Abstract
High-throughput RNA sequencing methods coupled with specialized bioinformatic analyses have recently uncovered tens of thousands of unique circular (circ)RNAs, but their complete sequences, genes of origin and functions are largely unknown. Given that circRNAs lack free ends and are thus relatively stable, their association with microRNAs (miRNAs) and RNA-binding proteins (RBPs) can influence gene expression programs. While exoribonuclease treatment is widely used to degrade linear RNAs and enrich circRNAs in RNA samples, it does not efficiently eliminate all linear RNAs. Here, we describe a novel method for the isolation of highly pure circRNA populations involving RNase R treatment followed by Polyadenylation and poly(A)+ RNA Depletion (RPAD), which removes linear RNA to near completion. High-throughput sequencing of RNA prepared using RPAD from human cervical carcinoma HeLa cells and mouse C2C12 myoblasts led to two surprising discoveries: (i) many exonic circRNA (EcircRNA) isoforms share an identical backsplice sequence but have different body sizes and sequences, and (ii) thousands of novel intronic circular RNAs (IcircRNAs) are expressed in cells. In sum, isolating high-purity circRNAs using the RPAD method can enable quantitative and qualitative analyses of circRNA types and sequence composition, paving the way for the elucidation of circRNA functions.
INTRODUCTION
Given that coding (messenger) RNAs comprise <5% of the eukaryotic transcriptome, most expressed transcripts are non-coding RNAs (ncRNA) (1). Ribosomal (r)RNAs and transfer (t)RNAs are among the most abundant ncRNAs and they generally control gene expression programs by carrying out housekeeping functions. Other ncRNAs such as microRNAs (miRNAs) and long non-coding (lnc)RNAs can elicit specific gene expression programs and are more diverse, although they are less abundant overall (2). Circular (circ)RNAs, another large and heterogeneous class of ncRNAs, are among the least-well characterized non-coding transcripts. CircRNAs are covalently closed via mechanisms that involve the splicing machinery, and therefore lack 5΄ or 3΄ ends, as shown in viroid and mitochondrial RNA decades ago and in higher eukaryotes more recently (3–8). High-throughput RNA sequencing (RNA-Seq) analyses have identified vast numbers of circRNAs (9). They can range in length from <100 nt to several kb and can arise from any genomic region (10–14). Most circRNAs reported to-date originate from exons of coding mRNAs, although some are derived from ncRNAs (11,14). Recent reports described circRNAs originated from intronic sequences, named circular intronic RNAs (ciRNAs) (12). CiRNAs are proposed to arise from the intronic lariats that form during splicing and fail to debranch due to the presence of RNA sequence motifs close to the 5΄splice site and branch point (12,15). In addition, some circRNAs have retained introns between exons and are termed exon–intron circRNAs, or EIcircRNAs (16).
Although thousands of circRNAs have been reported in recent years, only a few of them have been shown to influence physiologic or disease processes. Given that the circRNAs lack free ends and are resistant to exonucleases, they can have long half-lives and thus function as effective sponges or decoys for specific molecules interacting with them. There are a few recent examples of circRNAs that sponge microRNAs, in turn modulating the expression of mRNAs regulated by the microRNAs in question (11,17–19). CircRNAs have also been reported to act as decoys for RNA-binding proteins (RBPs). For example, CircMbl interacts with the splicing protein muscleblind (MBL) and alters splicing of the linear MBL mRNA by sponging MBL (20). Similarly, circ-Foxo3 altered cell cycle progression by forming a ternary complex with proteins CDK2 and p21/CDKN1A leading to cell cycle arrest (21).
Due to their great variability of length and their genesis from linear RNA, it is not possible to isolate circRNAs from other RNA species by size or sequence. Instead, circRNAs are typically identified by the presence of exons arranged out of order, forming ‘backsplice’ junctions. CircRNAs were first detected by electron microscopy, which did not differentiate circRNAs from RNA lariats (4). The recent development of bioinformatic tools (e.g. circ_finder, find_circ, CIRCexplorer and CIRI) and methods of enrichment of circRNAs by digesting linear RNAs with exoribonucleases (e.g. the 3΄→5΄ exonuclease RNase R) and depleting ribosomal RNA (rRNA) has helped to identify circRNAs in RNA-Seq datasets (11). These analytical methods rely on the alignment of fusion RNA-Seq reads to backsplice sequences for the identification of start-and-end coordinates of circRNAs, but cannot determine their full-length sequence. CircRNAs can also be detected by reverse transcription (RT) followed by conventional or quantitative (q) polymerase chain reaction (PCR) to amplify the circRNA backsplice junction using divergent primers and by Northern blot analysis if they are abundant (17,22). Unfortunately, current methods of rRNA depletion and RNase R digestion leave substantial amounts of linear RNAs intact, particularly linear RNAs with extensive secondary structures, hindering the quantitative and qualitative analysis of circRNAs.
Considering the rising interest in characterizing circRNAs comprehensively and elucidating their function, it is critical that superior methods be developed to isolate circRNA populations. Here, we describe a novel procedure to isolate highly pure circRNAs from total RNA by first depleting the linear RNA with RNase R, then polyadenylating the remaining RNAs bearing free 3΄-OH ends and finally depleting the poly(A)-containing RNAs that were initially resistant to RNase R digestion. This method, which we have termed (RPAD) for ‘RNase R treatment followed by polyadenylation and poly(A)+ RNA depletion’ yields circRNA populations of very high purity through the sequential depletion of linear RNAs. After sequencing RPAD-prepared RNA samples, the body of each circRNA can be assembled by joining the sequences that span the backsplice site. Using the RPAD method and high-throughput RNA-Seq analysis with paired-end reads, we identified full-length circRNA sequences that included many novel circRNA isoforms. As proof-of-principle, we identified full-length sequences of exonic and intronic circRNAs expressed in human and mouse cell lines (cervical carcinoma HeLa cells and C2C12 myoblasts, respectively). We termed the exonic and intronic circRNAs EcircRNAs and IcircRNAs, respectively.
MATERIALS AND METHODS
Cell culture and RNA isolation
Human cervical carcinoma HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) containing 10% fetal bovine serum (FBS, Gibco) and antibiotics (Life Technologies). Mouse C2C12 myoblasts were cultured in DMEM supplemented with 20% FBS and antibiotics. Both Hela and C2C12 cells were maintained at 37°C in a humidified atmosphere of 95% air, 5% CO2. Total RNA from cultured cells was isolated using the miRNeasy Mini Kit (#217004, Qiagen Inc.) following the manufacturer's protocol.
