tif51A-1 and tif51A-3 derivatives of the BY4741 strain (42) were grown in liquid YPD (1% yeast extract, 2% peptone, 2% glucose) media at 25°C until the exponential phase. Then cells were transferred to 37°C and were incubated for 4 h. Cells were harvested and pellets were frozen in liquid nitrogen and kept at –80°C until further 5PSeq processing. From the same cultures, the cells grown at 25°C, and also after 3 and 4 h of incubation at 34 and 37°C, were collected for subsequent protein analyses by Western blot. To check growth at permissive and restrictive temperatures, the yeast cells from fresh plates were streaked in YPD solid media and plates were incubated at 25 and 37°C for 2 days.
Western blot
Protein extraction and Western blot analyses were performed as previously described (43,44). A specific anti-eIF5A polyclonal antibody (kindly provided by S.R. Valentini, Sao Paulo State University) (45) was used to visualize the Tif51A protein. As described, the tif51A-3 protein showed slightly lower electrophoretic mobility than the wild-type protein due to the G118D mutation (45).
RNA extraction
Total RNA was extracted from yeast cells by phenol:chloroform extraction, as described in (46). Briefly, cell pellets were resuspended in LETS buffer (100 mM LiCl, 10 mM EDTA, 10 mM Tris–HCl pH 7.5, 0.2% SDS) and acid phenol–chloroform (5:1) and acid-washed glass beads (0.5 mm, Sartorius) were added. Cells were broken in Fast-Prep (MP-Biomedicals) and cell debris was pelleted. The supernatant was extracted with chloroform:isoamyl alcohol (24:1). The aqueous phase was precipitated with LiCl and cold EtOH, and then stored at –20°C. RNA pellets were washed with 70% (v/v) EtOH and then resuspended in the desired volume of RNase free water. The amount of RNA was quantified in a Nanodrop 2000 Spectrophotometer (Thermo Scientific) and RNA integrity was checked by agarose electrophoresis.
The 5PSeq method
The 5PSeq method was performed as previously described (40) with minor modifications. Specifically, 1 μg of total RNA was treated with DNAse and the 5΄ phosphorylated mRNA ends were ligated to a DNA/RNA oligo that contained a unique molecular identifier. After single-stranded RNA ligation, RNA samples were purified using Agencourt AmpureXP beads (Beckman Coulter) and RNA integrity was confirmed by running agarose gel electrophoresis. The rest of the sample was subjected to partial rRNA depletion using the RiboCop rRNA depletion kit (Lexogen). The rRNA-depleted sample was fragmented, reverse-transcribed and subjected to a one-cycle PCR reaction to generate a library that contained the cDNA which originated from the 5΄ phosphorylated mRNA molecules. Biotinylated cDNA fragments were captured using magnetic streptavidin beads, subjected to end-repair, dA addition and ligation of the P7MPX adapter compatible with illumina sequencing, as previously described (40). The streptavidin bound libraries were PCR-amplified (18 cycles), pooled and size-selected (300–500 nt). Samples were sequenced using a NextSeq500 to produce single-end 75 reads, and were demultiplexed using Illumina BaseSpace servers.
Sequence analysis
For each read we trimmed the first 8 nt (UMI, unique molecular identifier) and aligned the rest to the S. cerevisiae genome (version R64-1-1) using Hisat2 with default parameters (except when limiting the maximum intron size to 2 kb). The reads with the same 5΄mapping site and UMI were considered PCR duplicates, and were omitted from the analysis. We calculated differential ribosome pausing for each motif counting 5PSeq reads at a defined distance from the motifs genome-wide and computed their differential expression using DESeq2 (47). Briefly, a raw read count table was generated for the genome-wide aggregate of each motif in each replicate (Supplementary Dataset). The differential expression was computed across strains by taking into account the variability of biological replicates. An adjusted P-value was computed using Benjamin–Hochberg and shrunken fold-changes as implemented by DESeq2 (47). For the tripeptide analysis, all the possible combinations of motifs were analysed. For pentateptides, we restricted the analysis to those motifs that resulted from extending the 241 identified significant regulated tripeptides. For visualization purposes, the metagenes of the motifs were drawn using the raw abundance 5΄P intermediates in reads per million (rpm, Figure 1). Alternatively, to facilitate the visualization of codon-specific regulation, the total number of reads per sample in each metagene window (e.g. –80 to +20 from a motif) was normalized to the same total number of reads (normalized rpm). Independently of visualization, P-values and significance were computed with a raw number of unique identified molecules. The hierarchical cluster between the identified motifs was computed using Euclidean distances as defined by Biostrings (R package 2.40.2) and the complete linkage method. Weblogos were constructed using the online tool at http://weblogo.berkeley.edu/
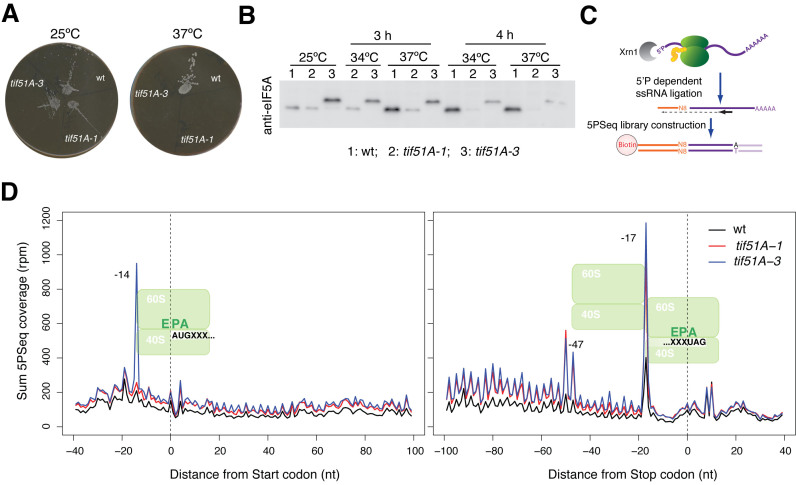
Figure 1.
eIF5A depletion yields a marked pause at the stop codon. (A) Growth of wild-type BY4741 and eIF5A temperature-sensitive mutant cells tif51A-1 and tif51A-3 in YPD media at permissive (25°C) and restrictive (37°C) temperatures. (B) Western blot analysis of eIF5A depletion. Cells were grown in YPD until the exponential phase and were then transferred to the indicated temperatures. Proteins were extracted and analysed by Western blot. The eIF5A protein was visualized with a specific anti-eIF5A antibody. (C) Outline of the 5PSeq method. (D) The metagene analysis displaying the abundance 5΄P intermediates in reads per million (rpm) in relation to the ORF start (left panel) and stop (right panel) codons.
