Abstract
Synthetic biology has great potential for future therapeutic applications including autonomous cell programming through the detection of protein signals and the production of desired outputs. Synthetic RNA devices are promising for this purpose. However, the number of available devices is limited due to the difficulty in the detection of endogenous proteins within a cell. Here, we show a strategy to construct synthetic mRNA devices that detect endogenous proteins in living cells, control translation and distinguish cell types. We engineered protein-binding aptamers that have increased stability in the secondary structures of their active conformation. The designed devices can efficiently respond to target proteins including human LIN28A and U1A proteins, while the original aptamers failed to do so. Moreover, mRNA delivery of an LIN28A-responsive device into human induced pluripotent stem cells (hiPSCs) revealed that we can distinguish living hiPSCs and differentiated cells by quantifying endogenous LIN28A protein expression level. Thus, our endogenous protein-driven RNA devices determine live-cell states and program mammalian cells based on intracellular protein information.
INTRODUCTION
Cell states are controlled by variety of biomacromolecules, including RNA, proteins and their complexes. Proteins are central to control gene expression, cell signaling and cell-fate regulation. In fact, protein expression levels determine cell fate (1) and human health (2,3). Thus, a synthetic system that can detect endogenous proteins and control gene expression in a living cell provides a useful tool for biological and therapeutic applications. The existing techniques to detect-specific proteins, including western blotting, immunostaining, LC-MS (4), chemical probes (5), proximity ligation (6) and tagging the protein of interest with reporter signals (7) (e.g. fluorescent proteins), enable us to analyze protein expression levels and cell states. However, it is difficult to apply these methods to the detection of endogenous proteins (i.e. without protein-modifications) in living cells.
Mammalian synthetic circuits delivered by RNA rather than DNA may provide a safer means to control cell behavior because synthetic RNA reduces the risk of genomic damage (8). A protein-driven mRNA device that detects a particular target protein and regulates post-transcriptional expression of exogenous genes can be used to build complex and sophisticated gene circuits, because the output protein from the device can serve as the input protein of other circuits (9–12). So far, several protein-responsive RNA devices, based on the conjugation of a specific protein-binding motif (aptamer) with messenger RNA (mRNA) (13) or short-hairpin RNA (shRNA) (14), have been reported. However, previous reports have either relied on exogenous RNA-binding proteins (e.g. MS2 coat protein or L7Ae ribosomal protein) that have to be overexpressed in the cells or required the use of DNA (plasmid DNA or viral vector) for circuit delivery. In addition, the number of available RNA devices is limited due to the difficulty in the sensitive recognition of endogenous proteins within the cell. Thus, the detection of endogenous proteins (e.g. marker proteins that represent cell state) and distinguish living cells by RNA-delivered devices remains a challenge.
In this article, we report a design strategy to construct mRNA devices that with improved sensitivity detect endogenous proteins in living human cells and transmit the information to synthetic translational regulatory systems (Figure 1). We engineered aptamer modules to protect and stabilize their active conformations in mRNA, while the original aptamers were insensitive to endogenous target proteins in cells. In addition, using a mRNA-delivery approach, we can distinguish human induced pluripotent stem cells (hiPSCs) from differentiated cells by quantifying the differential protein expression level of endogenous LIN28A.
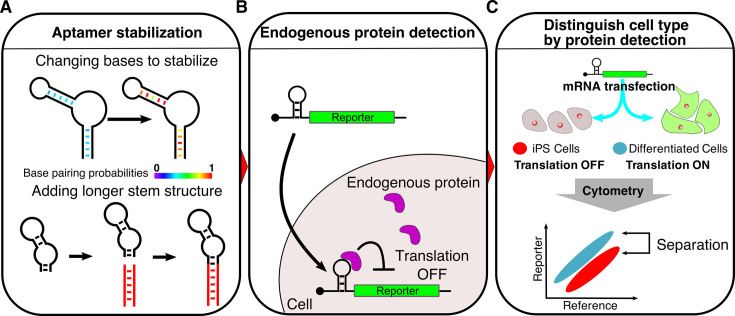
Figure 1.
Schematic illustration of detecting endogenous proteins and distinguishing mammalian cells via designed mRNA devices. (A) Stabilization of RNA secondary structures improves the sensitivity of protein-responsive mRNA devices. The RNA devices were stabilized by base-pair substitutions or elongation of the stem structure. Base pairs in red correspond to high base pairing probabilities. Red stems represent additional stem structures. (B) Detection of human endogenous proteins by mRNA devices. The mRNA devices bind to target proteins through RNA–protein interactions in the 5΄-UTR of the mRNA and repress translation of the reporter fluorescent protein, which enables the detection of native target proteins in living cells. (C) Distinction of cell types via mRNA devices. The mRNA-delivered device that responds to a marker protein expressed in human iPS cells can be used to distinguish iPS cells and differentiated cells after analysis of the translation level in each cell type.
MATERIALS AND METHODS
Plasmids construction
Device plasmids were derived from kt-EGFP as previously reported (13). To prepare pAptamerCassette-EGFP, kt-EGFP was digested by NheI and AgeI restriction enzymes and had inserted double strand oligo DNA (dsDNA), which was prepared by synthesized oligo DNAs, KWC0041 and KWC0042. The sequences of KWC0041 and KWC0042 were shown in Supplementary Table S1.
To construct each device plasmid, pAptamerCassette-EGFP or kt-EGFP were digested by AgeI and BamHI or AgeI and BglII, respectively, and had inserted dsDNA that contains an aptamer sequence shown in Supplementary Table S1. Oligo DNA and the multiple aptamer device (stbC x 2) were designed according to Ref: (15).
pTAPmyc-2A-tagRFP was newly constructed based on pIRES2-DsRed-Express (Clontech Laboratories). Inserted trigger proteins were cloned or constructed as shown in Supplementary Table S2. All plasmids used in this study are shown in Supplementary Table S3. iRFP670 ORF was originally obtained from piRFP670-N1 (16) (a gift from Vladislav Verkhusha; Addgene plasmid # 45457).
mRNA preparation
A template DNA for in vitro transcription (IVT) was generated by polymerase chain reaction (PCR) using appropriate primer sets, and template oligo DNA and/or plasmids (Supplementary Table S4). The PCR fragments of the 5΄UTR, ORF and 3΄UTR were fused to generate IVT template of reference, no aptamer and trigger mRNAs by using a primer set, T7Fwd and Rev120A. Plasmids in a PCR reaction were digested by 1 μl of DpnI (TOYOBO). The IVT template of device mRNAs was generated with the appropriate sets of forward primer, spacer, hmAG1 ORF, 3΄UTR and Rev120A (Supplementary Table S4). All PCR products were purified by MinElute PCR Purification Kit (QIAGEN). Synthetic mRNAs were transcribed by using MEGAScript T7 Kit (Ambion) as previously described (8,17). In general, modified nucleotides were used for IVT to avoid immune response (18–20). However, because we speculated that these unnatural bases might impair the interaction between protein and aptamer, we used natural nucleotides when transcribing the device mRNAs or control mRNA (without aptamer). Thus, except for device mRNAs, pseudouridine-5΄-triphosphate (Ψ) and 5-methylcytidine-5΄-triphosphate (m5C) (TriLink BioTechnologies) were used instead of uridine triphosphate and cytosine triphosphate, respectively. Five-fold diluted guanosine-5΄-triphosphate was used with Anti-Reverse Cap Analog (New England Biolabs). Reaction mixtures were incubated at 37°C for 6 h. After the incubation, TURBO DNase (Ambion) was added to the mixture and further incubated at 37°C for 30 min to remove the template DNA. The generated mRNAs were purified using a FavorPrep Blood/Cultured Cells total RNA extraction column (Favorgen Biotech) and incubated with Antarctic Phosphatase (New England Biolabs) at 37°C for 30 min. The reaction mixtures were purified again using an RNeasy MinElute Cleanup Kit (QIAGEN) according to the manufacturer's protocols.
Cell culture
293FT cells (Invitrogen) were cultured at 37°C in Dulbecco's modified Eagle's Medium (DMEM) medium (Nacalai tesque) supplemented with 10% fetal bovine serum (FBS) (#JBS020265; Japan Bio Serum), 2 mM L-Glutamine (Invitrogen), 0.1 mM Non-Essential Amino Acids (Invitrogen) and 1 mM Sodium Pyruvate (Sigma). HeLa cells (originally obtained from ATCC) were cultured in DMEM-F12 containing 10% FBS. Human iPS cells (201B7, a kind gift from Dr Masato Nakagawa, Kyoto University) were cultured in feeder-free condition with StemFit AK03 (Ajinomoto) as previously described (21). To prepare differentiated cells derived from iPSCs (D14), the cells were cultured with StemFit lacking bFGF for 2 weeks.