Targets and PCR primers
Ten circRNAs were selected from the circular RNA database circBase (http://www.circbase.org/) (9). All the divergent primers for circRNA detection were designed using the CircInteractome web tool (https://circinteractome.nia.nih.gov/Divergent_Primers/divergent_primers.html) (23). Convergent primers for detection of linear RNAs were designed using the NCBI primer tool, microRNAs were selected from the database miRBase (http://www.mirbase.org/), and microRNA sequences were used for forward primer design (24). All sequences are available (Supplementary Table S1).
Depletion of linear RNA
A 20-μl reaction was set up with 2 μg of Hela RNA, 20 U of RiboLock RNase inhibitor (Thermo Fisher Scientific), 1 × RNase R buffer and 20 U of RNase R (RNR07250, Epicentre) and incubated for 30 min at 37°C. Control reactions were carried out in the same conditions, but without RNase R. The RNA from control and RNase R-treated samples were isolated using miRNeasy Mini Kit following the manufacturer's instructions and eluted with 40 μl of nuclease-free water. RNase R was removed during the RNA isolation step using the miRNeasy Kit, which eliminates all protein from the sample. A 40-μl polyadenylation reaction was prepared with 20 μl of RNase R-treated RNA, 1 × E-PAP buffer, E-PAP (AM1350, Thermo Fisher Scientific), 1 mM ATP solution, 2.5 mM MnCl2 and 40 U of RiboLock RNase inhibitor, and incubated for 30 min at 37°C. The control RNA was processed using the same conditions, but without addition of E-PAP. Oligo-dT Dynabeads (10 μl) from Poly(A)Purist™ MAG Kit (AM1922) were washed three times with the 1 × binding buffer provided in the kit and dissolved in 40 μl of 2 × binding buffer. The Oligo-dT Dynabeads in 2 × binding buffer were added into the poly(A)-tailing reaction mix of RNase R-treated samples and incubated for 5 min at 75°C followed by 20 min at 25°C with shaking. The control RNA reaction was also incubated as above without the Oligo-dT Dynabeads. The RNA sample with Oligo-dT Dynabeads was kept for 2 min on the magnetic stand and the supernatant was collected for RNA isolation. The miRNeasy Mini Kit was used to prepare RNA from the control and RPAD samples, and dissolved in 40 μl of nuclease-free water.
Real-time reverse transcription-PCR
Total RNA or processed RNA was isolated using miRNeasy Mini Kit following the manufacturer's instructions. For regular cDNA synthesis, reverse transcription (RT) was performed in a 20-μl reaction containing RNA, 1 × RT buffer, 0.5 mM of each dNTP, 150 ng of random hexamers (11034731001, Roche) and maxima reverse transcriptase (RT) (EP0741, Thermo Fisher Scientific), and incubated at 25°C for 10 min followed by 30 min at 50°C; the RT enzyme was inactivated by heating at 85°C for 5 min (25). For qPCR analysis of circRNAs and mRNAs, 20-μl PCR reactions were set up with 0.1 μl of cDNA, 1 × KAPA SYBR® FAST qPCR mix (ABI Prism) (KK4605, KAPA Biosystems), and 250 nM gene-specific primers. The RT and PCR reactions were performed on Veriti® 96-Well Thermal Cycler (#4375786, Thermo Fisher Scientific, USA). QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific) with a cycle setup of 2 min at 95°C and 40 cycles of 2 s at 95°C plus 10 s at 60°C was used for RT-qPCR followed by calculation of the RNA fold change using the 2−ΔΔCT method (26).
Real-time reverse transcription-PCR analysis of microRNAs
RNA was reverse-transcribed using the Mir-X™ microRNA First Strand Synthesis Kit (#638315, Clontech) according to the manufacturer's protocol. The microRNAs and snRNAs were quantified by real-time PCR analysis using 20-μl reaction volumes containing 0.1 μl of cDNA, 1 × KAPA SYBR® FAST qPCR mix (ABI Prism), 250 nM miRNA-specific forward primers (Supplementary Table S1) and a universal reverse primer. RT-qPCR was performed on QuantStudio 5 Real-Time PCR System with a cycle setup consisting of 2 min at 95°C and 40 cycles of 2 s at 95°C plus 10 s at 60°C; the fold change in abundance was calculated using the 2−ΔΔCT method (26).
RT-PCR and circRNA sequencing
Backsplice sequence (junction) of circRNAs was amplified by PCR in a 50-μl reaction containing 0.1 μl of cDNA, 1 × KAPA SYBR® FAST qPCR mix (ABI Prism) and 250 nM circRNA-specific divergent primers. PCR was performed on a thermal cycler with a cycle setup consisting of 3 min at 95°C and 35 cycles of 5 s at 95°C plus 5 s at 60°C; the RT-PCR products were resolved by electrophoresis through 2% agarose gels stained with ethidium bromide and visualized on an ultraviolet transilluminator. The RT-PCR product was purified using the QIAquick Gel Extraction Kit (Cat# 28704, Qiagen) and the amplified PCR products were sequenced with forward or reverse primers (MCLAB, USA).
Circular RNA sequencing and annotation
After isolation of total RNA and RPAD-prepared RNA using miRNeasy Mini Kit, ribosomal (r)RNA was removed by rRNA Depletion Nano kit (Qiagen), whereupon cDNA was prepared and amplified using the Ovation RNA-Seq System V2 (NuGEN) kit following the manufacturer's instructions. The amplified cDNA was fragmented using a Bioruptor (Diagenode) followed by ligation of adaptors using TruSeq ChiP Sample Preparation kit (Illumina, San Diego, CA, USA), and DNA fragments ranging 300–350 bp were isolated from a 2.5% agarose gel and subjected to 17 cycles of PCR amplification. A Bioanalyzer 2100 instrument was used to analyze the quality of the cDNA libraries, which were sequenced using an Illumina HiSeq 2500 instrument (and deposited in GSE92632).
For identification of circRNAs in the RNA-Seq, adapter contamination was removed from the raw fastq files and TopHat2 (v2.1.0) was used to align the sequences to the human genome (hg19) and mouse genome (mm9). The aligned reads were used to identify the body of circular RNA and residual linear RNA using Cufflinks (v2.2.1) and default parameters (with the exception of assembled transfrags supported by five or more reported fragments). Both known (Ensembl GRCh37 Release 82 for human and Ensembl mm9 Release 67 for mouse) and novel transcripts were identified. The reads which did not align to the genome were used to identify fusion junctions using TopHat2. The CIRCexplorer program (v1.10) was used with the fusion junctions obtained from TopHat2 using the identified transcripts from the previous step to identify both the circularizing junction and the spliced sequence of circRNAs in HeLa and C2C12 cells, as described previously (22). Combined circRNA junction read numbers from two C2C12 samples and one HeLa sample with or without RPAD processing are shown (Supplementary Tables S2 and 3).