RESULTS
eIF5A depletion increases ribosome accumulation at start and stop codons
We aimed to analyse genome-wide the in vivo stalls of ribosomes in eIF5A-deficient S. cerevisiae cells. For this purpose we used the recently developed 5PSeq technique, which measures at a codon resolution the in vivo ribosome footprints generated by cellular co-translational 5΄-3΄ mRNA degradation (39). Since eIF5A is an essential protein in yeast, we used two eIF5A temperature-sensitive strains that contained a single Pro83 to Ser mutation (tif51A-1), and the double Cys39 to Tyr and Gly118 to Asp mutations (tif51A-3), in the highly expressed gene TIF51A (HYP2), which encodes the eIF5A protein (42,45). Both mutants grew well at 25°C and were unable to grow at 37°C. tif51A-3 showed a more severe growth defect than tif51A-1 at intermediate temperatures (Figure 1A and (12)). We incubated the wild-type and the two eIF5A mutant cells at 37°C for 4 h, conditions under which almost no drop in cell viability for both mutants was noted, but a significant reduction in the eIF5A protein took place (eIF5A protein levels, compared to the wild type ones, lowered to 0.17±0.13 and 0.18±0.10, respectively, in the tif51A-1 and tif51A-3 mutants, n = 3) (Figure 1B and (12)). We performed three independent 5PSeq experiments (40) with each tif51A-1 and tif51A-3 strain, and compared to two independent experiments with the isogenic wild-type BY4741 strain (see Methods and Figure 1C for details, and Supplementary Figure S1 for experiment reproducibility).
As we previously described, 5΄P mRNA degradation intermediates produced in vivo ribosome footprinting that peaked 17 nt upstream of the first nucleotide of the codon at the A position in the ribosome, and presented a clear three nucleotides periodicity (39) (Figure 1D). Surprisingly, we also observed a clear sharp accumulation of the 5PSeq reads at position -14 of the start codon (corresponding to a ribosome paused at the P site during initiation) in the tif51A-3 mutant, but not in tif51A-1 cells. This ribosome pausing was reminiscent of the general ribosome accumulation at the translation start site caused by the addition of cycloheximide or the naturally-occurring one in the stationary phase (Supplementary Figure S2A). Although eIF5A has been firstly described as a translation initiation factor (20,21), later reports have clearly established a role in translation elongation (22,23). The fact that this ribosome accumulation at the start codon was observed only in the tif51A-3 mutant, which showed a more defective growth phenotype than tif51A-1, suggests that this pause could be a consequence of a stronger inhibition of translation elongation in the tif51A-3 mutant, which could limit the availability of free ribosomes for initiation (48). As saturated yeast cultures (in stationary phase, Supplementary Figure S2A) also produce a clear accumulation at the translation initiation level, we cannot rule out that this pause could be caused by slower growth in the tif51A-3 mutant. However, as that phenotype is not observed in slower cultures grown in synthetic defined minimum media (SD, Supplementary Figure S2A) or in the alternative tif51A-1 mutant, a severe effect associated to the mutation present in the tif51A-3 strain is likely.
The ribosomes that paused at the stop codon in the wild-type yeast cells produced a clear peak, situated 17 nucleotides upstream of the stop codon (39). Here we found greater ribosomal footprinting at that position for both mutants tif51A-1 and tif51A-3, which suggests that the eIF5A factor is necessary for the termination of translation (Figure 1D). Ribosome accumulation at the stop codon also caused the appearance of a secondary peak (–47 and –50 nt), which is consistent with the protection of a second ribosome stalled in close proximity to that paused at the termination step. Similar disome patterns have been documented for pauses, such as that provoked in wild-type cells during oxidative stress at the stop codon, or more extreme pauses like those induced by the inhibition of histidine biosynthesis by 3-AT (39) or in the dom34Δ mutant (49). Presence of a proline residue immediately before the stop codon has been described to result in slower termination (50,51). However, in tif51 mutants, the strong pause observed at the stop was independent of presence of proline (Supplementary Figure S2B).
eIF5A depletion pauses the ribosome at the proline, glycine and charged amino acid codons at specific positions in the ribosome
Different features of codons and encoded amino acids have been suggested as being responsible for ribosomal velocity (2,4,6–8,41,52). We investigated the ribosomal footprinting signals at the 61 sense codons and the three non-sense stop codons in eIF5A-deficient strains, which we compared to the wild-type cell signals. We studied the 5΄P footprints that corresponded to the specific locations of the investigated codon (or the corresponding amino acid) in the ribosome: site A at position –17, site P at position –14, positions –11 and –8 that corresponded to the second and third amino acids upstream of site P and moved into the peptide exit tunnel, and position –20, the adjacent downstream position of site A (Figure 2A and (39)). Our results showed that presence of proline, glycine, and the charged amino acids arginine, lysine and aspartate, induced significantly higher ribosome footprinting signals in the eIF5A-deficient cells (Figure 2B, Supplementary Figure S3A and Supplementary dataset). The disparity in the intensity of the ribosome footprints between the eIF5A mutants and wild type was greater for the strain that carried two point mutations in eIF5A (tif51A-3) than was for that which carried only one-point mutation (tif51A-1). We observed similar 5PSeq footprints for different synonymous codons for proline (Supplementary Figure S3B), which suggests that the observed ribosome accumulation is an amino acid and not a codon effect.
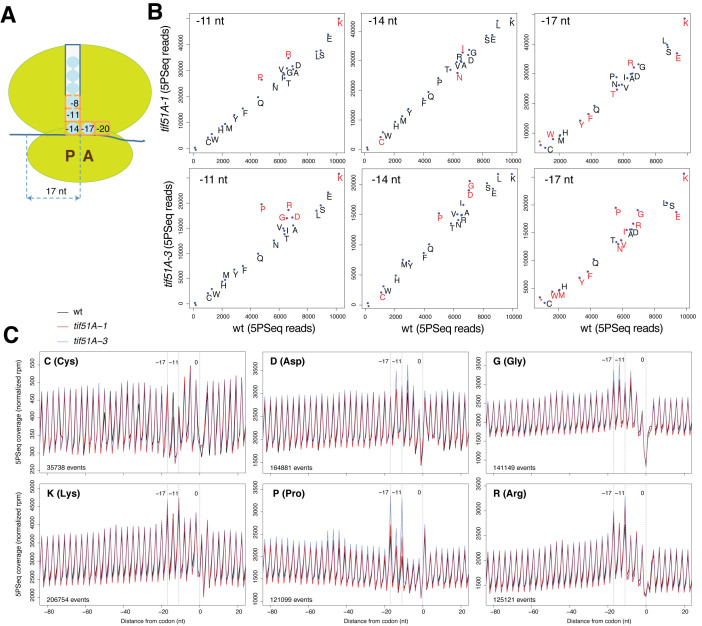
Figure 2.
eIF5A depletion reveals increased pauses at proline, glycine and charged amino acids. (A) The correspondence between the ribosomal sites occupied by codons and sequencing reads, as measured by 5PSeq. (B) The differential ribosome pausing for all amino acids at –11, –14 and –17 nt from the selected codon. Significantly regulated differences (adjusted P-value < 0.01) in red. (C) The metagene that represents the 5PSeq intermediates for the indicated amino acids and positions. To facilitate the identification of codon-specific regulation and sample comparison, the total number of reads for each metagene (–80 to +20 window) was normalized.
