Abstract
AGRO100 (also known as AS1411) is a G-rich oligonucleotide that has long been established as a potent anti-cancer aptamer. However, the structure of AGRO100 remained unresolved, due to the co-existence of multiple different G-quadruplex conformations. We identified a DNA sequence named AT11, derived from AGRO100, which formed a single major G-quadruplex conformation and exhibited a similar anti-proliferative activity as AGRO100. The solution structure of AT11 revealed a four-layer G-quadruplex comprising of two propeller-type parallel-stranded subunits connected through a central linker. The stacking between the two subunits occurs at the 3΄-end of the first block and the 5΄-end of the second block. The structure of the anti-proliferative DNA sequence AT11 will allow greater understanding on the G-quadruplex folding principles and aid in structural optimization of anti-proliferative oligonucleotides.
INTRODUCTION
Guanine-rich oligonucleotides are capable of folding into four-stranded structures called G-quadruplexes in the presence of cations (1). A folded intramolecular G-quadruplex consists of two main elements: core and loops (2–4). The core comprises of stacked G·G·G·G tetrad (or G-tetrad) layers, while the loops are linker sequences connecting the strands of the G-tetrad core. G-quadruplex structures are highly polymorphic, depending on the relative orientations of strands and types of loops (1–4). G-quadruplex polymorphism was also recently extended to motifs with bulges, duplex stems and missing guanines (5–7). In many cases, a single oligonucleotide could adopt multiple G-quadruplex conformations (4).
Over the past decades, numerous G-quadruplex-forming oligonucleotides have been reported as aptamers for a wide range of targets, such as thrombin (8–10), HIV (11–19) and cancer cells (20–25). The crystal structure of the complex between thrombin and a DNA aptamer (10) showed that the G-quadruplex formation was required for the recognition and biological activity of the oligonucleotide.
Most notably, AGRO100 (also known as AS1411), a G-rich oligonucleotide (sequence, GGTGGTGGTGGTTGTGGTGGTGGTGG), first discovered by Bates et al. (20), has garnered considerable research interest in the field of anticancer therapeutics (26,27). AGRO100 was reported to have inhibition activity against a large number of malignant cell lines with GI50 in the low micromolar range (26) and underwent Phase II clinical trials on renal cell carcinoma (28). The binding of AGRO100 to nucleolin, a protein participating ribosomal biogenesis (29), was proposed to be crucial for the anticancer activity of this oligonucleotide (30). Recently, AGRO100 has been reported to possess inhibitory activity against HIV (18,19).
Previous reports showed that AGRO100 adopted a mixture of multiple G-quadruplexes in solution, hindering structural studies (31). Using size-exclusion chromatography, Dailey et al. suggested the presence of at least eight species in this mixture (31). We previously determined the structure of a DNA sequence modified from AGRO100, revealing the formation of a left-handed G-quadruplex conformation (32).
In this study, we have identified a DNA sequence named AT11, derived from AGRO100, which formed a single major G-quadruplex conformation and exhibited a similar anti-proliferative activity as AGRO100. The solution structure AT11 revealed a novel G-quadruplex fold consisting of two propeller-type subunits. The structure of AT11 provides the basis for rational structural design and optimization of anti-proliferative oligonucleotides.
MATERIALS AND METHODS
DNA sample preparation
Unlabeled and site-specific labeled DNA oligonucleotides were chemically synthesized on an ABI 394 DNA/RNA synthesizer using products from Glen Research and Cambridge Isotope. DNA was purified by a reverse-phase Poly-Pak cartridge (Glen Research). Samples were then dialyzed successively against 25 mM KCl solution and against water. DNA oligonucleotides were dissolved in solution containing 70 mM KCl and 20 mM potassium phosphate (pH 7.0). DNA concentrations were determined by measuring the UV absorbance at 260 nm.
Cell proliferation assay
The following sequences, AT11, d(TGG TGG TGG TTG TTG TGG TGG TGG TGG T), AGRO100, d(GGT GGT GGT GGT TGT GGT GGT GGT GG) and a control sequence CRO (20), d(CCT CCT CCT CCT TCT CCT CCT CCT CC), were used in the cell proliferation assay. Oligonucleotides were purchased from Integrated DNA Technologies (Singapore), then dialyzed against water, followed by freeze-drying. The dried samples were dissolved in a buffer containing 70 mM KCl and 100 mM potassium phosphate (pH 7.0) to a final DNA stock concentration of 0.5 mM and stored at −30 °C for further usage.
Human cancer cell line A549 and human fibroblast cell line WI-38 were obtained from American Type Culture Collection (ATCC, Rockville, MD, USA). Cells were cultured in complete medium containing Dulbecco's modified Eagle's medium (high glucose, GlutaMAX™ supplement, pyruvate) (Gibco), 10% fetal bovine serum (Gibco), 1% Penicillin-Streptomycin (Sigma). Cell proliferation assay kit was purchased from ATCC (Rockville, MD, USA) including MTT reagent (3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide) and stop detergent reagent solution.