Identification of sequence motifs
To analyze the shared sequence motif at the circular RNA junction, genomic sequences corresponding to 20-nt junction sequences (10 nt of the circRNA end-joined with 10 nt of the start) were obtained from the hg19 (HeLa) and mm9 (C2C12) UCSC genome assembly using the ‘table browser’. The sequences upstream or downstream of the circRNAs were also obtained from the hg19 (HeLa) and mm9 (C2C12) UCSC genome assembly using the ‘table browser’. The WebLogo 3 webtool (http://weblogo.threeplusone.com/create.cgi) was used to generate the probability of different nucleotides near the circRNA junction (27).
RESULTS
Identification of backsplice sequences
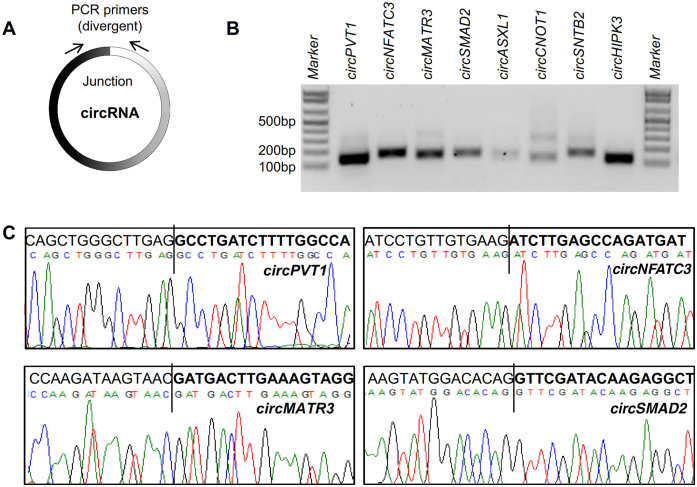
Although circRNAs can be detected by Northern blot analysis, this method has limitations, as the size of a circRNA is difficult to predict on a gel due to the presence of the secondary structure of the circRNA, the abundance of many circRNAs is too low for detection and this approach does not provide RNA sequence information. Alternatively, PCR analysis can be used to amplify a specific circRNA and identify its sequence, but this method is also challenging because the circRNA sequence is identical to that of the parent linear transcript. Primers can be designed (e.g. using the CircInteractome tool) to generate a PCR amplicon that spans the circRNA junction (Figure 1A). For example, RNA prepared from the human cervical carcinoma cell line HeLa was reverse-transcribed (RT) and amplified by PCR using divergent primers specific to several known circRNAs; the products were analyzed on 2% agarose gels stained with ethidium bromide (Figure 1B). All reactions produced amplicons of the expected size except for circCNOT1, which showed a larger band (∼300 bp) that could be a non-specific product or another circRNA isoform. The circRNAs used in validation experiments were named after their respective genes of origin (Table 1). After purification, the PCR products were sequenced, confirming that the circRNA junctions were indeed amplified and verifying the presence of backsplice sites in these transcripts (Figure 1C). Although this analysis suggests that circRNAs with these backsplice sites exist in the cell, it is possible that such backsplice sites might have been generated by other mechanisms like RNA transplicing, template switching during reverse transcription, or tandem duplication, as described previously (28). Thus, it is critical to deplete fully the linear RNAs from the total RNA population in order to analyze circRNAs quantitatively and qualitatively.
Figure 1.
Validation of backsplice sequences. (A) Schematic of the design of divergent primers used for the specific detection of circRNAs. (B) RT-qPCR products amplified with divergent primers resolved on ethidium bromide-stained, 2% agarose gels. (C) Representative PCR products purified and sequenced to confirm backsplice sequences.
Table 1. CircRNAs used for validation.
| CircRNA name | Chr location/CircRNA_junction_unique_IDs | circbase_circRNA ID | Exonic (E)circRNA | Gene_name | Length (nt) |
|---|---|---|---|---|---|
| circNFATC3 | hsa_chr16_68155889_68160513_F | hsa_circ_0000711 | EcircRNA | NFATC3 | 1298 |
| circMATR3 | hsa_chr5_138614015_138614818_F | hsa_circ_0008922 | EcircRNA | MATR3 | 161 |
| circSMAD2 | hsa_chr18_45391429_45423180_R | hsa_circ_0000847 | EcircRNA | SMAD2 | 783 |
| circHIPK3 | hsa_chr11_33307958_33309057_F | hsa_circ_0000284 | EcircRNA | HIPK3 | 1099 |
| circPVT1 | hsa_chr8_128902834_128903244_F | hsa_circ_0001821 | EcircRNA | PVT1 | 410 |
| circSNTB2 | hsa_chr16_69317950_69318147_F | hsa_circ_0004354 | EcircRNA | SNTB2 | 197 |
| circASXL1 | hsa_chr20_30954186_30956926_F | hsa_circ_0001136 | EcircRNA | ASXL1 | 195 |
| circCNOT1 | hsa_chr16_58594115_58594266_F | hsa_circ_0007079 | EcircRNA | CNOT1 | 151 |
| circUGP2 | hsa_chr2_64083439_64085070_F | hsa_circ_0001020 | EcircRNA | UGP2 | 236 |
| circANKRD17 | hsa_chr4_73950965_73958017_R | hsa_circ_0001417 | EcircRNA | ANKRD17 | 1832 |
| circPTK2 | hsa_chr8_141828375_141900868_R | hsa_circ_0005982 | EcircRNA | PTK2 | 899 |
| circBPTF | hsa_chr17_65941524_65972074_F | hsa_circ_0000799 | EcircRNA | BPTF | 2026 |
Depletion of linear RNAs and enrichment of circRNAs
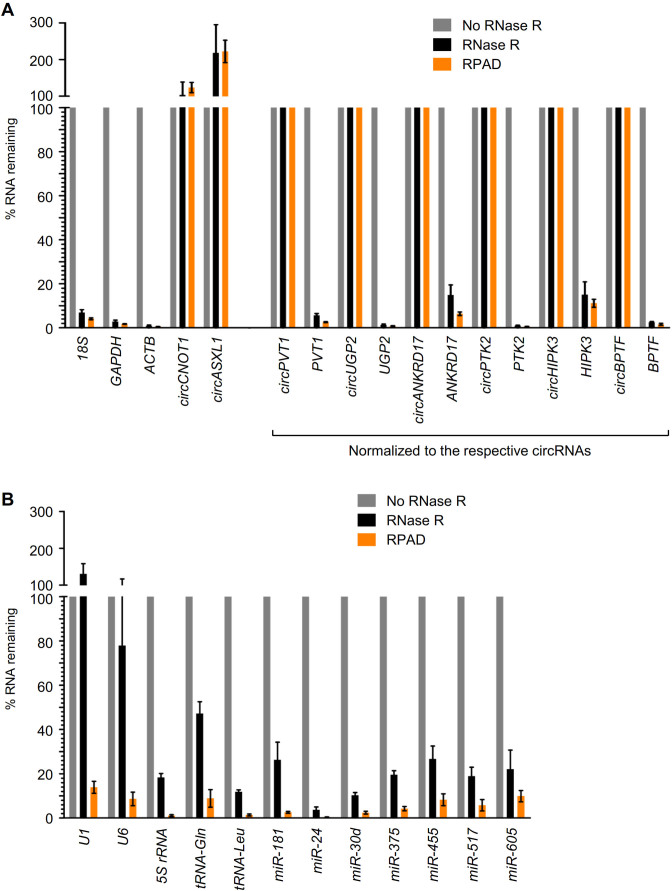
circRNAs can be enriched in the total RNA pool by degrading the linear RNAs using RNA exoribonucleases, which leaves circRNAs intact (29). Digestion of total HeLa RNA with the exonuclease RNase R degraded most of the linear RNA, leaving 15–20% of the RNA undigested (Supplementary Figure S1A), a fraction that likely includes circular RNAs, fragmented linear RNAs and linear RNAs resistant to RNase R treatment. To validate the digestion of linear RNAs with RNase R, we performed a limited screen of mRNAs and circRNAs by using RT-qPCR, employing convergent and divergent primers to detect linear and circular transcripts, respectively. Although mRNAs are presumed to be completely degraded by RNase R digestion, particularly after using high concentrations of RNase R and long incubation times, we were left with 2–20% of mRNAs (Figure 2A). Unlike mRNAs, circRNAs are highly resistant to RNase R digestion, in agreement with previous reports (Figure 2A). As double-stranded RNA and RNAs with extensive secondary structures are known to be refractory to RNase R digestion, we studied the extent of depletion of linear small RNAs such as 5S rRNA, tRNA, snRNAs and microRNAs. As expected, RNase R treatment was unable to degrade snRNAs U1 and U6, while other tested small RNAs were partially digested with RNase R, leaving 2–30% of the small RNAs intact (Figure 2B).