Next we examined the 5΄P profiles associated with the amino acids that caused higher ribosomal footprinting signals in the eIF5A mutants (Figure 2C). For proline, we detected strong accumulation signals at positions –11 and –17, and less intense ones at –14. This result agrees with the fact that eIF5A is necessary for the translation of polyproline motifs (i.e. PPP) and facilitates the formation of the peptide bond between the second and third proline (9,12). To study whether the proline-associated pauses at positions –11 and –17 were the result of these prolines forming part of a PPP motif (with P present at positions –11, –14 and –17), we studied the proline-associated pauses according to the neighboring amino acids. This analysis showed that the isolated prolines, when no other proline was immediately upstream or downstream (XPX motif, being X different of P), presented pauses at –14 and –8 (Supplementary Figure S3C). Our results supported the role of eIF5A in the formation of a peptide bond with the imino group of the proline situated at the P- and A-sites, and, more importantly, they suggest that eIF5A promotes peptide bond formation while proline enters the exit tunnel of the ribosome (proline at the second position upstream of the P-site; see the scheme in Figure 2A). In addition to this effect, presence of multiple adjacent prolines is associated with increased ribosome pausing (see below).
More intense 5΄P signals in the eIF5A mutants were also observed for glycine at position –14, and also at positions –17 and –8. For the basic amino acids, lysine and arginine, the most marked pause was detected at the –11 position, and at –8 for the acidic amino acid aspartic (Figure 2C). All these pauses suggest that eIF5A alleviates the inhibition of translation that takes place when these amino acids have been incorporated into the nascent polypeptide chain, and they move through the ribosomal exit tunnel. Indeed it is known in some cases that the interaction between the nascent peptide and the initial part of the ribosomal exit tunnel can lead to ribosome arrest (6,35,53).
Altogether, our results suggest that eIF5A alleviates the ribosome stalling produced by the incorporation of proline, glycine, and positively and negatively charged amino acids, into the nascent peptide chain.
eIF5A facilitates translation elongation at not only polyproline, but also at many other tripeptide motifs
The detected ribosome pauses associated with amino acids other than proline suggests that eIF5A plays additional roles than the previously described function to facilitate the synthesis of polyproline motifs (9,12). Studies in bacteria have shown that addition of basic, acidic and other specific amino acids in the diprolyl motif (PP) context increases translation pausing, which is alleviated by eIF5A homologous EF-P (31,32,35). In mammals, proteomic analyses have shown that eIF5A depletion causes the down-regulation of many proteins with no polyproline motif (36–38). To clarify which amino acid motifs provoke eIF5A-related pauses in yeast, we computed the differential 5΄P protection associated with all the possible tripeptide motifs (Figure 3). Many tripeptide motifs presented significant differential protection when comparing the eIF5A mutants with the wild-type strain (38 in tif51A-1 and 238 in tif51A-3, fold change>1.6 or <0.6, and P-value<0.01; and 151 tripeptide motifs at a more restrictive cutoff, fold change >2.0 or <0.5, and P-value <0.001) (Figures 3A and Supplementary dataset). We observed an increased disome signal (∼44 to 41 nt upstream) for many tripeptides, which indicates strong pause and ribosome stacking (Figure 3C G). Despite many motifs being associated with eIF5A-dependent pauses, the motifs at different positions of the ribosome clustered in a few groups and revealed specific shared features for the identified tripeptide sequences (Figure 3B, Supplementary Figure S4 and Supplementary Table S1).
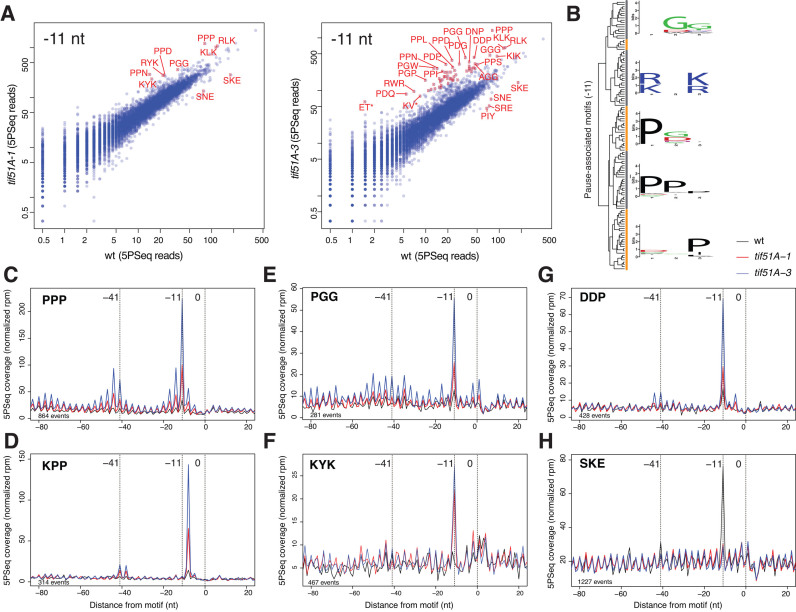
Figure 3.
Multiple tripeptide motifs influence eIF5A-dependent translation. (A) Differential ribosome pausing for all tripeptide motifs. Axes represent the raw number of reads for the metagene of each motif. Significantly regulated differences (adjusted P-value < 0.0001) in red, computed using shrunken log2 fold changes (see the Methods). (B) Hierarchical cluster depicting tripeptide motifs associated to eIF5A-dependent stall at –11nt. (C–H) The metagene that represents the 5PSeq intermediates for the selected tripeptide motifs. The total number of reads for each metagene (–80 to +20 window) was normalized.
Firstly, all except three of the motifs associated with the translation pauses in tif51A-1 were also identified in the tif51A-3 mutant. This confirmed our previous observation that cells with the double eIF5A mutation exhibit a similar (but stronger) phenotype to the single eIF5A mutant. From our 5PSeq, we also observed that a high percentage of the tripeptide motifs contained a diprolyl (PP) motif (43% in tif51A-1 and 15% in tif51A-3), and these motifs were mostly stronger and more significant (Supplementary Table S1 and Supplementary dataset). These prolyl-rich motifs rendered a high stalling signal at positions –11 for the PPX motifs and at –8 for the XPP motifs, where X indicated any amino acid (Figure 3C and Supplementary Figure S3C). These results indicated that the strongest ribosomal pauses occurred with peptidyl-Pro-Pro-tRNA at the P-site (that corresponded to 5΄P accumulation at –8 in relation to XPP, and at –11 in relation to PPX; see Figure 2A). This result agrees with similar phenomena described in Escherichia coli that lacked EF-P using ribosome profiling (32,35). In bacteria, ribosome profiling experiments have shown that stalling at PP dipeptides is enhanced by the presence of specific amino acids upstream and/or downstream of the motif. Remarkably, the pausing motifs that we identified in yeast contained all the tripeptides that have been previously described to cause strong stalling in bacteria (PPP, PPG, PPQ, PPR, PPD, PPW, PPN, DPP, EPP, GPP and APP) ((31–35) and reviewed in (54)).
Additionally, several of the identified eIF5A-dependent pause motifs contained a dipeptide GG (3% in tif51A-1 and 8% in tif51A-3), with peptidyl-Gly-tRNA at the P-site and Gly-tRNA at the A-site. We also observed that a significant number of motifs (40% in tif51A-1 and 12% in tif51A-3) contained positively charged amino acids at positions 5΄ and 3΄, thus rendering a motif (R/K)X(R/K). In these tripeptides, the 5΄ basic amino acid was situated mostly at –11. Motifs with dipeptides PG and GP were also abundant with glycine situated mostly at –14 (P-site) (8% and 6%, respectively). Finally, 13% of the tripeptide motifs contained negatively charged amino acid D at the first 5΄ position and proline and/or glycine in the downstream positions (Figure 3, Supplementary Figures S4 and S5 and Supplementary Table S1). These DXX motifs were found at –8, –11 and –14, which indicates that the pause was caused when aspartic was placed at the beginning of the peptide exit channel. All these data indicate that eIF5A alleviates the translational elongation stalling at the mRNAs which encode for many different specific tripeptide motifs, and may explain the previous puzzling results which indicated a strong impact in global translation by eliminating eIF5A (13,14).