Plasmids transfection
Cells were plated into appropriate multi-well plates. After 1-day incubation, cells were transfected with plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols. A total of 500 or 125 ng DNA was mixed with Opti-MEM (Life Technologies) in 24-well or 96-well format, respectively. After 4–6 h of transfection, the medium was replaced with fresh medium. Transfection details for each experiment are shown in Supplementary Table S5.
mRNA transfection
All transfections were carried out in a 24-well format. Transcribed mRNAs were transfected into the cells using StemFect (Stemgent) according to the manufacturer's protocols. CBG68Luc was used as control mRNA in the experiment of Figure 6. After 4–6 h of transfection, the medium was replaced with fresh medium. In the experiment of Figure 7, the medium was replaced with fresh medium at least 1 h before transfection. Transfection details for each experiment are shown in Supplementary Table S6. The mRNA sequences used in this study are shown in Supplementary sequences.
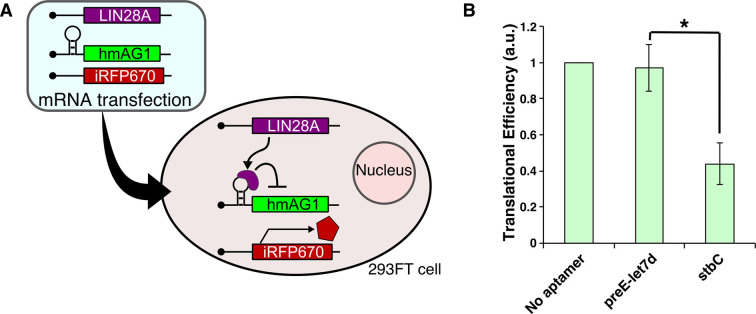
Figure 6.
RNA delivery of LIN28A-coding mRNA and LIN28A-responsive devices. (A) Schematic representation of RNA delivery of LIN28A-coding mRNA, LIN28A-responsive mRNA and reference mRNA. 293FT cells were co-transfected with three synthetic mRNAs, trigger mRNA (LIN28A or CBG68Luc mRNA), LIN28A-responsive hmAG1 mRNA and iRFP670 mRNA (reference mRNA) as a transfection control. (B) Translational efficiencies of LIN28A-responsive mRNAs. Transfection was performed in a 24-well format. Error bars represent mean ± SD (n = 3 independent experiments, each performed in triplicate). *P < 0.05 (Welch's t-test).
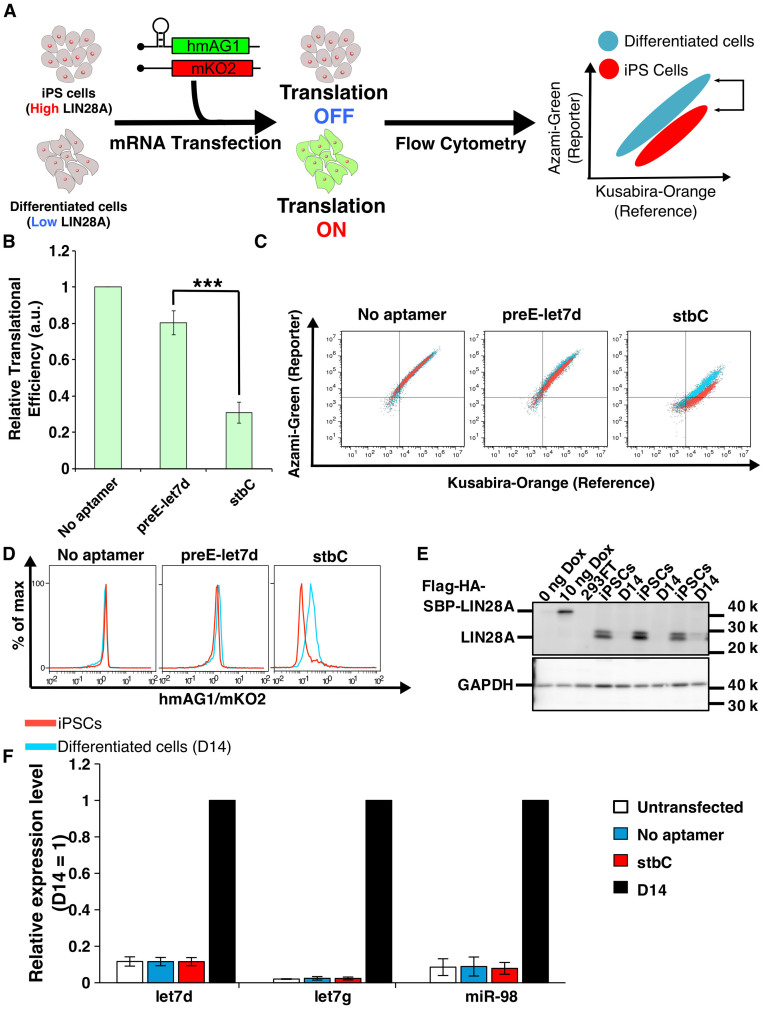
Figure 7.
Distinction of living cell types using LIN28A-responsive mRNA devices. (A) Distinction of human iPSCs with differentiated cells by detecting endogenous LIN28A expression level. LIN28A is highly expressed in iPSCs, and its expression level is decreased after differentiation. LIN28A in iPSCs reduces the translation level from LIN28A-responsive hmAG1 mRNA. In differentiated cells, LIN28A expression level is low, resulting in normal translation (left and middle panels). After co-transfection of the LIN28A-responsive hmAG1 mRNA and reference mRNA encoding mKO2, the cells typically show belt-like distribution in a two-dimensional plot by flow cytometry analysis (right panel). A shift of the population is observed as the result of translational repression of hmAG1 in iPSCs but not in differentiated cells, resulting in the separation of iPSCs from differentiated cells based on LIN28A detection. (B) Relative translational efficiency of transfected mRNAs in iPSCs and D14. Relative translational efficiency = (N.I. of iPSCs)/(N.I. of D14) (See ‘Flow cytometry data analysis’ in ‘Materials and Methods’ section). Error bars represent mean ± SD (n = 3 independent experiments, each performed in duplicate). ***P < 0.0005 (Welch's t-test). (C) Representative overlaid dot plots of iPSCs (red) and differentiated cells (D14, blue). (D) Representative histograms related to Figure 7C. Red, hiPSCs; blue, D14. (E) Western blot analysis of the LIN28A expression. A total of 0 ng Dox, 10 ng Dox and 293FT were applied as controls, and GAPDH was used as internal control. The samples used in these experiments were also used for the data in Figure 5C. iPSCs and D14 samples were collected in different experiments. LIN28A was detected by anti-LIN28A. (F) Endogenous miRNA expression levels by LIN28A-responsive mRNA transfection. Mature let7d, let7g and miR-98 levels were analyzed by qRT-PCR. We used differentiated cells (D14, black bar) as a control, because the expression levels of these miRNAs increased in differentiated cells compared with iPSCs. Thus, the expression levels of miRNAs in untransfected (white), mRNA (without aptamer)-transfected (blue) and stbC-containing mRNA-transfected (red) iPSCs were normalized by the expression level of the miRNAs in D14. Error bars indicate SD for three independent experiments. Significant differences were not observed among untransfected, no aptamer and stbC.
Flow cytometry measurements
One day after transfection, cells were washed with phosphate buffered saline (PBS). HeLa cells and 293FT cells were incubated in 100 μl of 0.25% Trypsin–EDTA (GIBCO) at 37°C. After the addition of 200 μl medium, cells were passed through the mesh and analyzed by flow cytometry. iPSCs and D14 cells were incubated in 200 μl of Accumax (Innovative Cell Technology) at 37°C, passed through the mesh and analyzed by flow cytometry. In experiments using a 24-well plate, we used Accuri C6 (BD Biosciences). EGFP and hmAG1 were detected by FL1 (533/30 nm) filters. tagRFP and mKO2 were detected by the FL2 (585/40 nm) filters. iRFP670 was detected by FL4 (675/25 nm) filters. In the experiments using a 96-well plate, we used BD LSRFortessa (BD Bioscuence). EGFP was detected by blue laser with an FITC filter (525/50 nm). tagRFP was detected by a green laser with a PE filter (582/15 nm).