Figure 2.
Validation of RPAD. (A) Subsets of HeLa circRNAs and mRNAs were quantified by RT-qPCR analysis with (black) or without (gray) RNase R treatment, as well as following RPAD (orange). (B) Subsets of HeLa snRNAs and miRNAs were quantified by RT-qPCR analysis with (black) or without (gray) RNase R treatment, as well as following RPAD (orange). In (A) since the relative circRNA levels rise when linear RNAs are depleted by RNase R, mRNA levels were normalized to the levels of the corresponding circRNAs, which were not affected by RNase R treatment or poly(A)-RNA depletion. The data represent the means ± SEM from five or more experiments.
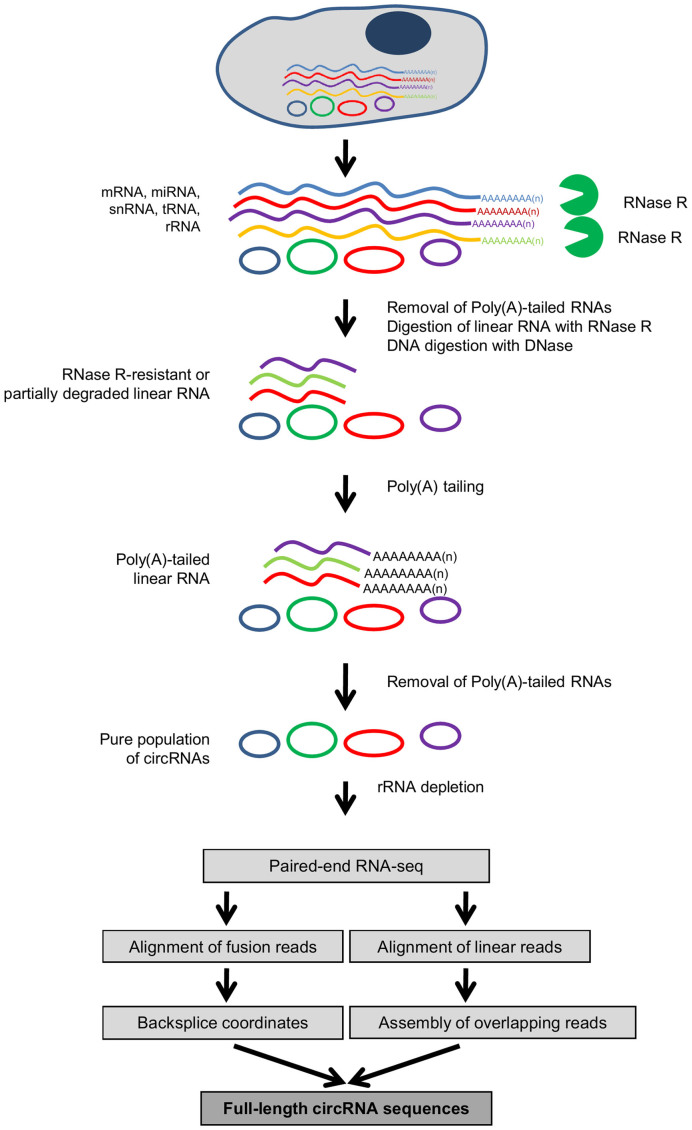
As the levels of circRNAs are often lower than those of the linear counterpart RNAs, even a small fraction of linear RNA left after RNase R treatment may surpass the levels of the cognate circRNA. To further deplete the linear RNAs after RNase R treatment, we devised a method that we called ‘RPAD’, outlined in Figure 3. Following RNase R digestion, depletion of poly(A)+ RNA using oligo(dT) beads (‘Materials and Methods' section) enriched the RNA population in circRNAs by depleting the poly(A)-bearing endogenous mRNAs. The depletion of poly(A) mRNA is moderately efficient, as it leaves ∼10–20% of mRNAs behind in the sample (Supplementary Figure S1B). As RNase R and Poly(A) depletion alone did not remove linear RNAs efficiently (30), we included a step in which a poly(A) tail was added to linear RNAs left in the sample, using Escherichia coli poly(A) polymerase (E-PAP, ‘Materials and Methods' section), followed by an additional round of depletion of polyadenylated RNAs that significantly eliminated mRNAs relative to circRNAs (Figure 2A, orange bars). The relative levels of circRNAs appeared to rise when linear RNAs were depleted using RNase R digestion, as reported (31), likely because in the absence of linear RNAs, circRNA cDNA synthesis was more efficient and thus circRNAs appeared to be more abundant (not shown); similarly, linear RNAs that were poorly digested by RNase R appeared overrepresented in RNase R-treated samples. To account for the variability introduced due to the efficiency of cDNA synthesis, the mRNA levels (Figure 2A) were normalized to their corresponding circRNAs, since circRNAs are not affected by RNase R treatment or poly(A)-RNA depletion. For example, as shown in Figure 2A, 15% of ANKRD17 mRNA was left after RNase R treatment, but only ∼7% of ANKRD17 mRNA remained after RPAD. Importantly, RPAD effectively depleted small RNAs, as miRNAs and snRNAs were also found to be extremely reduced using the RPAD method (Figure 2B, orange bars).