Unexpectedly, we also found tripeptide motifs (eight in tif51A-1 and the same motifs, plus five other motifs in tif51A-3) with fewer 5΄P intermediate reads in the eIF5A mutants than in the wild type. This finding suggests that these motifs produce a ribosome pause only in the presence of eIF5A (Figure 3 and Supplementary Table S1). Four (SKE, SNE, SPE and SRE) of these 13 motifs contained sequence SXE. When we re-analyzed our previous work (39), we found that wild-type cells also exhibited a ribosome pause associated with those motives. Interestingly, the ribosome pausing at those locations was highly regulated according to the physiological state of the cell, e.g. the SKE and SRE associated pauses increased dramatically in cells in the stationary phase, while the PIY associated pause was lost in the stationary phase, but increased for the cells grown in minimum media (SD) (Supplementary Figure S5B). This result suggests that eIF5A may play a role in regulating the translation elongation of these motifs during environmental adaptation and stress. However, more research is needed to fully understand the translation elongation mechanism that acts on SXE motifs.
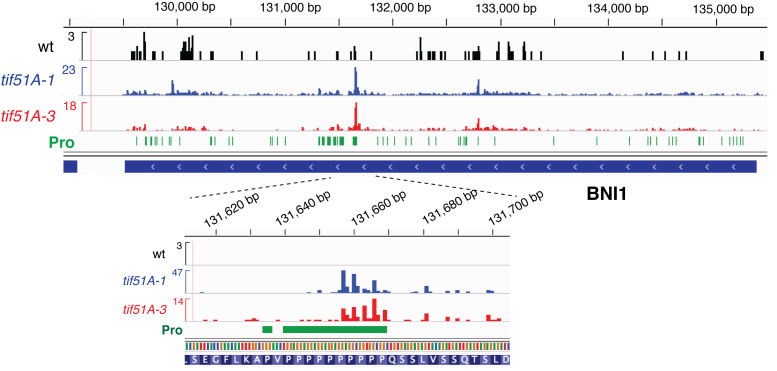
Having described the specific tripeptide motifs associated with differential translation regulation, we studied the impact of eIF5A depletion on individual genes. 5PSeq, as well as any other genome-wide approach to study ribosome footprints, produces relative sparse coverage at an individual gene level. For this reason, we focused our analysis on the genes that contain polyproline motifs (PPP). In line with our and other previous works (9,12), we detected the accumulation of 5΄P intermediates in close proximity to the polyproline motifs encoded in genes BNI1, VRP1 and EAP1 (Figure 4 and Supplementary Figure S6). These results confirm that 5PSeq can identify previously validated pauses, and can be used to identify ribosome stalling at the single codon level.
Figure 4.
5PSeq signals identify eIF5A-dependent ribosome stalling at particular genes. Coverage of the 5PSeq reads (collapsed to the single 5΄nucleotide) for the BNI1 gene in wild type (black), tif51A-1 (blue) and tif51A-3 (red). Proline codons are depicted in green.
Influence of the amino acids that precede the eIF5A-regulated tripeptide motifs in ribosome stalling
Polyproline, and the other tripeptides motifs associated with ribosome pausing in eIF5A mutants, are widely distributed in the yeast proteome, which means a vast number of putative eIF5A-translational regulated mRNAs. In bacteria, the sequence context of the polyproline motif can modulate translational stalling (34,35), and the residues upstream of the stalling motif can enhance or suppress this effect (55). To gain more information about eIF5A-dependent stall sites, we analysed the contribution of two additional upstream residues for all the tripeptide motifs presenting the above-identified eIF5A-dependent differential ribosome stalling. We calculated the pauses associated with the selected pentapeptides (i.e. all possible combinations of two random upstream amino acids, followed by the previously identified 241 significant regulated tripeptides) in relation to 5΄P positions –17, –14, –11, –8, –5 and –2, i.e. with ribosomal A-site occupied by the first, second, and up to the fifth, amino acid in the pentapeptide motif. As we can see in Supplementary Figure S7A, the number of motifs with significant differences (P-value < 0.01) between mutants tif51A-1 and tif51A-3 and the wild type was larger at positions –8, –5 and –2, which once again indicates the effect of eIF5A on the elongation of the selected tripeptide motifs as these amino acids move into the ribosomal exit tunnel. We then selected the pentapeptide motifs that gave the most significant pauses, P-value <0.001, at positions –11, –8, –5 and –2, and observed 38 pentapeptide motifs that yielded pauses in mutants tif51A-1 and/or tif51A-3 versus the wild type, and 11 pentapeptide motifs with higher 5΄P intermediates in the wild type than in the eIF5A mutants (Figure 5 and Supplementary Figure S7B-C). Interestingly, several pentapeptides were coincident in the amino acids at the specific positions in relation to the A-site of the ribosome (Supplementary Figure S7B and C), so they can be considered extended motifs (e.g. TAKPPPNS). To further dissect the influence of the polypeptide context for the prolyl-associated pauses, we also analyzed PPP motifs with respect to the observed pause. We noted that stalling at PPP was favored in some upstream peptide contexts, but was disadvantaged in others (Supplementary Figure S7D).
Figure 5.
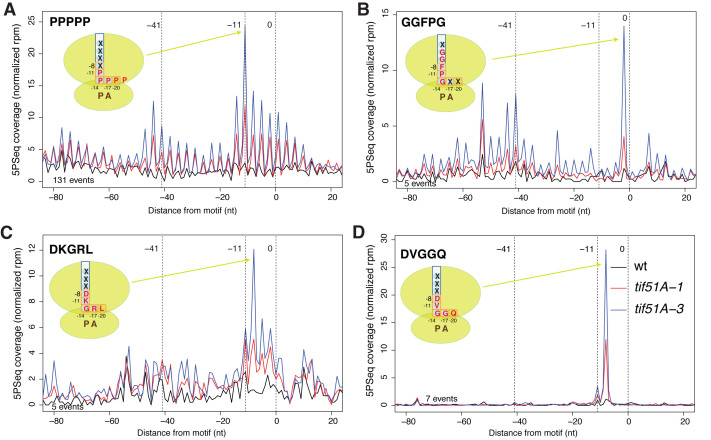
Extended context of peptide motifs influence eIF5A-dependent pausing. (A–D) The metagene that represents the 5PSeq intermediates for the selected pentapeptide motifs, and a scheme of their location in the ribosome exit tunnel. The total number of reads for each metagene (–80 to +20 window) was normalized.