In cell proliferation assay, cells were cultured in a 96-well plate at a low density of 1000 and 2000 cells per well for A549 and WI-38, respectively. Each well contained 100 μl complete medium and incubated at 37 °C, 5% CO2 for 18–20 h to allow cell adherence and growth. The medium was then replaced with a new complete medium containing an oligonucleotide at different concentrations (1, 5, 10 and 20 μM) followed by 5 days incubation at 37 °C, 5% CO2. Complete medium without additional oligonucleotides was used as control. In cell proliferation assay, 10 μl MTT was added to each well and incubated in 37 °C for 4 h. The reaction was stopped by adding 100 μl stop detergent reagent solution and incubated at 37 °C overnight to solubilize formazan crystal. Infinite 200 PRO Microplate Reader (Tecan, Switzerland) was used to record the absorbance at 570 nm, while the reference wavelength was 650 nm. Each experiment was repeated three times to have an average value of the measurements.
Circular dichroism
Circular dichroism (CD) spectra were recorded at 25 °C using a JASCO-815 spectropolarimeter with a 1-cm path length quartz cuvette containing a solution volume of 500 μl. Spectra (220–320 nm) were taken with a scan speed of 200 nm/min. DNA (∼5 μM) was dissolved in a buffer containing 70 mM KCl and 20 mM potassium phosphate (pH 7.0). For each measurement, an average of three scans was taken and the spectral contribution of the buffer was subtracted.
UV melting experiments
Melting experiments were performed on a JASCO V-650 UV-visible spectrophotometer (Tokyo, Japan). DNA concentration was ∼5 μM. Melting curves were monitored by the UV absorbance at 295 nm (33). Melting temperatures (Tm) reported represent an average over heating and cooling cycles.
NMR spectroscopy
Nuclear magnetic resonance (NMR) experiments were performed on 600 and 700 MHz Bruker spectrometers at 25 °C, unless otherwise stated. NMR spectra were recorded either directly or after following an annealing procedure (heating the sample at high temperature (∼95 °C) and slowly cooling down to room temperature). The concentration of DNA samples was typically 0.1–2.0 mM. Solution contained 70 mM KCl and 20 mM potassium phosphate (pH 7.0). Spectra analyses were performed using SpinWorks software (http://home.cc.umanitoba.ca/∼wolowiec/spinworks/), FELIX (Felix NMR, Inc.) and SPARKY (34) programs.
NMR-restrained structure calculation
The structure of AT11 was calculated using the XPLOR-NIH program (35). NMR-restrained computations were performed as described below. Structures were displayed using the PyMOL program (36).
NOE distance restraints
Distances between the non-exchangeable protons of AT11 were obtained from NOESY experiments at various mixing times (100 and 300 ms) recorded in D2O, those involving exchangeable protons were obtained from NOESY spectrum (mixing time, 200 ms) recorded in H2O solution. The NOE cross-peaks were classified as strong, medium and weak and given corresponding distance restraints as previously described (32).
Dihedral restraints
The glycosidic bonds of all 16 guanines were restrained according to their anti conformation. The glycosidic χ torsion angle for experimentally determined anti guanine residues were fixed at 240° ± 40°.
Hydrogen-bond and planarity restraints
Hoogsteen hydrogen bonds between guanines were restrained using H21–N7, N2–N7, H1–O6 and N1–O6 distances, which were set to 1.99 ± 0.1, 2.9 ± 0.1, 1.95 ± 0.1, 2.95 ± 0.1Å respectively. Planarity restrained was used for the G2·G5·G8·G11, G3·G6·G9·G12, G15·G18·G21·G24 and G17·G20·G23·G26 tetrads.
Repulsive restraints
Repulsive restraints (4.0–20.0 Å) were applied on pairs of protons that do not exhibit cross-peaks in a well-resolved region of the NOESY spectrum. A total of 17 repulsive restraints involving thymine residues were used: 9 between sugar protons and methyl protons, 1 between an aromatic proton and a methyl proton, 4 between sugar protons and T(H6) protons and 3 between sugar protons.
Distance geometry simulated annealing
An initial extended conformation of AT11 was generated using the XPLOR program. The system was then subjected to distance geometry simulated annealing by incorporating the hydrogen-bond, distance, dihedral, planarity and repulsive restraints. A total of 100 structures were generated and subjected to further refinements.
Distance-restrained molecular dynamics refinement
The 100 structures obtained from the simulated annealing step were refined with a distance-restrained molecular dynamics protocol incorporating all distance restraints. The system was heated from 300 to 1000 K in 5 ps and allowed to equilibrate for 1 ps, during which force constants for the distance restraints were kept at 2 kcal.mol−1.Å−2. The force constants for non-exchangeable proton, exchangeable proton and repulsive distance restraints were then increased to 64 kcal.mol−1.Å−2 in 26 ps. Next, the system was cooled down to 300 K in 14 ps, after which an equilibration was performed for 10 ps. The coordinates were saved every 0.5 ps during the last 4.0 ps and averaged. The average structure obtained was then subjected to minimization until the gradient of energy was <0.1 kcal.mol−1. Dihedral (50 kcal.mol−1.rad−2) and planarity (1 kcal.mol−1.Å−2 for tetrads) restraints were maintained throughout the course of refinement. Ten lowest-energy structures were chosen.