Flow cytometry data analysis
Flow cytometry datasets were analyzed using FlowJo (Tree Star), FlowCore (22) and Excel (Microsoft). Data were first screened to remove any events with non-positive forward scatter and side scatter. The remaining events were named P1 gate. P1 data were expanded to a histogram of reference (transfection control) count. This histogram was used to determine reference positive populations. In the following analysis, the median reporter/reference of each cell was calculated from the reference positive population by FlowJo.
Translational efficiency is defined using the following formulas.
Normalized Intensity (N.I.) = Median of the ratio (reporter intensity/reference intensity) of each cell.
Relative Intensity (R.I.) = (N.I. of Trigger +) / (N.I. of Trigger -).
Translational Efficiency = (R.I.) / (R.I. of No aptamer).
All values were normalized by No aptamer value.
Lin28A-expressing stable cell lines
Flp-In T-REx-293 cells were cultured in DMEM containing 10% FBS under a humidified 5% CO2 atmosphere at 37°C. A stable human cell line expressing streptavidin binding protein (SBP)-fused lin28A (SBP-lin28A) was established by Flp Recombinase-Mediated Integration (Flp-In system, Invitrogen) according to the manufacturer's instructions. The synthetic cDNA encoding human Lin28A was purchased from Eurofin Genomics, Japan and was cloned into the pcDNA5/FRT/TO-Flag-HA-SBP vector (kind gift from Dr Yamashita at Yokohama City University, Medical School). The resultant Lin28A expression plasmid, pcDNA5-SBP-lin28A, was co-transfected with pOG44 (Invitrogen) into the host Flp-In T-REx-293 cells (Invitrogen) using Lipofectamine 2000 (Invitrogen). The cell line harboring pcDNA5-SBP-lin28A was selected with hygromycin (100 mg/ml) and blasticidin (10 mg/ml) for 2 weeks according to the manufacturer's instructions.
Doxycycline induction analysis
Dox-LIN28A-293 cells were seeded in a 12-well cell culture plate. After 24 h, fresh media containing 0, 0.1, 0.2, 0.4, 0.8, 1.0, 3.0, 5.0 or 10 ng/ml doxycycline was added to the wells. After 1 h, cells were transfected with plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols. A total of 400 ng DNA was mixed with Opti-MEM (Life Technologies). After 4–6 h of transfection, the medium was replaced with fresh Dox-containing medium. Details about the transfections for each experiment are shown in Supplementary Table S5. One day after transfection, cells were washed with PBS. Cells were incubated in 300 μl of 0.25% Trypsin–EDTA (GIBCO) at 37°C. After the addition of 300 μl medium, cells were passed through the mesh. Cells in mixture were dispensed into 300 μl aliquots and collected for western blotting analysis or analyzed by Accuri C6 (BD Biosciences).
Knockdown assay
Cells were cultured in 24-well plates and transfected with 100 ng of device plasmid, 400 ng control plasmid and 5 pmol shRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol (see also ‘Plasmids Transfection’ in ‘Materials and Methods’ section and Supplementary Table S5). After 1 day incubation, cells were washed with PBS and incubated in 0.25% Trypsin–EDTA (GIBCO) at 37°C. After the addition of medium, cells were passed through the mesh and analyzed by Accuri C6 (BD Biosciences). The shRNA sequences were as follows:
Control-shRNA (23): 5΄- GCCUAAGGUUAAGUCGCCCUCGCAGCAUAGGCGAGGGCGACUUAACCUUAGGCAG -3΄
U1A-shRNA: 5΄- GAUCAAGAAGGAUGAGCUAAAAAAGAGCAUAGCUUUUUUAGCUCAUCCUUCUUGAUCAG -3΄
Aliquots of analyzed samples were collected for western blotting analysis.
Western blotting analysis
Collected cells were washed with 200 μl of PBS and lysed in 50 μl of RIPA buffer. The concentration of total protein was measured by Pierce BCA Protein Assay Kit (Thermo scientific). Samples (5 μg of protein) were applied to sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred into a PDVF membrane using iBlot (Invitrogen) according to the manufacturer's protocol. Membranes were incubated with specific primary antibodies. Anti-SNRPA (Santa Cruz Biotechnology, sc-101149) and Anti-human LIN28A (R and D Systems, AF3757) were used at 200-fold dilution and 10 000-fold dilution, respectively. GAPDH antibody (Santa Cruz Biotechnology, sc-25778) was used at 500-fold dilution. Then, the blot was incubated with secondary antibodies. Goat Anti-Mouse IgG (H+L)-HRP conjugate (BIO-RAD, 170–6516), Rabbit anti-Goat IgG (H+L) Secondary Antibody, HRP conjugate (Life Technologies, 81–1620) and Goat anti-Rabbit IgG (H+L)- HRP conjugate (BIO-RAD, 170–6515) were used at 2000-fold dilution. The blots were detected with ECL Prime Western Blotting Detection Reagent (GE Healthcare) and ImageQuant LAS 4000 (GE Healthcare). Protein expression level was calculated from bands intensities with ImageJ (NIH).
Quantitative RT-PCR
Cells for RNA analysis were pelleted, frozen and stored at −80°C until use. To quantify mature miRNAs, total RNA purification, reverse transcription and quantitative PCR were performed by using miRNA Cells-to-CT Kit (Applied Biosystems). Expression levels of mature miRNA were measured by using miRNA TaqMan probe (Applied Biosystems) and normalized using U6 snRNA (RNU6B) expression levels. The TaqMan probes are shown in Supplementary Table S7. To quantify the expression levels of immune response-related genes, pellets were thawed and then rapidly resuspended in Trizol (Thermo Fisher Scientific) according to the manufacturer's protocol. Extracted RNAs were treated with TURBO DNase (Ambion) and purified with Phenol/Chloroform/Isoamyl alcohol. Reverse transcription was carried out using ReverTra Ace qPCR RT Master Mix (TOYOBO) following the manufacturer's protocol. The primers for real-time amplification are listed in Supplementary Table S8. Target mRNA was normalized to GAPDH. The resulting cDNAs were amplified with THUNDERBIRD SYBR qPCR Mix (TOYOBO) using StepOne (Applied Biosystems). Relative expression levels were calculated using the ΔΔCt method.
WST-1 assay
Cell viability was measured with Cell Proliferation Reagent WST-1 (Roche). At one day after transfection of natural or modified hmAG1 mRNA, WST-1 substrate was added and cells were incubated for 2.5 h. The absorbance was measured at 440 nm with a reference wavelength at 600 nm on TECAN microplate reader (Infinite M1000).
Statistical analysis
Data of translational efficiency and relative translational efficiency are presented as means ± standard deviation (SD). Significant differences between means were determined by two-tailed Welch's t-test. The levels of significance are denoted as *P < 0.05, **P < 0.005 and ***P < 0.0005. Values of mean, SD and coefficient of determination were obtained, and Welch's t-test was performed by using Excel (Microsoft) or R.
RESULTS
Protein-responsive mRNA devices with improved sensitivity
Messenger RNA devices that have an aptamer module in the 5΄-UTR of reporter genes are capable of detecting target proteins, because the translation of reporter genes can be repressed through the protein–aptamer interaction (Supplementary Figure S1). The mRNA devices are also applicable to a RNA-only delivery approach, because translational regulation is independent of RNA nuclear history or splicing. Previous reports have generated protein-responsive mRNA devices that efficiently reduce the translational level of the mRNA in the presence of cognate proteins (13,24–28) and succeeded in delivering the devices into mammalian cells without DNA (8). However, the detection of endogenous proteins, including cytoplasmic marker proteins expressed in living cells has not been successfully implemented.
Thus, we aimed to create protein-responsive mRNA devices that efficiently repress translation by detecting target proteins in living cells. It has been reported that the presence of a protein-binding motif is not generally sufficient for effective RNA–protein interaction within a cell, and local RNA secondary structures affect the ability of binding (29,30). In fact, we observed that the simple insertion of protein (U1A and NF-κB p50)-binding sequences (aptamers) in the 5΄-UTR of reporter genes did not efficiently repress translation upon protein binding (Figure 2 and Additional text for Supplementary Figure S8). We hypothesized that the aptamers embedded in the 5΄-UTR of the mRNA may form unstable secondary structures that reduce binding ability to the target protein in cells. To investigate this possibility, we generated a set of synthetic mRNA devices that contain RNA aptamers with more stabilized secondary structures that maintain active conformations for protein binding and analyzed their ability to repress translation in the presence of cognate protein (Figure 2).