The number of proteins that contained each specific pentapeptide motif, and the number of proteins that contained the pentapeptide motifs which corresponded to each extended motif, was small, except for the group of proteins with five consecutive prolines (PPPPP). However, we found that gene ontology (GO) functional categories were over-represented in several pentapeptide protein groups (Supplementary Table S2). As expected, the GO categories ‘actin cytoskeleton organization’ and ‘mating projection tip’ were overrepresented in the proteins that contained the PPPPP motif (Supplementary Table S2). This result agrees with the physiological role of eIF5A in shmoo formation and fertility (12). We also found that the five proteins which contained motif GGFPG, encoded by genes SSA1-3, SIS1 and YDJ1, were involved in ‘protein folding’ (P-value = 4.6E–8) and, all except SIS1, were also involved in the ‘targeting of proteins to the endoplasmic reticulum (ER)’ (P-value = 5.2E–7) (Supplementary Table S2). In particular, we observed a clear increase in the 5΄P reads at position –2 of the GGFPG motif, which indicates pauses with the fifth amino acid, glycine, at the P-site and the previous four amino acids, GGFP, entering into the exit tunnel (Figure 5B). Another eIF5A-dependent pentapeptide motif, DKGRL, was present in the chaperone ATPase proteins encoded by SSA1-4 and KAR2, involved in ‘SRP-dependent cotranslational protein targeting to the membrane’ (P-value = 2.5E–12) (Supplementary Table S2). In this case, the stalling at the DKGRL motif was observed at position –8, with glycine at the P-site, arginine at the A-site and the charged amino acids aspartic and lysine in the ribosomal exit tunnel (Figure 5C). The discovery of stalled ribosomes upon eIF5A depletion at messenger-encoding proteins involved in protein folding and coupling translation to the ER may be related to the previous phenotypes observed in S. cerevisiae and mammalian cells that lack functional eIF5A (30,36). In yeast, eIF5A function loss resulted in defects in the co-translational translocation of the proteins in the ER (30). Additionally, a proteomic study in HeLa cells has identified ‘protein folding’ as the major cellular process affected in eIF5A-depleted cells, and has shown that lack of eIF5A leads to ER stress (36). Finally, we observed that five proteins (encoded by ARF1-3, ARL3 and GPA2), of the nine that contained the DVGGQ motif, were involved in ‘GTPase activity’ (P-value = 6.5E–9) (Supplementary Table S2). In this case the glycines of the pentapeptide were situated at P- and A-sites, and at the negatively charged aspartic residue in the exit tunnel (Figure 5D).
In short, our results indicate that specific combinations of the upstream amino acids in the eIF5A-dependent tripeptide motifs context determine the strength of the ribosomal stall, and that the identified pentapeptide motifs are enriched in groups of functionally related proteins.
Abundance and impact of yeast eIF5A-regulated motifs on the human proteome
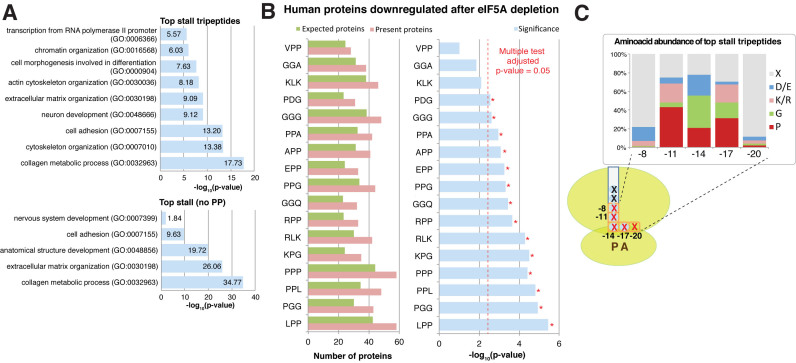
Having identified many new tripeptide and pentapeptide motifs associated with eIF5A depletion in S. cerevisiae, we studied the impact of these motifs on the human proteome. First we determined the abundance of the 43-top tripeptide, which caused the most marked ribosome stalls in the eIF5A mutants, and of all the identified eIF5A-dependent pentapeptide motifs in the human proteome. For both motif types, those with consecutive prolines were the most abundant (e.g. LPP, PPP and PPPPP motifs, rendered 24.5%, 23.3% and 3.6% of the human proteins, respectively). This result agrees with previously reports which have indicated that polyproline motifs have been selected through evolution (56,57). We next determined the number of total motifs per protein of the 43-top eIF5A-dependent tripeptides (and excluded all the eIF5A-dependent pentapeptides because of their generally poor representation in the human proteome) and searched for specific GO functional categories enriched in the proteins whose amino acid sequence contained more than 25 motifs. We observed significant enrichment in four groups of related GO terms (Figure 6A). The first related to extracellular matrix organization and collagen metabolism. A second group was related to differentiation and neurogenesis. The third was related to cytoeskeleton and actin organization. A fourth group included ‘chromatin organization’ and ‘RNA polymerase II transcription’. Since collagens, some transcription factors and proteins involved in actin organization are rich in proline and/or polyproline sequences (12,57), we repeated the gene ontology analysis by excluding the PPP motif. However, we obtained similar GO terms, except for ‘cytoskeleton and actin organization’, with similar significance (Figure 6A and Supplementary Figure S8A). We also obtained similar results, collagen, development and neurogenesis GO terms, when we excluded PP motifs, or even the tripeptides that contained proline (Figure 6A and Supplementary Figure S8A). These results suggest that, independently of the presence of proline, eIF5A-dependent tripeptides share common chemical or structural characteristics that could make the translation of functional related proteins dependent on presence of eIF5A.
Figure 6.
Conservation of the identified motifs in the human proteome. (A) Selected gene ontology terms significantly enriched in the human proteins with a high content (>25) of eIF5A-dependent motifs, including those tripeptides that contained PP or not. (B) Down-regulated proteins after eIF5A depletion (37) are enriched for the motifs that causer an eIF5A-dependent pause. Randomly expected proteins (green), identified (pink) and significance (according to the hypergeometric distribution, blue). Significance level for Bonferroni-adjusted multiple testing is depicted in red and indicated by *. Only the motifs present in at least 40 proteins identified by Fujimura et al. (37) were considered for the analysis. (C) Amino acid abundance in relation to the ribosome structure for those motifs associated with eIF5A-dependent pausing in S. cerevisiae.
To obtain an independent confirmation of our findings and to study to what degree eIF5A-dependent ribosome stalling is evolutionary conserved, we re-analyzed recent works in human cells. We particularly used proteomic analyses which investigated the effect of eIF5A depletion (36,37). Fujimura et al. identified eIF5A-dependent proteins that both contain and exclude polyproline motifs (37). Meanwhile, the down-regulated proteins identified by Mandal et al. were not enriched in the polyproline motifs (36). The 147 down-regulated proteins in the human pancreatic ductal adenocarcinoma (PDAC) cells under eIF5A depletion (37), but not the up-regulated proteins, were enriched in the PPPPP pentapeptide (P-value = 0.018) and in 14 of the 17 analysed motifs (by restricting the analysis to the 43-top tripeptide motifs or to the pentapeptide yeast motifs associated with pauses that are present in at least 40 proteins in the dataset; adjusted P-value < 0.05) (Figure 6B). Fujimura et al. showed that proteins RhoA, with no polyprolines, and its downstream target ROCK2, which contains two polyproline motifs, are regulated by eIF5A to mediate cell migration and invasion (37). Here, we observed that ROCK2 and RhoA were enriched in eIF5A-dependent motifs (14 and seven eIF5A-tripeptide motifs, respectively). In both cases, several of the motifs present in proteins do not contain diprolyl sequences. In the proteomic study by Mandal et al. conducted in HeLa cells, eIF5A depletion changed the levels of 104 proteins (36). As reported in that study, the identified eIF5A-dependent down-regulated proteins are no enriched in the polyproline motifs. However, we observed enrichment for motifs DDP (P-value = 1.7E–4) and RYK (P-value = 1.1E–3) (Supplementary Figure S8B).