Data deposition
The coordinates for the AT11 G-quadruplex have been deposited in the Protein Data Bank (accession code 2N3M).
RESULTS
Favoring a single G-quadruplex conformation from AGRO100 by sequence modifications
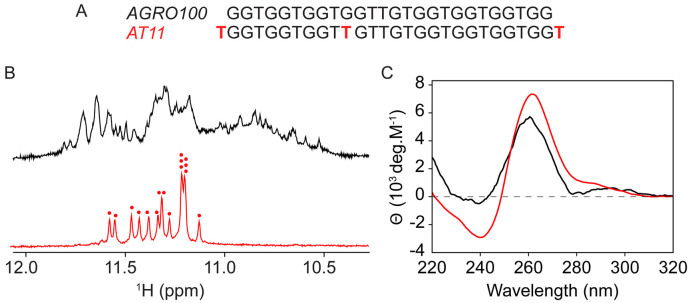
We recorded NMR and CD spectra of AGRO100 in K+ containing solution (Figure 1). The imino proton NMR spectrum of AGRO100 displayed multiple humps and peaks at 10.5–12.0 ppm (Figure 1A), indicating the presence of multiple G-quadruplex species, consistent with a previous report showing that AGRO100 could adopt at least eight different G-quadruplex conformations (31). CD spectrum of AGRO100 showed a minimum at 240 nm and a positive peak at 260 nm (Figure 1B), characteristic of the formation of parallel-stranded G-quadruplexes.
Figure 1.
(A) Sequences of the AGRO100 and AT11 oligonucleotides. (B) Imino proton NMR and (C) CD spectra of AGRO100 (black) and AT11 (red) in solution containing 70 mM KCl and 20 mM potassium phosphate (pH 7). Each red dot indicates an imino proton peak.
The complexity of NMR spectra of AGRO100 prevented its further structural elucidation. Sequence modifications were performed to favor a single G-quadruplex conformation and simplify the NMR spectra. The sequence AT11, derived from AGRO100 by an addition of thymine nucleotides to both 5΄- and 3΄-ends of the DNA sequence and a single G-to-T modification at position 11, exhibited a well-resolved NMR spectrum consistent with the formation of a single major G-quadruplex conformation (Figure 1). Addition of a thymine nucleotide to the 5΄- and 3΄-ends of the sequence was aimed to prevent stacking of G-quadruplexes at the 5΄/3΄-end surfaces which might lead to aggregation (15,37–40), while a G-to-T substitution reduced the number of guanines in the sequence from 17 to 16 with the aim to favor a single G-quadruplex fold (Supplementary Figure S1). Indeed, the imino proton spectrum of AT11 showed 16 peaks at 11.0–11.7 ppm, consistent with the formation of a G-quadruplex containing four G-tetrad layers. The same major conformation was formed by AT11 in the presence of varying K+ concentration (Supplementary Figure S2). The CD profile of AT11 was similar to that of AGRO100, showing a minimum at 240 nm and a positive peak at 260 nm (Figure 1C), suggesting the formation of a right-handed parallel-stranded G-quadruplex.
AT11 exhibits anti-proliferative activity
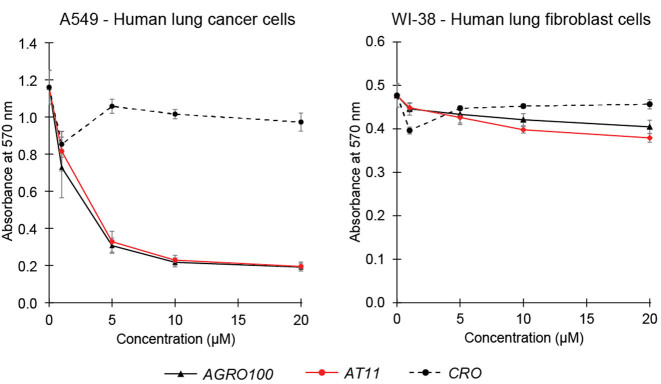
We tested the effect of different oligonucleotides on the proliferation of the human lung cancer cells A549. Both G-quadruplex-forming sequences AGRO100 and AT11 inhibited the proliferation of the A549 cells over a range of oligonucleotide concentrations from 5 to 20 μM, displaying similar anti-proliferative activities (Figure 2 and Supplementary Figure S3), while the control oligonucleotide CRO (20) did not inhibit the proliferation of the A549 cells in the same conditions. In comparison, none of the tested oligonucleotides showed anti-proliferative activity on the normal human cell line WI-38 (Figure 2).
Figure 2.