Figure 3.
Schematic of the RPAD method. Linear RNAs are first depleted by RNase R digestion. After the remaining RNA is polyadenylated, a second round of depletion of poly(A)+ RNAs using oligo(dT) beads leaves a highly enriched population of circular RNAs.
Identification of true exonic circRNAs (EcircRNAs) in HeLa cells and C2C12 myoblasts
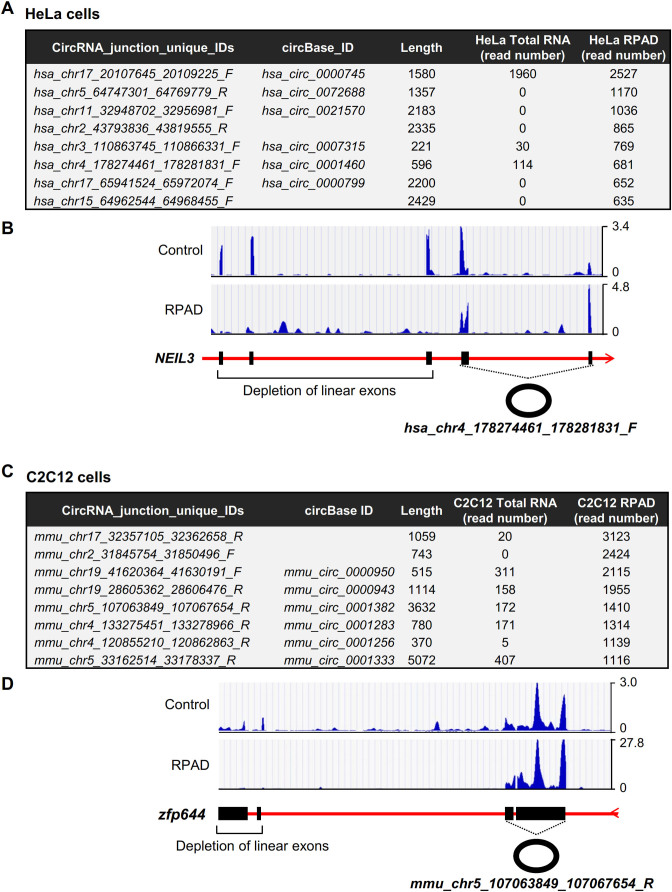
To identify exonic circRNAs (EcircRNAs) in Hela cells and C2C12 myoblasts, we used the RPAD method to deplete linear RNAs and enrich the circRNA population (both intronic and exonic circRNAs) present in total RNA, followed by high-throughput RNA-Seq analysis (GSE92632). By this approach, we identified ∼49,000 and ∼38,000 circRNAs in HeLa and C2C12 cells, respectively (Supplementary Tables S2 and S3), out of which ∼10,000 and ∼20,000, respectively, were only detected in control samples (untreated, no RNase R digestion, no polyadenylation and no poly(A)+ RNA depletion) and were thus deemed to be likely backsplice sites (Supplementary Figure S1C and data not shown). We detected 1374 EcircRNAs generated from backsplicing of exons in HeLa cells (Figure 4A and B; Supplementary Table S2) and 573 EcircRNAs from the exons in C2C12 cells (Figure 4C and Supplementary Table S3). Analysis of the sequencing reads revealed that the exons corresponding to circRNAs were protected while the linear exons were effectively depleted, showing only minimal levels remaining after processing of RNA from HeLa cells and C2C12 cells using RPAD (Figure 4B and D).
Figure 4.
Enrichment of EcircRNA. (A) Table of circRNAs in HeLa cells and their enrichment using the RPAD method. (B) Representative hg19 UCSC genome browser views of exons corresponding to hsa_chr4_178274461_178281831_F. (C) Table of circRNAs in C2C12 cells and their enrichment after RPAD. (D) Representative mm9 UCSC genome browser views of exons corresponding to mmu_chr5_107063849_107067654_R.
Full-length sequence of circRNA
To identify the full-length sequence of circRNAs, we generated paired-end RNA-Seq reads from RPAD-generated and rRNA-depleted RNA samples from HeLa and C2C12 cells. Assembly of RNA-Seq reads between the backsplice site coordinates in the genome were aligned to known and novel transcripts to find the sequence of the circRNA body (‘Materials and Methods’ section and Figure 3). Following this bioinformatic pipeline, we identified full-length sequences of 38,651 and 17,341 circRNAs in RPAD samples from HeLa and C2C12 cells, respectively (Supplementary Table S4).
Identification of EcircRNA isoforms with identical backsplice sequence
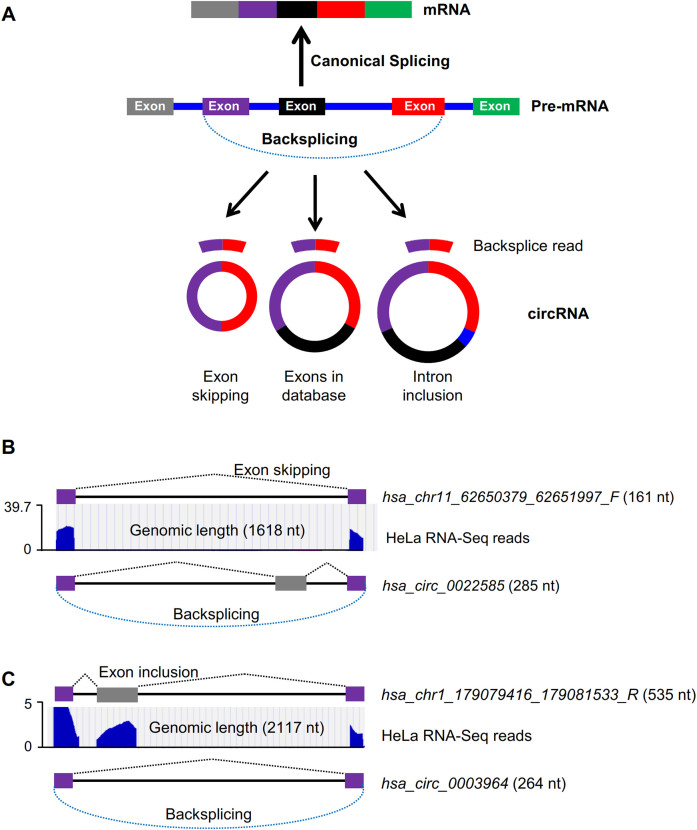
RNA-Seq analysis of RPAD samples revealed that 591 out of 1374 and 421 out of 573 EcircRNAs in HeLa and C2C12 cells, respectively, were novel EcircRNAs not reported by circBase (download 8/4/15). Surprisingly, we found that the lengths of ∼100 circRNAs identified were quite different from those previously reported in circBase (Supplementary Table S5). Although EcircRNAs are identified and validated only based on the backsplice site, our data suggested that the body of the EcircRNAs may vary depending on the inclusion or exclusion of exons during backsplicing (Figure 5A), as previously reported (32,33). For instance, our sequencing data revealed that circRNA hsa_chr11_62650379_62651997_F has an identical backsplice sequence as that in the previously reported hsa_circ_0022585, but one exon is skipped in hsa_chr11_62650379_62651997_F (Figure 5B). In another example, the EcircRNA hsa_chr1_179079416_179081533_R identified here has the same backsplice site as that in hsa_circ_0003964, but contains one extra exon (Figure 5C).