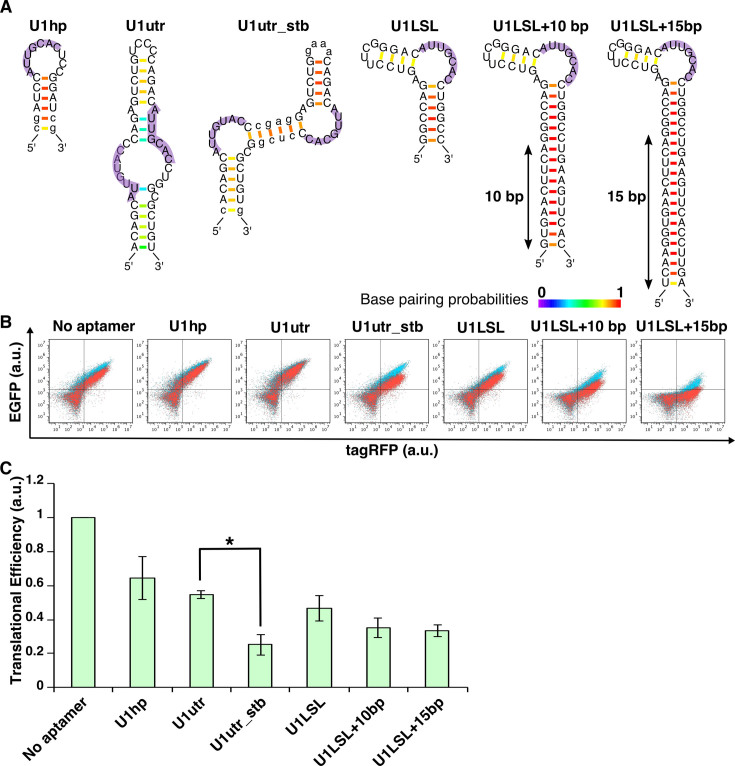
Figure 2.
Construction of U1A-responsive devices. (A) U1A-binding aptamers used in this study. Secondary structures were predicted by CentroidFold software (Ref:(35)). Small letters represent the nucleotides changed from U1hp or U1utr in an original report (Ref:(32)) to stabilize the aptamers. Purple shadows represent the U1A recognition sequences. Color scale represents base pairing probabilities calculated by CentroidFold software. (B) Overlaid dot plots from flow cytometry analysis. Red, cells co-transfected with EGFP reporter plasmid and trigger plasmid encoding U1A and tagRFP. Blue, cells co-transfected with EGFP reporter plasmid and control plasmid without U1A. (C) Translational efficiency of U1A-responsive devices. All values were normalized by cells transfected with reporter plasmid lacking aptamer (No aptamer). Transfection was performed in a 24-well format. Error bars represent mean ± SD (n = 3 independent experiments, each performed in triplicate). *P < 0.05 (Welch's t-test).
We first engineered U1A-responsive mRNA devices. U1A is a spliceosomal protein, and its binding aptamers (U1hp) are well characterized (31,32) (Figure 2A). U1A also binds to the 3΄UTR region of its own mRNA (U1utr) and regulates the polyadenylation process (33,34). We extracted the U1hp and U1utr sequences and inserted one into the 5΄-UTR of reporter plasmids that encode EGFP (Supplementary Figure S2). We also constructed a ‘trigger plasmid’ that expresses the U1A N-terminal RNA-recognition domain (14) conjugated with tagRFP via 2A peptide (Supplementary Figure S3). Twenty-four hours after co-transfection of these plasmids, we measured EGFP and tagRFP expression level by flow cytometry (Figure 2B) or fluorescence microscopy (Supplementary Figure 4) and analyzed translational repression efficiency (Figure 2C). The mRNA devices with U1hp or U1utr weakly repressed the translation in the presence of the trigger plasmid (∼30% repression) (Figure 2C). We speculated that the U1utr forms a less-stabilized secondary structure that may affect the U1A–mRNA interaction, because CentroidFold software (35) predicted that U1utr did not form the expected secondary structure reported previously (31) (Supplementary Figure S5). Thus, we replaced nucleotides of U1utr and newly designed the U1utr_stb aptamer that exhibits a more stabilized and expected secondary structure with single-stranded, two U1A-binding regions (shown in purple, Figure 2A and Supplementary Figure S5). Notably, mRNA containing the U1utr_stb aptamer repressed translation more efficiently (∼75% repression) compared with the original U1utr (Figure 2C and Supplementary Figure S4). We also designed alternative U1A-binding aptamers with a stabilized loop-stem-loop structure (U1LSL) and combined them with longer stem lengths (U1LSL + 10 bp and U1LSL + 15 bp) to reinforce the aptamer structures (the U1A-binding AUUGCAC sequences are located at the single stranded-loop region flanked by two stem regions). In the presence of the trigger plasmid, we observed that mRNA devices with these aptamers repressed translation more efficiently than with the original U1hp or U1utr, whereas the devices with defective U1LSL variants (U1LSLmut) that disrupt U1A binding did not repress translation (Figure 2C and Figure 3A, B). These data indicate that we can improve the sensitivity of protein-responsive mRNA devices by stabilizing RNA secondary structures with active conformations.
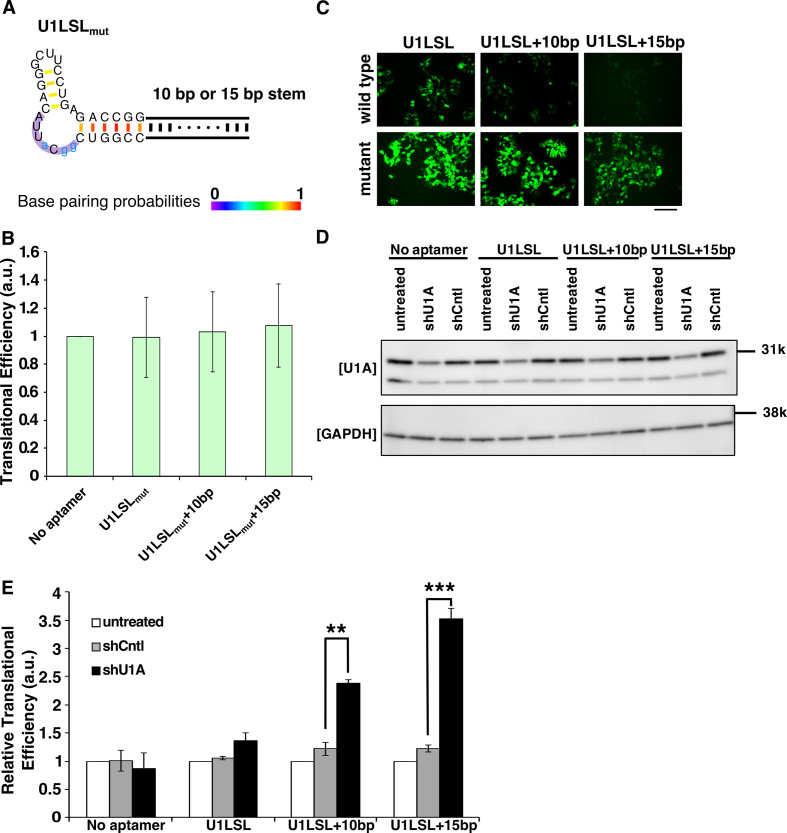
Figure 3.
U1LSL series-containing mRNA devices detect endogenous U1A. (A) Secondary structures of the mutant U1LSL series. Blue small letters represent the nucleotides changed from the original U1LSL to construct defective motif sequences (U1LSLmut). Purple shadow represents the U1A recognition sequences. Color scale represents base pairing probabilities calculated by CentroidFold software. (B) Translational efficiency of defective U1LSLmut series-reporter plasmids. The same experiment in Figure 2C was performed. Error bars represent mean ± SD (n = 3 independent experiments, each performed in triplicate). All values are normalized by cells transfected with a plasmid lacking aptamer sequence (No aptamer). (C) Fluorescent microscopic images of cells co-transfected wild-type U1LSL or mutant (U1LSLmut) series-reporter plasmids with control plasmid. EGFP was used as the reporter. The cells were observed at 24 h after transfection. Scale bar, 200 μm. wild-type, mRNA devices containing wild-type U1LSL sequence; mutant, mRNA devices containing U1LSLmut sequence. (D) Western blotting analysis of U1A knockdown after shRNA treatments. GAPDH was used as control. (E) Relative translational efficiency of EGFP intensities after endogenous U1A knockdown by shRNA. Relative translational efficiency = (N.I.)/(N.I. of each shRNA-untreated control) (See ‘Flow cytometry data analysis’ in ‘Materials and Methods’ section). The fluorescence intensities were normalized by cells transfected with only plasmids (untreated). Transfection was performed in a 24-well format. Error bars represent mean ± SD (n = 3 independent experiments, each performed in triplicate). **P < 0.005, ***P < 0.0005 (Welch's t-test).