Effect of different oligonucleotides on cell proliferation. Cell proliferation assays performed on A549 human lung cancer cells (left) and WI-38 human lung fibroblast cells (right) after incubation with different concentrations of DNA oligonucleotides (AGRO100, AT11 and CRO). Each measurement was repeated three times.
NMR structure of AT11 revealed a G-quadruplex with two parallel-stranded subunits
Spectral assignment
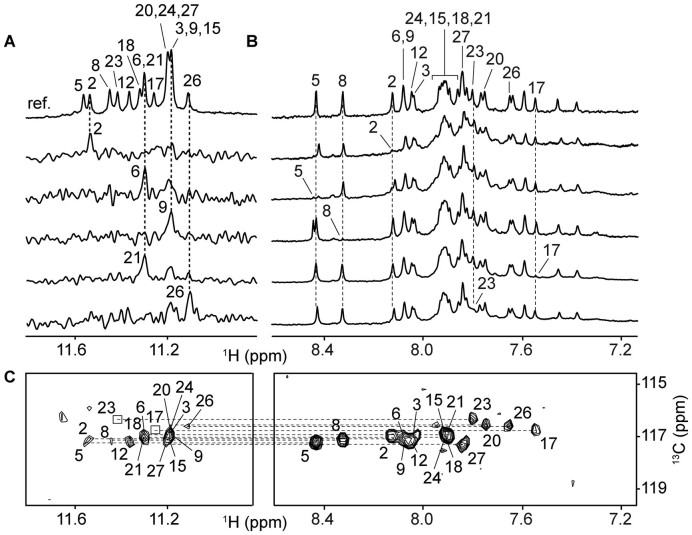
Unambiguous assignment of the imino and aromatic protons of AT11 was performed using site-specific low-enrichment (4 or 2%) 15N-labeling of guanines and site-specific deuteration of guanines, and through-bond correlation experiment (1H-13C-HMBC) (Figure 3 and Supplementary Figure S4). The remaining protons of AT11 were assigned accordingly using data obtain from through-bond (TOCSY, COSY, 1H-13C-HSQC and through-space (NOESY) correlation experiments (Supplementary Figure S5).
Figure 3.
NMR spectral assignments of AT11. (A) Examples of the assignment of guanine imino protons through site-specific 15N-labeling. (B) Examples of the assignment of guanine aromatic protons through site-specific deuterium labeling. Labeled guanine residues were indicated with their corresponding residue number. (C) Through-bond correlations between guanine imino and aromatic protons at natural abundance, using long-range J-couplings. Weak/missing signals are framed in boxes. DNA concentration was 0.2–0.5 mM for (A and B) and ∼1 mM for (C).
Folding topology of AT11
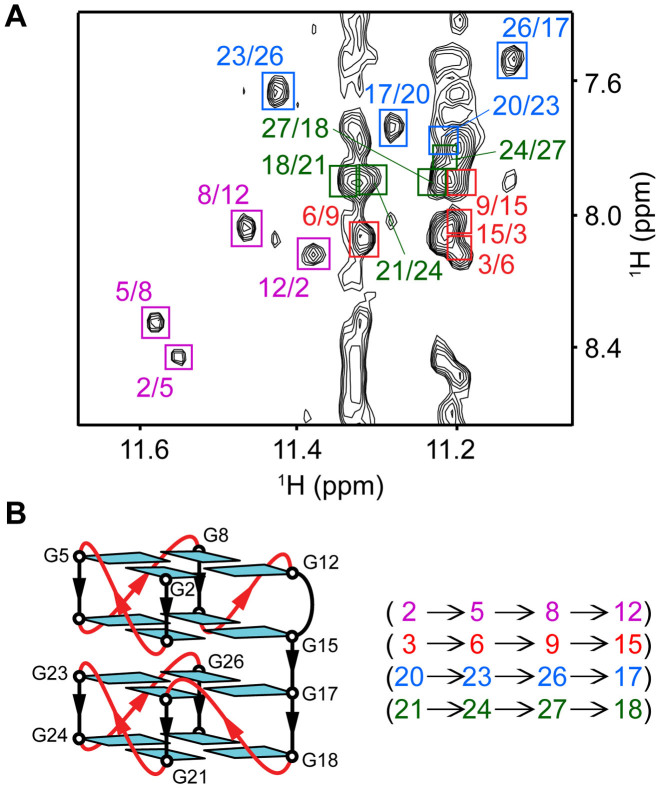
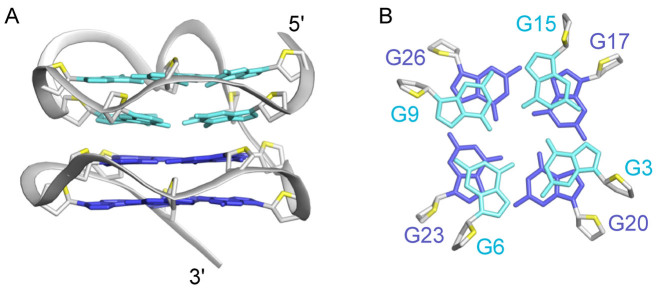
The folding topology of AT11 was determined by analyzing NOE cross-peaks between the imino (H1) and aromatic (H8) protons of the guanine bases (Figure 4A). From H1-H8 NOEs, the alignments of four G-tetrads, G2·G5·G8·G12, G3·G6·G9·G15, G17·G20·G23·G26 and G18·G21·G24·G27, were established (Figure 4). Solvent exchange data showed that imino proton peaks of the G3·G6·G9·G15 and G17·G20·G23·G26 tetrads were more protected than the counterparts from the other G-tetrads (Supplementary Figure S6). All together the data indicated the formation of a four-layer G-quadruplex structure as shown in Figure 4. The structure comprises of two 2-layered propeller-type parallel-stranded subunits connected by a single-residue linker (T16). H8/H6-H1΄ NOE sequential connectivity could be traced from T1 to T28, including the T1-G2-G3, G5-G6, G8-G9, T11-G12, G17-G18, G20-G21, G23-G24 and G26-G27-T28 segments, with several broken connections due to the presence of propeller loops, bulge and linker (Supplementary Figure S5). All guanines show weak intra-residue H8-H1΄ NOEs comparing to strong intra-residue thymine H6-CH3 NOEs, reflecting their anti glycosidic conformation. A two-residue bulge forms between G12 and G15 of the first subunit. The two subunits exhibit the 3΄-to-5΄ stacking mode (Figure 4B). Such an arrangement is supported by solvent exchange data and several NOEs observed at the interface between the two subunits, including: G20(H8)-G3(H1΄), G23(H8)-G6(H1΄), G26(H8)-G9(H1΄) and G17(H8)-G15(H1΄) (Supplementary Figures S5 and S7). The parallel-stranded folding topology of the AT11 G-quadruplex is consistent with its CD data (see above).
Figure 4.
Determination of G-quadruplex folding topology of AT11 in K+ solution. (A) NOESY spectrum (mixing time, 200 ms) showing the guanine H1-H8 NOE connectivity. Cross-peaks between the imino (H1) protons and aromatic (H8) protons are framed and labeled with the H1 and H8 proton assignment in the first and second position respectively. The first, second, third and fourth G-tetrad layers are colored in magenta, red, blue and forest green respectively. (B) Schematic structure showing the G-tetrad alignments and folding topology of AT11.
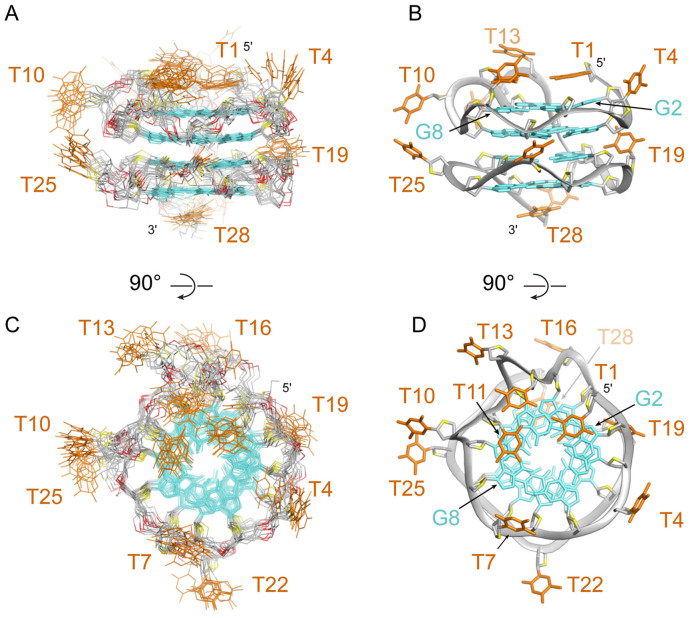
Solution structure of AT11
Following the unambiguous assignment of AT11 resonances, the solution structure of AT11 complex was computed (Table 1). The lowest-energy refined structures are presented in Figure 5. AT11 consists of two parallel-stranded propeller-type G-quadruplex blocks linked via a thymine residue (T16). The top G-quadruplex block contains three propeller loops (T4, T7 and T10–T11) and a bulge (T13–T14), while the bottom G-quadruplex block contains three single-nucleotide propeller loops (T19, T22 and T25). All guanine residues adopt the anti conformation. Partial overlap between the five- and six- membered rings of the guanine bases across the interface of the two G-quadruplex blocks (G3/G20, G6/G23, G9/G26 and G15/G17) (Figure 6) was observed to be similar to that between the two continuous G-tetrad layers.