Altogether, these results indicate the possibility that different eIF5A-regulated tripeptide motifs identified in yeast, apart from the polyproline motif, can cause translation defects in specific human proteins whose protein levels have been previously reported to be eIF5A-dependent.
DISCUSSION
eIF5A is an essential protein in eukaryotic cells that has been clearly related to translation elongation in the last few years. Studies on eIF5A in eukaryotes, and on the functional and structural homologous EF-P in bacteria, have shown that these factors are needed for the translation elongation of stretches of consecutive prolines. Here we extend these studies and determine the ribosome stalls along the S. cerevisiae transcriptome in eIF5A-depleted cells. Our results indicate clear ribosome stalls upon eIF5A depletion, provoked by presence of both single specific amino acids and tri- and pentapeptide motifs at specific positions in ribosomes. eIF5A depletion also provokes strong ribosome stalls in stop codons. We observed translation elongation stalls for more than 200 different tripeptides in the eIF5A mutants. Many of these tripeptides contain diprolyl motifs, which confirms previous reports on the effect of eIF5A on the translation of polyproline motifs. We identified in budding yeast all the diprolyl-containing motifs previously classified as EF-P-dependent in bacteria. Although our results agree with previous ribosome profiling studies in prokaryotes (31–35), we cannot rule out specific differences owing to the study of subpopulations of ribosome-associated co-translational degradation (39). We also identified many other eIF5A-regulated tripeptides independently of proline. eIF5A motifs are also enriched in diglycyl motifs, and in basic (arginine and lysine) and acidic (aspartic and glutamic) amino acids. From our 5Pseq data, we reach a consensus stalling peptide sequence with the relative frequency of every amino acid at each position when the ribosome stalls in eIF5A-depleted cells (Figure 6C).
One main question is how eIF5A alleviates ribosome stalls during elongation at these specific motifs. Our in vivo study cannot rule out that the observed ribosome stalls could be provoked by the down regulation of other translation elongation factors. However, the direct binding of eIF5A to the 80S ribosome, and a role in peptide bond formation demonstrated by previous crystallography and biochemical studies (9,24–27), suggest that the effects we describe herein are also direct. The frequency of specific amino acids at the P- and A-sites in the eIF5A-regulated motifs suggests that eIF5A facilitates the formation of peptide bonds in situations of slow reactivity when glycine, proline, or an acidic amino acid, acts as a donor and when proline, glycine or a basic amino acid acts as an acceptor (Figure 6C). Our results are in line with previous studies which have described that most of these amino acids act badly during peptide bond formation, e.g. presence of a secondary amino group in proline has been associated with slow peptide bond formation and translation (1–6,27). Previous studies have also detected poor reactivity for glycine and for basic amino acids arginine and lysine (4,58). Our data also support ribosome stalling in the absence of eIF5A when proline, lysine/arginine, glycine or aspartic/glutamic is/are immediately upstream of the aminoacyl tRNA at the P-site (position –11), and when acidic to basic amino acids are two positions upstream of the P-site (position –8) (Figure 6C). This extends previously described observations which have indicated that in wild-type cells, the proline located at the beginning of the exit tunnel can slow down ribosomes (4), and that positively charged amino acids in nascent chains slow down the elongation speed through their interaction with the negatively charged ribosome exit tunnel in wild-type cells (4,6–8). Studies into the mode of translation inhibition by antibiotics have pointed out the existence of functional interplay between the interactions of the nascent chain in the ribosomal exit tunnel and the formation of a peptide bond at the peptidyl transference center (59,60). Interestingly, it was shown that macrolide antibiotics erythromycin and telithromycin induced translation arrest at the consensus (R/K)X(R/K), with the X residue located at the P-site (59), which is coincident with one of the strong stall tripeptide consensus motifs that we found upon eIF5A depletion. These results suggest that peptide bond formation at this motif is poorly efficient, so it becomes rate-limiting in bacteria upon the addition of antibiotics and in yeast upon eIF5A depletion. Therefore, our ribosome footprinting suggests that presence of eIF5A near the tRNA exit site (E-site) facilitates peptide bond formation when there is a combination of amino acids with bad reactivity at the peptidyl transference center and when there is a specific nascent peptide at the beginning of the ribosome exit tunnel which negatively affects peptide bond formation. Interestingly, in addition to the positive effects in translation elongation alleviated by presence of eIF5A, we also identify its potential regulatory role for the translation of the proteins that contain SXE motifs.
During the review process of this work, an independent study was published that supports a global role of eIF5A in translation termination (61). Using ribosome profiling in a different experimental design, the results of Sculler et al. are coincident with ours in that they find a general role of eIF5A in the termination of translation and they also partially overlap our findings of new non-polyproline peptides producing ribosome stalls upon eIF5A depletion (of the top 29 tripeptide eIF5A-dependent motifs described by Schuller et al., 10 of 11 polyproline motifs and 15 of 18 non proline motifs have also been found in our study). In vitro translation assays validated a direct role of eIF5A in the translation elongation of the new stall motifs and showed that eIF5A hypusination is critical for the efficient translation of PPP motifs, but not for other non-proline stalling motifs (61).
The work presented herein predicts a considerable effect of eIF5A-depeletion on protein levels. We confirm at a single codon resolution the effect of eIF5A depletion on the synthesis of yeast proteins with polyproline stretches. Our analysis of pentapeptide motifs also indicates a role of eIF5A in the translation of the mRNAs that encode the specific chaperones involved in protein folding and ER-coupled translation, and in the translation of GTPases. Two previous studies conducted in yeasts and mammals have implicated eIF5A in the translocation of proteins to the ER (30) and ER-stress (36), respectively. However, both studies detected a rise in several chaperone levels when depleting activated-eIF5A. Our data suggest the opposite effect. A thorough analysis of the protein levels of all the implicated chaperones is necessary to clarify the precise role of eIF5A in ER-coupled translation. The enrichment in eIF5A-tripeptides observed in specific groups of functionally related human proteins suggests the possibility that several relevant cellular processes (cytoskeleton organization and RNA polymerase transcription), and also animal development processes (collagen metabolism and differentiation), require efficient translation by eIF5A. Interestingly, the motifs identified in budding yeast can explain previous proteomics observations in human cells, which suggest the significant conservation of its mechanism of action across eukaryotes. Although the eIF5A protein is highly expressed in all the eukaryotic cells, its expression is not always constant and can be regulated at transcriptional and post-transcriptional levels (reviewed in (18)). Due to the predicted high demand of eIF5A in translation, changes in eIF5A availability may render the modulation of the levels of its target proteins in the cell.