We next investigated whether the designed RNA devices can respond to endogenous U1A. We observed that the basal expression levels of EGFP from U1LSL (+10, +15 bp) aptamer-containing mRNAs were downregulated even in the absence of the trigger plasmid that overexpresses U1A compared with those from defective U1LSLmut aptamer-containing mRNAs (Figure 3A and C). These results suggest that the U1LSL series of mRNA devices are capable of detecting endogenous U1A expression. To prove this possibility, we carried out knockdown experiments of endogenous U1A and analyzed the effect on the translational regulation from the devices. We co-transfected U1A-targeting shRNA and the U1A-responsive reporter plasmids into 293FT cells. Twenty-four hours after transfection, EGFP expression levels of cells transfected with shU1A and reporter plasmids with U1LSL aptamers were higher than that transfected with control shRNA, indicating that knockdown of endogenous U1A increased EGFP expression from the reporter plasmids with U1LSL (+10, +15 bp) aptamers (Figure 3D and E). Thus, our U1A-responsive mRNAs can repress translation by detecting endogenous U1A protein.
Design and construction of human LIN28A-responsive mRNA devices
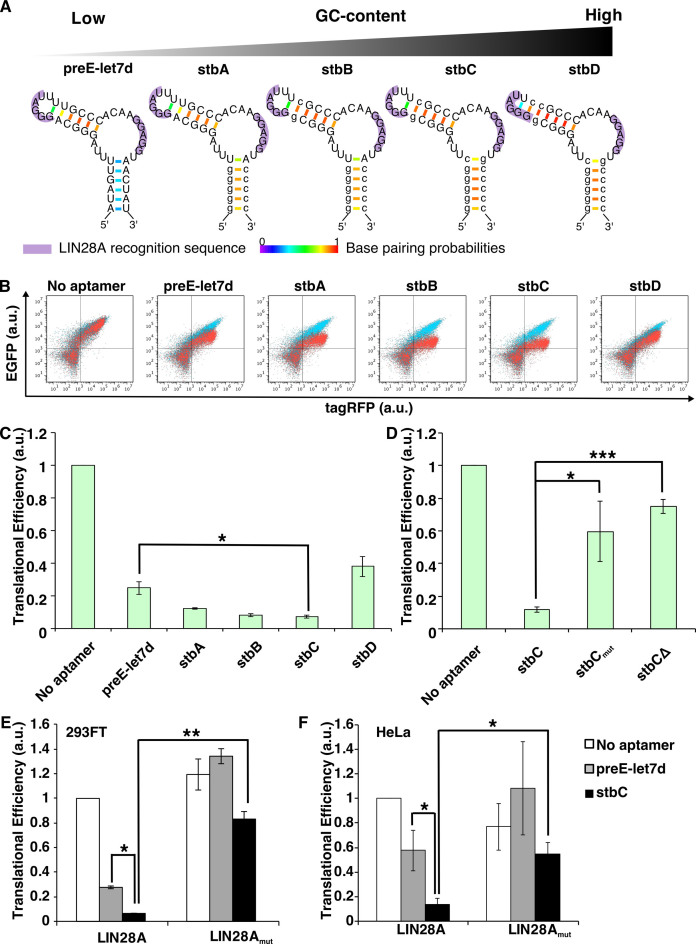
Next, we chose human LIN28A as a target protein to make new mRNA devices that distinguish living cell types. LIN28A is a marker protein that binds to a certain class of RNAs (36,37) and it is highly expressed in human pluripotent stem cells (38). For example, LIN28A binds to let-7 microRNA (miRNA) precursors, and the interaction inhibits the biogenesis of mature let-7 miRNAs (39,40), which is important to maintain stem cell states. We first extracted LIN28A-binding RNA sequences from pre-let-7d miRNA (a let-7 family miRNA precursor) (Figure 4A left; termed ‘preE-let7d’). Prediction of the secondary structure by CentroidFold software suggested the bottom stem region of preE-let7d with lower base-pairing probabilities may affect conformation of the aptamer inside the cell. We therefore stabilized the secondary structure of the LIN28A aptamers by increasing the number of GC base pairs in the stem region (Figure 4A, stbA-D), although prediction of the secondary structures suggested that stbD may disrupt one of the single-stranded loops (GGGAU) required for LIN28A-binding (41). Based on the designed stbA-D aptamers, we constructed a set of LIN28A-responsive devices that contain each aptamer in the 5΄-UTR of EGFP mRNAs (Supplementary Figure S2B). We also designed a trigger plasmid that expresses LIN28A (Supplementary Figure S3) and was co-transfected with the LIN28A-responsive reporter to analyze whether reporter EGFP expression could be repressed by LIN28A (Figure 4B and Supplementary Figure S6). Interestingly, in the presence of LIN28A, the mRNA devices with three designed aptamers (stbA-C) showed greater translational repression compared with the original preE-let7d aptamer (Figure 4C). The mRNA with stbD did not repress translation efficiently, probably due to disruption of the terminal loop structure (GGGAU) (Figure 4A, stbD). The mRNA with stbC aptamer showed the strongest translational repression (∼ 93%), whereas the mRNAs with mutated stbC (stbCmut) or deletion mutant (stbCΔ) (Supplementary Figure S2B), which lacks LIN28A-binding ability, recovered the translational efficiency (Figure 4D). The amino acid substitution (W46A) of LIN28A (LIN28Amut), which is known to interfere with binding to the terminal GGGAU loop of pre-let-7d miRNA (41,42), also inhibited the translational repression ability of stbC (Figure 4E), indicating that LIN28A–stbC interaction is indispensable for the translational repression. Moreover, the mRNA devices responded to LIN28A-expressing plasmid in HeLa cells in a manner similar to 293FT cells, suggesting that the functions of the mRNA devices are independent of the cell-line used (Figure 4F). Thus, LIN28A-responsive mRNA devices can be constructed in living cells by increasing the stability of aptamer-containing RNA modules that maintain an active secondary structure for efficient binding. We then focused on the mRNA device with stbC for further study.
Figure 4.
Construction of LIN28A-responsive devices. (A) Predicted secondary structures of the LIN28A aptamers used in this study. Small letters represent the nucleotides changed from the original preE-let7d sequence. Purple shadows represent the LIN28A recognition sequences. Color scale represents base pairing probabilities calculated by CentroidFold software. (B) Overlaid dot plots of the flow cytometry analysis. Red, cells co-transfected with EGFP reporter plasmid and trigger plasmid encoding LIN28A and tagRFP. Blue, cells co-transfected with EGFP reporter plasmid and control plasmid without LIN28A. (C) Translational efficiency of LIN28A-responsive devices. All values were normalized by cells transfected with a plasmid lacking aptamers (No aptamer). Transfection was performed in a 24-well format. Error bars represent mean ± SD (n = 3 independent experiments, each performed in triplicate). (D) Translational efficiency of defective LIN28A-responsive devices (sequences are described in Supplementary Figure S2). All values were normalized by cells transfected with No aptamer. Transfection was performed in a 96-well format. (E) Translational efficiency using mutant LIN28A (W46A, LIN28Amut), which weakens RNA binding. All values were normalized by cells co-transfected with No aptamer and trigger plasmid coding for wild-type LIN28A. Transfection was performed in a 24-well format. Error bars represent mean ± SD (n = 3 independent experiments, each performed in triplicate). (F) Translational efficiency of LIN28A-responsive devices in HeLa cells. Transfection was performed in a 24-well format. Error bars represent mean ± SD (n = 3 independent experiments, each performed in triplicate). All values were normalized as described in D. *P < 0.05, **P < 0.005, ***P < 0.0005 (Welch's t-test).