Table 1. NMR restraints and structure statistics.
| A. NMR restraints | ||
| Distance restraints | Exchangeable | Non-exchangeable |
| DNA restraints | ||
| Intra-residue | 0 | 141 |
| Inter-residue | 47 | 171 |
| Other Restraints | ||
| Hydrogen-bond restraints | 64 | |
| Torsion angle restraints | 16 | |
| Repulsive restraints | 17 | |
| B. Structure statistics | ||
| NOE violations | ||
| Numbers (>0.2 Å) | 0.000 ± 0.000 | |
| Deviations from standard geometry | ||
| Bond length (Å) | 0.004 ± 0.000 | |
| Bond angle (°) | 0.709 ± 0.016 | |
| Impropers (°) | 0.378 ± 0.007 | |
| Pairwise all heavy atom r.m.s.d. values (Å) | ||
| All heavy atoms | 1.611 ± 0.256 | |
| G-tetrad core | 1.010 ± 0.210 | |
Figure 5.
(A and C) Ten superimposed lowest-energy solution structures and (B and D) a representative structure of AT11. (A and B) Side view. (C and D) Top view.
Figure 6.
(A) AT11 consists of two propeller-type G-quadruplex subunits. (B) Stacking interface between the two subunits of AT11.
Effects of bulge and linker size on structure and thermal stability
Linker/bulge/loop elements in the AT11 G-quadruplex include five single-residue (T4, T7, T19, T22 and T25) and one two-residue (T10–T11) propeller loops, a two-residue bulge (T13–T14) and a one-residue linker between the two subunits (T16). To analyze the relationship between bulge/linker size and the thermal stability of AT11, modifications were performed on AT11 by varying the bulge/linker size from zero to two thymines (Table 2). It was observed that by decreasing either the bulge or linker size of AT11, an increase in thermal stability was achieved and vice versa, consistent with previous studies (5). For example, the sequence AT11-B0, a derivative of AT11 with no bulge (or bulge size zero), exhibits an increase in the melting temperature by 16 °C compared to that of AT11.
Table 2. Thermal stability of AT11 and variant sequences with different bulges and linkers.
| Sequence (5΄-3΄) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Bulge | Linker | T m (°C) | |||||||||||||||||
| AT11 | T | GG | T | GG | T | GG | TT | G | TT | G | T | GG | T | GG | T | GG | T | GG | T | 43 |
| AT11-B1 | T | GG | T | GG | T | GG | TT | G | T | G | T | GG | T | GG | T | GG | T | GG | T | 45 |
| AT11-B0 | T | GG | T | GG | T | GG | TT | G | G | T | GG | T | GG | T | GG | T | GG | T | 59 | |
| AT11-L0 | T | GG | T | GG | T | GG | TT | G | TT | G | GG | T | GG | T | GG | T | GG | T | 51 | |
| AT11-L2 | T | GG | T | GG | T | GG | TT | G | TT | G | TT | GG | T | GG | T | GG | T | GG | T | -a |
aUnable to detect fully-folded state at 20 °C
DISCUSSION
5΄-3΄ versus 5΄-5΄ stacking between propeller-type G-quadruplex subunits
In a previous study by Matsugami et al. (41), the NMR structure of a G-quadruplex adopted by the d(GGA)8 sequence in K+ solution was reported. Both the d(GGA)8 (41) and AT11 sequence (this work) fold into similar G-quadruplexes, comprising of two parallel-stranded blocks connected by a single-residue linker. However, in the d(GGA)8 structure, the stacking of two blocks occurs between two 5΄-end surfaces, while in AT11, the stacking occurs between the 3΄-end of the first block and the 5΄-end of the second block. Formation of a G-A-G-A-G-A-G heptad at the stacking interface of d(GGA)8 may be the reason for this difference in the stacking mode.
Right-handed versus left-handed G-quadruplexes
Recently, we reported on a left-handed G-quadruplex called Z-G4 (32) formed by another sequence derived from AGRO100. Like AT11, Z-G4 also forms a structure consisting of two parallel-stranded two-layer G-quadruplex subunits. However, unlike AT11, Z-G4 involves the stacking between the 5΄-ends of the two subunits and, particularly, the strand progression in the left-handed orientation.
Structure-activity relationship
It has been suggested that the cancer-selective anti-proliferative activity of G-rich oligonucleotides is associated with their interaction with proteins such as nucleolin, a protein abundant on the surface of many cancer cells (30). On the other hand, a recent study suggested that the anti-proliferative activity of G-rich oligonucleotides is mainly contributed by the guanine-containing degradation products (42). Further studies will be required to answer the question of whether and how the G-quadruplex structures of G-rich oligonucleotides contribute to their cellular uptake and anti-proliferative activity.
CONCLUSION
The AT11 sequence, derived from AGRO100 by a simple base substitution, was shown to favor a single major G-quadruplex conformation and exhibit a similar anti-proliferative activity as AGRO100. We have determined the structure of AT11, which adopts a four-layer G-quadruplex, which comprises of two propeller-type parallel-stranded subunits. The stacking between the two subunits occurs at the 3΄-end of the first block and the 5΄-end of the second block.
ACCESSION NUMBER
PDB ID: 2N3M.
Supplementary Material
Footnotes
Present addresses:
Ngoc Quang Do, Hanoi University of Pharmacy, Vietnam.