To summarize, our ribosome stalling results in eIF5A-deficient cells uncover a considerable effect on the translation elongation of many mRNAs and a clear role in termination. This work shows that eIF5A makes a major quantitative contribution to global cellular translation, which thus accounts for its essential role in the cell.
ACCESSION NUMBERS
The raw and processed sequencing data are deposited at GEO with accession number GSE91064.
AVAILABILITY
The computed abundance of the motifs identified in the human proteome are available at: http://data.pelechanolab.com/Publications/2017_eIF5A/
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Fany Carrasco for technical assistance, and also to Alice Stanciu and Elena Garre for their help while processing samples. We also thank the helpful discussions with all the members of the Alepuz and Pelechano laboratories, to Wu Wei and Manuel Sánchez del Pino for discussion and support, and to José-E. Pérez-Ortín for his critical review of the manuscript and useful suggestions. The computational analysis was performed at UPPMAX using the resources provided by the Swedish National Infrastructure for Computing (SNIC).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish MiNECO [BFU2013-48643-C3-3-P and BFU2016-77728-C3-3-P to P.A.]; Regional Valencian Government [PROMETEO II 2015/006]; European Union funds [FEDER]; Karolinska Institutet [SciLifeLab Fellowship (SFO) to V. P.]; Swedish Research Council (VR 2016-01842) [starting grant to V.P.]; Wallenberg Academy Fellowship (KAW) and the Swedish Foundations’ Starting Grant—Ragnar Söderberg Foundation (to V.P.). Funding for open access charge: Spanish MiNECO [BFU2013-48643-C3-3-P and BFU2016-77728-C3-3-P to P.A.]; Regional Valencian Government [PROMETEO II 2015/006]; European Union funds [FEDER]; Karolinska Institutet [SciLifeLab Fellowship (SFO) to V. P.]; Swedish Research Council (VR 2016-01842) [starting grant to V.P.]; Wallenberg Academy Fellowship (KAW) and the Swedish Foundations’ Starting Grant—Ragnar Söderberg Foundation (to V.P.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Wohlgemuth I., Brenner S., Beringer M., Rodnina M.V.. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J. Biol. Chem. 2008; 283:32229–32235. [DOI] [PubMed] [Google Scholar]
- 2. Artieri C.G., Fraser H.B.. Accounting for biases in riboprofiling data indicates a major role for proline in stalling translation. Genome Res. 2014; 24:2011–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gamble C.E., Brule C.E., Dean K.M., Fields S., Grayhack E.J.. Adjacent codons act in concert to modulate translation efficiency in yeast. Cell. 2016; 166:679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gardin J., Yeasmin R., Yurovsky A., Cai Y., Skiena S., Futcher B.. Measurement of average decoding rates of the 61 sense codons in vivo. Elife. 2014; 3, doi:10.7554/eLife.03735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pavlov M.Y., Watts R.E., Tan Z., Cornish V.W., Ehrenberg M., Forster A.C.. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sabi R., Tuller T.. A comparative genomics study on the effect of individual amino acids on ribosome stalling. BMC Genomics. 2015; 16(Suppl. 10):S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Charneski C.A., Hurst L.D.. Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol. 2013; 11:e1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu J., Deutsch C.. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 2008; 384:73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gutierrez E., Shin B.S., Woolstenhulme C.J., Kim J.R., Saini P., Buskirk A.R., Dever T.E.. eIF5A promotes translation of polyproline motifs. Mol. Cell. 2013; 51:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doerfel L.K., Wohlgemuth I., Kothe C., Peske F., Urlaub H., Rodnina M.V.. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science (New York, NY). 2013; 339:85–88. [DOI] [PubMed] [Google Scholar]
- 11. Ude S., Lassak J., Starosta A.L., Kraxenberger T., Wilson D.N., Jung K.. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science (New York, NY). 2013; 339:82–85. [DOI] [PubMed] [Google Scholar]
- 12. Li T., Belda-Palazon B., Ferrando A., Alepuz P.. Fertility and polarized cell growth depends on eIF5A for translation of polyproline-rich formins in Saccharomyces cerevisiae. Genetics. 2014; 197:1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kang H.A., Hershey J.W.. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J. Biol. Chem. 1994; 269:3934–3940. [PubMed] [Google Scholar]
- 14. Henderson A., Hershey J.W.. Eukaryotic translation initiation factor (eIF) 5A stimulates protein synthesis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:6415–6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park M.H., Nishimura K., Zanelli C.F., Valentini S.R.. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids. 2010; 38:491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dever T.E., Gutierrez E., Shin B.S.. The hypusine-containing translation factor eIF5A. Crit. Rev. Biochem. Mol. Biol. 2014; 49:413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakanishi S., Cleveland J.L.. Targeting the polyamine-hypusine circuit for the prevention and treatment of cancer. Amino Acids. 2016; 48:2353–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mathews M.B., Hershey J.W.. The translation factor eIF5A and human cancer. Biochim. Biophys. Acta. 2015; 1849:836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Caraglia M., Park M.H., Wolff E.C., Marra M., Abbruzzese A.. eIF5A isoforms and cancer: two brothers for two functions. Amino Acids. 2013; 44:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Benne R., Brown-Luedi M.L., Hershey J.W.. Purification and characterization of protein synthesis initiation factors eIF-1, eIF-4C, eIF-4D, and eIF-5 from rabbit reticulocytes. J. Biol. Chem. 1978; 253:3070–3077. [PubMed] [Google Scholar]
- 21. Schreier M.H., Erni B., Staehelin T.. Initiation of mammalian protein synthesis. I. Purification and characterization of seven initiation factors. J. Mol. Biol. 1977; 116:727–753. [DOI] [PubMed] [Google Scholar]
- 22. Saini P., Eyler D.E., Green R., Dever T.E.. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009; 459:118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Patel P.H., Costa-Mattioli M., Schulze K.L., Bellen H.J.. The Drosophila deoxyhypusine hydroxylase homologue nero and its target eIF5A are required for cell growth and the regulation of autophagy. J. Cell Biol. 2009; 185:1181–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zanelli C.F., Maragno A.L., Gregio A.P., Komili S., Pandolfi J.R., Mestriner C.A., Lustri W.R., Valentini S.R.. eIF5A binds to translational machinery components and affects translation in yeast. Biochem. Biophys. Res. Commun. 2006; 348:1358–1366. [DOI] [PubMed] [Google Scholar]
- 25. Jao D.L., Chen K.Y.. Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J. Cell. Biochem. 2006; 97:583–598. [DOI] [PubMed] [Google Scholar]
- 26. Schmidt C., Becker T., Heuer A., Braunger K., Shanmuganathan V., Pech M., Berninghausen O., Wilson D.N., Beckmann R.. Structure of the hypusinylated eukaryotic translation factor eIF-5A bound to the ribosome. Nucleic Acids Res. 2016; 44:1944–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Doerfel L.K., Wohlgemuth I., Kubyshkin V., Starosta A.L., Wilson D.N., Budisa N., Rodnina M.V.. Entropic contribution of elongation factor P to proline positioning at the catalytic center of the ribosome. J. Am. Chem. Soc. 2015; 137:12997–13006. [DOI] [PubMed] [Google Scholar]