Figure 5.
EcircRNA isoforms in HeLa cells. (A) Schematic of biogenesis of circRNA isoforms with identical backsplice sequence. (B) UCSC genome browser views of RNA-Seq reads from HeLa RNA prepared using RPAD to generate hsa_chr11_62650379_62651997_F, and the previously reported hsa_circ_0022585. (C) UCSC genome browser views of RNA-Seq reads from HeLa RNA prepared using RPAD to generate hsa_chr1_179079416_179081533_R, and the previously reported hsa_circ_0003964.
Enrichment of intronic circular RNAs (IcircRNAs) in RPAD samples
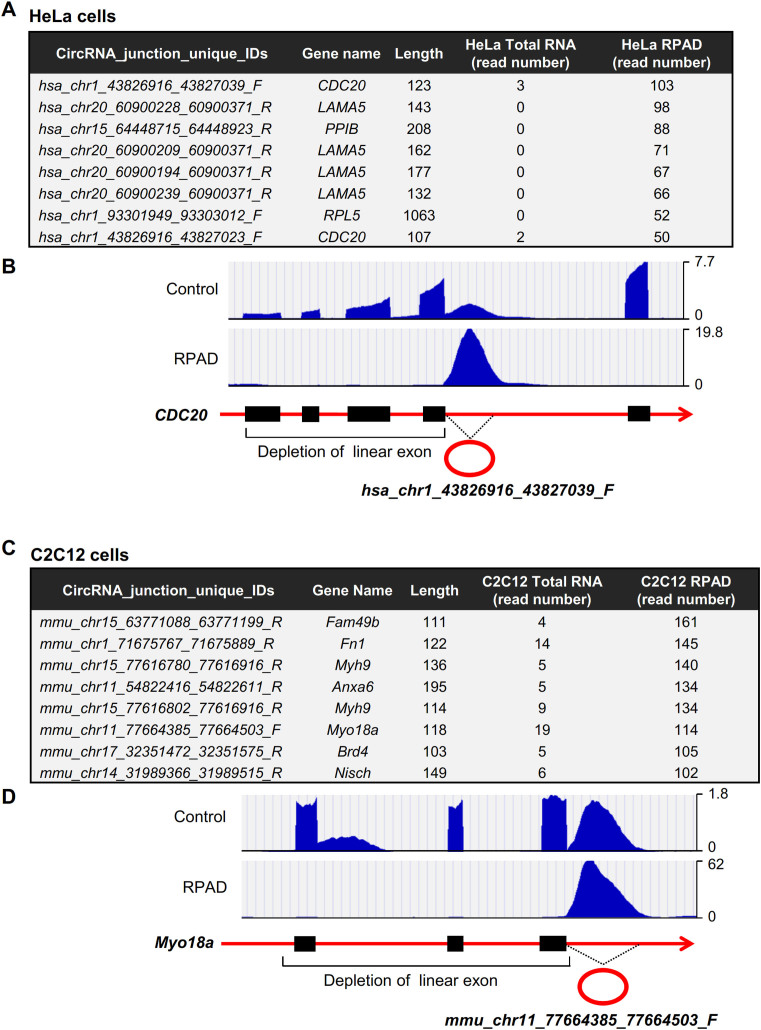
Although hundreds of EcircRNAs were enriched after RPAD (with a few of them shown in Figure 4), RPAD-generated circRNAs unexpectedly contained thousands of circRNAs generated from intronic sequences (Supplementary Figure S1C); we termed them IcircRNAs. HeLa RPAD analysis identified 37,277 novel IcircRNAs, while it only identified 1374 EcircRNAs; likewise, C2C12 RPAD analysis identified 16,768 novel IcircRNAs but only 573 EcircRNAs. The IcircRNA read numbers were greatly enriched in RPAD samples due to the depletion of linear exons, as illustrated in examples from HeLa cells (Figure 6A and B) and C2C12 cells (Figure 6C and D).
Figure 6.
Enrichment of IcircRNA. (A) Table of highly enriched IcircRNAs in HeLa cells prepared using the RPAD method. (B) Representative hg19 UCSC genome browser views of IcircRNA corresponding to hsa_chr1_43826916_43827039_F. (C) Table of highly enriched IcircRNAs in C2C12 cells prepared using the RPAD method. (D) Representative mm9 UCSC genome browser views of IcircRNA corresponding to mmu_chr11_77664385_77664503_F.
EcircRNAs with gt/ag splicing signatures
Attempts to validate the junction sequences in IcircRNAs by cloning and sequencing of amplified PCR fragments (PCR-Seq) revealed that the junction analysis by CIRCexplorer did not always faithfully identify the precise nucleotides immediately surrounding the junction. We discovered two main problems: (i) the annotation of splice junctions is incomplete at present, and (ii) many splice junctions have incorrect sequences likely due to biases in the bioinformatic analysis methods (as programs try to conform to classic splice junction sequences, typically GT-AG). Given the discrepancies between the IcircRNA junction sequences identified by RNA-Seq and those identified by PCR-Seq (typically 1–8 nt at the junction, Supplementary Figure S2), a global analysis of signature motifs for IcircRNAs junctions could not be performed.