Brahim Heddi, Laboratoire de Biologie et de Pharmacologie Appliquée, UMR 8113 CNRS, ENS Paris-Saclay, Université Paris Saclay, France
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Singapore Ministry of Education Academic Research Fund Tier 2 [MOE2015-T2-1-092]; National Research Foundation Investigatorship [NRF-NRFI2017-09]; Nanyang Technological University (to A.T.P.). Funding for open access charge: Singapore Ministry of Education Academic Research Fund Tier 2 [MOE2015-T2-1-092].
Conflict of interest statement. None declared.
REFERENCES
- 1. Davis J.T. G-quartets 40 years later: from 5΄-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. 2004; 43:668–698. [DOI] [PubMed] [Google Scholar]
- 2. Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S.. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006; 34:5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Patel D.J., Phan A.T., Kuryavyi V.. Human telomere, oncogenic promoter and 5΄-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007; 35:7429–7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Phan A.T. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010; 277:1107–1117. [DOI] [PubMed] [Google Scholar]
- 5. Mukundan V.T., Phan A.T.. Bulges in G-quadruplexes: broadening the definition of G-quadruplex-forming sequences. J. Am. Chem. Soc. 2013; 135:5017–5028. [DOI] [PubMed] [Google Scholar]
- 6. Lim K.W., Phan A.T.. Structural basis of DNA quadruplex-duplex junction formation. Angew. Chem. Int. Ed. 2013; 52:8566–8569. [DOI] [PubMed] [Google Scholar]
- 7. Heddi B., Martin-Pintado N., Serimbetov Z., Kari T.M., Phan A.T.. G-quadruplexes with (4n - 1) guanines in the G-tetrad core: formation of a G-triad.water complex and implication for small-molecule binding. Nucleic Acids Res. 2016; 44:910–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bock L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J.. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992; 355:564–566. [DOI] [PubMed] [Google Scholar]
- 9. Schultze P., Macaya R.F., Feigon J.. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG). J. Mol. Biol. 1994; 235:1532–1547. [DOI] [PubMed] [Google Scholar]
- 10. Russo Krauss I., Merlino A., Randazzo A., Novellino E., Mazzarella L., Sica F.. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012; 40:8119–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wyatt J.R., Vickers T.A., Roberson J.L., Buckheit R.W. Jr, Klimkait T., DeBaets E., Davis P.W., Rayner B., Imbach J.L., Ecker D.J.. Combinatorially selected guanosine-quartet structure is a potent inhibitor of human immunodeficiency virus envelope-mediated cell fusion. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:1356–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rando R.F., Ojwang J., Elbaggari A., Reyes G.R., Tinder R., McGrath M.S., Hogan M.E.. Suppression of human immunodeficiency virus type 1 activity in vitro by oligonucleotides which form intramolecular tetrads. J. Biol. Chem. 1995; 270:1754–1760. [DOI] [PubMed] [Google Scholar]
- 13. de Soultrait V.R., Lozach P.Y., Altmeyer R., Tarrago-Litvak L., Litvak S., Andreola M.L.. DNA aptamers derived from HIV-1 RNase H inhibitors are strong anti-integrase agents. J. Mol. Biol. 2002; 324:195–203. [DOI] [PubMed] [Google Scholar]
- 14. Phan A.T., Kuryavyi V., Ma J.B., Faure A., Andreola M.L., Patel D.J.. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:634–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Do N.Q., Lim K.W., Teo M.H., Heddi B., Phan A.T.. Stacking of G-quadruplexes: NMR structure of a G-rich oligonucleotide with potential anti-HIV and anticancer activity. Nucleic Acids Res. 2011; 39:9448–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mukundan V.T., Do N.Q., Phan A.T.. HIV-1 integrase inhibitor T30177 forms a stacked dimeric G-quadruplex structure containing bulges. Nucleic Acids Res. 2011; 39:8984–8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Musumeci D., Riccardi C., Montesarchio D.. G-quadruplex forming oligonucleotides as anti-HIV agents. Molecules. 2015; 20:17511–17532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Metifiot M., Amrane S., Mergny J.L., Andreola M.L.. Anticancer molecule AS1411 exhibits low nanomolar antiviral activity against HIV-1. Biochimie. 2015; 118:173–175. [DOI] [PubMed] [Google Scholar]
- 19. Perrone R., Butovskaya E., Lago S., Garzino-Demo A., Pannecouque C., Palu G., Richter S.N.. The G-quadruplex-forming aptamer AS1411 potently inhibits HIV-1 attachment to the host cell. Int. J. Antimicrob. Agents. 2016; 47:311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bates P.J., Kahlon J.B., Thomas S.D., Trent J.O., Miller D.M.. Antiproliferative activity of G-rich oligonucleotides correlates with protein binding. J. Biol. Chem. 1999; 274:26369–26377. [DOI] [PubMed] [Google Scholar]