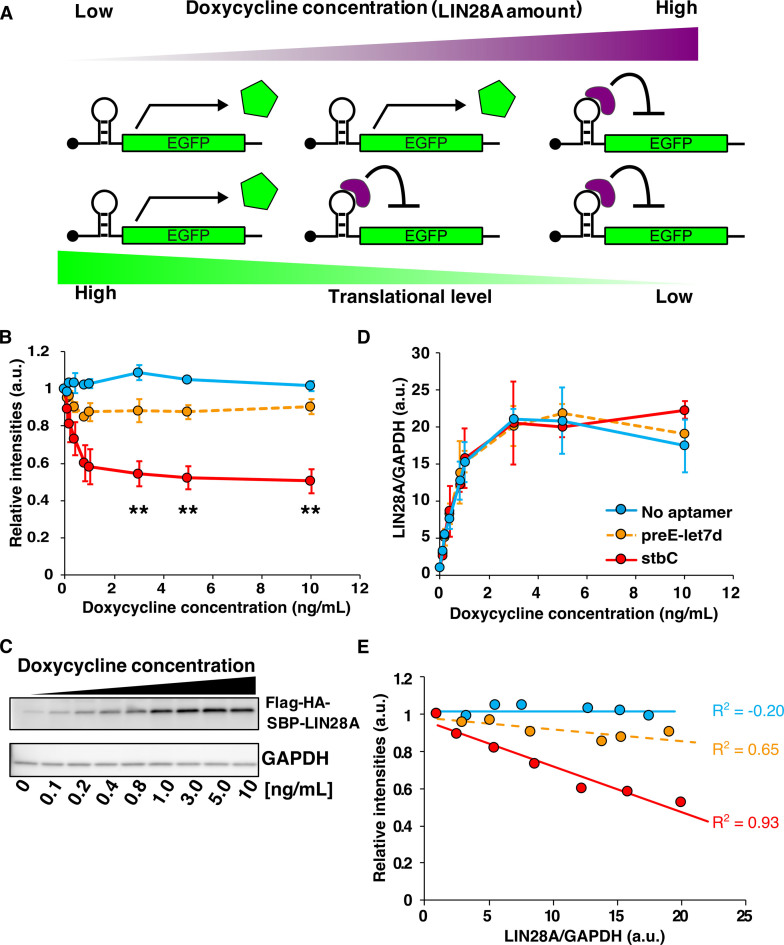
We next investigated whether the LIN28A-responsive mRNA device could quantify the LIN28A expression level. To control LIN28A expression level, we prepared a doxycycline (Dox)-inducible HEK293 cell line (Dox-LIN28A-HEK) that controls the level of LIN28A in a Dox-dependent manner (Figure 5). We transfected either the control (No aptamer) or LIN28A-responsive reporter plasmid into Dox-LIN28A-HEK cells. To analyze the correlation between the efficiency of translational repression and the level of LIN28A expression, we performed flow cytometry and western blotting to measure the intensity of EGFP and the amount of LIN28A, respectively (Figure 5B and C). Western blot analysis found that the addition of Dox at low dose (0–1 ng/ml) produced LIN28A in a Dox-concentration dependent manner, but LIN28A expression level reached plateau at high doses (more than 3 ng/ml) (Figure 5C and D). This result corresponded to the translational repression level of the mRNA device with stbC as analyzed by flow cytometry (Figure 5B). In fact, the intensity of EGFP produced from the stbC device was correlated with LIN28A expression level (R2 = 0.93, Figure 5E), indicating that the mRNA device responds to LIN28A in a concentration-dependent manner. However, the mRNA device with the original preE-let7d did not detect LIN28A efficiently (Figure 5B and E). Thus, our designed mRNA device with stbC showed higher sensitivity and its translation repression level depends on the LIN28A expression level, suggesting that the mRNA device can act as a LIN28A detector in living cells.
Figure 5.
Quantitative detection of LIN28A expression by mRNA devices. (A) Expected behavior of the LIN28A device in response to LIN28A protein expression. LIN28A is generated in a Dox-dependent manner, and the translation efficiency of the device relies on the expression level of intracellular LIN28A. At low Dox concentration, the expression level of LIN28A is also low and the device does not repress EGFP translation (left side). At high Dox concentration, LIN28A expression is also high and the intensity of EGFP decreases because of translational repression (right side). (B) Relationship between relative fluorescence intensity (translation of mRNA) and Dox concentration. Relative intensities = (N.I.)/(N.I. without Dox induction). (The relative intensities were normalized to the intensity at 0 ng Dox. See ‘Flow cytometry data analysis’ in ‘Materials and Methods’ section.) Error bars are mean ± SD. The statistical analysis was performed between No aptamer and stbC. **P < 0.005 (Welch's t-test). (C) Western blot analysis of LIN28A expression at 24 h after transfection. Tag sequence-fused LIN28A was induced by Dox. LIN28A was detected by anti-LIN28A. GAPDH was used as internal control. The concentration of Dox ranged between 0 and 10 ng/ml. (D) Relationship between relative LIN28A amount calculated from the western blotting analysis in Figure 5C and Dox concentration. Reporter expression from the device was controlled by the Dox concentration. Error bars are mean ± SD. (E) Relationship between the relative intensity of EGFP produced from the device and LIN28A amount. Plotted data are same as in Figure 5B and D. The maximum value of LIN28A/GAPDH in Figure 5E was taken from the minimum value of LIN28A/GAPDH between 3 and 10 ng/ml of Dox condition in Figure 5D. Blue, No aptamer; orange, preE-let7d; red, stbC. R2, coefficient of determination.
mRNA delivery of LIN28A-responsive devices to distinguish cell types
Endogenous protein-responsive RNA devices have so far relied exclusively on the introduction of foreign DNA (27,43). To investigate whether our LIN28A-responsive mRNA device can function for mRNA delivery, we in vitro transcribed mRNA that contained the stbC aptamer in 5΄-UTR and a humanized Azami-Green (hmAG1) ORF, respectively. We also prepared LIN28A-coding mRNA, and co-transfected them into 293FT cells (Figure 6A). Twenty-four hours after the transfection, we observed that the mRNA with stbC repressed translation in the presence of LIN28A, whereas the mRNA with the original preE-let7d did not respond to LIN28A expression (Figure 6B and Supplementary Figure S7). The results indicate that our LIN28A-responsive device functions by RNA-delivery.
Finally, we aimed to detect endogenous LIN28A for the purpose of distinguishing hiPSCs from differentiated cells (Figure 7A). LIN28A is highly expressed in hiPSCs, and its expression level is decreased as differentiation proceeds (36,38,44). To induce differentiation, we cultured hiPSC in medium lacking bFGF for 14 days (D14). We transfected LIN28A-responsive mRNA encoding hmAG1 and a reference mRNA encoding Kusabira-Orange (mKO2) into a hiPSC line (201B7) and D14. Notably, the LIN28A-responsive mRNA device with stbC efficiently reduced translation levels in hiPSCs compared with control mRNA without stbC or preE-let7d (Figure 7B). Moreover, an overlaying dot plot of hiPSCs and D14 showed that both populations were separated only when the stbC-mRNA device was transfected (Figure 7C and D). Western blotting analysis confirmed that LIN28A expression was detected in hiPSC lysates, but not in differentiated cell lysates (Figure 7E, iPSCs versus D14). In addition, we analyzed whether the LIN28A-responsive mRNA device could affect endogenous miRNA expression levels that are controlled by LIN28A. qRT-PCR analysis from hiPSCs revealed that the LIN28A-responsive mRNA device did not change the expression level of endogenous miRNAs (let7d, let7g and miR-98), which are known to be regulated by LIN28A and were also upregulated after differentiation (37) (Figure 7F). Thus, we conclude that our LIN28A-responsive device delivered by mRNA is able to distinguish hiPSCs from differentiated cells by detecting endogenous LIN28A without affecting the expression profile of its target miRNAs.