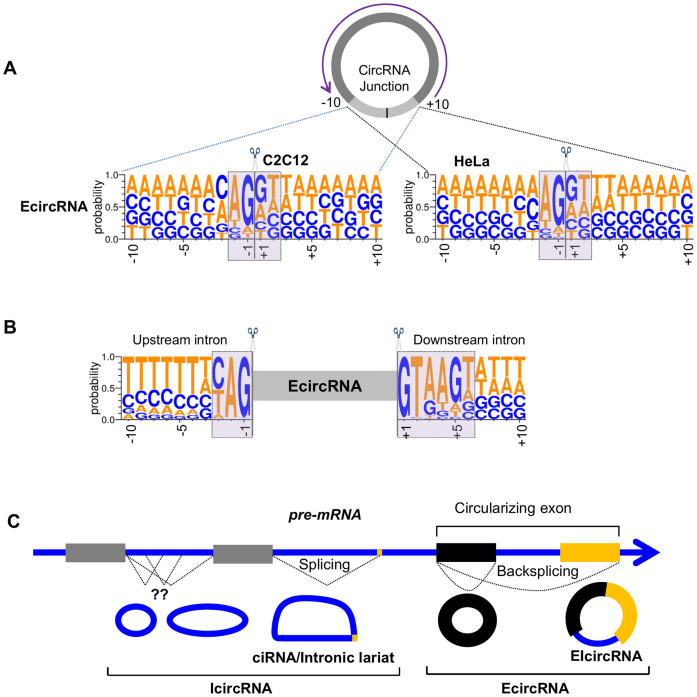
By contrast, it was possible to analyze the splicing pattern of the EcircRNA junctions by studying sequences 10 nt upstream and downstream of the backsplice site. As shown, the EcircRNA junctions showed a consensus of ‘GT’ and ‘AG’ at the 5΄ and 3΄ ends of circularizing exons (Figure 7A). The same observation was made with a similar motif analysis of circRNAs reported in circBase, with consensus sequences ‘GT’ and ‘AG’ at the 5΄ and 3΄ ends of circularizing exons (Supplementary Figure S3).
Figure 7.
Characteristics of circRNAs. (A) WebLogo representations of 20 nt backsplice sequences of EcircRNAs identified in HeLa and C2C12 cells. (B) WebLogo representations of upstream and downstream sequences of EcircRNAs identified in HeLa cells. (C) Schematic representation of different types of circular RNAs generated from a pre-RNA transcript. The RNA sequence is written in DNA format (T instead of U) following the WebLogo style.
Several studies have suggested that the biogenesis of circRNAs requires the endogenous spliceosome machinery. Almost all exonic annotated circRNAs reported in circBase are flanked by ‘ag’ and ‘gt’ dinucleotide pair (Supplementary Figure S3), supporting a biogenesis model involving the major U2-spliceosome complex which requires the A/CAG|gta/ggt at the 5΄ splice site and CAG|gt motif at the 3΄ splice site (34). Analysis of the flanking splice signal sequence immediately upstream or downstream of the exonic EcircRNAs identified using the RPAD method showed the presence of ‘ag’ at the end of upstream intron sequence and ‘gt’ at the start of downstream intron in almost every EcircRNA (Figure 7B), in agreement with previous reports (34).
DISCUSSION
Despite the decades that have elapsed since the discovery of circRNAs (3–6,35,36), the characterization of these interesting RNAs has progressed slowly. Recent high-throughput RNA-Seq analyses have begun to identify the vast abundance of circRNAs (10,14), uncovering their tissue specificity and evolutionary conservation (10,11,14). Although tens of thousands of circRNAs have been discovered to-date, their physiologic relevance has only been shown for a handful of them. The few circRNAs known in mechanistic detail to-date are described as regulators of gene expression by acting as decoys for regulatory miRNAs or proteins (reviewed in Ref. 37).
There is an increasing need to identify and characterize circRNAs comprehensively, as this ability could lead to a better understanding of the roles of circRNAs in cell physiology and disease processes. Current methods to detect and quantify circRNAs include circRNA microarray, RT-PCR/qPCR and Northern blot analyses, although these methods can only examine a limited subset of circRNAs and often have low sensitivity and/or specificity. Standard high-throughput RNA-Seq can identify circRNAs widely, but the body of the circRNA and the linear cognate RNA are indistinguishable, so one can only identify backsplicing junctions, not the entirety of the circRNAs. Given this limitation, interventions to deplete the linear RNA are particularly promising. Treatment of total RNA with exonucleases, particularly RNase R, has improved circRNA enrichments, but does not fully eliminate all linear RNAs (Figure 2). The method described here, RPAD (Figure 3), provides a practical solution for eliminating the substantial remaining linear RNA population, as it depletes linear RNA far more efficiently than exonuclease treatment alone. This method is particularly helpful for linear RNAs with extensive secondary structure, such as U1 and U6, which are highly resistant to RNase R digestion. Thus, RPAD can be used to isolate highly pure circRNAs from total RNA pools, increasing the chances of detecting novel circRNAs by RNA-Seq and enabling studies to elucidate their sequence, relative abundance and function.
Conventional circRNA identification is based on the presence of the backsplice sequence (the circRNA junction). By this approach, the body of the circRNA is predicted from the transcriptome considering all of the exons present between the backsplice sequences. Although most backsplice sequences identified are the same as those reported in circBase (9), several of them were found to have different sequences. Aligning RNA-Seq reads from RPAD-generated samples to known and novel transcripts helped assemble full-length circRNA sequences (Supplementary Table S4), which can then be used for elucidating their function. The RPAD assay quickly and conveniently isolates circRNAs from the total RNA population, allowing the validation of circRNA body sequences, which otherwise may only be predicted bioinformatically (Figure 5). As circular RNAs are known to regulate cell metabolism by interacting with gene regulatory factors (RBPs and miRNAs), we analyzed their interaction with RBPs using the CircInteractome web tool. Surprisingly, IcircRNAs and EcircRNAs were predicted to interact with many RBPs (Supplementary Figure S4 and Table S6), suggesting that the novel circRNAs discovered here may be important for cell physiology by acting as decoys for splicing factors and other RBPs. We further propose that some of the intronic CLIP tags found in RBP CLIP datasets might have originated from IcircRNAs which were previously thought to be derived from introns of pre-mRNAs.
CircRNA identification algorithms including CircRNAseq, find_circ, circRNA_ finder, CIRI and DCC use the splice donor and acceptor sequence for the U2 spliceosome (ag-gt) at the flanking introns to enhance the identification of true backspliced exons (38). These analyses exclude the identification of circRNAs generated from introns or by the minor spliceosome (utilizing U12 snRNA). We previously described thousands of intronic circular RNAs in monkey muscle as well as in human fibroblasts (22,39). In the present study, we used CIRCexplorer with modifications to identify thousands of intronic IcircRNAs and many exonic EcircRNAs with isoforms that were not previously reported (Figure 7C).
Several reports suggest that EcircRNAs are generated from the backsplicing of exons with flanking inverted complementary sequences and involve the spliceosome complex (7,8,15). Nearly all of the reported introns have ‘gt-ag’ dinucleotide sequences at the intron boundaries and spliced by the major spliceosome complex (40). Analysis of flanking intronic sequences (Figure 7B) of exonic EcircRNAs detected in HeLa cells shows the conservation of ‘ag’ and ‘gt’ dinucleotide at the end of upstream and start of downstream introns respectively. These data suggest that they may be generated by backsplicing of circularizing exons and removal of flanking introns by the spliceosome machinery. However, the vast majority of circular RNAs detected using RPAD RNA-Seq were intronic (IcircRNAs). In HeLa cells, 299 circular intronic RNAs (ciRNAs) have been reported to be generated from inefficient debranching of the intronic lariats (12). Interestingly, RNA-Seq analysis after RPAD did not detect any of the previously reported ciRNAs, possibly because of the polyadenylation and depletion of lariats due to their free 3΄-OH. Given that validation of IcircRNA junction sequences following RT-PCR analysis revealed poorly conserved upstream and downstream sequences (not shown), it is plausible that IcircRNAs are generated by other, as-yet unknown mechanisms. We propose that IcircRNAs were missed in previous studies due to technical challenges with the recognition of this class of circRNAs.