- 21. Simonsson T., Henriksson M.. C-myc suppression in Burkitt's lymphoma cells. Biochem. Biophys. Res. Commun. 2002; 290:11–15. [DOI] [PubMed] [Google Scholar]
- 22. Jing N., Li Y., Xiong W., Sha W., Jing L., Tweardy D.J.. G-quartet oligonucleotides: a new class of signal transducer and activator of transcription 3 inhibitors that suppresses growth of prostate and breast tumors through induction of apoptosis. Cancer Res. 2004; 64:6603–6609. [DOI] [PubMed] [Google Scholar]
- 23. Qi H., Lin C.P., Fu X., Wood L.M., Liu A.A., Tsai Y.C., Chen Y., Barbieri C.M., Pilch D.S., Liu L.F.. G-quadruplexes induce apoptosis in tumor cells. Cancer Res. 2006; 66:11808–11816. [DOI] [PubMed] [Google Scholar]
- 24. Choi E.W., Nayak L.V., Bates P.J.. Cancer-selective antiproliferative activity is a general property of some G-rich oligodeoxynucleotides. Nucleic Acids Res. 2010; 38:1623–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Edwards S.L., Poongavanam V., Kanwar J.R., Roy K., Hillman K.M., Prasad N., Leth-Larsen R., Petersen M., Marusic M., Plavec J. et al. Targeting VEGF with LNA-stabilized G-rich oligonucleotide for efficient breast cancer inhibition. Chem. Commun. 2015; 51:9499–9502. [DOI] [PubMed] [Google Scholar]
- 26. Bates P.J., Laber D.A., Miller D.M., Thomas S.D., Trent J.O.. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009; 86:151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bates P.J., Reyes-Reyes E.M., Malik M.T., Murphy E.M., O’Toole M.G., Trent J.O.. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms. Biochim. Biophys. Acta. 2016; doi:10.1016/j.bbagen.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 28. Rosenberg J.E., Bambury R.M., Van Allen E.M., Drabkin H.A., Lara P.N. Jr, Harzstark A.L., Wagle N., Figlin R.A., Smith G.W., Garraway L.A. et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Invest. New Drugs. 2014; 32:178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ginisty H., Amalric F., Bouvet P.. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998; 17:1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bates P.J., Choi E.W., Nayak L.V.. G-rich oligonucleotides for cancer treatment. Methods Mol. Biol. 2009; 542:379–392. [DOI] [PubMed] [Google Scholar]
- 31. Dailey M.M., Miller M.C., Bates P.J., Lane A.N., Trent J.O.. Resolution and characterization of the structural polymorphism of a single quadruplex-forming sequence. Nucleic Acids Res. 2010; 38:4877–4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chung W.J., Heddi B., Schmitt E., Lim K.W., Mechulam Y., Phan A.T.. Structure of a left-handed DNA G-quadruplex. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:2729–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mergny J.L., Phan A.T., Lacroix L.. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998; 435:74–78. [DOI] [PubMed] [Google Scholar]
- 34. Goddard T.D., Kneller D.G.. SPARKY 3. San Francisco: University of California. [Google Scholar]
- 35. Schwieters C.D., Kuszewski J.J., Tjandra N., Clore G.M.. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003; 160:65–73. [DOI] [PubMed] [Google Scholar]
- 36. Schrodinger LLC. The PyMOL Molecular Graphics System. 2015; Version 1.8 ed.
- 37. Kato Y., Ohyama T., Mita H., Yamamoto Y.. Dynamics and thermodynamics of dimerization of parallel G-quadruplexed DNA formed from d(TTAGn) (n=3-5). J. Am. Chem. Soc. 2005; 127:9980–9981. [DOI] [PubMed] [Google Scholar]
- 38. Smargiasso N., Rosu F., Hsia W., Colson P., Baker E.S., Bowers M.T., De Pauw E., Gabelica V.. G-quadruplex DNA assemblies: loop length, cation identity, and multimer formation. J. Am. Chem. Soc. 2008; 130:10208–10216. [DOI] [PubMed] [Google Scholar]
- 39. Martadinata H., Phan A.T.. Structure of propeller-type parallel-stranded RNA G-quadruplexes, formed by human telomeric RNA sequences in K+ solution. J. Am. Chem. Soc. 2009; 131:2570–2578. [DOI] [PubMed] [Google Scholar]
- 40. Sket P., Plavec J.. Tetramolecular DNA quadruplexes in solution: insights into structural diversity and cation movement. J. Am. Chem. Soc. 2010; 132:12724–12732. [DOI] [PubMed] [Google Scholar]
- 41. Matsugami A., Ouhashi K., Kanagawa M., Liu H., Kanagawa S., Uesugi S., Katahira M.. An intramolecular quadruplex of (GGA)4 triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction. J. Mol. Biol. 2001; 313:255–269. [DOI] [PubMed] [Google Scholar]
- 42. Zhang A., Bing T., Liu X., Qi C., Shen L., Wang L., Shangguan D.. Cytotoxicity of guanine-based degradation products contributes to the antiproliferative activity of guanine-rich oligonucleotides. Chem. Sci. 2015; 6:3831–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.