DISCUSSION
Here, we developed endogenous protein (LIN28A and U1A)-responsive mRNA devices by stabilizing the active conformation (i.e. secondary structure) of aptamers. The original aptamers (preE-let7d, U1utr), which mediate the binding of LIN28A and U1A, respectively, did not efficiently respond to target proteins, indicating that their structures are less stable in cells compared with those of engineered aptamers. Our design strategy is simple and versatile for the creation and improvement of protein-responsive RNA devices that function in living cells. In fact, LIN28A-responsive mRNA distinguished hiPSCs and differentiated cells by detecting endogenous LIN28A expression levels. To our knowledge, no previous study has shown that synthetic RNA-delivered devices can detect endogenous proteins, control translation and distinguish mammalian cells. Additionally, given that aptamers that bind proteins of interest are generated by in vitro selection, our design strategy can be applied not only to naturally occurring protein-binding RNA motifs, but also to synthetic RNA aptamers. In fact, we designed alternative protein-responsive mRNA devices based on the interaction between nuclear factor-κB (NF-κB) p50 protein and the in vitro selected p50-binding aptamer (p50A) (Supplementary Figure S8 and Additional text for Supplementary Figure S8). The mRNA with engineered aptamer (p50A-stb) efficiently detected p50, whereas that with the original aptamer was insensitive to p50, indicating that we are able to design a variety of mRNA devices using both natural and synthetic aptamers. However, it is noteworthy that the basal expression levels of mRNAs that contain stabilized aptamers at the 5΄-UTR region tend to be low in the absence of target proteins (Figure 3C, Supplementary Figures S4 and S6). Although the U1LSL series with long stem structures showed reduced reporter expression levels (Supplementary Figure S4), this repression was likely caused by endogenous U1A (Figure 3). To increase the basal expression levels of mRNA devices with stabilized aptamers, the position of the aptamers can be shifted to the downstream region (13) (Supplementary Figure S9A). Thus, the position of the stabilized aptamers is an important factor in the target sensitivity and basal expression level.
It has been reported that several protein-responsive RNA devices were designed based on the insertion of aptamers into RNA. Examples include the regulation of mRNA translation (24,25,27), shRNA/miRNA processing (14,45), alternative splicing (43), nonsense-mediated mRNA decay (46) and ribozyme activity. However, these devices were exclusively delivered by plasmid DNA, which may raise the risk of random genomic integration. Messenger RNA delivery may reduce the risk of genomic damage, providing an avenue for future therapeutic applications, such as live-cell detection or purification for regenerative medicine (17), based on intracellular protein information. Indeed, a recent synthetic biology study showed that synthetic circuits with RNA binding proteins can function in mammalian cells when delivered by RNA (8), however, these circuits also require specific exogenous RNA-binding proteins (L7Ae or MS2-CNOT7) for their construction. In contrast, our mRNA devices for RNA delivery are able to detect endogenous LIN28A protein in living cells. It is known that higher expression of LIN28A is correlated with several diseases and cancers, including glioblastoma (47). Thus, LIN28A-responsive mRNA devices may distinguish not only pluripotent stem cells but also LIN28A-related cancer cells within a heterogeneous cell population.
In this study, we used unmodified, native mRNA-delivered devices to study the function of LIN28A-binding aptamers (Figures 6 and 7). Although unmodified mRNAs may induce an immune response in several cell types, we used 293FT cell lines and hiPSCs that show attenuated responses to interferon signaling (48,49). Indeed, our mRNA-delivered devices did not increase the expression levels of interferon signaling-related OAS1 and IFIT1 genes, nor did they induce a loss of cell viability (Supplementary Figure S10). These results indicate that the synthetic mRNA devices did not cause an innate immune response under our experimental condition. It is also noteworthy that the use of modified mRNAs may affect the function of the devices. In fact, we found that modified mRNA devices with stbC aptamer did not respond to target LIN28A protein efficiently (Supplementary Figure S9B), indicating that some modified bases in the mRNA may affect binding ability to target proteins and stability of the RNA structures in cells. To apply our mRNA devices to many cell types, we expect that generation of mRNA devices with modified bases will also be important for future study.
Synthetic mRNA devices with RNA-binding proteins have been applied to construct cellular logic gates and to control cell fate (25,46). The direct detection of human endogenous proteins by synthetic RNAs may open the door to program cell fate autonomously based on intracellular protein information. Additionally, endogenous protein-responsive RNA devices can be used to measure the protein expression level in living cells. Indeed, reporter expression levels from the LIN28A-responsive device were correlated with LIN28A amount in the cell (Figure 5). Thus, we may be able to monitor the amount of target protein expressed in living cells without the need for tagging the protein. Moreover, combining with current gene regulation technologies, including miRNA responsive switches (8,17,50), RNA inverter modules (46), cell classifier circuits (50), ribozyme devices (51,52) and two dimensional tuning (optimized positioning and multiple insertion of RNA motifs in mRNA) (13) will enable us to build more sensitive, precise and increasingly sophisticated systems to program cell fate. In fact, the tandem insertion of stbC motif enabled to construct more effective mRNA device (Supplementary Figure S11). Thus, we believe endogenous protein-responsive RNA devices with greater safety (i.e. RNA-delivery) and sensitivity will expand the field of biomedicine and synthetic biology.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Peter Karagiannis and Dr Callum Parr (Kyoto University) for critical reading of the manuscript, Dr Kei Endo (The University of Tokyo) and Dr Shunnich Kashida (Ecole Normale Superieure) for critical discussions and some materials preparation during the early stages of the research. Dr Takuya Yamamoto (Kyoto University) for advice about statistical analysis, and Karin Hayashi and Moe Hirosawa (Kyoto University) for supporting experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
JSPS KAKENHI [15H05722, 24104002 to H.S.]; Naito Foundation (to H.S.); Canon Foundation (to H.S.); Nakatani Foundation (to H.S.). Funding for open access charge: JSPS KAKENHI [15H05722].
Conflict of interest statement. Kyoto University has filed a patent application broadly relevant to this work. S. K. and H. S. are the investigators of record listed on the patent application.
REFERENCES
- 1. Niwa H., Miyazaki J., Smith A.G.. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000; 24:372–376. [DOI] [PubMed] [Google Scholar]
- 2. Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011; 144:646–674. [DOI] [PubMed] [Google Scholar]
- 3. López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G.. The hallmarks of aging. Cell. 2013; 153:1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walker J.M. Walker JM. The Protein Protocols Handbook. 2002; 2nd edn, NJ: Humana Press. [Google Scholar]
- 5. Tsukiji S., Hamachi I.. Ligand-directed tosyl chemistry for in situ native protein labeling and engineering in living systems: from basic properties to applications. Curr. Opin. Chem. Biol. 2014; 21:136–143. [DOI] [PubMed] [Google Scholar]
- 6. Greenwood C., Ruff D., Kirvell S., Johnson G., Dhillon H.S., Bustin S.A.. Proximity assays for sensitive quantification of proteins. Biomol. Detect. Quantif. 2015; 4:10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lo C.-A., Kays I., Emran F., Lin T.-J., Cvetkovska V., Chen B.E.. Quantification of protein levels in single living cells. Cell Rep. 2015; 13:2634–2644. [DOI] [PubMed] [Google Scholar]
- 8. Wroblewska L., Kitada T., Endo K., Siciliano V., Stillo B., Saito H., Weiss R.. Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat. Biotechnol. 2015; 33:839–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hsu H.-T., Lin Y.-H., Chang K.-Y.. Synergetic regulation of translational reading-frame switch by ligand-responsive RNAs in mammalian cells. Nucleic Acids Res. 2014; 42:14070–14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cao J., Arha M., Sudrik C., Mukherjee A., Wu X., Kane R.S.. A universal strategy for regulating mRNA translation in prokaryotic and eukaryotic cells. Nucleic Acids Res. 2015; 43:4353–4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stapleton J.A, Endo K., Fujita Y., Hayashi K., Takinoue M., Saito H., Inoue T.. Feedback control of protein expression in mammalian cells by tunable synthetic translational inhibition. ACS Synth. Biol. 2012; 1:83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ausländer S., Ausländer D., Müller M., Wieland M., Fussenegger M.. Programmable single-cell mammalian biocomputers. Nature. 2012; 487:123–127. [DOI] [PubMed] [Google Scholar]