Further investigation is required to elucidate the mechanism of IcircRNAs biogenesis and function. Methods such as RPAD will enable rapid progress toward characterizing circRNAs in their vast heterogeneity, so that we may investigate comprehensively their involvement in physiology and disease.
Supplementary Material
ACKNOWLEDGEMENTS
We thank William H. Wood III and Elin Lehrmann for their help with circRNA sequencing and data deposition in GEO.
Footnotes
Present address: Amaresh C. Panda, Department of Biochemistry and Molecular Biology, University of Miami Miller School of Medicine, Miami, FL 33136, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute on Aging Intramural Research Program of the National Institutes of Health [Z01-AG000511-19]. Funding for open access charge: NIH Intramural Research Program.
Conflict of interest statement. None declared.
REFERENCES
- 1. Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. et al. . Landscape of transcription in human cells. Nature. 2012; 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beermann J., Piccoli M.T., Viereck J., Thum T.. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol. Rev. 2016; 96:1297–325. [DOI] [PubMed] [Google Scholar]
- 3. Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K.. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U.S.A. 1976; 73:3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arnberg A.C., Van Ommen G.J., Grivell L.A., Van Bruggen E.F., Borst P.. Some yeast mitochondrial RNAs are circular. Cell. 1980; 19:313–319. [DOI] [PubMed] [Google Scholar]
- 5. Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., Goodfellow P., Lovell-Badge R.. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993; 73:1019–1030. [DOI] [PubMed] [Google Scholar]
- 6. Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D., Kinzler K.W., Vogelstein B.. Scrambled exons. Cell. 1991; 64:607–613. [DOI] [PubMed] [Google Scholar]
- 7. Szabo L., Morey R., Palpant N.J., Wang P.L., Afari N., Jiang C., Parast M.M., Murry C.E., Laurent L.C., Salzman J.. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015; 16:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hudson A.J., Stark M.R., Fast N.M., Russell A.G., Rader S.D.. Splicing diversity revealed by reduced spliceosomes in C. merolae and other organisms. RNA Biol. 2015; 12:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Glazar P., Papavasileiou P., Rajewsky N.. circBase: a database for circular RNAs. RNA. 2014; 20:1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E.. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013; 19:141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. et al. . Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013; 495:333–338. [DOI] [PubMed] [Google Scholar]
- 12. Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L.. Circular intronic long noncoding RNAs. Mol. Cell. 2013; 51:792–806. [DOI] [PubMed] [Google Scholar]
- 13. Guo J.U., Agarwal V., Guo H., Bartel D.P.. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014; 15:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rybak-Wolf A., Stottmeister C., Glazar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R. et al. . Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015; 58:870–885. [DOI] [PubMed] [Google Scholar]
- 15. Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016; 17:205–211. [DOI] [PubMed] [Google Scholar]
- 16. Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L. et al. . Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015; 22:256–264. [DOI] [PubMed] [Google Scholar]
- 17. Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J.. Natural RNA circles function as efficient microRNA sponges. Nature. 2013; 495:384–388. [DOI] [PubMed] [Google Scholar]
- 18. Li F., Zhang L., Li W., Deng J., Zheng J., An M., Lu J., Zhou Y.. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015; 6:6001–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. et al. . Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016; 7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S.. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014; 56:55–66. [DOI] [PubMed] [Google Scholar]
- 21. Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B.. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016; 44:2846–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abdelmohsen K., Panda A.C., De S., Grammatikakis I., Kim J., Ding J., Noh J.H., Kim K.M., Mattison J.A., de Cabo R. et al. . Circular RNAs in monkey muscle: age-dependent changes. Aging. 2015; 7:903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dudekula D.B., Panda A.C., Grammatikakis I., De S., Abdelmohsen K., Gorospe M.. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016; 13:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kozomara A., Griffiths-Jones S.. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014; 42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Panda A.C., Abdelmohsen K., Martindale J.L., Di Germanio C., Yang X., Grammatikakis I., Noh J.H., Zhang Y., Lehrmann E., Dudekula D.B. et al. . Novel RNA-binding activity of MYF5 enhances Ccnd1/Cyclin D1 mRNA translation during myogenesis. Nucleic Acids Res. 2016; 44:2393–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schmittgen T.D., Livak K.J.. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008; 3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 27. Crooks G.E., Hon G., Chandonia J.M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jeck W.R., Sharpless N.E.. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014; 32:453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suzuki H., Zuo Y., Wang J., Zhang M.Q., Malhotra A., Mayeda A.. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006; 34:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vincent H.A., Deutscher M.P.. Substrate recognition and catalysis by the exoribonuclease RNase R. J. Biol. Chem. 2006; 281:29769–29775. [DOI] [PubMed] [Google Scholar]
- 31. You X., Vlatkovic I., Babic A., Will T., Epstein I., Tushev G., Akbalik G., Wang M., Glock C., Quedenau C. et al. . Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015; 18:603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang X.O., Dong R., Zhang Y., Zhang J.L., Luo Z., Zhang J., Chen L.L., Yang L.. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016; 26:1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gao Y., Wang J., Zheng Y., Zhang J., Chen S., Zhao F.. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat. Commun. 2016; 7:12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang M.Q. Statistical features of human exons and their flanking regions. Hum. Mol. Genet. 1998; 7:919–932. [DOI] [PubMed] [Google Scholar]
- 35. Kos A., Dijkema R., Arnberg A.C., van der Meide P.H., Schellekens H.. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986; 323:558–560. [DOI] [PubMed] [Google Scholar]
- 36. Cocquerelle C., Daubersies P., Majerus M.A., Kerckaert J.P., Bailleul B.. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992; 11:1095–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Panda A.C., Grammatikakis I., Munk R., Gorospe M., Abdelmohsen K.. Emerging roles and context of circular RNAs. Wiley Interdiscip. Rev. RNA. 2016; 8:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Szabo L., Salzman J.. Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 2016; 17:679–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Panda A.C., Grammatikakis I., Abdelmohsen K., Martindale J.L., Kim K.M., De S., Yang X., Gorospe M.. Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence supressor CircPVT1. Nucleic Acids Res. 2016; 45:4021–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sheth N., Roca X., Hastings M.L., Roeder T., Krainer A.R., Sachidanandam R.. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006; 34:3955–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.