- 28. Hanauske-Abel H.M., Park M.H., Hanauske A.R., Popowicz A.M., Lalande M., Folk J.E.. Inhibition of the G1-S transition of the cell cycle by inhibitors of deoxyhypusine hydroxylation. Biochim. Biophys. Acta. 1994; 1221:115–124. [DOI] [PubMed] [Google Scholar]
- 29. Chatterjee I., Gross S.R., Kinzy T.G., Chen K.Y.. Rapid depletion of mutant eukaryotic initiation factor 5A at restrictive temperature reveals connections to actin cytoskeleton and cell cycle progression. Mol. Genet. Genomics. 2006; 275:264–276. [DOI] [PubMed] [Google Scholar]
- 30. Rossi D., Galvao F.C., Bellato H.M., Boldrin P.E., Andrews B.J., Valentini S.R., Zanelli C.F.. eIF5A has a function in the cotranslational translocation of proteins into the ER. Amino Acids. 2014; 46:645–653. [DOI] [PubMed] [Google Scholar]
- 31. Peil L., Starosta A.L., Lassak J., Atkinson G.C., Virumae K., Spitzer M., Tenson T., Jung K., Remme J., Wilson D.N.. Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:15265–15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Woolstenhulme C.J., Guydosh N.R., Green R., Buskirk A.R.. High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep. 2015; 11:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hersch S.J., Wang M., Zou S.B., Moon K.M., Foster L.J., Ibba M., Navarre W.W.. Divergent protein motifs direct elongation factor P-mediated translational regulation in Salmonella enterica and Escherichia coli. MBio. 2013; 4:e00180–00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Starosta A.L., Lassak J., Peil L., Atkinson G.C., Virumae K., Tenson T., Remme J., Jung K., Wilson D.N.. Translational stalling at polyproline stretches is modulated by the sequence context upstream of the stall site. Nucleic Acids Res. 2014; 42:10711–10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Elgamal S., Katz A., Hersch S.J., Newsom D., White P., Navarre W.W., Ibba M.. EF-P dependent pauses integrate proximal and distal signals during translation. PLoS Genet. 2014; 10:e1004553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mandal A., Mandal S., Park M.H.. Global quantitative proteomics reveal up-regulation of endoplasmic reticulum stress response proteins upon depletion of eIF5A in HeLa cells. Sci. Rep. 2016; 6:25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fujimura K., Choi S., Wyse M., Strnadel J., Wright T., Klemke R.. Eukaryotic translation initiation factor 5A (EIF5A) regulates pancreatic cancer metastasis by modulating RhoA and Rho-associated kinase (ROCK) protein expression levels. J. Biol. Chem. 2015; 290:29907–29919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Memin E., Hoque M., Jain M.R., Heller D.S., Li H., Cracchiolo B., Hanauske-Abel H.M., Pe’ery T., Mathews M.B.. Blocking eIF5A modification in cervical cancer cells alters the expression of cancer-related genes and suppresses cell proliferation. Cancer Res. 2014; 74:552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pelechano V., Wei W., Steinmetz L.M.. Widespread Co-translational RNA Decay Reveals Ribosome Dynamics. Cell. 2015; 161:1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pelechano V., Wei W., Steinmetz L.M.. Genome-wide quantification of 5΄-phosphorylated mRNA degradation intermediates for analysis of ribosome dynamics. Nat. Protoc. 2016; 11:359–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science (New York, NY). 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li Z., Vizeacoumar F.J., Bahr S., Li J., Warringer J., Vizeacoumar F.S., Min R., Vandersluis B., Bellay J., Devit M. et al. . Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat. Biotechnol. 2011; 29:361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zuzuarregui A., Li T., Friedmann C., Ammerer G., Alepuz P.. Msb2 is a Ste11 membrane concentrator required for full activation of the HOG pathway. Biochim. Biophys. Acta. 2015; 1849:722–730. [DOI] [PubMed] [Google Scholar]
- 44. Garre E., Romero-Santacreu L., De Clercq N., Blasco-Angulo N., Sunnerhagen P., Alepuz P.. Yeast mRNA cap-binding protein Cbc1/Sto1 is necessary for the rapid reprogramming of translation after hyperosmotic shock. Mol. Bol. Cell. 2012; 23:137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Valentini S.R., Casolari J.M., Oliveira C.C., Silver P.A., McBride A.E.. Genetic interactions of yeast eukaryotic translation initiation factor 5A (eIF5A) reveal connections to poly(A)-binding protein and protein kinase C signaling. Genetics. 2002; 160:393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Garre E., Romero-Santacreu L., Barneo-Munoz M., Miguel A., Perez-Ortin J.E., Alepuz P.. Nonsense-mediated mRNA decay controls the changes in yeast ribosomal protein pre-mRNAs levels upon osmotic stress. PLoS One. 2013; 8:e61240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shah P., Ding Y., Niemczyk M., Kudla G., Plotkin J.B.. Rate-limiting steps in yeast protein translation. Cell. 2013; 153:1589–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guydosh N.R., Green R.. Dom34 rescues ribosomes in 3΄ untranslated regions. Cell. 2014; 156:950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hayes C.S., Bose B., Sauer R.T.. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J. Biol. Chem. 2002; 277:33825–33832. [DOI] [PubMed] [Google Scholar]
- 51. Sunohara T., Abo T., Inada T., Aiba H.. The C-terminal amino acid sequence of nascent peptide is a major determinant of SsrA tagging at all three stop codons. RNA (New York, NY). 2002; 8:1416–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lareau L.F., Hite D.H., Hogan G.J., Brown P.O.. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. Elife. 2014; 3:e01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ito K., Chiba S.. Arrest peptides: cis-acting modulators of translation. Annu. Rev. Biochem. 2013; 82:171–202. [DOI] [PubMed] [Google Scholar]
- 54. Lassak J., Wilson D.N., Jung K.. Stall no more at polyproline stretches with the translation elongation factors EF-P and IF-5A. Mol. Microbiol. 2016; 99:219–235. [DOI] [PubMed] [Google Scholar]
- 55. Woolstenhulme C.J., Parajuli S., Healey D.W., Valverde D.P., Petersen E.N., Starosta A.L., Guydosh N.R., Johnson W.E., Wilson D.N., Buskirk A.R.. Nascent peptides that block protein synthesis in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E878–E887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mandal A., Mandal S., Park M.H.. Genome-wide analyses and functional classification of proline repeat-rich proteins: potential role of eIF5A in eukaryotic evolution. PLoS One. 2014; 9:e111800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Morgan A.A., Rubenstein E.. Proline: the distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PLoS One. 2013; 8:e53785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Johansson M., Ieong K.W., Trobro S., Strazewski P., Aqvist J., Pavlov M.Y., Ehrenberg M.. pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kannan K., Kanabar P., Schryer D., Florin T., Oh E., Bahroos N., Tenson T., Weissman J.S., Mankin A.S.. The general mode of translation inhibition by macrolide antibiotics. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:15958–15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Marks J., Kannan K., Roncase E.J., Klepacki D., Kefi A., Orelle C., Vázquez-Laslop N., Mankin A,S.. Context-specific inhibition of translation by ribosomal antibiotics targeting the peptidyl transferase center. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:12150–12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schuller A.P., Wu C.C., Dever T.E., Buskirk A.R., Green R.. eIF5A Functions globally in translation elongation and termination. Mol. Cell. 2017; 66:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.