- 13. Endo K., Stapleton J.A, Hayashi K., Saito H., Inoue T.. Quantitative and simultaneous translational control of distinct mammalian mRNAs. Nucleic Acids Res. 2013; 41:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kashida S., Inoue T., Saito H.. Three-dimensionally designed protein-responsive RNA devices for cell signaling regulation. Nucleic Acids Res. 2012; 40:9369–9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Endo K., Saito H.. Engineering protein-responsive mRNA switch in mammalian cells. Methods Mol. Biol. 2014; 1111:183–196. [DOI] [PubMed] [Google Scholar]
- 16. Shcherbakova D.M., Verkhusha V.V. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods. 2013; 10:751–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miki K., Endo K., Takahashi S., Funakoshi S., Takei I., Katayama S., Toyoda T., Kotaka M., Takaki T., Umeda M. et al. Efficient detection and purification of cell populations using synthetic microRNA switches. Cell Stem Cell. 2015; 16:699–711. [DOI] [PubMed] [Google Scholar]
- 18. Andreev D.E., Terenin I.M., Dmitriev S.E., Shatsky I.N.. Pros and cons of pDNA and mRNA transfection to study mRNA translation in mammalian cells. Gene. 2016; 578:1–6. [DOI] [PubMed] [Google Scholar]
- 19. Antony J.S., Dewerth A., Haque A., Handgretinger R., Kormann M.S.D.. Modified mRNA as a new therapeutic option for pediatric respiratory diseases and hemoglobinopathies. Mol. Cell. Pediatr. 2015; 2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Warren L., Manos P.D., Ahfeldt T., Loh Y.-H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A. et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010; 7:618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nakagawa M., Taniguchi Y., Senda S., Takizawa N., Ichisaka T., Asano K., Morizane A., Doi D., Takahashi J., Nishizawa M. et al. A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci. Rep. 2014; 4:3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ellis B., Haaland P., Hahne F, Le Meur N, Gopalakrishnan N, Spidlen J., Jiang M.. flowCore: Basic structures for flow cytometry data. 2016; R Package Version 1.40.0.https://bioconductor.org/packages/release/bioc/html/flowCore.html. [Google Scholar]
- 23. Sarbassov D.D., Guertin D.A, Ali S.M., Sabatini D.M.. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005; 307:1098–1101. [DOI] [PubMed] [Google Scholar]
- 24. Saito H., Kobayashi T., Hara T., Fujita Y., Hayashi K., Furushima R., Inoue T.. Synthetic translational regulation by an L7Ae-kink-turn RNP switch. Nat. Chem. Biol. 2010; 6:71–78. [DOI] [PubMed] [Google Scholar]
- 25. Saito H., Fujita Y., Kashida S., Hayashi K., Inoue T.. Synthetic human cell fate regulation by protein-driven RNA switches. Nat. Commun. 2011; 2:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nie M., Htun H.. Different modes and potencies of translational repression by sequence-specific RNA-protein interaction at the 5΄-UTR. Nucleic Acids Res. 2006; 34:5528–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu Y., Huang W., Zhou D., Han Y., Duan Y., Zhang X., Zhang H., Jiang Z., Gui Y., Cai Z.. Synthesizing oncogenic signal-processing systems that function as both ‘signal counters’ and ‘signal blockers’ in cancer cells. Mol. Biosyst. 2013; 9:1909–1918. [DOI] [PubMed] [Google Scholar]
- 28. Stripecke R., Oliveira C.C., McCarthy J.E., Hentze M.W.. Proteins binding to 5΄ untranslated region sites: a general mechanism for translational regulation of mRNAs in human and yeast cells. Mol. Cell. Biol. 1994; 14:5898–5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taliaferro J.M., Lambert N.J., Sudmant P.H., Dominguez D., Merkin J.J., Alexis M.S., Bazile C.A., Burge C.B.. RNA sequence context effects measured in vitro predict in vivo protein binding and regulation. Mol. Cell. 2016; 64:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van Nostrand E.L., Pratt G.A., Shishkin A.A., Gelboin-Burkhart C., Fang M.Y., Sundararaman B., Blue S.M., Nguyen T.B., Surka C., Elkins K. et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods. 2016; 13:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagai K., Oubridge C., Ito N., Avis J., Evans P.. The RNP domain: a sequence-specific RNA-binding domain involved in processing and transport of RNA. Trends Biochem. Sci. 1995; 20:235–240. [DOI] [PubMed] [Google Scholar]
- 32. Jovine L., Oubridge C., Avis J.M., Nagai K.. Two structurally different RNA molecules are bound by the spliceosomal protein U1A using the same recognition strategy. Structure. 1996; 4:621–631. [DOI] [PubMed] [Google Scholar]
- 33. Boelens W.C., Jansen E.J.R., van Venrooij W.J., Stripecke R., Mattaj I.W., Gunderson S.I.. The human U1 snRNP-Specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell. 1993; 72:881–892. [DOI] [PubMed] [Google Scholar]
- 34. Gunderson S.I., Beyer K., Martin G., Keller W., Boelens W.C., Mattaj I.W.. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell. 1994; 76:531–541. [DOI] [PubMed] [Google Scholar]
- 35. Sato K., Hamada M., Asai K., Mituyama T.. CentroidFold: a web server for RNA secondary structure prediction. Nucleic Acids Res. 2009; 37:277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mayr F., Heinemann U.. Mechanisms of Lin28-mediated miRNA and mRNA regulation—a structural and functional perspective. Int. J. Mol. Sci. 2013; 14:16532–16553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thornton J.E., Gregory R.I.. How does Lin28 let-7 control development and disease?. Trends Cell Biol. 2012; 22:474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang J., Ratanasirintrawoot S., Chandrasekaran S., Wu Z., Ficarro S.B., Yu C., Ross C.A., Cacchiarelli D., Xia Q., Seligson M. et al. LIN28 regulates stem cell metabolism and conversion to primed pluripotency. Cell Stem Cell. 2016; 19:66–80. [DOI] [PubMed] [Google Scholar]
- 39. Heo I., Joo C., Kim Y.K., Ha M., Yoon M.J., Cho J., Yeom K.H., Han J., Kim V.N.. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009; 138:696–708. [DOI] [PubMed] [Google Scholar]
- 40. Piskounova E., Polytarchou C., Thornton J.E., Lapierre R.J., Pothoulakis C., Hagan J.P., Iliopoulos D., Gregory R.I.. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011; 147:1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nam Y., Chen C., Gregory R.I., Chou J.J., Sliz P.. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011; 147:1080–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Loughlin F.E., Gebert L.F.R., Towbin H., Brunschweiger A., Hall J., Allain F.H.-T.. Structural basis of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28. Nat. Struct. Mol. Biol. 2011; 19:84–89. [DOI] [PubMed] [Google Scholar]
- 43. Culler S.J., Hoff K.G., Smolke C.D.. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science. 2010; 330:1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tano K., Yasuda S., Kuroda T., Saito H., Umezawa A., Sato Y.. A novel in vitro method for detecting undifferentiated human pluripotent stem cells as impurities in cell therapy products using a highly efficient culture system. PLoS One. 2014; 9:e110496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bloom R.J., Winkler S.M., Smolke C.D.. A quantitative framework for the forward design of synthetic miRNA circuits. Nat. Methods. 2014; 11:1147–1153. [DOI] [PubMed] [Google Scholar]
- 46. Endo K., Hayashi K., Inoue T., Saito H.. A versatile cis-acting inverter module for synthetic translational switches. Nat. Commun. 2013; 4:2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mao X., Hütt-Cabezas M., Orr B.A., Weingart M., Taylor I., Rajan A.K.D., Odia Y., Kahlert U., Maciaczyk J., Nikkhah G. et al. LIN28A facilitates the transformation of human neural stem cells and promotes glioblastoma tumorigenesis through a pro-invasive genetic program. Oncotarget. 2013; 4:1050–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Andries O., Filette M.De, De Smedt S.C., Demeester J., Poucke M.Van, Peelman L., Sanders N.N.. Innate immune response and programmed cell death following carrier-mediated delivery of unmodified mRNA to respiratory cells. J. Control. Release. 2013; 167:157–166. [DOI] [PubMed] [Google Scholar]
- 49. Hong X.-X., Carmichael G.G.. Innate immunity in pluripotent human cells: attenuated response to interferon-β. J. Biol. Chem. 2013; 288:16196–16205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xie Z., Wroblewska L., Prochazka L., Weiss R., Benenson Y.. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011; 333:1307–1311. [DOI] [PubMed] [Google Scholar]
- 51. Ausländer S., Stücheli P., Rehm C., Ausländer D., Hartig J.S., Fussenegger M.. A general design strategy for protein-responsive riboswitches in mammalian cells. Nat. Methods. 2014; 11:1154–1160. [DOI] [PubMed] [Google Scholar]
- 52. Kennedy A.B., Vowles J.V., D’Espaux L., Smolke C.D.. Protein-responsive ribozyme switches in eukaryotic cells. Nucleic Acids Res. 2014; 42:12306–12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.